Abstract
The structure specific flap endonuclease 1 (FEN1) plays an essential role in long-patch base excision repair (BER) and in DNA replication. We have generated a fluorescently tagged FEN1 expressing mouse which allows monitoring the localization and kinetics of FEN1 in response to DNA damage in living cells and tissues. The expression of FEN1, which is tagged at its C-terminal end with enhanced yellow fluorescent protein (FEN1-YFP), is under control of the endogenous Fen1 transcriptional regulatory elements. In line with its role in processing of Okazaki fragments during DNA replication, we found that FEN1-YFP expression is mainly observed in highly proliferating tissue. Moreover, the FEN1-YFP fusion protein allowed us to investigate repair kinetics in cells challenged with local and global DNA damage. In vivo multi-photon fluorescence microscopy demonstrates rapid localization of FEN1 to local laser-induced DNA damage sites in nuclei, providing evidence of a highly mobile protein that accumulates fast at DNA lesion sites with high turnover rate. Inhibition of poly (ADP-ribose) polymerase 1 (PARP1) disrupts FEN1 accumulation at sites of DNA damage, indicating that PARP1 is required for FEN1 recruitment to DNA repair intermediates in BER.
INTRODUCTION
All cells are confronted with tens of thousands of DNA lesions each day by endogenous cellular processes. Generally, these DNA lesions severely affect DNA metabolism. Base excision repair (BER) is the major pathway for removing oxidative DNA damage (1,2). In brief, BER is initiated by one of many DNA glycosylases, which recognizes and removes the damaged base. The apurinic/apyrimidinic (AP) sites generated are further processed by downstream enzymes that carry out strand incision, gap filling and ligation in a coordinated manner. Two sub-pathways of BER have been characterized in vitro, and are classified according to the length of the repair patch produced in the reaction: (i) short-patch (SP, one nucleotide replaced) and (ii) long-patch (LP, more than one nucleotide replaced) BER (2).
The structure specific flap endonuclease 1 (FEN1) plays a crucial role in maintaining genome integrity of cells and cleaves 5′ DNA flaps formed as intermediates in LP-BER and during DNA replication (3–7). In addition to flap removal in DNA repair and replication, FEN1 has been implicated to play a role in recombination (8,9), apoptosis (10), non-homologous end joining (11), repeat expansion (12,13) and telomere maintenance (14–17), demonstrating the indispensable role of FEN1 in genome maintenance and cell viability.
A large panel of proteins required for genome stability have been reported to interact with FEN1. These include proliferating cell nuclear antigen (PCNA), replication protein A (RPA), DNA polymerases β and δ, AP endonuclease 1 (APE1), RecQ helicases WRN and BLM, endonuclease/helicase DNA2, endonuclease G (ENDO G), HIV integrase and p300 (18). In mammalian cells, FEN1 is widely expressed. Highly proliferative tissues like bone marrow, testis and thymus show particularly high expression of FEN1 (19–22). In agreement with the many important roles of FEN1, mouse null-mutant FEN1 causes embryonic lethality (23,24). It has also been shown that the conserved amino-acids of the PCNA binding site of FEN1 are required for viability (25,26). In contrast, mice carrying mutations in the nuclease domain of FEN1 are viable and cancer-prone (25,27). Recently, another 5′flap cleaving helicase/nuclease, human DNA2, originally identified in yeast as a nuclear DNA replication and repair factor, was found to predominantly locate to mitochondria, suggesting that FEN1 could be the main 5′flap endonuclease in the nucleus (28,29).
The BER pathway has been extensively studied in vitro, yet, the kinetics of BER proteins in vivo is less well known. Live cell FEN1 kinetic studies could answer questions on FEN1 mobility and mode of action in different cell types. Mechanisms of FEN1 activity in replication and LP-BER, and mechanism of interaction with other proteins, throughout the cell cycle and during aging under varying physiological conditions can be revealed by in vivo imaging. Moreover, it is difficult to address the relative contribution of SP-BER and LP-BER in different cell types, although many in vitro studies imply that SP-BER is the predominant sub-pathway (30–32). To repair oxidative damage, a cell will either exploit SP- or LP-BER, and several hypotheses have been formulated regarding which pathway will be used. Briefly, when an oxidized or reduced AP-site cannot be processed by polymerase β, as the polymerase can not eliminate the modified sugar by 5′dRP lyase activity, a DNA strand displacement and flap formation will occur, inducing the LP-BER (5). Alternatively, the ATP concentration near the AP-site could direct the pathway choice (33). LP-BER, resulting from XRCC1 promoting DNA strand displacement by polymerase β, is predominant during energy depletion, and is required for ATP generation from poly (ADP-ribose) (PAR). SP-BER is preferred during energy abundance, when Ligase III prevents strand displacement synthesis and promotes ligation.
