Abstract
Accumulation of misfolded proteins in the cell at high temperature may cause entry into a nonproliferating, heat-shocked state. The imino acid analog azetidine 2-carboxylic acid (AZC) is incorporated into cellular protein competitively with proline and can misfold proteins into which it is incorporated. AZC addition to budding yeast cells at concentrations sufficient to inhibit proliferation selectively activates heat shock factor (HSF). We find that AZC treatment fails to cause accumulation of glycogen and trehalose (Msn2/4-dependent processes) or to induce thermotolerance (a protein kinase C-dependent process). However, AZC-arrested cells can accumulate glycogen and trehalose and can acquire thermotolerance in response to a subsequent heat shock. We find that AZC treatment arrests cells in a viable state and that this arrest is reversible. We find that cells at high temperature or cells deficient in the ubiquitin-conjugating enzymes Ubc4 and Ubc5 are hypersensitive to AZC-induced proliferation arrest. We find that AZC treatment mimics temperature up-shift in arresting cells in G1 and represses expression of CLN1 and CLN2. Mutants with reduced G1 cyclin-Cdc28 activity are hypersensitive to AZC-induced proliferation arrest. Expression of the hyperstable Cln3–2 protein prevents G1 arrest upon AZC treatment and temperature up-shift. Finally, we find that the EXA3–1 mutation, encoding a defective HSF, prevents efficient G1 arrest in response to both temperature up-shift and AZC treatment. We conclude that nontoxic levels of misfolded proteins (induced by AZC treatment or by high temperature) selectively activate HSF, which is required for subsequent G1 arrest.
Eukaryotic cells respond to heat shocks by the induction of a conserved set of proteins, the heat shock proteins, many of which function as molecular chaperones (1). In the budding yeast Saccharomyces cerevisiae, these inductions are caused by increased transcription of the corresponding genes (2). Two transcriptional control systems appear to be responsible for the majority if not all of the profound gene expression changes that occur upon heat shock, that involving heat shock factor (HSF) and that involving Msn2 and Msn4 (3). HSF binds to heat shock elements (HSEs) found upstream of many heat-inducible genes (4). The thermal induction of these genes depends on the activation of HSF and on the presence of the HSEs. It recently has become clear that activation of HSF is also required for repression of the ribosomal protein genes, although the mechanism is unclear (5). Msn2 and Msn4 are transcription factors that bind to the stress response element (STRE), also found upstream of many heat-inducible genes. The STRE and HSF regulons appear to account for most, if not all, of the transcriptional inductions caused by mild heat shock (6).
Heat-shocked yeast cells display a number of characteristic phenotypes in addition to their transcriptional program. For example, cells treated with a mild heat shock accumulate the storage carbohydrates glycogen and trehalose (a process that requires Msn2 and Msn4) (7), acquire thermotolerance [a process dependent on activation of the protein kinase C–mitogen-activated protein (Pkc1-MAP) kinase pathway] (8), and become transiently arrested in the G1 phase of the cell cycle by an unknown mechanism (9, 10).
How does heat shock trigger these changes? The activation of HSF by heat shock is likely to result from some thermal misfolding of cellular protein. Such misfolding proteins bind avidly to cytoplasmic/nuclear Hsp70 chaperones (11), and the resulting titration of “free” Hsp70s has been proposed to activate HSF, which is already bound to its cognate promoters (12).
Activation of the STRE regulon by heat shock requires Msn2 and Msn4, but the mechanism of activation of these trancription factors by heat shock remains obscure (3). Whatever the primary trigger, it is unlikely that misfolded proteins cause activation of this system (see below).
Activation of the Pkc1-MAP kinase pathway at high temperature appears to be due to thermal effects on the cell surface (8). The cell surface proteins Hcs77/Wsc1 (13, 14) and Mid2 (15–17) are required for activation of the Pkc1-MAP kinase pathway by heat shock and have been proposed to be sensors of the cell surface state. Activation of this pathway by heat shock is a slow process, taking up to 45 min, and requires protein synthesis (8). It is therefore possible that activation of the HSF regulon by misfolding of nascent proteins is required to generate the cell surface defects that are sensed by Hcs77 and Mid2. Alternatively, defects generated by the thermal misfolding of newly synthesized membrane proteins may directly cause the cell surface defects to which the Pkc1 pathway responds.
Cells subjected to temperature up-shift (e.g., 23°C to 36°C) become transiently arrested in the G1 phase of the cell cycle (9, 10). It is not known what parameter of the cell is monitored by this system (e.g., misfolded proteins? membrane damage?), nor is the mechanism by which the signal is transduced to the cell cycle machinery understood. The G1 arrest appears to be due to a loss of G1 cyclin function: expression of CLN1 and CLN2 is transiently repressed by temperature up-shift, and expression of hyperstable forms of Cln3 or Cln2 can prevent the G1 arrest (10).
