Abstract
CCR5, a cell surface molecule critical for the transmission and spread of HIV-1, is dynamically regulated during T cell activation and differentiation. The molecular mechanism linking T cell activation to modulation of CCR5 expression remains undefined. KLF2 is a transcription factor that promotes quiescence, survival, and in part by modulating chemokine receptor levels, induces homing to secondary lymphoid organs. Given the relationship between T cell activation and chemokine receptor expression, we tested whether the abundance of KLF2 following T cell activation regulates CCR5 expression and thus susceptibility of a T cell to CCR5 dependent HIV-1 strains (R5). We observed a strong correlation between T cell activation, expression of KLF2 and CCR5, and susceptibility to infection. To directly measure how KLF2 affects CCR5 regulation, we introduced siRNA targeting KLF2 expression and demonstrated reduced KLF2 expression also resulted in less CCR5. Chromatin immunoprecipitation assays identified KLF2 bound to the CCR5 promoter in resting but not CD3/28 activated T cells, suggesting that KLF2 directly regulates CCR5 expression. Introduction of KLF2 under control of a heterologous promoter could restore CCR5 expression and R5 susceptibility to CD3/28 costimulated T cells and some transformed cell lines. Thus, KLF2 is a host factor that modulates CCR5 expression in CD4 T cells and influences susceptibility to R5 infection.
Introduction
There is great variability within the population regarding susceptibility to HIV-1 infection and the rate at which those infected progress to AIDS. A large number of host factors including HLA, TRIM5α, APOBEC3G, CCR5 and CXCR4 have been shown to play a role in either determining susceptibility to HIV-1 infection or in the progression to AIDS (1). While much study has been devoted to identifying particular alleles associated with resistance to infection or altered progression to AIDS, less progress has been made identifying factors regulating these susceptibility and progression factors. CCR5, which is highly regulated during T cell activation and differentiation, is particularly interesting because there is a direct correlation with the amount of CCR5 on the cell surface and susceptibility to R5 infection (2). It has long been noted that there is an inverse relationship between the strength of a T cell activation signal and the amount of CCR5 expression (3–6); however, to date no molecular mechanism has been identified which would explain how the strength of T cell activation would regulate CCR5 expression.
Given the predominant role CCR5 plays in HIV-1 acquisition and progression, there have been many attempts to understand the factors that regulate CCR5 expression (7). CCR5 transcription is initiated at multiple sites via two promoters (8, 9). Interestingly, Promoter 1 (Pr1) is more active in transformed cell lines whereas Pr2 appears to be the dominant promoter used in activated primary human T cells (8, 10). Although many factors including NF-κB/Rel, NF-AT, YY1, GATA-1, CREB-1, Oct1 and Oct-2 have been shown to regulate CCR5 expression (8, 9, 11–17), the spatial-temporal coordination of these factors modulating CCR5 expression in response to various activation signals is unclear. Moreover, because T cell lines largely have lost the ability to express CCR5 and structure/function promoter studies in primary T cells are challenging to perform, it has been difficult to pinpoint the factors that control CCR5 expression in primary human T cells in response to developmental, environmental and antigen signals.
Kruppel-like factors are a family of at least 17 zinc-finger transcription factors that play key roles in differentiation, quiescence, and homeostasis of many cell lineages (18). Krueppel-like factor 2, KLF2, highlights the importance of this family of transcription factors to the immune system as mice lacking KLF2 showed an activated T cell phenotype with severely reduced T cells in the periphery (19). KLF2 plays an active role in keeping T cells in a resting state, in part by reducing cMyc expression (20). KLF2 has also been shown to regulate T cell migration by controlling the expression of key regulators of T cell trafficking such as CD62-L and S1P1 (21, 22), explaining in part the lack of T cells in the lymph nodes and peripheral blood of KLF2-deficient mice. Further investigation demonstrated KLF2 deficient murine T cells have augmented mRNA levels of the inflammatory chemokine receptors CXCR3, CCR3 and CCR5 (23) ; however, subsequent studies indicated that loss of KLF2 lead to a cell-non-autonomous increase in CCR5 expression (24). This suggests that current mouse models of KLF2 deficiency will not be informative to study cell-intrinsic effects of KLF2 loss in T cells. Based upon the correlation amongst KLF2, chemokine receptor expression and T cell activation, we speculated that KLF2 may regulate a human T cell’s susceptibility to HIV-1 infection. Our studies indicate that KLF2 does indeed regulate CCR5 expression in primary human T cells in a manner which alters a cell’s susceptibility to HIV-1 infection.
Materials and Methods
Lentiviral vectors, transduction and cell culture
KLF2 and TRIM22 cDNAs were obtained from Open Biosystems (Huntsville, AL) and cloned into a lentiviral expression system that linked expression to either GFP or mRFP via a 2A sequence. Murine CD28 with an introduced stop codon placed right after the transmembrane domain (aa 167) was also inserted into a lentiviral vector backbone. Construction of huTRIM5α lentiviral vector, high titer vector production, and transduction into primary T cells was performed as previously described (25).
siRNA electroporation
Electroporation of primary human T cells was performed as described (26), using 20 µg siRNA comprising a pool of 4 control or KLF2-specific siRNAs (Dharmacon, Lafayette, CO).
FACS Analysis, Cell Sorting and Quantitative RT-PCR
Cell-surface staining for CCR5 was performed as previously described (27) using a BD LSR II. Cell sorting was performed by Flow Cytometry & Cell Sorting Core at the University of Pennsylvania as previously described (25). KLF2, CCR5 and GAPDH expression levels were quantitated by real time PCR using a 7900HT thermocycler (Life Technologies) using FAM-based inventoried assays Hs00360439_g1, Hs99999149_s1, and Hs00266705_g1 (Life Technologies), as recommended by the manufacturer and relative expression was calculated as previously described (28).
Chromatin Immunoprecipitation
ChIP was performed using the EZ-MagnaChIP G kit (Millipore) as previously described (29) with anti-KLF2 monoclonal antibody (M09, Abnova). Real time PCR primers are identified at their extreme ends in Fig. 2A and full length sequences are available upon request.
Figure 2.