Here, we have examined the FEN1 kinetics at sites of DNA damage in vivo by measuring enhanced yellow fluorescent protein (eYFP) from a novel mouse model where FEN1 is fused to eYFP (in the following denoted as YFP). The FEN1-YFP model enabled us to characterize expression levels and distribution of FEN1-YFP in cultured mouse cells and living tissues. We also studied FEN1-YFP repair kinetics in cells when challenged with local and global DNA damage and following poly (ADP-ribose) polymerase (PARP) inhibition. Moreover, the dual expression of two fluorescently tagged proteins, FEN1-YFP and mCherry-PCNA, allowed us to visualize the dynamics of FEN1 and its interacting partner PCNA in DNA replication foci and at DNA repair sites over time.
MATERIALS AND METHODS
General methods used (mice, organs, RNA and protein isolation and western blot analysis) is found in Supplementary Data.
Ethics statement
Mice were maintained in the minimal disease unit in an animal facility under barrier conditions. Animal experiments were performed in accordance with institutional guidelines and national legislation, and according to the 3R animal welfare rules.
Generation of the targeting construct
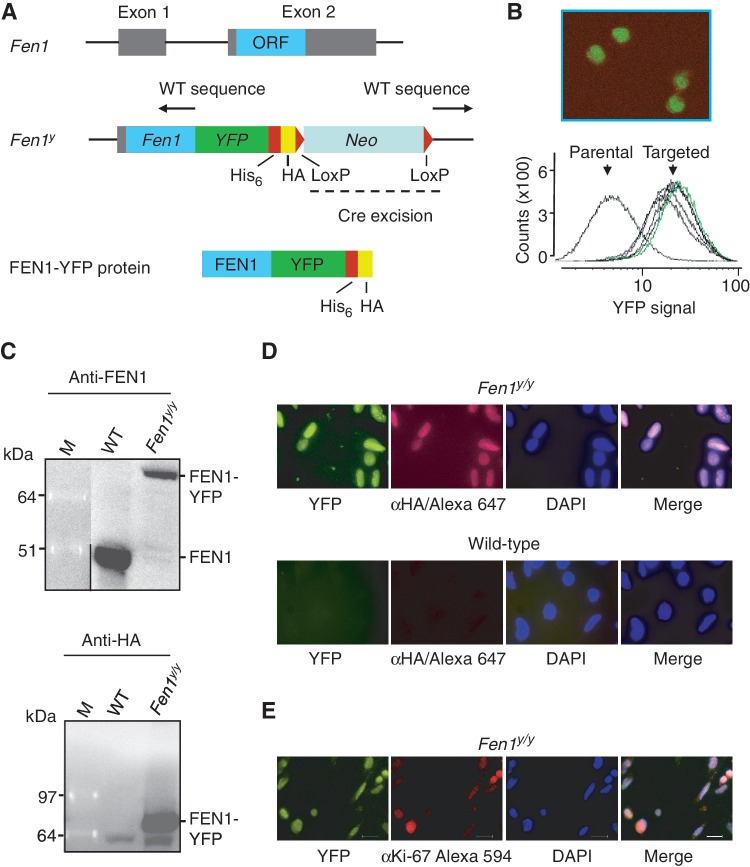
The knock-in targeting vector consisted of an ∼12 kb mouse genomic DNA, containing the Fen1 locus. A plasmid vector containing ATG-less YFP, His6 and HA epitope tags and a Neomycin (Neo) expression cassette flanked by two LoxP sites, was cloned in frame to the C-terminal genomic part of Fen1 to generate a FEN1-YFP fusion gene with the aim to express FEN1-YFP protein. The YFP vector was produced as previously described (34). To make this Fen1-YFP targeting vector, the genomic Fen1 was PCRed from 129/SvJ mouse genomic DNA with SacII and BamHI restriction enzyme sites in the primers. The Polymerase chain reaction (PCR) was designed to generate a 2.1 kb homologous arm fragment from Fen1 intron1-2, covering the Fen1 translated part of exon 2 and to exclude the stop codon of Fen1. Following PCR, restriction enzyme digestion and blunting of the BamHI site, the 2.1 kb fragment containing Fen1 was cloned into the YFP vector, using SacII and SmaI in the YFP vector multiple cloning site (MCS). From the Fen1 E160D knock-in targeting vector (25), the Fen1 and Neo genes were cut out using HpaI and AscI. The remaining vector, containing the homologous arm with the 3′ sequence downstream Fen1, was blunted and ligated with the SacI and NotI digested blunted Fen1-YFP fragment, obtaining the Fen1-YFP-His6-HA-LoxP-Neo-LoxP vector (Figure 1A).
