Abstract
Although a network of transcription factors that specifies neural crest identity in the ectoderm has been defined, expression of neural crest transcription factors does not guarantee eventual migration as a neural crest cell. While much work has gone into determining regulatory relationships within the transcription factor network, the ability of protein modifications like phosphorylation to modulate the function of neural crest regulatory factors and determine when and where they are active also has crucial implications. Paladin, which was previously classified as a phosphatase based on sequence similarity, is expressed in chick neural crest precursors and is maintained throughout their epithelial to mesenchymal transition and migration. Loss of Paladin delays the expression of transcription factors Snail2 and Sox10 in premigratory neural crest cells, but does not affect accumulation of FoxD3, Cad6B or RhoB, indicating that Paladin differentially modulates the expression of genes previously thought to be coregulated within the neural crest gene regulatory network. Both gain and loss of Paladin function result in disrupted neural crest migration, reinforcing the importance of precisely regulated phosphorylation for neural crest migration. Mutation of critical, catalytic cysteine residues within Paladin’s predicted phosphatase active site motifs did not abolish the function of Paladin in the neural crest. Collectively, these data indicate that Paladin is an antiphosphatase that modulates the activity of specific neural crest regulatory factors during neural crest development. Our work identifies a novel regulator of phosphorylation status that provides an additional layer of regulation in the neural crest.
Keywords: neural crest, phosphorylation, migration
Introduction
Neural crest cells are a multipotent, migratory cell type that is unique to vertebrate embryos. Neural crest precursors originate as epithelial cells within the dorsal neural tube, then undergo an epithelial to mesenchymal transition and delaminate from the neural tube. Once motile, neural crest cells migrate to the periphery and eventually differentiate into a variety of cell types, including neurons and glia of the peripheral nervous system, craniofacial bone and melanocytes (LeDouarin and Kalcheim, 1999).
While a gene regulatory network (GRN) of transcription factors that regulate neural crest specification has been established (Betancur et al., 2010; Sauka-Spengler and Bronner-Fraser, 2008), cells of the dorsal neural tube are not uniquely fated to give rise to neural crest cells (Collazo et al., 1993; Selleck and Bronner-Fraser, 1995). In particular, expression of neural crest transcription factors does not guarantee that a cell will migrate (Chizhikov and Millen, 2004b; Duband, 2006; Linker et al., 2000). For example, dorsal neural tube cells co-express Snail2, a neural crest marker, and Lmx1a, a roof plate marker, and become either cell type (Chizhikov and Millen, 2004a). Because expression of transcription factors that specify neural crest cell identity does not ensure neural crest development, we postulate that modulation of protein activity provides an additional layer of control during neural crest development.
Phosphorylation is a well-established mechanism for regulating protein activity. A phosphate group is added to a serine, threonine or tyrosine residue within the targeted protein by a kinase, and the process is reversible through the activity of a phosphatase (Sefton and Shenolikar, 2001). Phosphorylation status is also regulated by a third class of proteins, the antiphosphatases, sometimes referred to as pseudophosphatases. Antiphosphatases contain incomplete or divergent inactive phosphatase domains and bind to phosphorylated residues on target proteins to protect them from dephosphorylation (Cui et al., 1998; Gingras et al., 2009; Wishart and Dixon, 1998). While kinase and phosphatase inhibitor treatments show that phosphorylation in general regulates neural crest adhesion and migration (Brennan et al., 1999; Minichiello et al., 1999; Monier-Gavelle and Duband, 1995; Newgreen and Minichiello, 1995), only recently have specific regulators of phosphorylation status been identified. A growing list of kinases active in neural crest cells (for example Banerjee et al., 2011; Jin et al., 2012; Murphy et al., 2011; Ossipova and Sokol, 2011) reinforces the idea that phosphorylation is important for neural crest development. Meanwhile, mutation of the phosphatase Shp2 was recently shown to cause abnormalities in neural crest derivatives (Nakamura et al., 2009; Stewart et al., 2010) while inhibition of PP2A phosphatase activity results in increased cranial neural crest migration (Latta and Golding, 2012). While target protein phosphorylation status can determine biological activity, for example phosphorylation of the neural crest transcription factor Twist affects binding partner choice and transcriptional outcome (Firulli and Conway, 2008), a neural crest antiphosphatase that can sustain target phosphorylation has never been described.
Paladin (Pald) is a protein that contains two phosphotyrosine phosphatase consensus active site motifs but lacks other domains characteristic of phosphatases. Found only in vertebrates, Pald was first identified in a large-scale cDNA sequencing project (Nagase et al., 1999) and was subsequently identified in a screen for genes upregulated as a consequence of neural crest induction (Adams et al., 2008; Gammill and Bronner-Fraser, 2002). Recent work has shown that Pald inhibits insulin-stimulated phosphorylation of AKT; however, in vitro assays have failed to detect phosphatase activity for Pald (Huang et al., 2009). As no in vivo functional assays have assessed the importance of Pald’s phosphatase domains, it is unclear whether to categorize Pald as a phosphatase or antiphosphatase (Alonso et al., 2004).
We have examined the role that Pald plays in chick neural crest development. Pald is expressed in premigratory and migratory neural crest cells, consistent with a role in the neural crest. Loss of Pald results in reduced expression of some neural crest transcription factors, but not others, indicating Pald differentially regulates pathways in the neural crest GRN. Later in development, both gain and loss of Pald result in disrupted neural crest migration, indicating Pald levels are strictly regulated during neural crest migration. Additionally, mutation of critical cysteines in Pald’s putative phosphatase active site motifs does not abolish Pald function. As these mutations should eliminate Pald activity if it is a phosphatase, and because Pald does not exhibit phosphatase activity in vitro (Huang et al., 2009), we propose that Pald is an antiphosphatase that regulates the phosphorylation and activity of target proteins during neural crest development.
Materials and Methods
Chicken Embryo Culture
Fertile chicken embryos were obtained from local sources and incubated in a humidified incubator at 37–38°C (G. Q. F. Manufacturing; Savannah, GA). Embryos were staged according to Hamburger and Hamilton (Hamburger and Hamilton, 1951) or by counting somite pairs.
Morpholinos and DNA Constructs
FITC- or lissamine-tagged morpholinos (MOs) were obtained from GeneTools, LLC (Philmouth, OR) with the following sequences: translation blocking cPald MO TGCACGCTCTCAAAGGCGGAGGCTG; 5 bp mismatch control mmcPald MO TGgACcCTCTgAAAGcCGGAcGCTG; splice blocking spcPald MO CAACAAGCAGCAATGCCAACCTGCA; 5 bp mismatch control mmspcPald MO CAAgAAcCAcCAATcCCAACCTcCA. For overexpression, full-length Pald was subcloned using standard methods into pMES (Swartz et al., 2001) with GFP replaced with mcherry (pMESmcherry) using PCR driven overlap extension (Heckman and Pease, 2007). Phosphatase active site mutations were made using a QuikChange II Site-Directed Mutagenesis Kit (Agilent; Santa Clara, CA).
Electroporation
Embryos at HH stage 4–5 were electroporated with morpholino and DNA as previously described (Gammill and Krull, 2011). Morpholinos were used at 1.0 mM with or without DNA. DNA for overexpression was electroporated at 1–5 mg/ml as indicated along with 1.0 mM standard control MO. After electroporation, embryos were reincubated to the appropriate stages, fixed with 4% paraformaldehyde at RT for one hour, and imaged using fluorescent microscopy. Embryos were screened for appropriately targeted, bright MO incorporation prior to analysis because the MO fluorescent tag often does not persist after in situ hybridization. Embryos were then dehydrated into methanol and stored at −20° C for in situ hybridization or embedded immediately for sectioning.
In vitro translation
In vitro translation was performed using the FluoroTect GreenLys in vitro Translation labeling system and Rabbit Reticulocyte Lysate (Promega; Madison, WI) according to the manufacturer’s instructions. Briefly, full length chick Paladin, including 44 bp of 5’ UTR, was cloned from a chick embryo expression library (Gammill and Bronner-Fraser, 2002) and subcloned into CS2+myc (Turner and Weintraub, 1994). A construct containing a 5 base pair mismatch in the morpholino target sequence (mmcPald) was generated by PCR using primers containing the mismatch target site. mRNA for Pald or mmcPald was in vitro transcribed and used as template for in vitro translation. 0.5 mM or 1.0 mM cPald MO was added to selected samples. Reactions were run out on a 10% Tris- HCl SDS PAGE gel, and results visualized on an FLA-5000 imager (FujiFilm; Stamford, CT). Band intensity was measured and compared using Image Gauge software (FujiFilm).