In our previous work, we have exploited the imino acid analog azetidine 2-carboxylic acid (AZC) (E.W.T., G.A.P., and J.V.G., unpublished work). AZC is an analog of proline that is incorporated into cellular protein competitively with proline and can misfold proteins into which it is incorporated (18, 19). We found that the AZC addition to yeast, at sublethal concentrations sufficient to inhibit proliferation, selectively and strongly induces the expression of genes containing heat shock elements via activation of a heat shock factor (HSF) mutant (E.W.T., G.A.P., and J.V.G., unpublished work). We found that AZC treatment selectively and strongly represses expression of the ribosomal protein genes. This repression and that caused by mild heat shock are independent of known regulators of ribosomal protein gene expression but depend on the proper activation of HSF. We found that the STRE regulon is weakly activated, if at all, by AZC addition. Thus, to a first approximation, AZC treatment selectively activates only the HSF regulon. Misfolded proteins are thus competent to be intermediates in the heat shock activation of the HSF regulon but not the STRE regulon.
AZC affords us a unique opportunity to determine the cellular phenotypes that result from the protein misfolding and subsequent activation of HSF. In this paper, we examine a number of phenotypes of cells treated with AZC. We find that AZC treatment does not cause Msn2/4 and Pkc1 pathway-dependent phenotypes, but that cells prearrested in AZC can acquire these phenotypes in response to a subsequent heat shock. Misfolded proteins are thus unlikely to be involved in the thermal activation of the STRE regulon or the Pkc1 pathway but are not detrimental to viability and responsiveness. We find that AZC arrests proliferation in the G1 phase of the cell cycle by the same mechanism as temperature up-shift. Appropriate activation of HSF is required to trigger proliferation arrest.
Materials and Methods
Chemicals, Media, and Manipulations.
All chemicals were purchased from Sigma. Components of growth media were from Difco. Liquid medium was prepared as described by Ogas et al. (21). Solid medium contained an additional 2% agar.
For drug treatments, cells were grown at 30°C to an OD600 of 0.05, unless stated otherwise. AZC was dissolved in water to a stock concentration of 500 mM and added to growth medium to achieve the final concentrations specified in individual experiments.
For temperature up-shift experiments, cells were grown at 23°C to an OD600 of 0.2 and added to an equal volume of yeast extract/peptone/dextrose (YPD) in a conical flask preheated at 36°C in a water bath. Incubation was continued at 36°C with agitation for the time course of each experiment.
All optical density measurements were made on a Milton Roy Spectronic 601 spectrophotometer.
Strains and Plasmids.
Strains W303–1A (wild type, WT: MATa.ura3–1.lys2.trp1–1.leu2–3.112.his3–11.can1–100.ade2–1), JVG961 (S288c WT: MATα.ura3–52.lys2–801.his3-Δ200.leu2.ade2–101.ho∷LacZ), and JVG917 (EG123 WT: MATa.trp1.leu2.ura3.his4.can1) were from the laboratory collections. Strains MH297 (EXA3–1 strain: MATα.leu2–3, 112.lys2,ura3–52, his3–11, 15, trp1-Δ1,EXA3–1) and a corresponding WT, DS10 (MATa.leu2, 112, lys2,ura3–52,his3–11, 15,trp1-Δ1,lys1), were kind gifts from E. Craig (University of Wisconsin, Madison). GAP510 (MATa.ubc4Δ1∷HIS3.ubc5Δ1∷LEU2.leu2–3, 112.ura3–52.trp1–1.his3-Δ200.lys2–801) and a corresponding WT GAP512 (MATα.leu2–3, 112.ura3–52.trp1–1.his3-Δ200.lys2–801) were kind gifts from S. Jantsch (Max-Planck-Gesellschaft, Tubingen, Germany).
The strain MH297 (EXA3–1) contains a mutation in the HSP104 gene (22). The plasmid pYS104 containing HSP104 in pRS316 was kindly supplied by E. Craig. Transformants containing pYS104 were grown in SD-ura overnight and transferred to YPD 3–4 h before heat shock or AZC treatment.
The following mutants in the S288C strain background were kindly provided by J. P. Ogas (University of California, San Francisco). The genotypes were as for JVG961 (see above), except as follows: JVG1052 (trp1Δ63.cln1Δ∷TRP1.cln2Δ∷LEU2), JVG1022 (cdc28–1.HIS3.tyr1), JVG1023 (cdc28–9), and JVG1041 (cdc28–1N.HIS3).
pJO96 (CLN3–2 in YCp50) was a kind gift from J. P. Ogas.
Glycogen and Trehalose Assays.
Extraction and estimation of carbohydrate levels were performed as described by Sullivan et al. (22). For both glycogen and trehalose determinations, cells were chilled quickly by the addition of crushed ice, collected by centrifugation at 4°C, and washed three times with distilled water at 4°C. The cell pellets were frozen and stored at −20°C.
For trehalose measurements, the pellets were thawed in 3 vol of 500 mM trichloroacetic acid and extracted for 40–60 min at room temperature. After centrifugation and a second extraction of the pellets, the combined supernatants were assayed for anthrone-positive material.