Figure 1.
Generation and characterization of mice expressing FEN1-YFPHis6-HA fusion protein. (A) Physical map of the genomic DNA containing the mouse Fen1 gene. Exon 2 contains the entire ORF (in turquoise) of the Fen1 gene. Genomic (WT) sequences, indicated with dotted lines and black arrows, were subcloned upstream and downstream of the genes encoding enhanced YFP and Neo. The Fen1 ORF was included in the targeting construct, excluding the stop codon, to allow in-frame fusion with the modified fluorescent ATG-less YFP, with an additional stretch of six histidines and an HA-epitope tag. The dominant selectable marker, Neo, was flanked by two LoxP sites to enable Cre-excision. Details of the targeting strategy are presented in section ‘Materials and Methods’ and in Supplementary Data. (B) YFP expression in ES cells. Left: FACS analysis of parental and five targeted Fen1yNeo/WT ES cell clones. The positive clones all display histograms with YFP-signal after homologous recombination of the targeting construct, which is lacking in the parental ES cells. Indicated in green is the ES cell line taken further for blastocyst injection, which resulted in chimeric mice used for generation of Fen1yNeo/WT mice. Visualization of YFP in the superior ES cell line is shown to the right (Zeiss LSM 510 microscope). (C) Expression of the FEN1-YFPHis6-HA fusion protein in knock-in mice. Representative western blots with 40 μg of protein whole cell extracts (WCE) from thymus of a wild-type and a Fen1y/y knock-in mouse. Both anti-FEN1 and anti-HA antibodies detect a 70 kDa FEN1-YFPHis6-HA fusion protein. Moreover Anti-FEN1 detects the 43 kDa FEN1 protein from wild-type thymus. Anti-HA also binds to a smaller, unspecific band. (D) Endogenous expression and IF demonstrating FEN1-YFPHis6-HA fusion protein expression in primary MEF cells. Wild-type and Fen1y/y primary MEFs were stained with anti-HA (recognizing FEN1-YFPHis6-HA, red) and DAPI (staining nuclear DNA, blue). In Fen1y/y primary MEFs both endogenous YFP signal and anti-HA (IF) from the fusion protein were detected, whereas neither could be observed in wild-type primary MEFs, as expected. (E) FEN1 and Ki-67 expression in proliferating Fen1y/y primary MEF cells. Fen1y/y primary MEFs were stained with an anti-Ki-67 antibody (proliferation marker, red) and DAPI (staining nuclear DNA, blue). In Fen1y/y primary MEFs both endogenous YFP (green) signal from the fusion protein and Ki-67 immunostaining were detected in the nucleus. The scale bar is 20 μm. The cells in D. and E. were analysed using an Axio Observer.Z1 fluorescence microscope, with a Plan-Apochromat 63x/1.40 Oil DIC M27 objective and AxioCamMR3 camera.
Embryonic stem cell culture and gene targeting
About 25 μg of NotI linearized targeting vector was electroporated into approximately 1.6 × 107 embryonic stem (ES) cells (129, substrain R1), in 650 μl FBS-ES cell medium. Positive selection of Neo-resistance with G418 (300 μg/ml) was started 24 h after electroporation of ES cells. Negative selection agent FIAU (1-2′-deoxy-2′-fluoro-ß-D-arabinofuranosyl-5-iodouracil, 0.2 μm) was added 48 h after electroporation. FIAU selects against random integration events in the ES cells. The herpes simplex virus thymidine kinase (Tk) gene is present outside the region of homology in the targeting construct. After 8–9 days, resistant single colonies were isolated, and those were further grown to screen for homologous recombinants by Southern blot analyses of BamHI-digested DNA with a 463 bp 3′ external probe (Supplementary Figure S1A and B, the probe was PCRed with primers 19 and 20 in Supplementary Table S1). Out of 288 G418 resistant clones, 27 ES clones were positive homologous recombinant (10% of recombination). After DNA sequence verification, 2 out of the 27 correctly targeted ES clones were injected into blastocysts of C57Bl/6J mice and transplanted into CD1 foster mothers. Chimeric mice were further crossed to C57Bl/6J mice, and germline transmission of the targeted allele to offspring was genotyped by PCR (Supplementary Figure S1B–E and Supplementary Table S1).