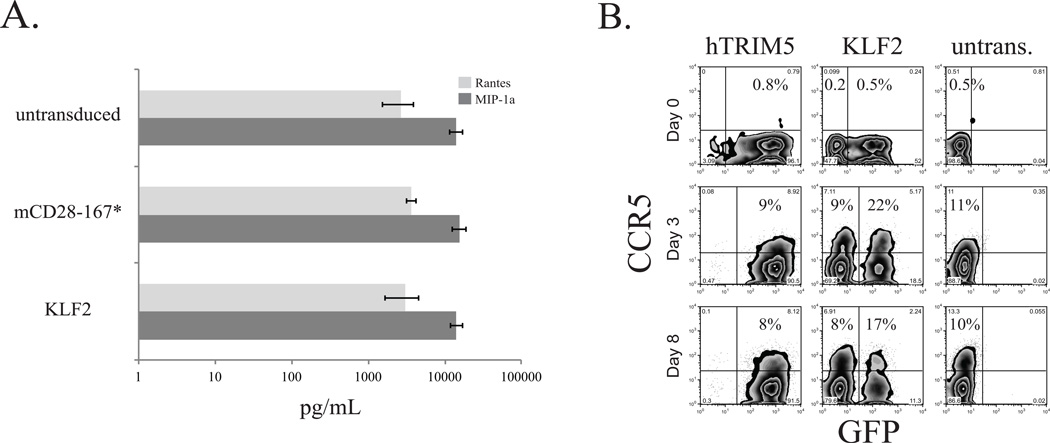
A. Resting CD4 T cells were stimulated with PHA + IL2 for 24 hours, and then transduced with KLF2-expressing or mCD28 167* lentivirus, or left untransduced. After 5 days, transduced cells were sorted and stimulated with CD3/CD28 coated beads for 24 hours and supernatants were collected. Concentration of MIP-1α (CCL3) and RANTES (CCL5) was measured by ELISA. Error bars reflect SEM from three independent experiments. B. CD3/CD28 coated bead stimulated CD4 T cells were transduced with KLF2 or huTRIM5α expression vectors. After 3 days, the αCD3/28 coated beads were removed (Day 0, top panel), and the T cells were cultured an additional 3 (middle panel) or 8 (bottom panel) days. Staining for cell surface CCR5 expression was monitored in parallel with GFP by FACS, and percentages of CCR5+ cells were calculated for both GFP+ and GFP- populations.
HIV-1 Challenge and Entry Assay
KLF2-, mCD28-167*- or untransduced CD4 T cells were costimulated with αCD3/CD28-coated beads at a 3:1 bead to cell ratio, and challenged 2 hours later with cell-free HIV-1BalHIV-1US1 or HIV-1BK132 at a multiplicity of infection of 0.1, or mock challenged as previously described (27). After 48 hours, 2–4 × 105 cells were harvested for DNA isolation using a QIAamp DNA Mini kit (Qiagen, Valencia, CA). The extent of infection in mock and HIV-challenged samples was quantified by TaqMan for HIV-1 gag and gapdhusing DNA from stably-infected Ach-2 cells to construct a gag standard curve (25). To perform HIV-1 pseudotype entry assay, PHA + IL2 stimulated primary human CD4 T cells were transduced with mRFP 2A KLF2- or CCR5-expressing lentivectors, or were left untransduced as a control. After 4 days, KLF2- and hTRIM22-expressing cells were sorted based on mRFP expression. The next day, cells were subjected to the 24-hour BlaM assay as previously described (30) and then assayed for HIV-1 fusion by FACS for cleaved CCF2.
Results
KLF2 and CCR5 expression are linked in primary human CD4 T cells
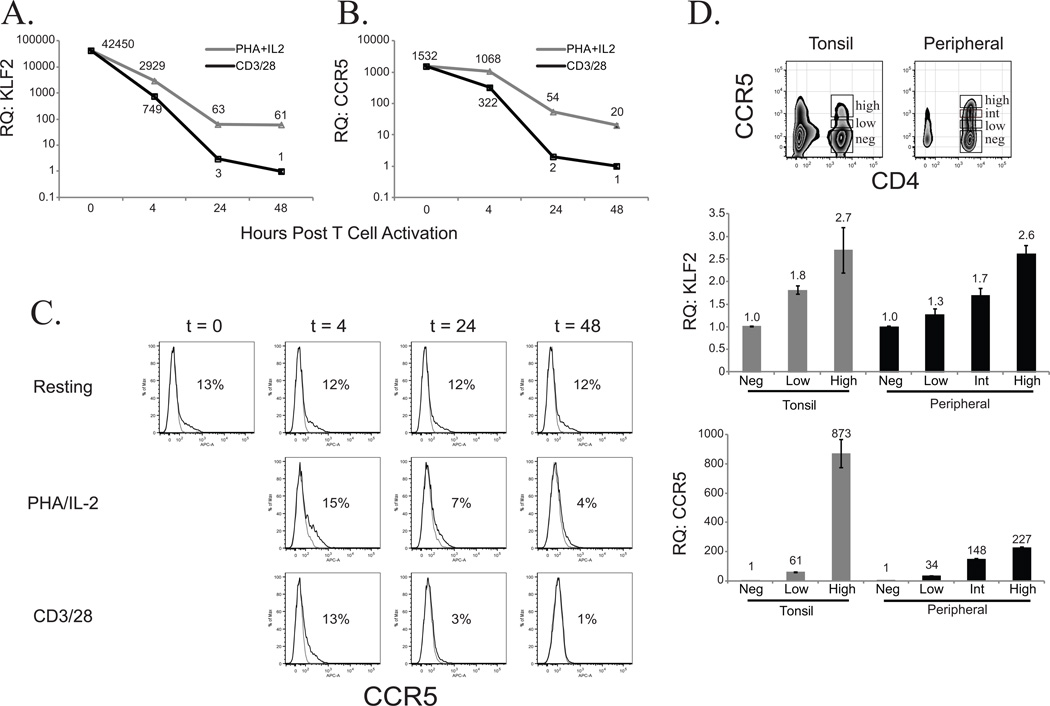
HIV-1 preferentially infects activated T cells (31), but the manner by which the T cell is activated influences its susceptibility to infection (32). We compared the expression levels of KLF2 and CCR5 in primary human CD4 T cells after being activated by either PHA + IL-2 or immobilized anti-CD3 and anti-CD28 Ab coated beads (CD3/28 bead). These two means of activating human CD4 T cells diverge in their ability to drive T cell expansion and support R5 infection. PHA + IL-2 stimulation results in limited T cell expansion and supports R5-dependent HIV-1 infection, whereas CD3/28 bead costimulation is not conducive to a productive R5 infection but enables robust and long term expansion of human CD4 T cells (5, 6, 33, 34). Consistent with previous findings, we observed high expression of KLF2 in resting T cells that was rapidly downregulated following T cell activation (28, 35). However, KLF2 downregulation was delayed in PHA + IL-2 blasts relative to CD3/28 bead stimulation and remained significantly higher even after the cells had started to expand (Fig 1A). The inability of PHA + IL-2 stimulation to completely downregulate KLF2 expression correlated well with higher CCR5 expression in PHA blasts relative to cells stimulated by CD3/28 coated beads (Fig 1B). Examination of CCR5 surface expression reveals a similar, albeit less dramatic, finding (Fig 1C); however, examination of CCR5 surface expression is complicated by the fact that β-chemokine binding to CCR5 forces CCR5 internalization (36, 37), making steady-state CCR5 mRNA expression levels a more accurate depiction of CCR5 regulation in activated primary human T cells. Thus, higher levels of KLF2 expression in PHA blasts also correlated with less cell expansion relative to CD3/28 bead stimulation (34), suggesting a link between KLF2 expression, T cell expansion and susceptibility to HIV-1 infection. We further examined the association of CCR5 and KLF2 expression using freshly isolated CCR5HIGHCCR5LOW and CCR5NEG populations from resting PBMC and tonsil tissue (Fig 1D). Again, we observed a positive correlation between KLF2 and CCR5 expression.
Figure 1. KLF2 and CCR5 expression are linked in primary human CD4 T cells.