Glycogen content was determined by enzymatic hydrolysis for the combined supernatants prepared as described above. Two milliliters of amyloglucosidase (1 mg/ml in 0.2 M acetate buffer, pH 4.8) and 0.3 ml of neutralized carbohydrate sample were incubated for 2 h at 37°C. The incubation was terminated by heating to 100°C for 10 min. Denatured protein was removed by centrifugation. Total sugar liberated was estimated spectrophotometrically with p-hydroxybenzoic acid hydrazide reagent, with the use of glucose as a standard. Amyloglucosidase is known to release more than 98% of the glucose from a glycogen sample when incubated as described.
Glucose standards were used for both trehalose and glycogen determinations. All determinations were made at least three times. Heat shocks (42°C for 30 min, 1 h, or 2 h) were used as positive controls.
Acquired Thermotolerance Assays.
Cells (JVG961: S288c WT) were grown in YPD medium at 23°C to midlogarithmic growth. Samples, either pretreated or not with 10 mM AZC in YPD for 3 h, were incubated at 23°C or 37°C for an additional hour before heat shock to 50°C by a 1:40 dilution into preheated medium at 50°C. Samples were incubated at 50°C for the times indicated in Table 1 before transfer to iced water. Cells were diluted into fresh YPD and immediately plated on YPD plates and incubated at 30°C for 3 days. The number of colonies on each plate was quantitated. The experiment was performed three times independently, yielding qualitatively similar results. The qualitative results for one replicate are shown in Table 1.
Table 1.
AZC does not induce thermotolerance
| Treatment before heat shock | # Colonies formed after heat shock to 50°C for:
|
||
|---|---|---|---|
| 0 min | 10 min | 30 min | |
| 23°C | 3,504 | 0 | 0 |
| 23°C + AZC (20 mM) for 3 h | 3,232 | 0 | 0 |
| 23°C to 37°C for 1 h | 3,600 | 2,160 | 308 |
| 23°C + AZC (20 mM) for 2 h then to 37°C for 1 h (still in AZC) | 3,040 | 936 | 244 |
Northern Probes.
Probes were amplified from yeast amplified ORFs (Research Genetics, Huntsville, AL) using universal yeast primers (Research Genetics) by PCR using Reddy-load PCR mix (Advanced Biotechnologies, Surrey, U.K.) according to the manufacturer's instructions. The genes that were probed (and their ORF numbers) are as follows: ACT1 (YFL039c), CLN1 (YMR199w), CLN2 (YPL256c), and CLN3 (YAL040c).
Amplified yeast ORFs were run on 1% agarose gels and purified with a Concert Rapid Gel Extraction System (Life Technologies, Grand Island, NY) according to the manufacturer's instructions. Amplified yeast ORFs were labeled with [32P]dCTP (New England Nuclear) with the use of a Prime-It II random primer labeling kit (Stratagene) and purified with NucTrap purification columns (Stratagene) according to the manufacturer's instructions.
Northern Analysis.
Extraction of total RNA was performed as described (20). For Northern analysis, 50 μg of total RNA was denatured at 65°C for 5 min before separation on a 1.3% agarose gel containing 10% formaldehyde. The RNA was transferred overnight to biodyne-b (0.45 μm) membrane (Pall) in 10× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7). Membranes were baked for 2 h at 200°C. Membranes were prehybridized in 50% formamide, 10× Denhardt's solution, 2% SDS, 5× SSC, and 100 μg/ml denatured salmon sperm DNA for 2 h. The labeled probe was boiled for 5 min before addition to the membrane in prehybridization mix. Membranes were hybridized overnight at 42°C. After stringent washing (2 × 15 min in 1% SDS, 0.25× SSC at room temperature; 15 min in 0.1% SDS, 0.1% SSC at room temperature; and 30 min in 0.1% SDS, 0.1% SSC at 65°C), membranes were exposed to x-ray film (Konica, Tokyo) at −70°C with intensifying screens for an appropriate length of time.
Flow Cytometry.
Cultures were fixed in ethanol (70%) for 1 h. Fixed cells were treated with RNase overnight, stained with propidium iodide, sonicated, and subjected to flow cytometry analysis, as described by Gray et al. (13).
Budding Indices.
Budding indices were determined on samples that had been fixed in ethanol (70%) for 1 h and resuspended in TE buffer (10 mM Tris/1 mM EDTA, pH 7.5). Samples were sonicated immediately before analysis, as described by Gray et al. (13). At least 200 cells were counted for each sample.
Results
AZC Does Not Cause Accumulation of Glycogen and Trehalose.