Mice
The knock-in Fen1y (resulting fusion gene between Fen1 and YFP, coding for FEN1-YFP) allele was maintained in C57Bl/6J background. The Cre-expressing mice used to excise Neo from the Fen1yNeo knock-in mice were B6.C-Tg (CMV-cre)1Cgn/J mice (stock number 006054) from Jackson laboratories.
Cell culture and specific treatments
Primary mouse embryonic fibroblasts (MEFs) were isolated from Fen1WT/WT, Fen1WT/y and Fen1y/y E13.5 dpc embryos after a heterozygous × heterozygous mating of Fen1WT/y mice. The primary MEFs were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS, 1% Pen/Strep and 1% L-glutamine, incubated at 37°C and in 5% CO2. Fen1y/y primary MEFs were treated with 10 μm PARP1-inhibitor 3,4-Dihydro-5[4-(1-piperindinyl)butoxy]-1(2H)-isoquinoline (DPQ) for 3 h at 37°C, 5% CO2, 3% O2 prior to multiphoton local damage experiment. Treatment of Fen1y/y primary MEFs with another PARP inhibitor, Nu1025, was performed at 150 μm for 3 h at 37°C, 5% CO2, 3% O2 prior to multiphoton local damage experiments. Individual treatments of primary Fen1y/y MEFs with 250μm MMS for 1 h, 4 mM KBrO3 for 30 min and 150 μm H2O2 for 12 min, followed by Phosphate buffered saline (PBS) wash, replacement in fresh media and immediate strip-FRAP measurements were performed. Strip-FRAP measurements were also performed after treatment of Fen1y/y MEFs with toxic concentrations of DNA damaging agents, 1.5 mM MMS for 1 h, 40 mM KBrO3 for 30 min, and 10 mM H2O2 for 10 min.
Double colour cell line expressing FEN1-YFP and mCherry-PCNA
Fen1y/y MEFs were transfected with mCherry-PCNA expression construct (35) to obtain FEN1-YFP and mCherry-PCNA double colour cell line. About 2 µg of the plasmid expressing mCherry-PCNA was transfected into primary MEFs endogenously expressing FEN1-YFP using the JetPEI transfection agent (Polyplus transfection), cells were allowed to incorporate the DNA and fresh medium was replaced 48 hours after transfection. Cells were then selected with Neomycin (100 µg/ml) containing medium. Time-lapse imaging on the multiphoton microscope was performed to study the spatio-temporal distribution of FEN1 and PCNA throughout the cell cycle. FEN1-YFP and mCherry-PCNA were monitored during S-phase to see whether FEN1 and PCNA colocalize in DNA replication foci. G1 or G2 phase FEN1-YFP and mCherry-PCNA double colour MEFs were monitored after induction of local laser damage and the fluorescence accumulation of the respective proteins to the lesion site was imaged over time.
Fluorescence activated cell sorting analysis
ES cells were trypsinized, collected in medium, spun down, washed with PBS, resuspended in PBS and measured with fluorescence activated cell sorting (FACS) using the 488 nm laser. FACS analysis was performed on one parental and five targeted Fen1yNeo/WT ES cell clones using a FACSVantage machine (Becton Dickinson).
Immunofluorescence
Fen1WT/WT and Fen1y/y primary MEFs were grown in 8-well chamber glass slides (BD Falcon), washed twice with PBS, fixed with 2% paraformaldehyde at room temperature for 15 min, washed with PBS, permeabilized with 0.1% Triton X-100 for 4–10 min at room temperature, washed with PBS and blocked in PBS+ (PBS, 2% BSA, 0.15% glycine, 5% goat serum) for 1–3 h. Primary antibodies used were: rat anti-HA (Roche) at 1:100 dilution, and rabbit anti-FEN1 (Abcam) at 1:500 dilution. Incubation was performed over night at 4°C. After three PBS washes, the secondary antibodies, goat anti-rat Alexa Fluor 647 and goat anti-rabbit Alexa Fluor 594, diluted 1:1000 in PBS+ were added and incubated for 1 h at room temperature. After three PBS washes, DAPI (1 μg/μl) was added and the cells were incubated for 5–10 min at room temperature, followed by