RT-PCR
Electroporated embryos were incubated to 6–8 somites. MO targeted hemi-heads were dissected from these embryos and RNA was isolated using Trizol (Life Technologies; Grand Island, NY) and reverse transcribed using SuperScript III (Life Technologies; Grand Island, NY) according to manufacturer’s instructions. cDNA was then used as template for PCR reactions using Choice Taq (Denville Scientific; Metuchen, NJ). No reverse transcriptase samples were used as negative controls. Primer sequences were as follows: Pald exon2 F-CCTGAGCATCCACTCCTTTC; Pald exon4 R-GCTGACCCATACCAAACACC; GAPDH F-GGACACTTCAAGGGCACTGT; GAPDH R-TCTCCATGGTGGTGAAGACA.
In situ hybridization
Digoxigenin-labeled RNA probes were translated using the following constructs as templates: Snail2 (Gammill and Bronner-Fraser, 2002); Sox10 (Cheng et al., 2000); FoxD3 (Kos et al., 2001); Cad6B (Gammill and Bronner-Fraser, 2002); RhoB (gift of Lisa Taneyhill); Msx1 (clone ChEST900p21; Boardman et al., 2002); and Sox2 (Uwanogho et al., 1995). In situ hybridization was performed as described previously (Wilkinson, 1992). Embryos were viewed in whole mount with a Zeiss Discovery V8 stereoscope and selected embryos were embedded and sectioned with a Leica CM1900 cryostat at 10–20 µm and imaged on a Zeiss AxioImager A1.
Phenotype Evaluation
Phenotype was evaluated at cranial axial levels well targeted with fluorescent MO signal. Effects on premigratory gene expression in whole mount were analyzed by comparing the relative intensity of in situ hybridization colorimetric signal on the targeted and untargeted sides of the embryo using the criteria shown in Fig. S1. In section images, the cross-sectional area of the expression domain on targeted and untargeted sides of the neural tube were compared in 5 sections of selected embryos using Image J (NIH). Effects on migration were scored by comparing the distance migrated on targeted and untargeted sides using the criteria shown in Fig. S1. Statistical analysis of phenotypic effects was performed using Fisher’s exact test in R (R Development Core Team, 2011).
Immunostaining and Cell Counting
Embryos were infiltrated with 5 and 15% sucrose, embedded in gelatin, frozen in liquid nitrogen and sectioned with a Leica CM1900 cryostat at 10-20 µm. Sections were stained with anti-HNK-1 (ATCC; Manassas, VA), anti-cleaved caspase3 (Cell Signaling; Danvers, MA) or anti-phospho-Histone H3 (Millipore; Billerica, MA) followed by the appropriate secondary antibody (Cy2 anti-Mouse IgM or Cy5 anti-Rabbit, Jackson; West Grove, PA) as indicated. Slides were mounted with PermaFluor (Thermo Fisher Scientific; Waltham, MA) containing DAPI and viewed on either a Zeiss AxioImager A1 or a Zeiss CARV spinning disc confocal microscope. Images were adjusted in Photoshop (Adobe). Nuclei and dividing or dying cells were counted in the dorsal quarter of the cranial neural tube on both the targeted and untargeted sides of the embryo for at least 5 sections per embryo and statistics were performed in Excel (Microsoft).
Results
Chick Paladin is expressed in premigratory and migratory neural crest cells
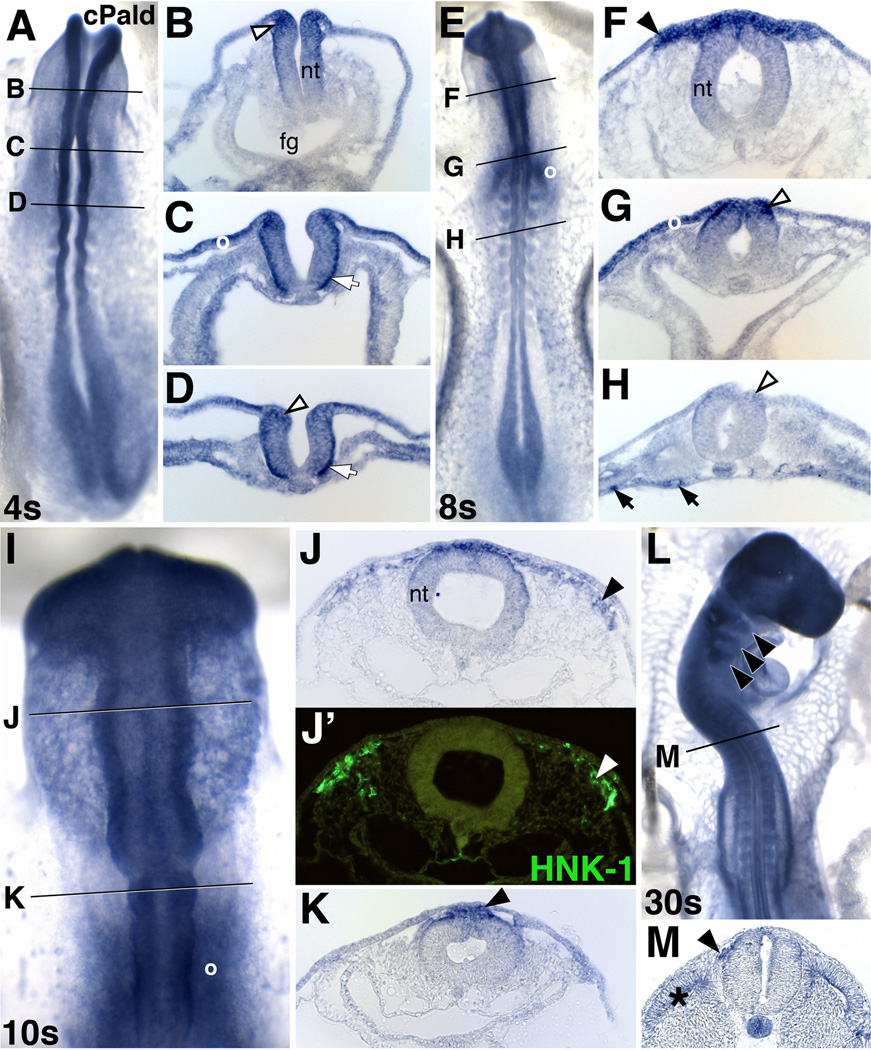
In order to better understand early neural crest development, previous work identified 176 genes that are upregulated as neural crest cells are preparing to migrate (Adams et al., 2008; Gammill and Bronner-Fraser, 2002). One of these genes, Paladin (incorrectly identified as Palladin in Gammill and Bronner-Fraser, 2002), is annotated as a putative phosphatase and its expression profile remains unreported. Chick Pald was expressed in premigratory neural crest cells at 4 somites in cranial (Fig.1B, open arrowhead) and trunk domains (Fig.1D, open arrowhead) as well as along the basal surface of the neural tube (Fig.1C,D, white arrows). This expression persisted in cranial and trunk premigratory neural crest precursors at 8 somites (Fig.1G,H, arrowheads), indicating that Pald is expressed throughout premigratory neural crest development.
Figure 1. Paladin is expressed in premigratory and migratory neural crest cells.
(A–D) Cranial (B, C) and trunk (D) premigratory neural crest cells (open arrowhead) express Pald mRNA at 4 somites (4s). (E–M) At 8s (E–H), 10s (I–K), and 30s (L, M), Pald expression persists in hindbrain (G) and trunk premigratory neural crest cells (H, open arrowheads) and is maintained during delamination and migration of midbrain (F,I,J) hindbrain (K,L) and trunk neural crest cells (M; arrowheads). Pald expressing cells (J, arrowhead) are positive by immunofluorescence for the migratory neural crest marker HNK-1 (J’, arrowhead), confirming their identification as neural crest cells. Pald is also expressed in the basal surface of the neural tube (white arrows), blood vessels (black arrows), and dermomyotome (asterisk). A, E, I, dorsal view; L, dorsolateral view; B–D, F–H, J, K, M, transverse section at the level indicated in the accompanying whole mount. I, higher magnification view. Fg, foregut; nt, neural tube; o, otic placode.
Both cranial and trunk neural crest cells maintained expression of Pald as they delaminated from the neural tube (Fig.1K,M, closed arrowheads) and migrated away (Fig.1F, J). Pald mRNA expressing cells outside of the neural tube (Fig.1J, arrowhead) were confirmed as neural crest cells by costaining with HNK-1 (Fig.1J’, arrowhead). Expression of Pald was still apparent in migrating neural crest cells and their derivatives within the branchial arches at 30 somites (Fig.1L, arrowheads). Additionally, Pald was expressed in otic placode and vesicle cells (Fig.1C,E,G,I, o) as well as nascent blood vessels (Fig.1H, black arrows). Expression of Pald transcripts in neural crest cells at multiple stages puts Pald in the right time and place to regulate neural crest development.