From our previous studies, we found that AZC treatment at concentrations sufficient to inhibit proliferation strongly activates HSF but does not robustly activate the Msn2/4-STRE regulon. It has recently been shown that the heat shock-induced accumulation of the storage carbohydrates glycogen and trehalose depends on Msn2 and Msn4 (7). If AZC treatment indeed fails to significantly activate the STRE regulon, then we expect AZC treatment not to induce accumulation of these storage carbohydrates (7). We find that AZC treatment for up to 5 h fails to stimulate accumulation of glycogen and trehalose (data not shown). However, the incorporation of AZC into cellular protein may somehow prevent proper regulation of the STRE regulon. We find that pretreatment with AZC does not preclude accumulation of these carbohydrates in response to a subsequent heat shock (data not shown). We conclude that AZC treatment does not activate the STRE regulon, even though the regulon remains inducible by a subsequent heat shock. Misfolded proteins are thus unlikely to trigger the STRE regulon.
AZC Does Not Induce Thermotolerance.
Activation of the Pkc1-MAP kinase pathway by heat shock has been proposed to result from thermal damage to the cell surface (8, 13). This heat shock activation of the pathway requires protein synthesis (8). It is possible that the trigger for heat shock activation of the Pkc1-MAP kinase pathway is the misfolding of proteins at the cell surface. In this case, AZC should induce the Pkc1 pathway. The acquisition of thermotolerance by incubation at 37°C before a subsequent, and otherwise lethal, heat shock has been shown to depend on the Pkc1 pathway (8). Indeed, activation of the MAP kinase branch of the pathway has been shown to be sufficient for the acquisition of thermotolerance (8). We therefore monitored thermotolerance as a sensitive readout of the state of activation of the Pkc1-MAP kinase pathway. As shown in Table 1, we find that AZC treatment does not lead to thermotolerance, indicating that AZC does not significantly induce the Pkc1 pathway. However, AZC may prevent activation of the Pkc1 pathway or the acquisition of thermotolerance by an indirect mechanism. As shown in Table 1, we find that AZC-arrested cells are competent to acquire thermotolerance. We conclude that AZC does not induce thermotolerance. We infer that AZC does not cause activation of the Pkc1-MAP kinase pathway.
In addition, these data indicate that the arrest in response to AZC is reversible upon washout of the compound. Indeed, we have found that the arrest is reversible even after 24 h in AZC (G.A.P., unpublished observation).
AZC Treatment and Temperature Up-shift Prevent Entry into the Cell Cycle by the Same Mechanism.
AZC inhibits cell proliferation at the concentrations used in this study. The arrested cells are clearly viable (see previous section). Is AZC causing proliferation arrest by the same mechanism as mild heat shock? As shown in Fig. 1A, we find that growth of a WT strain is hypersensitive to AZC at 37°C relative to 28°C. This observation is true for multiple strain backgrounds tested (data not shown). This finding is consistent with AZC and high temperature arresting proliferation by a common mechanism.
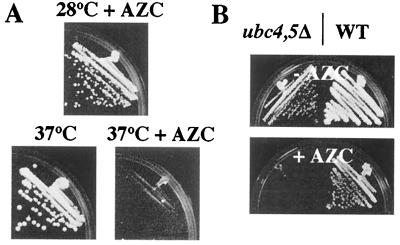
Figure 1.
AZC stops proliferation via incorporation into protein and may act by the same mechanism as mild heat shock. (A) JVG718 (EG123 WT) was streaked on YPD plates or on YPD plates containing 7 mM AZC. Plates were incubated at either 28°C or 37°C for 3 days. (B) GAP510 (WT) and a congenic ubc4Δ.ubc5Δ mutant (GAP512) were streaked on YPD plates or on YPD plates containing AZC (3 mM). Plates were incubated at 23°C for 3 days.
It is known that mutants deficient in the ubiquitin-conjugating enzymes Ubc4 and Ubc5 are unable to efficiently degrade misfolded and analog-containing proteins (23). ubc4Δ.ubc5Δ mutants are unable to proliferate at 37°C, indicating that they are hypersensitive to high temperature (23). As shown in Fig. 1B, we find that ubc4Δ.ubc5Δ double mutants are also hypersensitive to growth inhibition by AZC treatment. This finding is again consistent with high temperature and AZC treatment arresting proliferation by the same mechanism. Furthermore, this finding strongly suggests that AZC exerts its growth-inhibitory effect via incorporation into cellular protein.
Proliferating cells shifted from 23°C to 36°C (“temperature up-shift”) become transiently arrested in the G1 phase of the cell cycle (9, 10). Is AZC mimicking this effect? As shown in Figs. 2A and 3B, we find that AZC treatment causes WT cells to accumulate in an unbudded state with 1N DNA content. Thus AZC treatment and temperature up-shift arrest cells at the same stage of the cell cycle, namely G1.
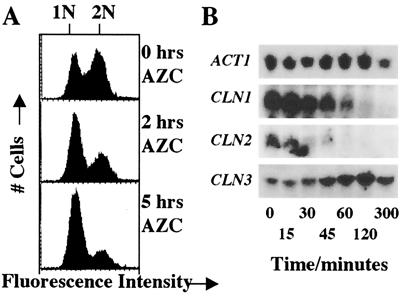
Figure 2.