three more PBS washes. The chamber wells were removed, and the stained cells mounted with Mowiol. For Ki-67-staining, cultured Fen1y/y primary MEFs were washed with PBS, fixed, permeabilized and blocked in PBS+ as described above. Primary antibody was mouse anti Ki-67 (Dako, clone MIB5) at 1:50 dilution, and secondary antibody was Alexa Fluor 594 goat anti-mouse (Invitrogen, Molecular Probes) at 1:500 dilution in PBS+. An Axio ObserverZ1 fluorescence microscope, with a Plan-Apochromat 63x/1.40 Oil DIC M27 objective and AxioCamMR3 camera was used to acquire fluorescent cell images. Prior to immunofluorescence staining of postnatal day (P) 7.5 mouse brains, 4 μm paraffin sections were deparaffinized followed by antigen retrieval in sodium citrate pH 6.0 buffer for 20 min. The subsequent steps were essentially the same as described for MEFs above, with the exception that a slightly longer permeabilization step (15 min) and a slightly shorter blocking step (30 min) was used for the paraffin sections. For the brain sections, primary antibodies used for incubation over night were mouse anti-PAR (Enzo Life Sciences, clone 10H) 1:100, rabbit anti-PARP1 (Abcam, Ab6079) 1:100, rabbit anti-FEN1 (Abcam, Ab17993) 1:500, rabbit anti-GFAP (glial fibrillary acidic protein) (Invitrogen, 18-0063) 1:1000, mouse anti-NeuN (Millipore, MAB377) 1:200 and mouse anti-Ki-67 (Dako, clone MIB5) 1:50 and the secondary antibodies were goat anti-rabbit Alexa Fluor 488 1:500 and goat anti-mouse Alexa Fluor 594 1:500. Prior to immunofluorescence on Fen1y/y primary MEFs after strip laser damage, the cells were grown on glass cover slips (24 mm), laser irradiated, fixed, permeabilized and blocked (described for MEFs in the beginning of this section). Primary antibodies used for strip laser damaged cells were rabbit anti-APE1 (Abcam, Ab82) 1:100, mouse anti-LIG1 (MBL, clone 5H5) 1:200, mouse anti-PAR (Enzo Life Sciences, clone 10H) 1:100, rabbit anti-PARP1 (Abcam, Ab6079) 1:100 and secondary antibodies were goat anti-mouse Alexa Fluor 633 1:500 and goat anti-rabbit Alexa Fluor 633 1:500. The laser irradiated and fluorescently stained MEFs were mounted with Vectashield containing DAPI.
Microscopy and laser-induced DNA damage
To locally induce DNA damage in living cells, we used a tuneable near-infrared pulsed laser (Cameleon Ultra II, Coherent Inc, USA) directly coupled to an inverted confocal microscope equipped with a 40x/1.3 oil objective and a thermostatic chamber maintained at 37°C with 5% CO2 (LSM 710-NLO, Zeiss, Germany). Typically, a small circular area (∼2 μm in diameter) within the nucleus of a live cell was targeted for 34 ms (single scan iteration at 800 nm, 10% power output). Image analysis was done via ImageJ (Rasband, W.S., National Institutes of Health, USA) and a custom-built macro. Briefly, after a time series image file was imported (LSM ToolBox plugin) and adjusted to compensate for cell movement (StackReg plugin), a region of interest (ROI) was defined outside the cell and was used to determine the background signal (to be subtracted from each of the quantitative image data). Next, another ROI spanning the total nucleus was defined to compensate for unwanted photobleaching during the acquisition of images, and finally a ‘local damage’ ROI was indicated to allow for the quantification of the total fluorescence due to the recruitment of FEN1-YFP at the laser damaged area. Data was then exported to Excel (Microsoft, USA) for plotting. To target a large number of nuclei for immunofluorescence (IF) (Figure 5A), several adjacent fields were scanned by the 800 nm multiphoton laser in a pattern of evenly spaced parallel lines.
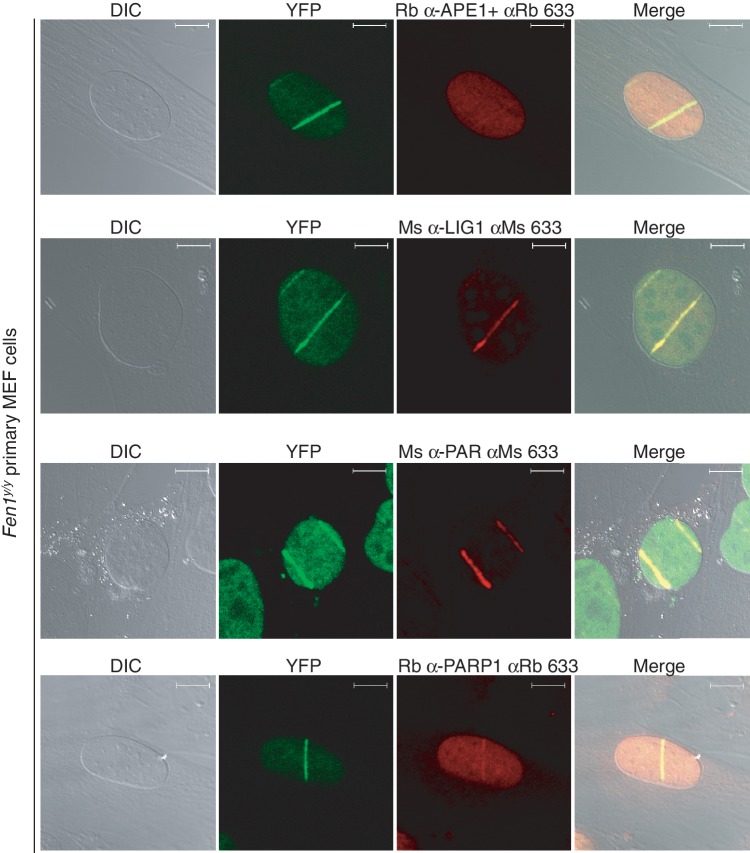
Figure 5.