Relative KLF2 (A.)and CCR5 (B.) expression levels were determined by real time RT-PCR using cDNA from resting CD4 T cells (T = 0 hrs), and at subsequent timepoints (T = 4, 24, 48 hrs) after PHA/IL2 activation or αCD3/CD28 coated bead costimulation. Expression levels were normalized to those in αCD3/CD28 stimulated CD4 T cells at assay endpoint (T = 48 hrs). C. FACS analysis of CCR5 expression on the cell surface in resting, PHA/IL2 or αCD3/CD28 stimulated CD4 T cells, relative to isotype control, during the first 48 hours after activation. D. CD4 T cells from tonsil or PBMC were FACS-sorted based on CCR5 expression, and their cDNAs were assayed by RT-PCR for relative KLF2 and CCR5 expression levels normalized to GAPDH.
KLF2 expression targets CCR5 but not β-chemokine expression
As shown above, analysis of CCR5 surface expression in primary human T cells is made challenging by inherently low expression. Further complicating analysis is the fact that CD3/28 costimulation results in secretion of high levels of the β-chemokines CCL3, CCL4, CCL5 which are also known as MIP-1α, MIP-1β and RANTES (6), leading to the internalization of CCR5 after binding. Thus, it is possible some of the differences in the CCR5 surface expression observed in Fig 1 are the result of KLF2 interfering with β-chemokine production rather than a direct effect on CCR5 protein expression. To test whether KLF2 expression altered β-chemokine production, we transduced PHA + IL-2 stimulated blasts with a lentiviral vector that co-expressed monomeric (m) RFP and KLF2 via a T2A sequence or a control vector that expressed mCD28 with a truncated cytoplasmic tail (mCD28 167*). Transduced cells were purified by cell sorting after which they were stimulated with CD3/28 coated beads and β-chemokine production was measured 24h later (Fig 2A). We observed no significant difference in β-chemokine production between the different cell populations indicating that enhanced expression of KLF2 does not regulate β-chemokine production (MIP-1α (p=0.511) and Rantes (p=0.831), by ANOVA), and differences in CCR5 expression observed in KLF2 expressing cells are likely due to direct effects of KLF2 on CCR5 regulation. To better understand how KLF2 controls CCR5 expression, we set up an assay to measure recovery of CCR5 expression after CD3/28 coated beads are removed, after which β-chemokine levels drop considerably following 8–10 days of culture (38). We transduced CD3/28 costimulated CD4 T cells with lentivirus vectors expressing either GFP T2A KLF2 or GFP T2A human TRIM5α. The TRIM5α expression vector was chosen as a control because it was constructed on the same backbone as the KLF2-expressing vector and is not thought to be involved in CCR5 regulation. After 3 days, the beads were removed and CCR5 expression was examined after an additional 3 and 8 days of culture. We observed approximately twice as many CCR5 expressing cells in KLF2 transduced cells as compared to untransduced or hTRIM5α-transduced cells (Fig 2B). These data further demonstrate that KLF2 expression augments CCR5 expression in primary human T cells. Moreover, this data shows the cell-intrinsic nature of this relationship between KLF2 and CCR5 expression, as the GFP-negative cells within the KLF2 expressing cultures exhibit the same amount of CCR5 as the TRIM5α and untransduced cell populations.
Targeting KLF2 expression using siRNA results in reduced CCR5 expression
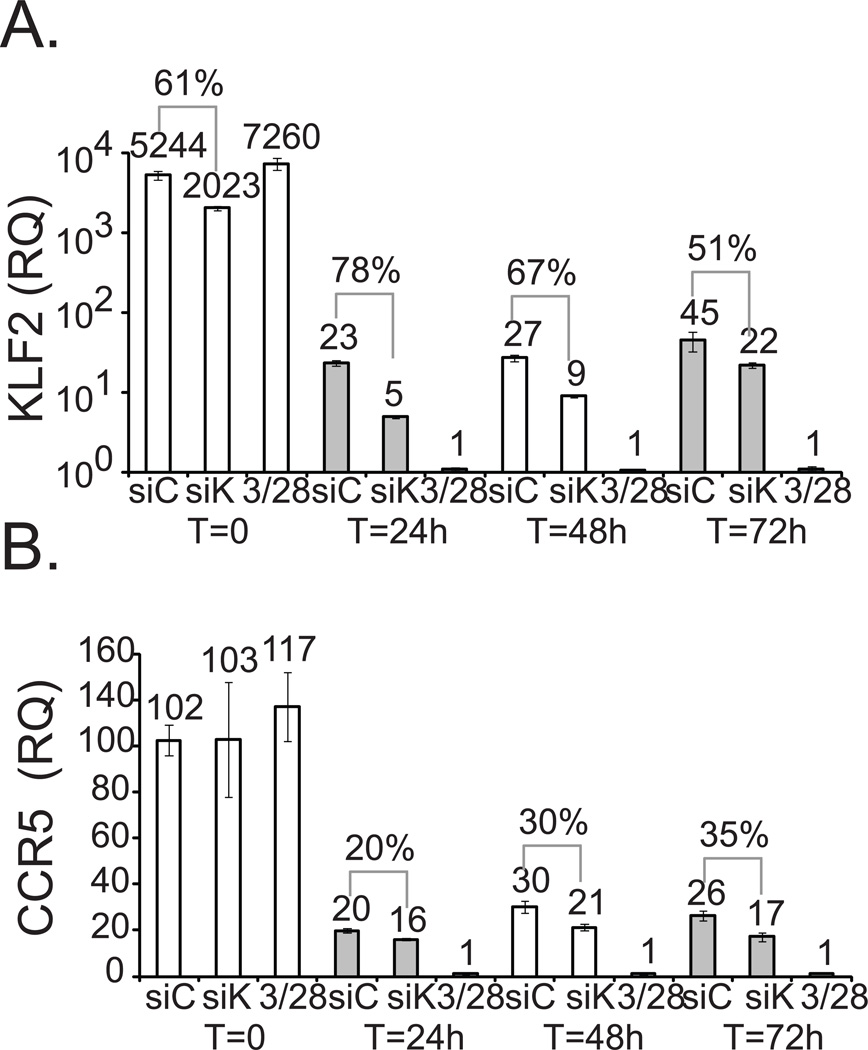
While CD3/28 bead stimulation potently downregulates expression of KLF2 and CCR5, it does not establish whether KLF2 directly regulates CCR5 expression. To do this, we transfected a pool of 4 control siRNAs, or a pool of 4 KLF2-specific siRNAs into primary human CD4 T cells, and the expression of both KLF2 and CCR5 mRNA levels were monitored by quantitative RT-PCR the next day (t = 0) and for the next 72 hours after PHA/IL-2 activation. KLF2-specific siRNAs reduced KLF2 mRNA levels by 61% in resting CD4 T cells incubated overnight following transfection (t = 0), relative to control siRNA transfection (Fig 3A). KLF2 knockdown was most pronounced 24 hours after PHA/IL-2 activation (t = 24) in which there was 78% less KLF2 mRNA present. After an additional 24 and 48 hours of culture, KLF2 expression gradually returned. Knockdown of KLF2 resulted in a 20% decrease in CCR5 after 24 hours, a 30% decline by 48 hours, and a 35% reduction after 72 hours (Fig 3B). Hence, specific knockdown of KLF2 using siRNAs subsequently reduced expression of CCR5 but not the control GAPDH, indicating KLF2 expression can regulate CCR5 levels in primary human T cells.