AZC treatment causes G1 arrest and repression of CLN1 and CLN2. (A) A midlogarithmic culture of JVG961 (S288C WT) growing in YPD at 30°C was treated with AZC (10 mM) at 0 h. Samples were stained with propidium iodide and analyzed by flow cytometry. Cells with unduplicated DNA or with duplicated DNA are indicated by the 1N and 2N labels, respectively. (B) A midlogarithmic culture of JVG961 (S288C WT) growing in YPD at 30°C was treated with AZC (10 mM) at 0 min. Total RNA was prepared from samples taken at the times indicated. Samples (50 μg RNA) were subjected to Northern analysis. Blots were probed for CLN1, CLN2, CLN3, and ACT1.
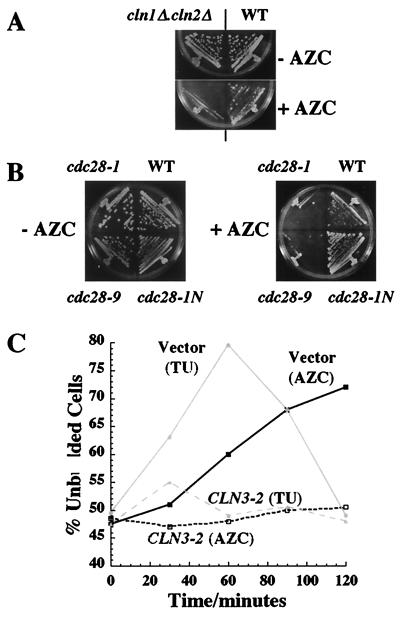
Figure 3.
AZC treatment limits G1 cyclin activity. (A) Strains JVG961 (WT) and JVG1052 (cln1Δ.cln2Δ) were streaked on YPD plates containing AZC (7 mM) or no AZC and incubated at 30°C for 3 days. (B) Strains JVG961 (WT), JVG1022 (cdc28–1), JVG1023 (cdc28–9), and JVG1041 (cdc28–1N) were streaked on plates containing AZC (5 mM) or no AZC and were incubated at 23°C for 4 days. (C) Midlogarithmic cultures of JVG961 (S288C WT) transformants containing pCLN3–2 (pJO96) or a vector control (YCp50) growing in SD-URA at 23°C or 30°C were subjected to a temperature up-shift (TU: 23°C to 37°C) or were treated with AZC (10 mM, 30°C), respectively. The percentage of unbudded cells was determined for each culture as a function of time after treatment was initiated.
Temperature up-shift is known to cause repression of the CLN1 and CLN2 genes coincident with the transient G1 arrest (10). Temperature up-shift does not repress expression of CLN3 (10). Does AZC also cause repression of CLN1 and CLN2? Unfortunately, these genes did not yield high-quality signals in our microarray analysis (E.W.T., unpublished observations). We therefore monitored the expression of CLN1 and CLN2 in response to AZC treatment by Northern analysis. As shown in Fig. 2B, we find that expression of CLN1 and CLN2 is severely reduced by AZC treatment. In contrast, CLN3 expression is not affected by AZC treatment (Fig. 2B). Thus both AZC treatment and temperature up-shift cause repression of CLN1 and CLN2.
Does AZC treatment therefore cause G1 arrest via limiting G1 cyclin activity? If so, we expect that mutants defective in the G1/S transition should be hypersensitive to proliferation arrest caused by AZC treatment. As shown in Fig. 3A, we find that growth of a mutant lacking CLN1 and CLN2 is indeed inhibited by a concentration of AZC that is insufficient to arrest its WT. We find that temperature-sensitive alleles of CDC28 that arrest in G1 at restrictive temperature (cdc28–1 and cdc28–9) are hypersensitive to AZC at permissive temperatures (Fig. 3B). In contrast, the cdc28–1N allele, which arrests in G2 at restrictive temperatures, is not hypersensitive to AZC at permissive temperatures (Fig. 3B). Hence, AZC-treated cells are limiting for G1 cyclin-Cdc28 activity. Expression of hyperstable forms of either Cln2 or Cln3 have been shown to prevent G1 arrest in response to temperature up-shift (10). As shown in Fig. 3C, we find that expression of the hyperstable Cln3–2 (Daf1–1) prevents G1 arrest in response to both temperature up-shift and AZC treatment. We conclude that AZC-treated cells are limited for G1 cyclin activity.
Taken together, these results strongly indicate that AZC treatment and temperature up-shift arrest proliferation by the same mechanism.
Proper Activation of HSF Is Required for G1 Arrest in Response to AZC Treatment and Temperature Up-Shift.
The mechanism by which temperature-up-shifted cells affect G1 cyclin activity is not well understood. In particular, we do not know what heat-sensitive sensor initiates the signal to cease proliferation. However, we have shown that AZC selectively and almost exclusively activates HSF-dependent transcriptional events (E.W.T., G.A.P., and J.V.G., unpublished work). Is proper activation of HSF required for G1 arrest in response to AZC treatment or temperature up-shift?