Accumulation of BER proteins at laser-induced DNA damage. Accumulation of FEN1-YFP, APE1, LIG1, PAR and PARP1 at laser-induced DNA damage in Fen1y/y primary MEF cells. In transmitted light (DIC, grey, left panels) part of the irradiated cell’s cytoplasm and its entire nucleus was visualized. Live imaging of FEN1-YFP (Multiphoton LSM 710 microscope) visualized FEN1 (YFP, green panels) accumulation in the irradiated strip. Immunofluorescent staining of BER proteins and the PAR polymer with the indicated antibodies visualized that APE1, LIG1, PAR and PARP1 all accumulated in the DNA damage site. The scale bar is 10 μm.
Time-lapse imaging of live cells on glass coverslips for up to 48 h was performed on the same inverted confocal microscope. To follow the movement of cells during their cycle, we acquired 2 × 2 tile scans (4096 × 4096 pixels) at minimum zoom (∼700 × 700 μm field of view) at 0.5 or 1 h intervals.
Fluorescence recovery after photobleaching
To determine the mobility of FEN1-YFP in nuclei of living cells a specialized fluorescence recovery after photobleaching (FRAP) procedure was applied: strip-FRAP (36). In these experiments a narrow strip spanning the nucleus of a cell was monitored 200 times every 20 ms at 1% laser intensity (25 mW Argon laser, 514 nm line) to allow the measured fluorescence to reach a steady state level (after ∼4 s). The same strip was then photobleached with three iterations at 100% laser intensity. Subsequently, the recovery of fluorescence within this strip was monitored (1% laser intensity) every 20 ms for 20 s. The shown FRAP data were corrected for background noise and normalized to pre-bleach values, without corrections for unwanted ‘monitor’ photobleaching.
FRAP on local damage was performed to determine the dwell time of FEN1-YFP within the damaged area and conducted as follows: Two to three minutes after DNA damage was induced by multiphoton laser (when assembly and dissociation are in equilibrium) within a MEF cell nucleus, a slightly larger circle encompassing the damaged area was photobleached. Photobleaching was induced by three successive, 100% laser power, iterations at 514 nm. Fluorescence recovery was monitored by measuring fluorescence (full image acquisition) every 2 s for 2 min, at 0.4% laser intensity to minimize unwanted photobleaching. FRAP on local damage data were corrected for background noise, normalized to pre-bleach and corrected for monitor photobleaching.
RESULTS
Generation of the FEN1-YFP knock-in mouse model
In order to visualize and study the kinetics of FEN1 within different cells and tissues, we created mice expressing endogenous mouse FEN1 tagged with YFP at its C-terminus. Gene-targeting constructs and strategy is shown in Figure 1A (see section ‘Materials and Methods’ and Supplementary Data for details). Expression of the fusion gene was under control of the endogenous mouse Fen1 promoter, thereby providing a high probability of physiological expression levels in all different cell types throughout development. Additional C-terminal His6- and HA-tags were added for convenient detection and purification of the FEN1-YFP fusion protein. ES cells transfected with the Fen1-YFP fusion construct were selected and analysed by Southern hybridization (Supplementary Figure S1A). FACS analysis of five selected positive clones revealed YFP expression in all cells of the five analysed targeted ES cell clones (Figure 1B). The YFP expression in ES cells containing the Fen1-YFP fusion gene was also visualized by confocal microscopy (Figure 1B). Chimeric offspring generated from the ES-clone with superior YFP-expression (green in Figure 1B) were further crossed to C57Bl/6J mice and shown to provide germ-line transmission of the targeted allele (Supplementary Figure S1B).