Paladin is required for premigratory expression of Snail2 and Sox10 but not other neural crest genes
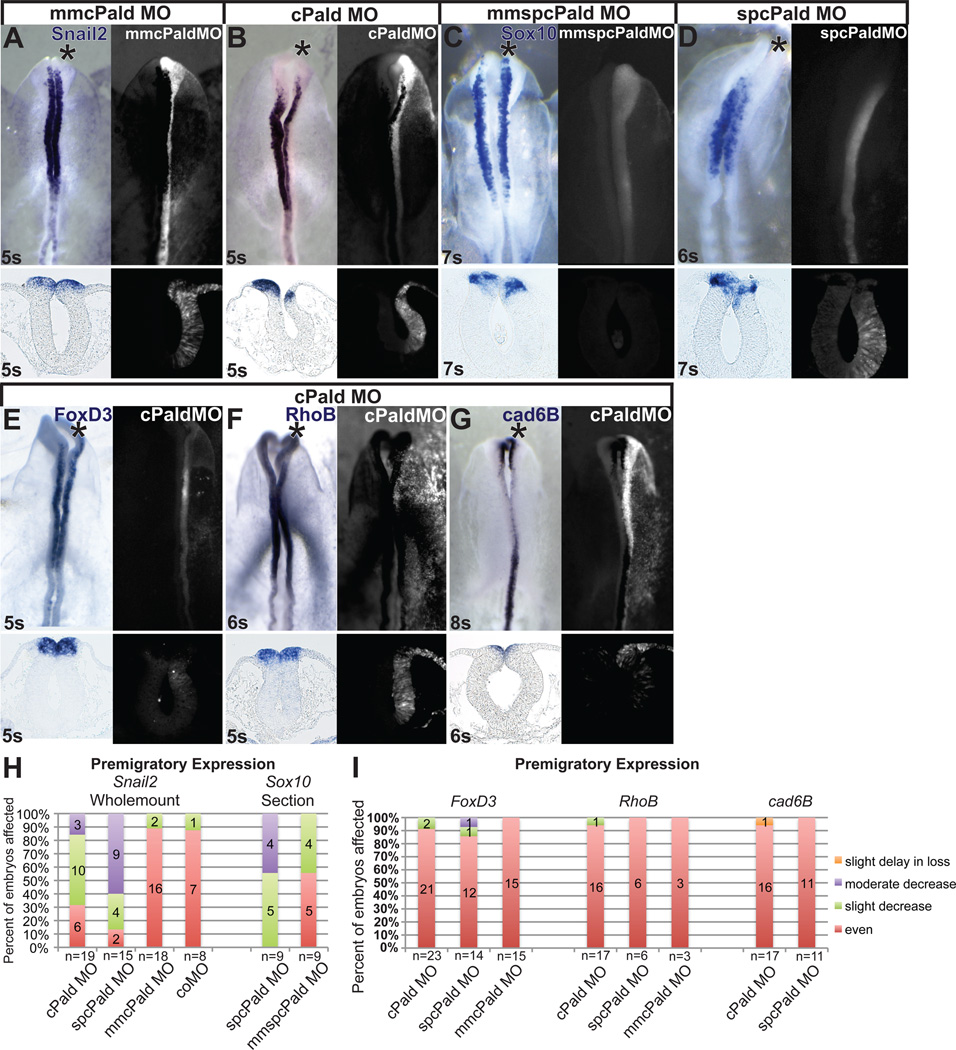
We designed a translation blocking morpholino against chick Pald (cPald MO) to examine the requirement for Pald during neural crest development. To test the ability of this morpholino to inhibit translation of PALD protein, we performed in vitro translation reactions. Inclusion of cPald MO in the translation reaction resulted in a dose-dependant decrease in the amount of PALD produced (Fig.S2A). cPald MO was ineffective when 5 base pairs of the morpholino target site were changed, indicating that cPald MO is specific. We also utilized a second morpholino designed against the splice donor site of intron 3 (spcPald MO). This morpholino altered Pald mRNA splicing when assessed using RT-PCR (Fig.S2B).
To determine whether Pald regulates neural crest development in chicken embryos, we electroporated the Pald MOs unilaterally into neural crest precursors at mid-gastrula, during neural crest specification (Basch et al., 2006; Gammill and Krull, 2011). Electroporated embryos were then analyzed for expression of Snail2 by in situ hybridization in whole mount and transverse sections when neural crest cells were still in the neural tube. Electroporation of a morpholino with a 5 base pair mismatch (mmcPald MO) did not affect premigratory expression of Snail2 on the targeted side of embryos (Figure 2A, asterisk; Fig.2H, n=18). However, knockdown of Pald using cPald MO resulted in a consistent decrease in the expression of Snail2 on the targeted side relative to the untargeted side of 68% of embryos analyzed (Fig.2B, asterisk; Fig.2H, p=1.1×10−3, n=19). A similar, but slightly stronger effect was observed in 87% of embryos electroporated with spcPald MO (Fig.2H, p=1.1×10−3, n=15). Thus, Pald is required for premigratory Snail2 expression in the neural crest.
Figure 2. Pald is required for premigratory expression of Snail2 and Sox10.
Embryos were unilaterally electroporated at HH stage 4–5 with mmcPald MO (A), cPald MO (B, E, F, G), mmspcPald MO (C) or spcPald MO (D). After reincubation to 4–8 somites (s), in situ hybridization was performed to assess expression of Snail2 (A, B), Sox10 (C, D), FoxD3 (E), RhoB (F), or Cad6B (G). (A–D) Representative examples of Snail2 and Sox10 expression inhibition following Pald knock down. Snail2 expression is unaffected in embryos electroporated with mmcPald MO (A) but reduced on the side of the neural tube targeted with cPald MO (B). Likewise, Sox10 expression is unaffected in embryos electroporated with mmspcPaldMO (C) but reduced on the side of the neural tube targeted with spcPald MO (D). (E–G) Representative examples of neural crest regulators unaffected by Pald knock down. Expression of the transcription factor FoxD3 (E) and neural crest effector genes RhoB (F) and Cad6B (G) is equivalent on cPaldMO targeted and untargeted sides of the neural tube. (H) Stacked bar graphs depicting the frequency and severity of Snail2 and Sox10 expression defects in embryos electroporated with cPald MO, spcPald MO, mmcPald MO, mmspcPald MO, or control (co) MO when assayed in whole mount (Snail2) or sections (Sox10). (I) Stacked bar graphs reveal the absence of FoxD3, Cad6B, and RhoB expression defects in nearly all embryos assayed. In A-G, top row: dorsal views of in situ hybridization in left panel, fluorescent MO targeting in right panel; bottom row: transverse sections at the level of the midbrain. Note that the section shown is not necessarily from the wholemount embryo pictured. Asterisk, targeted side of the embryo.
To assess whether Pald regulates neural crest cell specification generally or Snail2 expression specifically, we examined premigratory expression of other neural crest cell markers. While effects on Sox10 expression were subtle in whole mounts (Fig.2C, D top), defects were apparent in sectioned embryos (Fig.2C, D bottom). 44% of mmspcPald MO electroporated embryos displayed slightly decreased Sox10 expression (Fig.2C,H, n=9), indicating that the morpholino is at the upper limit of its effective- and specific-dose range (Moulton and Yan, 2008). Nevertheless, Sox10 expression was slightly to moderately decreased in all embryos electroporated with spcPald MO, with 44% exhibiting a moderate reduction (Fig.2D,H, p=8.5×10−3, n=9). While both Snail2 and Sox10 are transcription factors regulating neural crest cell fate, a third transcription factor, FoxD3, was unaffected upon Pald knockdown by either cPald (n=23) or spcPald (n=14) MO (Fig.2E,I). Furthermore, two neural crest cell effector genes, RhoB (Fig.2F,I) and the epithelial cadherin Cad6B (Fig.2G,I), were normally expressed as well following Pald MO electroporation. Importantly, Cad6B was downregulated on time in Pald-deficient embryos (Fig.2G). Notably, neural and dorsal neural tube makers were also expressed normally in Pald knock down embryos, revealing intact patterning of neural tube fates (Fig. S3). As in the case of Snail2 expression, both MOs elicited similar phenotypes for all neural crest markers analyzed (Fig.2H,I). These results indicate that Pald is required for expression of both Snail2 and Sox10, but not for neural crest specification generally.
Initial reductions in Snail2 and Sox10 transcripts could have been caused by increased rates of cell death or decreased cell proliferation in neural crest cells. To examine this possibility, we electroporated embryos with mmspcPald MO or spcPald MO and assessed cell proliferation and death markers in embryo sections at premigratory neural crest cell stages. Quantification of dying and proliferating cells revealed that electroporation of neither mmspcPald MO nor spcPald MO altered rates of either cell death or proliferation in the dorsal neural tube (Fig.S4). Similar results were observed with cPald MO (data not shown). Thus Paladin does not appear to regulate cell proliferation or death but expression of particular neural crest genes specifically.