Figure 3. Targeting KLF2 expression using siRNA results in reduced CCR5 expression.
Resting, freshly isolated CD4 T cells were electroporated with control (siC) or KLF2-specific siRNA (siK) pools. The next day, relative KLF2 (A.)and CCR5 (B.) expression levels were determined by real time RT-PCR (t = 0) for siRNA treated and untreated control cells (t = 0*). siRNA treated T cells were stimulated by PHA + IL-2, whereas untreated control cells were stimulated with CD3/28 coated beads. Relative CCR5 and KLF2 expression levels were measured by RT-PCR from cDNA isolated at the indicated time points. Similar results were obtained in three separate experiments.
KLF2 binds to the CCR5 promoter in resting but not CD3/28 activated T cells
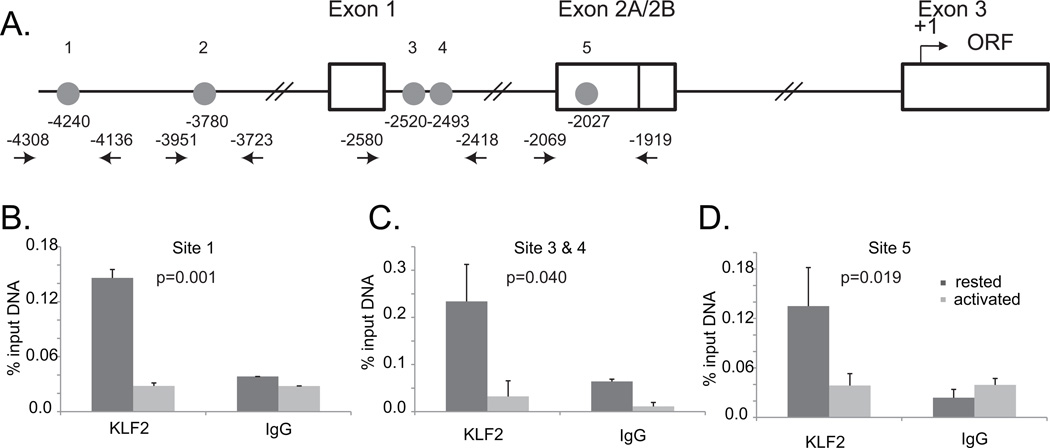
KLF2 is a zinc finger transcription factor that plays a direct role in regulating the expression of IL-2, CD62-L and S1P1 (22, 39). There are 5 putative KLF2 binding sites “CCACCC” within the first 4500 basepairs upstream of the CCR5 translational start site (Fig 4A). To determine whether KLF2 binds the CCR5 promoter, we performed chromatin immunoprecipitation (ChIP) assays using freshly obtained T cells that were stimulated with CD3/28 beads for 48 hours. We observed KLF2 occupancy on the CCR5 promoter in resting but not activated primary human CD4 T cells (Fig. 4B). Although this data does not eliminate the possibility that KLF2 can regulate the activity or expression of other factors involved in CCR5 regulation, it does show that KLF2 engages the CCR5 promoter region in a manner that correlates with changes in CCR5 expression, suggesting that it is a direct transactivator of gene expression for not only CD62-L and S1P1 (22), but CCR5 as well.
Figure 4. KLF2 binds to the CCR5 promoter and directly regulates its expression.
A. Schematic of the CCR5 promoter and exon/intron structure. Numbering is relative to the start site of translation. Each circle depicts a KLF2 binding site (CCACCC), and the arrows surrounding these sites indicate the primer sets with the number representing the 5’ base of the oligo primer used to for the chromatin immunoprecipation (ChIP) assay. B. C. and D. Data depict triplicate PCR reactions, with p values determined using Students t test. Background values obtained by using control IgG Ab were subtracted prior to statistical analysis. This data is representative of two independent experiments in which both pull down and quantitative PCR was performed. We were unable to get the assay to work for Site 2.
KLF2 expression induces CCR5 in some but not all transformed T cell lines and restores susceptibility to R5 HIV-1 infection
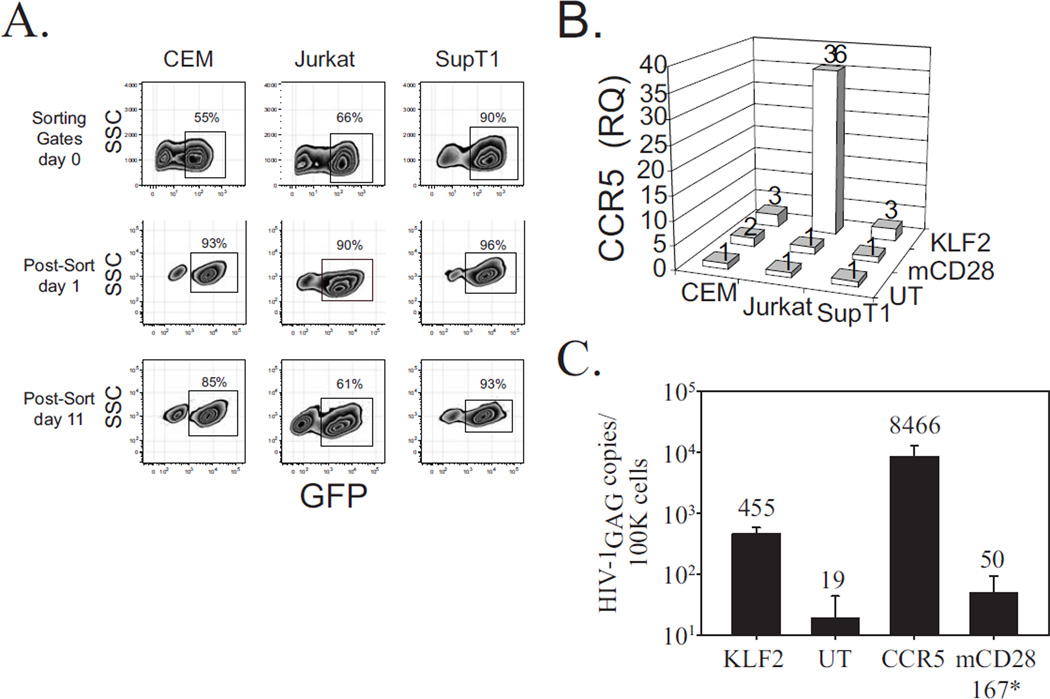
Given its ability to act as an anti-proliferative gene, we suspected KLF2 would likely be poorly expressed in continuously dividing T cell lines, potentially explaining why T cell lines are CCR5 deficient. To test this, we transduced three T cell lines used often in HIV-1 research: CEM, SupT1 and Jurkat with a GFP 2A KLF2 lentiviral vector. After transduction, these cells were sorted by GFP expression in near but not absolutely pure populations (Fig 5A). After an additional 10 days of culture, we observed that all three cell lines had fewer GFP positive T cells than on Day 1 suggesting that the introduced KLF2 was functionally active as an anti-proliferative agent and T cell lines not expressing KLF2 had a selective growth advantage (39). Of note, loss of KLF2 expressing cells was most pronounced in Jurkat T cells, suggesting that KLF2 has much functional activity in this cell line. Next, we examined CCR5 expression levels in KLF2-transduced and untransduced T cell lines and observed that KLF2 expression induced significantly higher levels of CCR5 expression in Jurkat T cells, with a more modest increase in CEM and SupT1 T cells (Fig. 5B). To determine if KLF2-mediated upregulation of CCR5 expression in Jurkat T cells now made these cells sensitive to CCR5 tropic (R5) HIV-1 strains, we challenged them with HIV-1BaL and then harvested the cells 48 hours later and measured HIV-1 gag copy-number by quantitative PCR. As a positive control, Jurkat T cells were transduced with a CCR5 expressing lentiviral vector and negative controls included the parent Jurkat cell line or those transduced with murine CD28 devoid of a cytoplasmic tail (mCD28 167*) (Fig. 5C). KLF2-expressing Jurkat cells were much more susceptible to HIV-1BaL infection compared to mCD28-167*-expressing or untransduced Jurkat T cells. Therefore, augmented KLF2 expression in a transformed T cell line can lead to a physiologically relevant induction of CCR5, suggesting that KLF2 is a key regulator of CCR5 expression and loss of KLF2 expression is why Jurkat T cells cannot support an R5 infection. Moreover, the ability of KLF2 to robustly restore CCR5 expression to some but not all transformed T cells lines suggests that KLF2 is necessary but not sufficient to drive CCR5 expression, and/or cell lines such as SupT1 or CEM are insensitive to KLF2 activity.