The selectable Neo marker gene, which might interfere with Fen1-YFP expression, was removed by breeding Fen1yNeo/WT and Fen1yNeo/yNeo mice with a ubiquitous Cre-recombinase-expressing mouse (Supplementary Figure S1B and D). The presence of the fluorescent tag did not significantly interfere with the activity of the FEN1 protein (Supplementary Figure S1H), and the Fen1y/y mice appeared healthy and fertile. Western blotting of whole cell extracts from the thymus of wild-type Fen1WT/WT mice and homozygous Fen1y/y mice using anti-FEN1 and anti-HA antibodies showed that the 43 kDa FEN1 was expressed in wild-type but not in Fen1y/y mice, while the HA-tagged 70 kDa FEN1-YFP protein was expressed exclusively in Fen1y/y mice (Figure 1C). FEN1-YFP was also readily detected in whole cell extracts from spleen, testis and lung (Supplementary Figure S1F and data not shown). Immunofluorescence performed on MEF cells confirmed the expression of the fusion protein in these cells (Figure 1D). Moreover, the YFP signal from FEN1-YFP protein could be detected by direct fluorescence microscopy of Fen1y/y MEFs, without antibody staining (Figure 1D). The FEN1-YFP protein was exclusively seen inside the cell nucleus (Figure 1D and E, Figure 5 and Supplementary Figure S1M). Wild-type Fen1 and Fen1y/y mRNA showed similar expression levels as determined by quantitative real-time PCR (Supplementary Figure S1G). Direct in vivo confocal imaging of live embryos (E.9.5, E.14.5) and adult mouse organs including brain, spleen, testis, thymus, kidney, intestine and skin, confirmed the FEN1-YFP-expression seen in the nuclei of cultured Fen1y/y MEFs (E9.5 embryo in Figure 2A, brain, skin, thymus, liver and intestine tissue images in Figure 2C and kidney, spleen and testis tissue in Supplementary Figure S1L). We stained Fen1y/y MEFs for the proliferative marker Ki-67, and indeed FEN1-YFP expression correlates with Ki-67 (Figure 1E and Supplementary Figure S1I), in accordance with FEN1’s role in DNA replication. Moreover, FEN1-YFP expression was detected in proliferative cells (stained by anti-Ki-67) in the external granule layer of Fen1y/y cerebellum sections (Figure 2B lower panel and Supplementary Figure S1J). Stained paraffin-sections did not reveal any significant difference in FEN1 versus FEN1-YFP expression between wild-type and Fen1y/y cerebellums (Supplementary Figure S1J). The wild-type and Fen1y/y cerebellums also displayed the same expression of the proliferative marker Ki-67 (Supplementary Figure S1J). Furthermore, immunofluorescence co-staining of Fen1y/y mouse brain sections with antibodies against PAR and FEN1 revealed a very similar distribution of PAR and FEN1 in the cerebellum (Figure 2B upper panel). NeuN (neuronal cell marker) and FEN1 co-staining showed that neuronal cells express FEN1 (Figure 2B middle panel). Ki-67 and GFAP (glial cell marker) co-staining (Figure 2B lower panel) showed the distribution of Ki-67 in the external granule cell layer of the cerebellum at 5x magnification, and the glial cell distribution in the cerebellum. Higher magnifications of the images in Figure 2B are shown in Supplementary Figure S1K, along with a 40x magnification of immunofluorescent co-staining of a Fen1y/y mouse brain section with antibodies against PAR and PARP1. As a whole, these results allow us to conclude that the fluorescent tag does not disturb FEN1’s expression, localization or activity significantly and therefore that our Fen1y/y mouse model should be ideal for in vivo studies of FEN1 functions, in particular during DNA repair via BER.