Neural crest migration is disrupted upon Paladin knockdown
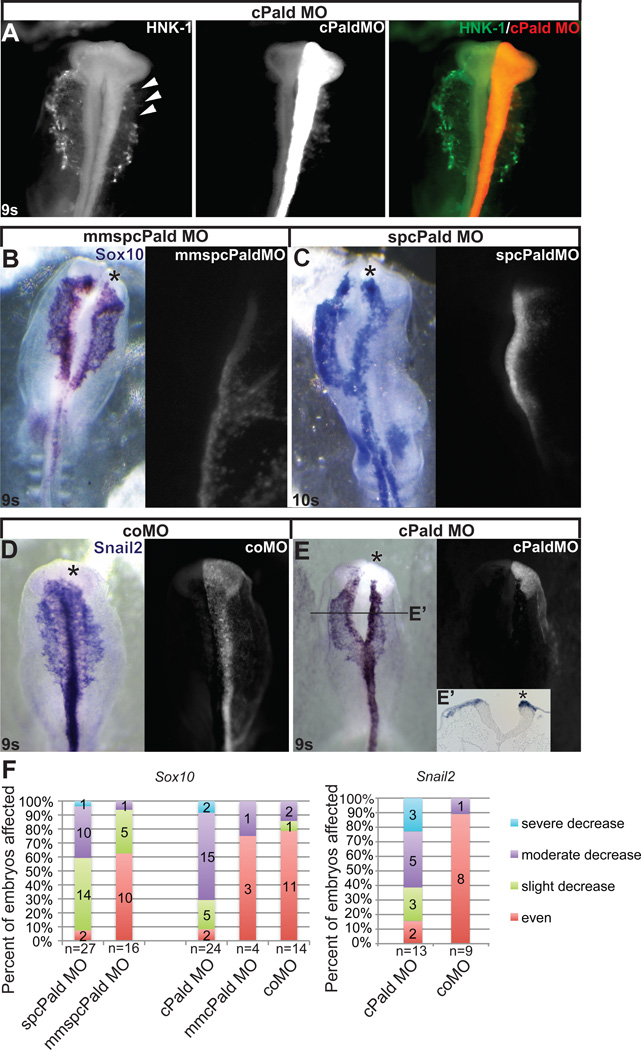
Because Pald was expressed throughout neural crest development (Fig.1), we wanted to determine if Paladin was also required for neural crest migration. We again knocked down Pald using unilateral morpholino electroporation at mid-gastrula, but instead let embryos develop to 8 to 11 somites. When cranial neural crest cells were visualized in these embryos by wholemount HNK-1 immunofluorescence, we noted that migration of cPaldMO-electroporated neural crest cells was curtailed (Fig.3A, arrowheads). To characterize this disruption, we performed Sox10 in situ hybridization, as Sox10 is a robust marker of neural crest cells throughout their emigration and active migration (Cheng et al., 2000), and because moderate levels of Sox10 expression are retained in Pald knock down embryos (Fig.2D). While a minority of mmspcPald, mmcPaldMO, and standard control MO electroporated embryos exhibited slight migratory defects on the targeted side (Fig.3B,F), electroporation of spcPald MO or cPald MO resulted in a slight to severe decrease in the distance migrated on the targeted side compared to the untargeted side in over 90% of embryos (Fig.3C,F, spcPald MO p=4.8×10−4, cPald MO p=6.3×10−5). Together, these results indicate that Pald is required for proper neural crest migration.
Figure 3. Loss of Pald impedes neural crest migration.
Embryos were unilaterally electroporated at HH stage 4–5 with cPaldMO, mmspcPald MO, spcPald MO, or standard control MO (coMO) and reincubated to 8-11 somites (s). (A) Neural crest migration defects visualized by HNK-1 immunofluorescence following Pald knock down. cPald MO-targeted neural crest cells (red; arrowheads) do not migrate as far as HNK-1 positive neural crest cells (green) on the untargeted side. (B, C) A representative example of neural crest migration defects visualized by Sox10 in situ hybridization following Pald knockdown. While mmspcPald MO does not affect neural crest migration (A), spcPald MO-targeted neural crest cells’ migration distance is reduced compared to the untargeted side (B). (D, E) A representative example of neural crest migration defects visualized by Snail2 in situ hybridization following Pald knockdown. While coMO does not affect neural crest migration (D), cPald MO-targeted neural crest cells’ migration distance is reduced compared to the untargeted side (E), transverse section at the level of the line indicated in E’. (B–E) Dorsal views of in situ hybridization in left panel, fluorescent MO targeting in right panel. Asterisk, targeted side of the embryo. (F) Stacked bar graphs depicting the severity and frequency of migration impairment in embryos electroporated with cPald MO, spcPald MO, mmcPald MO, mmspcPald MO, or coMO and visualized by Sox10 (left) or Snail2 (right) in situ hybridization. Migration of neural crest cells is mildly disrupted by electroporation of mmspcPald MO, indicating that 1.0mM spcPald MO is the upper limit of its effective and specific dose.
As Sox10 is expressed at relatively normal levels in Pald knock down embryos at migratory stages (compare Figs.2D and 3C), we determined whether Snail2 expression would also recover. Interestingly, when cPaldMO electroporated embryos were processed by Snail2 in situ hybridization at early migratory stages, elevated Snail2 expression was apparent in the targeted neural fold (Fig.3E,E’, asterisk). Moreover, cPaldMO-targeted migratory neural crest cells expressed Snail2 at levels equivalent to migratory neural crest cells on the untargeted side (Fig.3E), and once again revealed that Pald is required for neural crest migration (Fig.3F, p=4.8×10−3). As neural crest migration progressed, Pald-deficient neural crest cells appeared to “catch up” to the untargeted side regardless of the marker used (data not shown). These data suggest that absence of Snail2 is not preventing neural crest migration, and that Pald is necessary to coordinate early migration rather than active migration into the periphery.
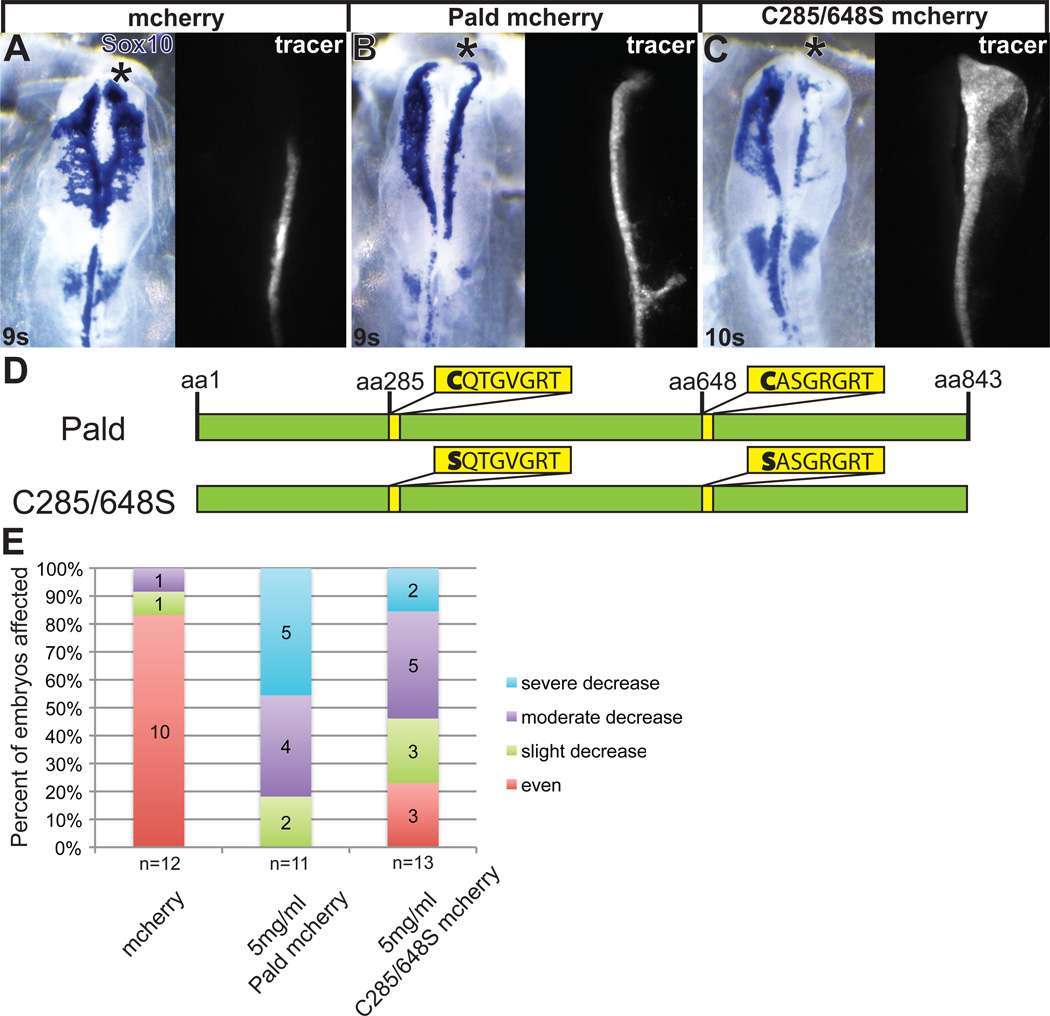
Excess Paladin impedes neural crest migration
Because loss of Pald disrupted neural crest migration, we next wanted to examine the effects of Pald excess. To achieve this, we cloned chick Pald into the bicistronic expression construct pMES-mcherry. We electroporated pMES-Pald-mcherry DNA unilaterally into midgastrula stage embryos and assessed neural crest cell migration. FITC-tagged control morpholino was included in these electroporations because we find coelectroporation of morpholino along with DNA results in a higher efficiency of DNA transfection and FITC fluorescence is brighter than mcherry, allowing for more accurate detection of properly transfected embryos. Surprisingly, overexpression of Pald-mcherry at 5 mg/ml yielded a decrease in neural crest migration distance (Fig.4B,E, p=5.6×10−5), similar to Pald knockdown, whereas electroporation of pMES-mcherry alone did not affect migration (Fig.4A,E). Importantly, the incidence and severity of Pald overexpression defects was dose dependent (data not shown). Pald overexpression outside of the neural crest domain had no apparent effect, suggesting Pald can affect active neural crest signaling pathways, but cannot trigger their activation. These results indicate that both loss and gain of Pald function disrupt neural crest migration, and Pald modulates neural crest development but does not initiate it.