Figure 5. KLF2 expression induces CCR5 in some but not all transformed T cell lines and restores susceptibility to R5 infection.
A. KLF2-expressing CEM, Jurkat and SupT1 cells were sorted by GFP expression GFP+ cells on day 0, and then monitored for stability of expression subsequently at day 1 and day 11. B. Relative CCR5 expression levels were measured by quantitative PCR using cDNA obtained from freshly (day 1) sorted KLF2-expressing CEM, Jurkat and SupT1 cells, as well as mCD28-167*-expressing and untransduced controls for each human CD4 T cell line. Relative KLF2 and CCR5 expression levels were normalized to those observed in untransduced cells for each T cell line. C. Untransduced and KLF2-, mCD28-167*-, or CCR5-expressing Jurkat T cells were challenged with cell-free HIV-1BaL. After 48 hours, cells were harvested and TaqMan was performed for HIV-1 gag to assess the extent of infection, using DNA from stably-infected Ach-2 cells as a standard. Samples were run in duplicate, and results are indicative of those obtained in three separate experiments.
KLF2 expression induces CCR5 in primary human CD4 T cells and renders them susceptible to R5-tropic HIV infection
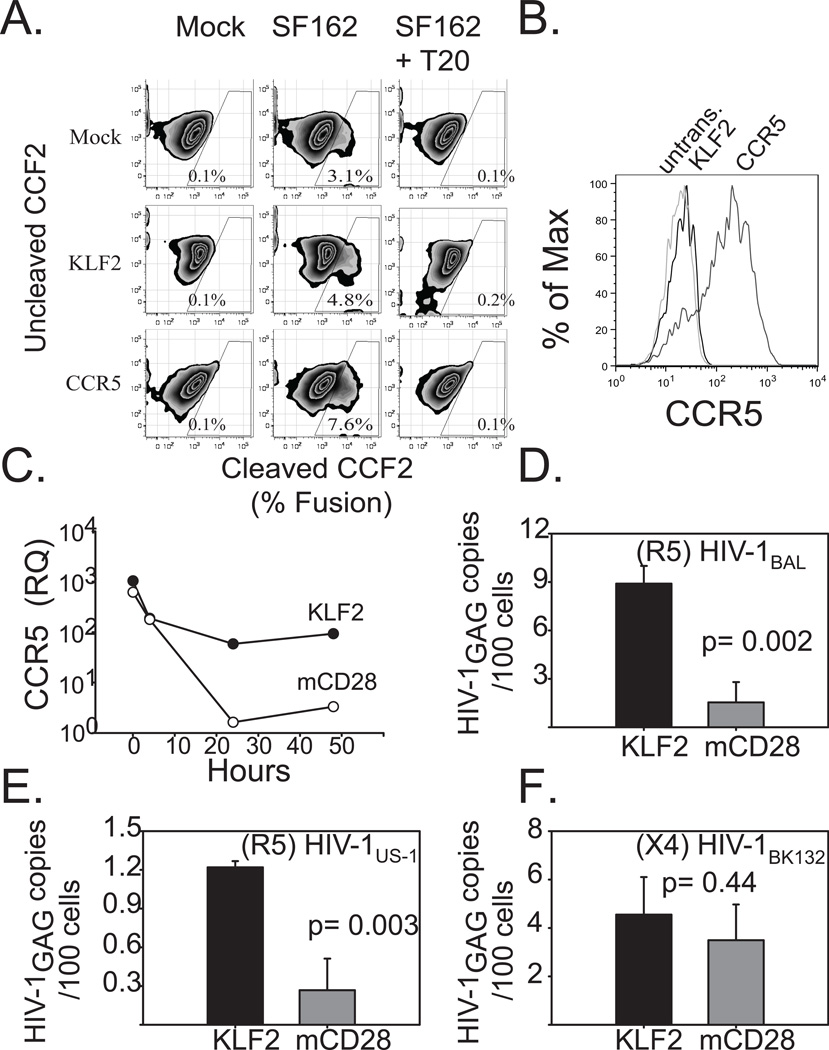
Having established a link between KLF2 and CCR5 expression and susceptibility to R5 infection in transformed T cell lines, we wished to determine whether modulation of KLF2 expression could alter susceptibility to HIV-1 infection in primary human T cells. Since CD3/28 costimulation proved to be a much more effective means of downregulating KLF2 expression than introduction of KLF2-specific siRNAs, we decided to determine whether enforced expression of KLF2 could overcome the previously described CD28 costimulation-mediated downregulation of CCR5 (37, 40, 41). First, we hypothesized that KLF2 expression would facilitate enhanced HIV-1 entry due to augmented CCR5 expression. PHA blasts were transduced with mCD28 167*, GFP T2A KLF2 or CCR5 expressing vectors and pure populations of transduced T cells were obtained by cell sorting. These cells were loaded with the fluorescent dye CCF2-AM and then infected with a R5 (SF162)ENV-HIV-1 viral pseudotype that incorportated a Vpr-β-lactamase fusion protein. Binding and fusion of the viral membrane with the host cell delivers the Vpr-β-lactamase fusion protein to the host cell cytoplasm where it cleaves CCF2-AM, resulting in a shift in fluorescence that can be easily monitored by FACS. Thus, this assay monitors virus entry into cells, which is the step of the virus life cycle impacted by CCR5 expression. We observed that KLF2-transduced T cells expressed slighlty more CCR5 than control cells and also permitted higher levels of viral entry (4.8% KLF2-transduced vs 3.1% for untransduced controls-Fig. 6A), while cells transduced with the CCR5 expression vector supported even higher levels of CCR5 (Fig. 6B) and virus entry (7.6%, Fig. 6A), suggesting that the differences observed between the KLF2-transduced and untransduced T cells are biologically relevant differences.