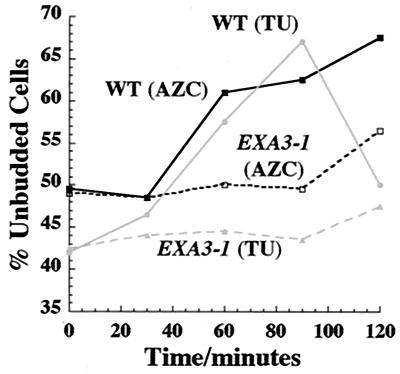
To address this question, we exploited a mutant form of the HSF1 gene encoding HSF, EXA3–1 (21). This mutation leads to aberrant timing and extent of activation of HSF-dependent transcripts in response to severe heat shocks, temperature up-shifts, and AZC treatment (5, 22). This mutant (and its WT equivalent) do not arrest permanently in AZC, but display a transient activation of HSF upon AZC treatment (ref. 6; data not shown). We tested the capacity of the WT and EXA3–1 strains to become arrested in G1 in response to both temperature up-shift and AZC treatment. As shown in Fig. 4, we find that the WT strain arrests in G1 in response to both treatments. The AZC-induced arrest is transient in this strain background (data not shown). In contrast, we find that the EXA3–1 mutant is defective in G1 arrest in response to temperature up-shift and AZC treatment.
Figure 4.
The EXA3–1 mutation in the gene encoding HSF prevents G1 arrest upon both temperature up-shift and AZC treatment. Midlogarithmic cultures of DS10 (WT) and the congenic EXA3–1 mutant (MH297) growing in YPD at 23°C were subjected to a temperature up-shift (TU: 23°C to 37°C). Midlogarithmic cultures of the same strains growing in YPD at 30°C were treated with AZC (50 mM) at t = 0 without a temperature change. The percentage of unbudded cells was determined for each culture as a function of time after treatment was initiated.
We conclude that proper activation of HSF is required for efficient G1 arrest in response to both temperature up-shift and AZC treatment. This finding indicates that the misfolding of cellular protein triggers G1 arrest upon temperature up-shift via activation of HSF.
[Note: The EXA3–1 strain is deficient in HSP104 (21). However, the introduction of HSP104 on a plasmid into this strain does not alter its capacity for G1 arrest in response to either temperature up-shift or AZC treatment (data not shown).]
Discussion
AZC Acts by Misfolding a Fraction of Cellular Protein.
AZC is known to be incorporated into cellular protein in organisms for which the analog is growth inhibitory (18, 19). The consequence of AZC incorporation is well understood. AZC is incorporated into protein competitively with proline. The four-membered ring of the analog (compared with the five-membered ring of proline) causes a distortion of the polypeptide backbone at the point of incorporation. The consequence is an altered protein fold, leading to reduced thermal stability or failure to successfully fold (24–26). AZC-containing proteins bind avidly to Hsp70 chaperones in vivo (11) and are prone to degradation by a ubiquitin-dependent mechanism involving the Ubc4 and Ubc5 ubiquitin-conjugating enzymes (23).
A number of lines of evidence suggest that the effect of AZC on the cell is via its incorporation into proteins (and consequently their misfolding). First, the expression changes induced by AZC are closely mimicked by ethanol treatment, another agent capable of misfolding protein (E.W.T., G.A.P., and J.V.G., unpublished work). Second, the relative capacity of AZC and canavanine (an arginine analog) to induce the heat shock transcripts correlates with their relative capacities to misfold proteins into which they are incorporated (AZC being more potent) (E.W.T., G.A.P., and J.V.G., unpublished work). Finally, ubc4Δ.ubc5Δ mutants defective in the degradation of analog-containing and misfolded proteins are hypersensitive to AZC (Fig. 1B), consistent with the AZC-generated signal originating from AZC-containing proteins.
To date, we have been unable to synthesize radioactive AZC to directly determine the extent of incorporation of the analog in our experiments. Despite this limitation, it is clear that the extent of protein misfolding caused by AZC in our experiments is relatively low. (Note: Proline is present in the media used for our experiments.) First, AZC-treated cells are viable and arrest is reversible upon washout of the compound. Second, AZC-treated cells remain responsive to subsequent heat shocks (accumulation of glycogen and trehalose, and acquisition of thermotolerance) and to treatment with rapamycin (induction of GAP1 and MEP2; E.W.T., G.A.P., and J.V.G., unpublished work). Third, AZC treatment induces a very specific subset of transcripts, as determined by genomewide microarray analysis (E.W.T., G.A.P., and J.V.G., unpublished work). Such specificity strongly argues against widespread cellular dysfunction. We conclude that AZC misfolds a small fraction of cellular protein to which HSF is exquisitely sensitive.
Misfolded Proteins Do Not Induce the STRE Regulon.