Figure 4. Pald overexpression disrupts neural crest migration.
Embryos were unilaterally electroporated at HH stage 4–5 with pMES-mcherry expression constructs mixed with 1.0 mM FITC-labeled standard control MO (coMO). The MO served as a lineage tracer and a carrier for the DNA. At 8-11 somites (s), embryos were harvested and neural crest cells visualized by in situ hybridization for Sox10. (A–C) Representative embryos showing inhibition of migration by Pald overexpression. Whereas neural crest migration is unaffected by pMES-mcherry electroporation (A), electroporation of Paldmcherry (B) or C285/648S phosphatase domain mutant Pald (C) disrupt neural crest migration on the targeted side. Dorsal views of in situ hybridization in left panel, fluorescent MO in right panel. Asterisk, targeted side of the embryo. (D) Schematic of Pald sequence. Phosphotyrosine phosphatase sites in yellow, active site cysteine is mutated in both domains in C285/648S. (E) Stacked bar graphs depicting the severity and frequency of migration defects in embryos overexpressing either wildtype or phosphatase mutant Pald.
Mutation of Paladin’s protein tyrosine phosphatase domains does not abolish Paladin function
While Paladin has been preliminarily classified as a protein tyrosine phosphatase (PTP), little has been done to characterize its biological function, likely because Pald does not exhibit phosphatase activity in vitro (Huang et al., 2009). To examine the activity of Pald, we assessed whether Pald’s PTP motifs were essential for proper neural crest migration. Pald contains two core PTP active site signature motifs, CX5R (Fig.4D, Tonks, 2006), but no other phosphatase accessory domains. Because the conserved cysteine is critical for catalytic activity and phosphatase function as it transiently accepts the phosphate during catalysis (Guan and Dixon, 1991), we mutated both of Pald’s putative active site cysteines (C285 and C648) to serines (Fig.4D). With other phosphatases, this strategy abolishes phosphatase activity, resulting in proteins that still recognize their substrates but no longer dephosphorylate and release them; cysteine mutants are thus called substrate-trapping mutations (Milarski et al., 1993). We therefore reasoned that if Pald was a phosphatase, these mutations (C285/648S) would eliminate Pald function, whereas if Pald was an antiphosphatase, Pald activity would be retained. Like wildtype Pald, electroporation of C285/648S disrupted neural crest migration (Fig.4C,E, p=0.016). The number of embryos affected by C285/648S overexpression was not significantly different compared to wildtype Pald overexpression (Fig.4E, p=0.31), however, the phenotypic severity was decreased. These data suggest that the conserved cysteines within the Pald PTP catalytic domains are required for complete Pald activity in regulating neural crest migration. However, because the migration disrupting activity of Pald was still intact, the active sites are unlikely to confer phosphatase catalytic activity.
Paladin active site mutants rescue paladin loss of function in neural crest cells
To fully assess the biological activity of Pald during neural crest development, we wanted to see if the C285/648S mutant protein could substitute for endogenous Pald. First, we needed to establish the ability of wildtype Pald to rescue spcPald MO electroporated embryos. We coelectroporated spcPald MO with varying amounts of Pald-mcherry DNA that is not targeted by either MO. While some rescue was observed at 3 and 4 mg/ml Paldmcherry (data not shown), optimal rescue was observed at 5 mg/ml and further experiments were carried out at this concentration (Fig.5B,D). Knockdown of Pald using spcPald MO once again disrupted neural crest migration by decreasing the distance migrated in almost 90% of embryos (Fig.5A,D), whereas coelectroporation of spcPald MO with Pald-mcherry resulted in normal migration in about 50% of embryos (Fig.5B,D, p=0.039). These data indicate that disrupted neural crest migration following spcPald MO electroporation is specifically due to loss of Pald function, as replacement with exogenously supplied Pald rescues this defect.
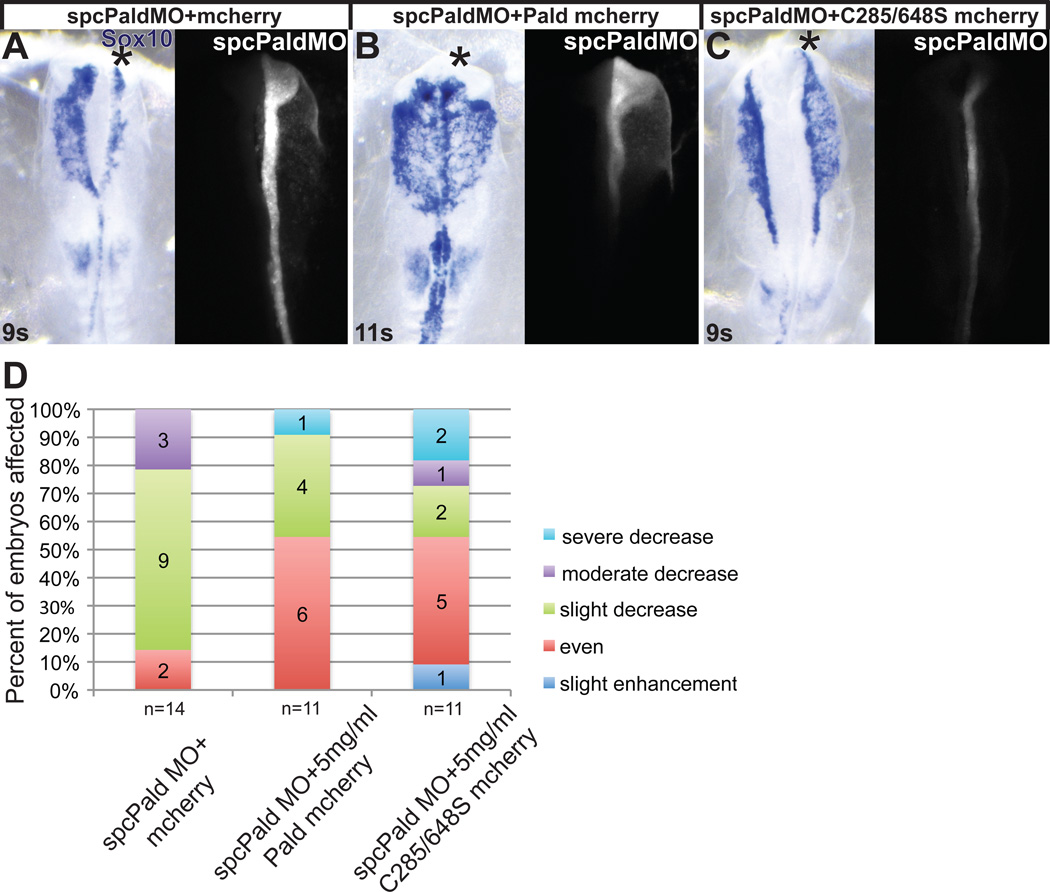
Figure 5. Phosphatase domain catalytic cysteine residues are not essential for Paladin activity in neural crest cells.