Figure 6. Forced expression of KLF2 in CD4s restores CCR5 expression during CD3/28 costimulation, and renders them susceptible to R5-tropic HIV.
A. PHA + IL2 stimulated primary human CD4 T cells were transduced with mRFP 2A KLF2-, or CCR5-expressing lentiviral vectors, or were left untransduced as a control. After 4 days of culture, KLF2- expressing cells were sorted based on mRFP expression. The following day cells were loaded with CCF2 and challenged with HIV-1 pseudotype expressing the HIVSF162 (R5) Env and Vpr-BlaM fusion in the presence or absence of 1μg of T20. Cleaved CCF2 was analyzed by flow cytometry 24 hours later. B. Untransduced and KLF2-, and CCR5-expressing CD4 T cells used in the BlaM assay were stained for cell surface CCR5 expression and assayed via flow cytometry. C. Untransduced, sorted KLF2-, or sorted mCD28-167*- expressing human CD4 T cells were activated with αCD3/CD28-coated beads. Relative CCR5 expression levels were determined by real time prior to (T = 0 hrs) and during αCD3/CD28 coated bead costimulation (T = 4, 24, 48 hrs). Expression levels were normalized to those in untransduced, αCD3/CD28 costimulated CD4 T cells at assay endpoint (T = 48 hrs). Similar results were obtained in five separate experiments, using cells from different donors. D,E.F KLF2- and mCD28-167*- expressing human CD4 T cells were prepared as above and activated with αCD3/CD28-coated beads and then challenged two hours later with CCR5-tropic or CXCR4-tropic cell-free HIV-1. After 48 hours, DNA was isolated from harvested cells, and real time PCR was performed for integrated HIV-1 gag DNA. DNA from stably-infected Ach-2 cells was used as a standard. D. CCR5-tropic HIV-1BaL challenge. E. CCR5-tropic HIV-1US1 challenge. F. CXCR4-tropic HIV-1BK132 challenge. Each experiment was performed in duplicate and data shown is representative of three independent studies. Error bars reflect the standard deviation of real time PCR assay of one representative experiment.
To determine whether a similar experimental plan could be used to measure susceptibility to infection by replication-competent HIV-1, we activated T cells that had been transduced with the mCD28 167*, GFP T2A KLF2 or CCR5 expression vectors and then measured CCR5 expression 4, 24 and 48 hours post CD3/28 costimulation (Fig 6C). We observed that CCR5 expression remained detectable in T cells transduced with the KLF2 expressing vector, whereas T cells transduced with mCD28 167* rapidly downregulated CCR5 expression after CD3/28 bead stimulation. We obtained pure populations of KLF2 and mCD28 167* expressing cells by transducing KLF2 or mCD28 167* expression vectors into PHA blasts and sorting them based on GFP and mCD28 expression. To determine whether KLF2 expression could block the CD3/28 antiviral effect, these KLF2 and mCD28 167* expressing T cells were stimulated with CD3/28 coated beads and two hours later challenged with one of two R5 viruses, HIV-1BAL or HIV-1US-1or the X4 tropic virus HIV-1BK132. We observed that KLF2 expression made CD3/28 costimulated T cells much more susceptible to R5 but not X4 infection (Fig 6D–F), demonstrating expression of the host factor KLF2 regulates susceptibility to CCR5-dependent HIV-1 strains.
Discussion
HIV-1 requires T cell activation to initiate a productive infection (31); therefore it was quite surprising that a supra-physiological stimulus like CD3/28 costimulation permitted the long term expansion of HIV-1 infected CD4 T cells in the absence of a spreading infection without the addition of antiretrovirals (5, 6). These studies and others (4, 27) highlight the “bell curve” relationship between T cell activation and susceptibility to CCR5-dependent HIV-1 infection. While this finding that strong stimulation leads to a robust antiviral state in primary human CD4 T cells has permitted testing of cell therapy approaches to treat HIV-1 infections (42), a molecular mechanism that would explain this relationship has yet to be described. Our data suggest KLF2 expression plays a pivotal role in determining how T cell activation affects receptiveness to R5 HIV-1 infection. Our findings support the view that KLF2 plays at least two roles in T cell biology that are, in part, regulated by its relative abundance in the cell. When expressed at high levels, KLF2 enforces T cell quiescence, and activating stimuli must sufficiently downregulate KLF2 expression in order to enable the transition from G0 to G1 (20, 43, 44). The second role for KLF2 is regulation of chemokine receptor expression in lymphocyte trafficking (45–48). Our data would indicate that much more modest levels of KLF2 are required to regulate chemokine receptor expression as opposed to T cell quiescence, as less than 1% of KLF2 present in resting T cells is still able to upregulate CCR5 expression in PHA blasts (Fig 2A). Moreover, we find KLF2 expression must be essentially undetectable to render T cells completely CCR5 negative. Thus, suboptimal modes of T cell activation such as use of soluble anti-CD28 Ab coupled with plate bound anti-CD3 Ab (4), lectin (PHA or ConA) +IL-2 (5), or by APCs that express both costimulatory and negative regulators of T cell activation (27) are insufficient to completely downregulate KLF2 expression, rendering T cells stimulated in this manner susceptible to R5 infection. Thus, agents that promote T cell activation such as CTLA-4 and PD-1 blocking antibodies (49) may provide a dual benefit to fight HIV-1 infection by first augmenting T cell activity, and second by further reducing KLF2 expression resulting in less CCR5 expression. Absolute downregulation of KLF2 by CD3/28 costimulation provides a molecular mechanism underlying why primary human CD4 T cells stimulated in this manner are resistant to R5 infection.
Clearly, KLF2 is not the only factor regulating CCR5. While KLF2 was necessary and sufficient to restore CCR5 expression to Jurkat T cells, it was not enough to restore high levels of CCR5 to CEM or SupT1 cells, suggesting these cell lines are deficient in other factors necessary to induce CCR5 expression. Moreover, with multiple promoters and start sites of transcription to which many transcription factors including NF-KB/Rel, NF-AT, YY1, GATA-1, CREB-1, Oct1 and Oct-2 have been shown to bind to the CCR5 promoter, (8, 9, 11–17) coupled with developmental, environmental, and antigenic signals that all regulate CCR5 expression, understanding the molecular mechanisms by which KLF2 modulates CCR5 expression will undoubtedly be complicated. Nonetheless, our data argues that KLF2 is a rate-limiting factor in regards to CCR5 expression following T cell activation, given the direct correlation observed between KLF2 and CCR5 expression. Interestingly, there is evidence suggesting that CCR5 may also regulate KLF2 expression, suggesting that there may be a positive regulation loop. Studies of CCR5 deficient mice demonstrate that the CCR5−/− CD4 SP T cells which preferentially accumulated in the thymus also lacked KLF2 and S1P1 expression compared to WT cells, and signaling via CCR5 in WT cells induced KLF2 expression, highlighting their potential co-regulation (50). Lastly, given the tight correlation of KLF2 and CCR5 expression we would predict that small differences in KLF2 expression (perhaps mediated by SNPs within the KLF2 gene) could translate into meaningful differences in an individual’s susceptibility to HIV-1 infection as well as the rate by which an individual would progress to AIDS.