From our previous analysis of genomewide expression changes induced by AZC, it is clear that the STRE regulon is not efficiently induced by AZC treatment (E.W.T., G.A.P., and J.V.G., unpublished work). Indeed, the STRE regulon is only very weakly induced by treatment with ethanol, another agent capable of misfolding cellular protein (27). These data suggest that misfolded proteins may not the primary signal for the heat shock activation of the STRE regulon. Our results here support this conclusion. We find that the accumulation of glycogen and trehalose (Msn2/4-dependent processes) does not occur upon AZC treatment (data not shown). Misfolded proteins are thus unlikely to be the primary trigger for heat shock activation of the STRE regulon. The nature of that trigger is unknown.
Misfolded Proteins Do Not Activate the Pkc1-MAP Kinase Pathway.
The Pkc1-MAP kinase pathway is inducible by elevated temperature (8). The MAP kinase branch of this pathway is required for acquired thermotolerance (8). Indeed, activation of the MAP kinase branch of the pathway is sufficient to confer acquired thermotolerance (8). Acquired thermotolerance is thus a very sensitive readout of the state of activation of the Pkc1-MAP kinase pathway.
We find that thermotolerance is not induced by AZC treatment, even though AZC-arrested cells competently acquire thermotolerance in response to a subsequent heat shock (Table 1). Indeed, the MAP kinase of the pathway is not significantly activated by AZC treatment, as determined by biochemical assay (S.A.K., unpublished observation). We conclude that misfolded proteins are not involved in generating the cell surface stresses that trigger the Pkc1 pathway.
AZC Treatment and Temperature Up-Shift Arrest Cell Proliferation by a Common Mechanism Dependent on HSF.
It is clear from the findings here that AZC treatment arrests cells in a viable state, that arrest is reversible upon washout of the compound, and that arrested cells are competent to respond normally to a variety of stimuli. We find that this arrest occurs by the same mechanism as that caused by mild heat shock. First, the transcript profiles of AZC-treated cells are remarkably similar to those of temperature-up-shifted cells when they are transiently inhibited for proliferation (E.W.T., G.A.P., and J.V.G., unpublished work). Second, cells at 37°C are more sensitive to growth inhibition by AZC than are the same cells at 28°C. Third, proliferation of cells deficient in Ubc4 and Ubc5 is hypersensitive to elevated temperature and to AZC-induced arrest. Fourth, both AZC treatment and temperature up-shift cause a G1 arrest. Fifth, both treatments lead to repression of CLN1 and CLN2 expression but do not affect CLN3 mRNA levels. Sixth, expression of a hyperstable form of the G1 cyclin Cln3 prevents G1 arrest by both temperature up-shift and AZC treatment. Finally, a mutated from of HSF prevents G1 arrest in response to both treatments.
We conclude that some misfolding of cellular proteins, either by AZC treatment or at elevated temperature, causes a G1 arrest by limiting G1 cyclin activity via an unknown mechanism dependent on the proper activation of HSF.
Note that the EXA3–1 mutation and expression of Cln3–2 prevent G1 arrest in response to AZC, but they do not make cells resistant to AZC (data not shown). Rather, these cells arrest randomly in the cell cycle upon AZC treatment (data not shown). We do not know the basis for this random arrest, nor do we know if HSF is required for it (inasmuch as the EXA3–1 allele is not a null).
The EXA3–1 mutation leads to a delay in the timing of induction of HSF-dependent transcripts in response to heat shocks and AZC treatment (5). Thus the failure of the EXA3–1 mutant to arrest in G1 in response to temperature up-shift and AZC treatment indicates that transcription induction by activated HSF is somehow required to halt cell cycle progression in G1 by limiting G1 cyclin activity. It should be noted that the extent of induction of transcripts by heat shock or AZC treatment is also affected by the EXA3–1 mutation: some transcripts are induced more strongly in the mutant, whereas others are not induced as well (5, 21). Thus, it is formally possible, if unlikely, that the mutation is preventing G1 arrest by causing the overexpression of some target gene(s). In any event, activation of HSF modulates the G1/S transition.
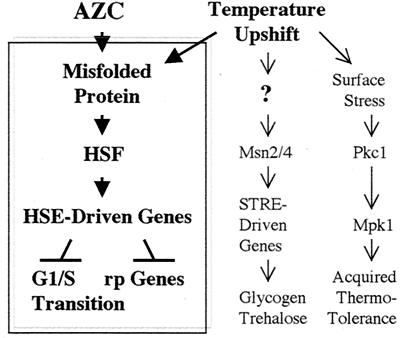
In summary, we propose that the misfolding of small amounts of cellular proteins by AZC treatment (or mild heat shock) selectively induces HSF-dependent events: activation of HSE-containing genes, repression of the ribosomal protein genes, and inhibition of the G1/S transition. HSF appears to be an exquisitely sensitive sensor of misfolded proteins (Fig. 5). To a first approximation, it appears that the cellular response to mild heat shock is the sum of the separate responses to individual heat-induced defects, of which we can identify misfolded proteins and cell surface stress. An unknown heat-sensitive parameter triggers activation of the STRE regulon (Fig. 5).
Figure 5.