Embryos were unilaterally electroporated at HH stage 4–5 with spcPald MO mixed with pMES-mcherry vector DNA, pMES-Pald-mcherry or pMES-C285/648S-mcherry and reincubated. At 8–11 somites (s) embryos were harvested and neural crest cells visualized by in situ hybridization for Sox10. (A–C) Representative examples of spcPald MO rescue. While neural crest migration is disrupted upon unilateral coelectroporation of empty pMES-mcherry (A), neural crest cells migrate normally following coelectroporation of 5 mg/ml pMES-Pald-mcherry (B) or 5mg/ml C285/648S-mcherry (C). Dorsal views of in situ hybridization in left panel, fluorescent MO in right panel. Asterisk, targeted side of the embryo. (D) Stacked bar graphs depicting the severity and frequency of migration defects in embryos coelectroporated with spcPald MO and wildtype or phosphatase mutant Pald.
Next, to assess of the importance of the putative active site cysteines, we examined the ability of C285/648S to rescue spcPald MO knockdown. While overexpression of C285/648S suggested that these cysteine residues, which are critical for function of a phosphatase active site, are important for Pald function during neural crest migration, we do not know the mechanism by which overexpression prevents neural crest migration. Knocking down endogenous Pald with spcPald MO and rescuing with C285/648S provides a more direct test of the importance of the putative active site cysteines. Following coelectroporation of spcPald MO with C285/648S, about 50% of embryos exhibited normal neural crest migration (Fig.5C,D, p=0.031). Although the proportion of embryos rescued by wildtype or C285/648S coelectroporation was similar and not significantly different (Fig.5D; p=0.76), those embryos that were affected exhibited more severe phenotypes with C285/648S rescue than those rescued with wildtype Pald. These results indicate that while putative active site cysteine residues are not essential for Pald function in the neural crest, these residues are necessary for maximal Pald activity during neural crest migration.
Discussion
The function of Pald, which was presumed to be a phosphatase, was unknown during embryogenesis. Here, we show that Pald is expressed in premigratory and migratory neural crest cells, and that this expression is maintained in later neural crest derivatives, suggesting that Pald plays a role throughout neural crest development. Pald knockdown demonstrates that Pald affects premigratory Snail2 and Sox10 expression but not neural crest specification generally. Additionally, loss and gain of function analysis reveals that Pald regulates early neural crest migration. Finally, our mutational analysis reveals that protein tyrosine phosphatase active site cysteine residues are not essential for Pald activity, indicating that Pald is an antiphosphatase that binds and protects phosphorylated residues during neural crest migration.
Paladin differentially affects members of the neural crest gene regulatory network
Genes involved in the early stages of neural crest development have been characterized in multiple species and their regulatory interactions organized into a gene regulatory network (Betancur et al., 2010). The neural crest specification transcription factors Snail2, Sox10, and FoxD3 are expressed in premigratory neural crest cells and downregulated at various points once migration commences. Two of these transcription factors, Snail2 and FoxD3, were found to be activated by similar sets of genes, namely Msx1 (Tribulo et al., 2003), Pax3/7 (Sato et al., 2005), and Hairy2 (Glavic et al., 2004; Wettstein et al., 1997). Interestingly, loss of Pald in premigratory neural crest cells results in differential effects on these genes with a disruption in Snail2 expression (Fig.2B,H) and no effect on FoxD3 (Fig.2E,I), indicating that these transcription factors have differential regulation that has not been appreciated by neural crest gene regulatory analysis to date. The third transcription factor, Sox10, is expressed at a slightly later stage in development than Snail2 and FoxD3 and its expression in premigratory neural crest cells is also disrupted by loss of Pald, though to a lesser extent than Snail2 (Fig.2D,H). These data indicate that Pald selectively impacts neural crest transcription factor expression.
One potential explanation for this differential effect on neural crest transcription factors is that Pald directly regulates Snail2 expression. Interestingly, SNAIL2 is a highly unstable protein in neural crest cells (Vernon and LaBonne, 2006) and in cancer cells, Snail stability is influenced by phosphorylation (Zhou et al., 2004). Furthermore, Snail positively autoregulates its own expression (Fuse et al., 1996). Nevertheless, we were unable to detect an interaction between PALD and SNAIL2 using directed yeast-two hybrid (data not shown). This implies that Pald does not directly regulate Snail2 expression by modulating SNAIL2’s phosphorylation status, and instead that Pald modulates a protein that regulates Snail2 but not FoxD3 expression. Efforts to identify such a target of Pald are currently underway.
Examination of neural crest effector genes upon Pald knockdown also revealed novel insights into previously characterized neural crest gene regulatory interactions. Cellular effectors of neural crest development act subsequent to neural crest transcription factor expression, promoting delamination and migration. RhoB activity is required for delamination from the neural tube (Liu and Jessell, 1998) and its expression was unaffected by Pald knockdown (Fig.2F,I). Cad6b is an epithelial cadherin whose expression is directly downregulated in cranial neural crest cells by Snail2 prior to delamination (Taneyhill et al., 2007). Interestingly, though Snail2 expression is disrupted in premigratory neural crest cells, we did not observe a delay in the downregulation of Cad6b transcripts as we would have expected given their direct regulatory relationship (Fig.2G,I). This unexpected result reveals that regulation of Cad6b expression can be uncoupled from Snail2, indicating that Cad6B transcriptional regulation is more complex than currently appreciated and warrants additional characterization. Altogether, our data reveal that Pald is required for expression of specific neural crest regulatory genes, but not neural crest specification in general.
Paladin regulates but is not essential for neural crest migration
Our data indicate that either loss or gain of Pald disrupts neural crest migration. Analysis during early stages of neural crest migration shows a decreased migration distance upon overexpression (Fig.4B) or knockdown (Fig.3A,C,E). However, when these embryos are allowed to develop to later stages, neural crest migration recovers (data not shown), indicating that migration is disrupted but not prevented by loss or gain of Pald. Interpretation of knock down effects is complex: because embryos were electroporated with morpholino at mid-gastrula stages, migration defects (Fig. 3) could be a secondary consequence of earlier effects on neural crest gene expression during specification (Fig. 2). Unfortunately we could not separate the role of Pald in premigratory and migratory neural crest development by timed electroporation as electroporation of Pald MOs later than HH stage 5 had no phenotype (data not shown). Given this is quite an early stage, we presume that Pald is a stable protein that accumulates to sufficient levels for normal development by stage 5. Nevertheless, three observations argue that Pald regulates migration directly. First of all, Snail2 and Sox10 expression in neural crest cells recovers by migratory stages although migration is still impaired (Fig. 3). This suggests specification defects are not preventing migration. Second, migration is more severely affected than specification following Pald knockdown (compare Figs. 2H and 3F), suggesting these outcomes are independent. Third, following Pald knock down Cad6B downregulates on schedule and RhoB is expressed normally, suggesting there is not a general block to neural crest development, but that specific regulators of specification and migration are affected. Importantly, temporally normal expression of Cad6B and RhoB also suggests that Pald targets likely do not include factors that regulate epithelial to mesenchymal transition. Taken together, these results suggest that Pald regulates early migration away from the neural tube, but its function is not critical for emigration and active migration into the periphery.
It is surprising that we observed similar phenotypes upon overexpression and knockdown of Pald. One explanation is that providing excess Pald causes it to be mislocalized and/or abnormally sequestered within a cell, removing it from its normal site of action and effectively causing a Pald loss of function phenotype. Another possibility is that a precise level of Pald is necessary to maintain the correct balance of target protein phosphorylation/dephosphorylation necessary for proper function. In other words, target dephosphorylation (absence of Pald, phosphorylated sites unprotected) and target hyperphosphorylation (excess of Pald, sites always phosphorylated) might equally prevent normal target protein activity and thus elicit the same phenotype.
Classification of Paladin as an antiphosphatase
Determining the mechanism by which Pald regulates neural crest development is difficult without knowing its biological activity. While Pald contains two core catalytic protein tyrosine phosphatase (PTP) active site motifs (CX5R, Fig.4D; Alonso et al., 2004; Fauman and Saper, 1996; Tonks, 2006), it is missing additional domains typically associated with tyrosine phosphatase activity, namely the majority of the 280 amino acid extended catalytic domain (Tonks, 2006). For these reasons Pald is frequently cited as an example of a divergent tyrosine phosphatase family member. Alternatively, antiphosphatases contain catalytically inactive PTP domains with altered critical catalytic residues (Alonso et al., 2004). Although the core phosphatase motifs are intact in Paladin, Pald does not exhibit phosphatase activity in vitro (Huang et al., 2009), increasing our suspicion that Pald may instead be an antiphosphatase.