Acknowledgments
We are gratefully acknowledge the CFAR/Cancer Center Immunology Core for providing the primary human T cells used in this study, Drs. Mike Betts and Laura McLane for help with ChIP assay; the CFAR Virology core for providing TaqMan primers and probes to measure HIV-1 infection. This manuscript is dedicated to the memory of Dr. Richard Carroll who first observed a correlation between CCR5 expression and T cell activation.
Footnotes
This work was funded in part by U19AI082628 and R01CA147795.
References
- 1.Rowland-Jones S, Pinheiro S, Kaul R. New Insights into Host Factors in HIV-1 Pathogenesis. Cell. 2001;104:473–476. doi: 10.1016/s0092-8674(01)00235-5. [DOI] [PubMed] [Google Scholar]
- 2.Wu L, Paxton WA, Kassam N, Ruffing N, Rottman JB, Sullivan N, Choe H, Sodroski J, Newman W, Koup RA, Mackay CR. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp.Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward SG, Bacon K, Westwick J. Chemokines and T lymphocytes: more than an attraction. Immunity. 1998;9:1–11. doi: 10.1016/s1074-7613(00)80583-x. [DOI] [PubMed] [Google Scholar]
- 4.Creson JR, Lin AA, Li Q, Broad DF, Roberts MR, Anderson SJ. The mode and duration of anti-CD28 costimulation determine resistance to infection by macrophage-tropic strains of human immunodeficiency virus type 1 in vitro. J Virol. 1999;73:9337–9347. doi: 10.1128/jvi.73.11.9337-9347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll RG, Riley JL, Levine BL, Feng Y, Kaushal S, Ritchey DW, Bernstein W, Weislow OS, Brown CR, Berger EA, June CH, St LDC. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science. 1997;276:273–276. doi: 10.1126/science.276.5310.273. [DOI] [PubMed] [Google Scholar]
- 6.Levine BL, Mosca JD, Riley JL, Carroll RG, Vahey MT, Jagodzinski LL, Wagner KF, Mayers DL, Burke DS, Weislow OS, St LDC, June CH. Antiviral effect and ex vivo CD4+ T cell proliferation in HIV-positive patients as a result of CD28 costimulation. Science. 1996;272:1939–1943. doi: 10.1126/science.272.5270.1939. [DOI] [PubMed] [Google Scholar]
- 7.renzana-Seisdedos F, Parmentier M. Genetics of resistance to HIV infection: Role of co-receptors and co-receptor ligands. Seminars in Immunology. 2006;18:387–403. doi: 10.1016/j.smim.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Mummidi S, Adams LM, VanCompernolle SE, Kalkonde M, Camargo JF, Kulkarni H, Bellinger AS, Bonello G, Tagoh H, Ahuja SS, Unutmaz D, Ahuja SK. Production of specific mRNA transcripts, usage of an alternate promoter, and octamer-binding transcription factors influence the surface expression levels of the HIV coreceptor CCR5 on primary T cells. J Immunol. 2007;178:5668–5681. doi: 10.4049/jimmunol.178.9.5668. [DOI] [PubMed] [Google Scholar]
- 9.Mummidi S, Ahuja SS, McDaniel BL, Ahuja SK. The human CC chemokine receptor 5 (CCR5) gene. Multiple transcripts with 5'-end heterogeneity, dual promoter usage, and evidence for polymorphisms within the regulatory regions and noncoding exons. The Journal of biological chemistry. 1997;272:30662–30671. doi: 10.1074/jbc.272.49.30662. [DOI] [PubMed] [Google Scholar]
- 10.Liu R, Zhao X, Gurney TA, Landau NR. Functional analysis of the proximal CCR5 promoter. AIDS research and human retroviruses. 1998;14:1509–1519. doi: 10.1089/aid.1998.14.1509. [DOI] [PubMed] [Google Scholar]
- 11.Kuipers HF, Biesta PJ, Montagne LJ, van Haastert ES, van der Valk P, van den Elsen PJ. CC chemokine receptor 5 gene promoter activation by the cyclic AMP response element binding transcription factor. Blood. 2008;112:1610–1619. doi: 10.1182/blood-2008-01-135111. [DOI] [PubMed] [Google Scholar]
- 12.Guignard F, Combadiere C, Tiffany HL, Murphy PM. Gene organization and promoter function for CC chemokine receptor 5 (CCR5) J Immunol. 1998;160:985–992. [PubMed] [Google Scholar]
- 13.Mummidi S, Bamshad M, Ahuja SS, Gonzalez E, Feuillet PM, Begum K, Galvis MC, Kostecki V, Valente AJ, Murthy KK, Haro L, Dolan MJ, Allan JS, Ahuja SK. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. The Journal of biological chemistry. 2000;275:18946–18961. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- 14.Moriuchi H, Moriuchi M, Fauci AS. Cloning and analysis of the promoter region of CCR5, a coreceptor for HIV-1 entry. J Immunol. 1997;159:5441–5449. [PubMed] [Google Scholar]
- 15.Moriuchi M, Moriuchi H. Octamer transcription factors up-regulate the expression of CCR5, a coreceptor for HIV-1 entry. The Journal of biological chemistry. 2001;276:8639–8642. doi: 10.1074/jbc.M008391200. [DOI] [PubMed] [Google Scholar]
- 16.Moriuchi M, Moriuchi H. YY1 transcription factor down-regulates expression of CCR5, a major coreceptor for HIV-1. The Journal of biological chemistry. 2003;278:13003–13007. doi: 10.1074/jbc.M204980200. [DOI] [PubMed] [Google Scholar]
- 17.Moriuchi M, Moriuchi H, Fauci AS. GATA-1 transcription factor transactivates the promoter for CCR5, a coreceptor for human immunodeficiency virus type 1 entry. Blood. 1999;93:1433–1435. [PubMed] [Google Scholar]
- 18.Hart GT, Hogquist KA, Jameson SC. Kruppel-like factors in lymphocyte biology. J Immunol. 2012;188:521–526. doi: 10.4049/jimmunol.1101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 20.Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc--dependent pathway. Nat.Immunol. 2001;2:698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- 21.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 22.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J.Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 23.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nature immunology. 2008;9:292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 24.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson MW, Carroll RG, Stremlau M, Korokhov N, Humeau LM, Silvestri G, Sodroski J, Riley JL. Mode of transmission affects the sensitivity of human immunodeficiency virus type 1 to restriction by rhesus TRIM5alpha. J Virol. 2008;82:11117–11128. doi: 10.1128/JVI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plesa G, Zheng L, Medvec A, Wilson CB, Robles-Oteiza C, Liddy N, Bennett AD, Gavarret J, Vuidepot A, Zhao Y, Blazar BR, Jakobsen BK, Riley JL. TCR affinity and specificity requirements for human regulatory T-cell function. Blood. 