Model for cellular response to temperature up-shift and AZC treatment. AZC misfolds cellular protein into which it is incorporated. Misfolded protein specifically activates HSF, causing induction of HSE-driven transcripts and (by unknown mechanisms) subsequent repression of the ribosomal protein (rp) genes and G1 arrest. Temperature up-shift causes protein misfolding and thereby triggers the HSF-dependent events. The thermal trigger for the STRE regulon is unknown (?) but is independent of protein misfolding. The Pkc1 pathway is activated by thermal stress to the cell surface independently of misfolded proteins.
Acknowledgments
We thank Ian Graham and members of the Gray and Petsko laboratories for their help and support throughout this work. Maria Giannakou contributed to this project at an early stage. We thank E. Craig, S. Jantsch, and J. P. Ogas for their generous gifts of strains and plasmids. This work was supported by funds from the Ellison Research Foundation (to G.A.P.) and grants from the Royal Society (RS19987) and the Wellcome Trust (054419) (to J.V.G.). E.W.T. was supported by a Ph.D. studentship from the Biotechnology and Biological Sciences Research Council.
Abbreviations
- AZC
azetidine-2-carboxylic acid
- HSF
heat shock factor
- HSE
heat shock element
- STRE
stress response element
- Pkc1
protein kinase C of Saccharomyces cerevisiae
- MAP
mitogen-activated protein
- YPD
yeast extract/peptone/dextrose
- WT
wild type
References
- 1.Parcell D A, Lindquist S. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 2.Lindquist S. Nature (London) 1981;293:311–314. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- 3.Ruis H, Schüller C. BioEssays. 1995;17:959–996. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 4.Wu C. Annu Rev Cell Dev Biol. 1993;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 5.Lopez N, Halliday J, Walter W, Craig E A. J Bacteriol. 1999;181:3136–3143. doi: 10.1128/jb.181.10.3136-3143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boy-Marcotte E, Lagiel G, Perrot M, Bussereau F, Boudsocq A, Jacquet M, Labarre J. Mol Microbiol. 1999;33:274–283. doi: 10.1046/j.1365-2958.1999.01467.x. [DOI] [PubMed] [Google Scholar]
- 7.Parrou J L, Teste R A, Francoise J. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- 8.Kamada Y, Jung U S, Piotrowski J, Levin D E. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 9.Johnston G C, Singer RA. Mol Gen Genet. 1980;178:357–360. doi: 10.1007/BF00270484. [DOI] [PubMed] [Google Scholar]
- 10.Rowley A, Johnston G C, Butler B, Werner-Washburne M, Singer R A. Mol Cell Biol. 1993;13:1034–1041. doi: 10.1128/mcb.13.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckmann R P, Mizzen L A, Welch W J. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- 12.Craig E A, Gross C A. Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 13.Gray J V, Ogas J P, Kamada Y, Stone M, Levin D E, Herskowitz I. EMBO J. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verna J, Lodder A, Lee K, Vagts A, Ballester R. Proc Natl Acad Sci USA. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ketela T, Green R, Bussey H. J Bacteriol. 1999;181:3330–3340. doi: 10.1128/jb.181.11.3330-3340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajavel M, Philip B, Bueher B M, Errede B, Levin D E. Mol Cell Biol. 1999;19:3969–3976. doi: 10.1128/mcb.19.6.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin H, Rodriguez-Pachon J M, Ruis C, Nombela C, Molina M. J Biol Chem. 2000;275:1511–1519. doi: 10.1074/jbc.275.2.1511. [DOI] [PubMed] [Google Scholar]
- 18.Fowden L, Richmond M H. Biochim Biophys Acta. 1963;71:459–461. [Google Scholar]
- 19.Fowden L, Lewis D, Tristram H. Adv Enzymol Relat Areas Mol Biol. 1967;29:89–163. doi: 10.1002/9780470122747.ch3. [DOI] [PubMed] [Google Scholar]
- 20.Ogas J, Andrews B J, Herskowitz I. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 21.Halladay J T, Craig E A. Mol Cell Biol. 1995;15:4890–4897. doi: 10.1128/mcb.15.9.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan P A, Yin C Y, Molloy C, Templeton M D, Shepard M. Can J Microbiol. 1983;29:1514–1525. doi: 10.1139/m83-233. [DOI] [PubMed] [Google Scholar]
- 23.Seufert W, Jantsch S. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zagari A, Nemethy G, Scheraga H A. Biopolymers. 1990;30:951–959. doi: 10.1002/bip.360300909. [DOI] [PubMed] [Google Scholar]
- 25.Zagari A, Nemethy G, Scheraga H A. Biopolymers. 1990;30:967–974. doi: 10.1002/bip.360300911. [DOI] [PubMed] [Google Scholar]
- 26.Zagari A, Nemethy G, Scheraga H A. Biopolymers. 1994;34:51–60. doi: 10.1002/bip.360340107. [DOI] [PubMed] [Google Scholar]
- 27.Schüller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]