Mutation of a PTP domain by substituting the active site cysteine with a serine inactivates tyrosine phosphatases (Fauman and Saper, 1996) and leads to sustained interactions between a phosphatase and its targets (Flint et al., 1997). We predicted that if Pald is a phosphatase, the cysteine to serine mutation would abolish Paladin function, whereas if Pald is an antiphosphatase the mutation would affect Pald activity minimally, if at all. Overexpression of either wildtype or C285/648S Pald disrupts neural crest migration without any statistically significant difference in the phenotypic outcome (Fig.4B,C,E). Furthermore, C285/648S is able to rescue Pald knockdown embryos (Fig.5C,D), indicating the mutation does not abolish Pald activity and implying that Pald is an antiphosphatase. Because the overall proportion of embryos affected by Pald overexpression and C285/648S overexpression is similar with a slight reduction in severity of phenotypes observed (Fig.4E), and while C285/648S is equally efficient at rescuing Pald knock down with a slight increase in the severity of those affected (Fig.5D), we conclude that PTP cysteine residues are required for optimal Pald activity.
Because we deduce that Pald is an antiphosphatase, this suggests that regulation of the phosphorylation status of Pald target proteins is required for expression of Snail2 and Sox10 during neural crest development. Due to the ability of antiphosphatases to bind to their phosphorylated targets and protect them from dephosphorylation, and the fact that phosphorylation is a known regulator of protein activity, we postulate that Pald acts a signal enhancer to reinforce the activity of phosphorylated target proteins that are necessary for neural crest specification and early steps in migration. The end result is that, although neural crest cells will eventually migrate without Pald activity, expression of Pald prolongs the intensity and duration of regulatory factor activation to promote/expedite early neural crest development. This hypothesis is supported by the fact that ectopic Pald can only affect neural crest development within the neural crest-forming domain: Pald can only exert its effects when target proteins are already phosphorylated, and Pald itself cannot initiate neural crest formation (Fig.4B). Further work is needed to identify Pald targets to fully explain the mechanism whereby Pald regulates neural crest development.
A Model for Paladin’s Role in Neural Crest Development
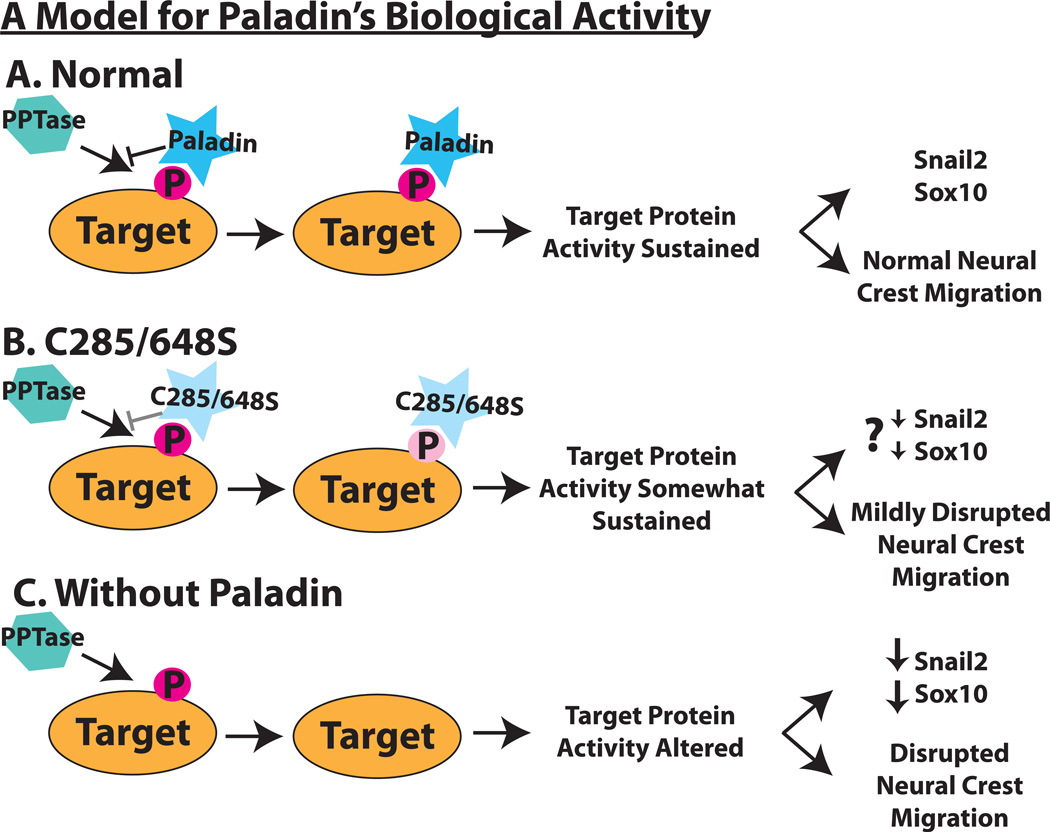
Based on the effects of Pald gain and loss of function, and the consequences of cysteine active site mutations, we propose that Pald is an antiphosphatase. Given that changes in phosphorylation state are essential for proper neural crest migration (Minichiello et al., 1999; Monier-Gavelle and Duband, 1995; Newgreen and Minichiello, 1995), we hypothesize that specific neural crest regulatory proteins are phosphorylated and that this phosphorylation status either positively or negatively affects protein activity during neural crest development depending on the identity of the protein. According to our working model, Pald binds to these phosphorylated proteins, protects their phosphotyrosine residues from dephosphorylation and sustains the activity necessary for proper neural crest gene expression and subsequent migration (Fig.6A). When Pald phosphatase domain active site cysteines are mutated to serines in C285/648S, the mutant protein does not protect its phosphorylated targets as well as wildtype Pald. This results in less stable phosphorylation and altered activity of these targets, leading to mild effects on migration and, we predict, gene expression (Fig.6B). Pald loss of function results in dephosphorylation of Pald targets, which leads to a reduction in both Snail2 and, to a lesser extent, Sox10, as well as disrupted neural crest migration (Fig.6C). Future identification of Pald targets and examination of their phospho-regulation within premigratory and migratory neural crest cells will shed light on an understudied mechanism to regulate neural crest development.
Figure 6. A model for Paladin’s role in neural crest development.
(A) Predicted mechanism of Paladin’s biological activity. A Paladin target protein (orange oval) is tyrosine-phosphorylated (dark pink P) during neural crest development. Paladin (blue star) binds to the phosphotyrosine, protecting it from removal by phosphatases (turquoise hexagon) and sustaining target protein activity. (B) In a C285/648S Paladin mutant, we predict that the loss of the critical cysteine reduces the affinity or specificity of mutant Paladin (light blue star) for the target phosphotyrosine, making protection from dephosphorylation less efficient (light pink P). We predict that expression of C285/648S mutant Paladin results in partial dephosphorylation of target protein(s), which we expect would lead to a slight decrease in Snail2 and Sox10 as well as the observed mild disruption of neural crest migration and less effective rescue. (C) Paladin knockdown results in unprotected target phosphotyrosines, allowing for dephosphorylation by a phosphatase. This alters target protein activity which impedes Snail2 and Sox10 expression and disrupts early phases of neural crest migration.
Supplementary Material
Highlights.
Paladin is an antiphosphatase that is expressed throughout neural crest development.
Paladin modulates expression of neural crest transcription factors Snail-2 and Sox10
Paladin does not regulate neural crest specification generally.
Paladin promotes neural crest migration away from the neural tube.