2012;119:3420–3430. doi: 10.1182/blood-2011-09-377051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley JL, Schlienger K, Blair PJ, Carreno B, Craighead N, Kim D, Carroll RG, June CH. Modulation of susceptibility to HIV-1 infection by the cytotoxic T lymphocyte antigen 4 costimulatory molecule. J.Exp.Med. 2000;191:1987–1997. doi: 10.1084/jem.191.11.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riley JL, Mao M, Kobayashi S, Biery M, Burchard J, Cavet G, Gregson BP, June CH, Linsley PS. Modulation of TCR-induced transcriptional profiles by ligation of CD28, ICOS, and CTLA-4 receptors. Proc.Natl.Acad.Sci.U.S.A. 2002;99:11790–11795. doi: 10.1073/pnas.162359999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funderburg NT, Jadlowsky JK, Lederman MM, Feng Z, Weinberg A, Sieg SF. The Toll-like receptor 1/2 agonists Pam(3) CSK(4) and human beta-defensin-3 differentially induce interleukin-10 and nuclear factor-kappaB signalling patterns in human monocytes. Immunology. 2011;134:151–160. doi: 10.1111/j.1365-2567.2011.03475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavrois M, Neidleman J, Bigos M, Greene WC. Fluorescence resonance energy transfer-based HIV-1 virion fusion assay. Methods Mol Biol. 2004;263:333–344. doi: 10.1385/1-59259-773-4:333. [DOI] [PubMed] [Google Scholar]
- 31.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 32.Carroll RG, Riley JL, Levine BL, Blair PJ, St Louis DC, June CH. The role of co-stimulation in regulation of chemokine receptor expression and HIV-1 infection in primary T lymphocytes. Semin Immunol. 1998;10:195–202. doi: 10.1006/smim.1998.0131. [DOI] [PubMed] [Google Scholar]
- 33.Riley JL, Levine BL, Craighead N, Francomano T, Kim D, Carroll RG, June CH. Naive and memory CD4 T cells differ in their susceptibilities to human immunodeficiency virus type 1 infection following CD28 costimulation: implicatip6s for transmission and pathogenesis. The Journal of Virology. 1998;72:8273–8280. doi: 10.1128/jvi.72.10.8273-8280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine BL, Bernstein WB, Connors M, Craighead N, Lindsten T, Thompson CB, June CH. Effects of CD28 costimulation on long-term proliferation of CD4+ T cells in the absence of exogenous feeder cells. J Immunol. 1997;159:5921–5930. [PubMed] [Google Scholar]
- 35.Wan Q, Kozhaya L, ElHed A, Ramesh R, Carlson TJ, Djuretic IM, Sundrud MS, Unutmaz D. Cytokine signals through PI-3 kinase pathway modulate Th17 cytokine production by CCR6+ human memory T cells. The Journal of experimental medicine. 2011;208:1875–1887. doi: 10.1084/jem.20102516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mack M, Luckow B, Nelson PJ, Cihak J, Simmons G, Clapham PR, Signoret N, Marsh M, Stangassinger M, Borlat F, Wells TN, Schlondorff D, Proudfoot AE. Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling: a novel inhibitory mechanism of HIV infectivity. The Journal of experimental medicine. 1998;187:1215–1224. doi: 10.1084/jem.187.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riley JL, Carroll RG, Levine BL, Bernstein W, St LDC, Weislow OS, June CH. Intrinsic resistance to T cell infection with HIV type 1 induced by CD28 costimulation. J.Immunol. 1997;158:5545–5553. [PubMed] [Google Scholar]
- 38.Levine BL, Bernstein WB, Aronson NE, Schlienger K, Cotte J, Perfetto S, Humphries MJ, Ratto-Kim S, Birx DL, Steffens C, Landay A, Carroll RG, June CH. Adoptive transfer of costimulated CD4+ T cells induces expansion of peripheral T cells and decreased CCR5 expression in HIV infection. Nat.Med. 2002;8:47–53. doi: 10.1038/nm0102-47. [DOI] [PubMed] [Google Scholar]
- 39.Wu J, Lingrel JB. KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene. 2004;23:8088–8096. doi: 10.1038/sj.onc.1207996. [DOI] [PubMed] [Google Scholar]
- 40.Levine BL, Mosca JD, Riley JL, Carroll RG, Vahey MT, Jagodzinski LL, Wagner KF, Mayers DL, Burke DS, Weislow OS, St Louis DC, June CH. Antiviral effect and ex vivo CD4+ T cell proliferation in HIV-positive patients as a result of CD28 costimulation. Science. 1996;272:1939–1943. doi: 10.1126/science.272.5270.1939. [DOI] [PubMed] [Google Scholar]
- 41.Carroll RG, Riley JL, Levine BL, Feng Y, Kaushal S, Ritchey DW, Bernstein W, Weislow OS, Brown CR, Berger EA, June CH, St Louis DC. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science. 1997;276:273–276. doi: 10.1126/science.276.5310.273. [DOI] [PubMed] [Google Scholar]
- 42.June CH, Blazar BR, Riley JL. Engineering lymphocyte subsets: tools, trials and tribulations. Nat.Rev.Immunol. 2009;9:704–716. doi: 10.1038/nri2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 44.Schober SL, Kuo CT, Schluns KS, Lefrancois L, Leiden JM, Jameson SC. Expression of the transcription factor lung Kruppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. J.Immunol. 1999;163:3662–3667. [PubMed] [Google Scholar]
- 45.Finlay D, Cantrell D. Phosphoinositide 3-kinase and the mammalian target of rapamycin pathways control T cell migration. Ann.N.Y.Acad.Sci. 2010;1183:149–157. doi: 10.1111/j.1749-6632.2009.05134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hart GT, Wang X, Hogquist KA, Jameson SC. Kruppel-like factor 2 (KLF2) regulates B-cell reactivity, subset differentiation, and trafficking molecule expression. Proc.Natl.Acad.Sci.U.S.A. 2011;108:716–721. doi: 10.1073/pnas.1013168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Zhang Y, Liu Z, Wu X, Zheng Y, Tao Z, Mao K, Wang J, Lin G, Tian L, Ji Y, Qin M, Sun S, Zhu X, Sun B. ECM1 controls T(H)2 cell egress from lymph nodes through re-expression of S1P(1) Nat.Immunol. 2011;12:178–185. doi: 10.1038/ni.1983. [DOI] [PubMed] [Google Scholar]
- 48.Winkelmann R, Sandrock L, Porstner M, Roth E, Mathews M, Hobeika E, Reth M, Kahn ML, Schuh W, Jack HM. B cell homeostasis and plasma cell homing controlled by Kruppel-like factor 2. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:710–715. doi: 10.1073/pnas.1012858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaufmann DE, Walker BD. PD-1 and CTLA-4 inhibitory cosignaling pathways in HIV infection and the potential for therapeutic intervention. J Immunol. 2009;182:5891–5897. doi: 10.4049/jimmunol.0803771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kroetz DN, Deepe GS., Jr An aberrant thymus in CCR5−/− mice is coupled with an enhanced adaptive immune response in fungal infection. J Immunol. 2011;186:5949–5955. doi: 10.4049/jimmunol.1003876. [DOI] [PMC free article] [PubMed] [Google Scholar]