Acknowledgements
We thank Carol Erickson, Paul Scotting, Andrea Streit, and Lisa Taneyhill for kind gifts of plasmids. Special thanks to Wuming Gong for assistance with statistical analyses. We are grateful to the members of the Gammill Lab and Yasuhiko Kawakami for their input over the course of this project. This work was supported by K22 DE15309 (to LSG), F32 DE019973 (to JRA), and a Minnesota Medical Foundation Research Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Adams MS, Gammill LS, Bronner-Fraser M. Discovery of transcription factors and other candidate regulators of neural crest development. Dev Dyn. 2008;237:1021–1033. doi: 10.1002/dvdy.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Gordon L, Donn TM, Berti C, Moens CB, Burden SJ, Granato M. A novel role for MuSK and non-canonical Wnt signaling during segmental neural crest cell migration. Development (Cambridge, England) 2011;138:3287–3296. doi: 10.1242/dev.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch M, Bronner-Fraser M, Garcia-Castro M. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman PE, Sanz-Ezquerro J, Overton IM, Burt DW, Bosch E, Fong WT, Tickle C, Brown WRA, Wilson SA, Hubbard SJ. A Comprehensive Collection of Chicken cDNAs. Current Biology. 2002;12:1965–1969. doi: 10.1016/s0960-9822(02)01296-4. [DOI] [PubMed] [Google Scholar]
- Brennan H, Smith S, Stoker A. Phosphotyrosine signalling as a regulator of neural crest cell adhesion and motility. Cell motility and the cytoskeleton. 1999;42:101–113. doi: 10.1002/(SICI)1097-0169(1999)42:2<101::AID-CM2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Cheng Y-C, Cheung M, Abu-Elmagd MM, Orme A, Scotting PJ. Chick Sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Developmental Brain Research. 2000;121:233–241. doi: 10.1016/s0165-3806(00)00049-3. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Millen KJ. Control of roof plate formation by Lmx1a in the developing spinal cord. Development (Cambridge, England) 2004a;131:2693–2705. doi: 10.1242/dev.01139. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Millen KJ. Mechanisms of roof plate formation in the vertebrate CNS. Nature reviews. 2004b;5:808–812. doi: 10.1038/nrn1520. [DOI] [PubMed] [Google Scholar]
- Collazo A, Bronner-Fraser M, Fraser SE. Vital dye labelling of Xenopus laevis trunk neural crest reveals multipotency and novel pathways of migration. Development (Cambridge, England) 1993;118:363–376. doi: 10.1242/dev.118.2.363. [DOI] [PubMed] [Google Scholar]
- Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary ML. Association of SET domain and myotubularin-related proteins modulates growth control. Nature genetics. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- Duband JL. Neural crest delamination and migration: integrating regulations of cell interactions, locomotion, survival and fate. Advances in experimental medicine and biology. 2006;589:45–77. doi: 10.1007/978-0-387-46954-6_4. [DOI] [PubMed] [Google Scholar]
- Fauman EB, Saper MA. Structure and function of the protein tyrosine phosphatases. Trends in biochemical sciences. 1996;21:413–417. doi: 10.1016/s0968-0004(96)10059-1. [DOI] [PubMed] [Google Scholar]
- Firulli AB, Conway SJ. Phosphoregulation of Twist1 provides a mechanism of cell fate control. Curr Med Chem. 2008;15:2641–2647. doi: 10.2174/092986708785908987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint AJ, Tiganis T, Barford D, Tonks NK. Development of "substrate-trapping" mutants to identify physiological substrates of protein tyrosine phosphatases. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse N, Hirose S, Hayashi S. Determination of wing cell fate by the escargot and snail genes in Drosophila. Development (Cambridge, England) 1996;122:1059–1067. doi: 10.1242/dev.122.4.1059. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Genomic analysis of neural crest induction. Development (Cambridge, England) 2002;129:5731–5741. doi: 10.1242/dev.00175. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Krull CE. Embryological and genetic manipulation of chick development. Methods in molecular biology (Clifton, N.J. 2011;770:119–137. doi: 10.1007/978-1-61779-210-6_5. [DOI] [PubMed] [Google Scholar]
- Gingras MC, Zhang YL, Kharitidi D, Barr AJ, Knapp S, Tremblay ML, Pause A. HD-PTP is a catalytically inactive tyrosine phosphatase due to a conserved divergence in its phosphatase domain. PLoS One. 2009;4:e5105. doi: 10.1371/journal.pone.0005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavic A, Silva F, Aybar MJ, Bastidas F, Mayor R. Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development (Cambridge, England) 2004;131:347–359. doi: 10.1242/dev.00945. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. The Journal of biological chemistry. 1991;266:17026–17030. [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. Journal of Morphology. 1951;88:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Huang SM, Hancock MK, Pitman JL, Orth AP, Gekakis N. Negative regulators of insulin signaling revealed in a genome-wide functional screen. PLoS One. 2009;4:e6871. doi: 10.1371/journal.pone.0006871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wu L-J, Jun J, Cheng X, Xu H, Andrews NC, Clapham DE. The channel kinase, TRPM7, is required for early embryonic development. Proceedings of the National Academy of Sciences. 2012;109:E225,ÄìE233. doi: 10.1073/pnas.1120033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development (Cambridge, England) 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- Latta EJ, Golding JP. Regulation of PP2A activity by Mid1 controls cranial neural crest speed and gangliogenesis. Mechanisms of development. 2012 doi: 10.1016/j.mod.2012.01.002. [DOI] [PubMed] [Google Scholar]
- LeDouarin N, Kalcheim C. The Neural Crest. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- Linker C, Bronner-Fraser M, Mayor R. Relationship between gene expression domains of Xsnail, Xslug, and Xtwist and cell movement in the prospective neural crest of Xenopus. Developmental biology. 2000;224:215–225. doi: 10.1006/dbio.2000.9723. [DOI] [PubMed] [Google Scholar]
- Liu JP, Jessell TM. A role for rhoB in the delamination of neural crest cells from the dorsal neural tube. Development (Cambridge, England) 1998;125:5055–5067. doi: 10.1242/dev.125.24.5055. [DOI] [PubMed] [Google Scholar]
- Milarski KL, Zhu G, Pearl CG, McNamara DJ, Dobrusin EM, MacLean D, Thieme-Sefler A, Zhang ZY, Sawyer T, Decker SJ, et al. Sequence specificity in recognition of the epidermal growth factor receptor by protein tyrosine phosphatase 1B. The Journal of biological chemistry. 1993;268:23634–23639. [PubMed] [Google Scholar]
- Minichiello J, Ben-Ya'acov A, Hearn CJ, Needham B, Newgreen DF. Induction of epithelio-mesenchymal transformation of quail embryonic neural cells by inhibition of atypical protein kinase-C. Cell Tissue Res. 1999;295:195–206. doi: 10.1007/s004410051225. [DOI] [PubMed] [Google Scholar]
- Monier-Gavelle F, Duband JL. Control of N-cadherin-mediated intercellular adhesion in migrating neural crest cells in vitro. Journal of cell science. 1995;108(Pt 12):3839–3853. doi: 10.1242/jcs.108.12.3839. [DOI] [PubMed] [Google Scholar]
- Moulton JD, Yan YL. Current protocols in molecular biology / edited by Frederick M. Ausubel ... [et al Chapter 26, Unit 26 28. 2008. Using Morpholinos to control gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DA, Diaz Ba, Bromann PA, Tsai JH, Kawakami Y, Maurer J, Stewart RA, Izpisua-Belmonte JC, Courtneidge SA. A Src-Tks5 Pathway Is Required for Neural Crest Cell Migration during Embryonic Development. PLoS One. 2011;6:e22499. doi: 10.1371/journal.pone.0022499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Kikuno R, Hirosawa M, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:337–345. doi: 10.1093/dnares/6.5.337. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Gulick J, Colbert MC, Robbins J. Protein tyrosine phosphatase activity in the neural crest is essential for normal heart and skull development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11270–11275. doi: 10.1073/pnas.0902230106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgreen DF, Minichiello J. Control of epitheliomesenchymal transformation. I. Events in the onset of neural crest cell migration are separable and inducible by protein kinase inhibitors. Developmental biology. 1995;170:91–101. doi: 10.1006/dbio.1995.1198. [DOI] [PubMed] [Google Scholar]
- Ossipova O, Sokol SY. Neural crest specification by noncanonical Wnt signaling and PAR-1. Development (Cambridge, England) 2011;138:5441–5450. doi: 10.1242/dev.067280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2011 [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development (Cambridge, England) 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Sefton BM, Shenolikar S. Overview of protein phosphorylation. Curr Protoc Protein Sci Chapter 13, Unit13. 2001:11. doi: 10.1002/0471140864.ps1301s00. [DOI] [PubMed] [Google Scholar]
- Selleck MA, Bronner-Fraser M. Origins of the avian neural crest: the role of neural plate-epidermal interactions. Development (Cambridge, England) 1995;121:525–538. doi: 10.1242/dev.121.2.525. [DOI] [PubMed] [Google Scholar]
- Stewart RA, Sanda T, Widlund HR, Zhu S, Swanson KD, Hurley AD, Bentires-Alj M, Fisher DE, Kontaridis MI, Look AT, Neel BG. Phosphatase-dependent and -independent functions of Shp2 in neural crest cells underlie LEOPARD syndrome pathogenesis. Developmental cell. 2010;18:750–762. doi: 10.1016/j.devcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz ME, Eberhart J, Pasquale EB, Krull CE. EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development (Cambridge, England) 2001;128:4669–4680. doi: 10.1242/dev.128.23.4669. [DOI] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development (Cambridge, England) 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Tribulo C, Aybar MJ, Nguyen VH, Mullins MC, Mayor R. Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development (Cambridge, England) 2003;130:6441–6452. doi: 10.1242/dev.00878. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes & development. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mechanisms of development. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Vernon AE, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development (Cambridge, England) 2006;133:3359–3370. doi: 10.1242/dev.02504. [DOI] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development (Cambridge, England) 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Wilkinson D. Whole mount in situ hybridization of vertebrate embryos. In: Wilkinson D, editor. In Situ Hybridization: A Practical Approach. Oxford: Oxford University Press; 1992. pp. 75–83. [Google Scholar]
- Wishart MJ, Dixon JE. Gathering STYX: phosphatase-like form predicts functions for unique protein-interaction domains. Trends in biochemical sciences. 1998;23:301–306. doi: 10.1016/s0968-0004(98)01241-9. [DOI] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nature cell biology. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.