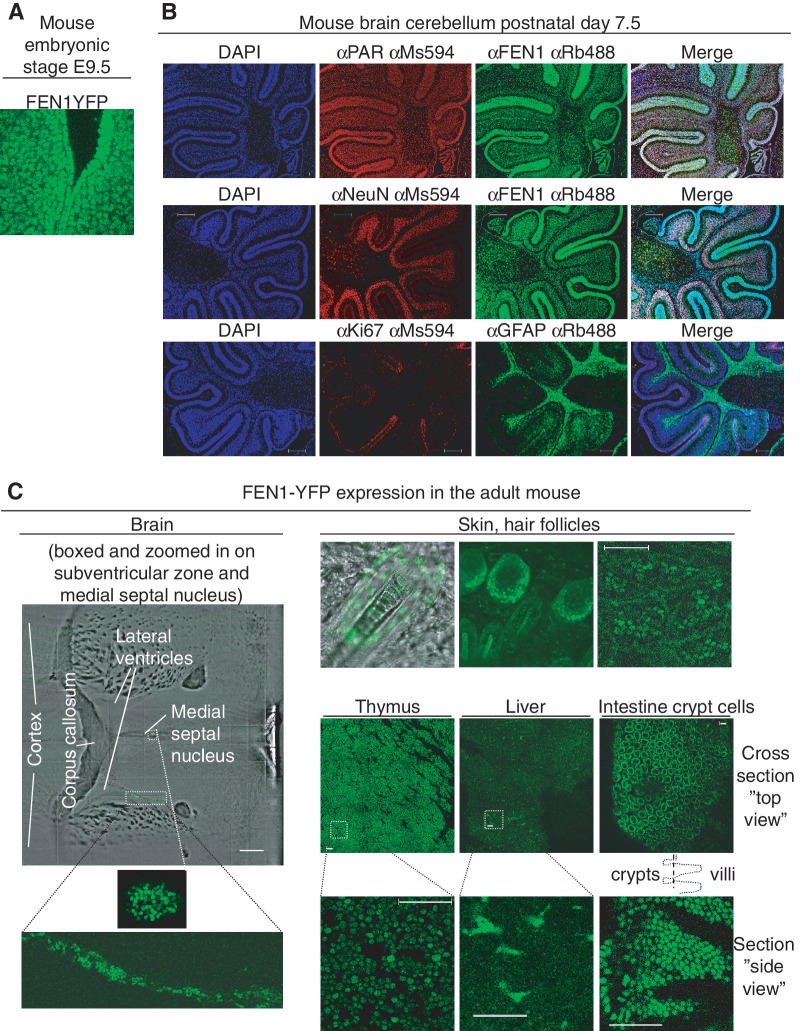
Figure 2.
FEN1-YFP expression in different mouse tissues. (A) In vivo imaging of FEN1-YFP expression. FEN1-YFP expression (green) is seen in the developing Fen1yNeo/yNeo mouse embryo (40×, embryonic stage E9.5, Confocal LSM 510 microscope). (B) Immunofluorescence staining of FEN1, poly (ADP-ribose) polymer (PAR, red in upper panel), brain cell markers (anti-NeuN stain neurons (red in middle panel) and anti-GFAP stain glia (green in lower panel)) and proliferation marker (anti-Ki-67, red in lower panel) in cerebellum from paraffin sections of Fen1y/y mouse brain 7.5 days after birth. Antibodies against the indicated polymer/proteins were used for co-staining of PAR and FEN1, NeuN and FEN1 and Ki-67 and GFAP. DAPI (blue) stained nuclear DNA. To the right, a merge of blue, red and green channel is shown. The scale bar is 200 μm. (C) Expression of FEN1-YFP in adult Fen1y/y mouse tissue. In brain from a 6 weeks old Fen1y/y mouse (left panel), FEN1-YFP cells could be detected in the subventricular zones and in the medial septal nucleus (Multiphoton LSM 710 microscope). Brain regions with FEN1-YFP expressing cells in the subventricular zone of one lateral ventricle and in the medial septal nucleus are enlarged (lower left panels). FEN1-YFP is expressed in skin (imaged with hair) from an adult Fen1yNeo/yNeo mouse (Confocal LSM 510 microscope), and in skin from an adult Fen1y/y mouse (Multiphoton LSM 710 microscope). In skin (three upper right panels), keratinocytes expressing FEN1-YFP could readily be detected in proliferative zones of the skin. FEN1-YFP expressing cells are readily found in thymus and intestine from a 6 weeks old Fen1y/y mouse (lower right panels). In liver from a 6 weeks old Fen1y/y mouse, FEN1-YFP expressing cells are found in a subpopulation of cells, which possibly could be Kuppfer cells, liver-specific macrophages (lower right panels). Kuppfer cells proliferate in the adult, and can have more than one nucleus. A Zeiss LSM 710 microscope with a 40× 1.3 oil objective was used for live imaging of FEN1-YFP in mouse tissue. The scale bar is 50 μm in images of live tissue, except for the 500 μm scale bar in brain.
FEN1 is expressed in adult mouse organs, including brain
The successful visualization of FEN1-YFP tempted us to further study FEN1 using live cell imaging on living tissues. Under normal non-exposed conditions FEN1 is mainly involved in DNA replication, and thus we expected to find FEN1 expression correlating with the proliferative status of a cell. We already knew that ES cells (Figure 1B), MEFs (Figure 1D and E) and cells in the developing embryo (E9.5 and E14.5, Figure 2A and data not shown) expressed FEN1-YFP at levels easily detected by microscopy. In the adult Fen1y/y mouse, we analysed the fusion gene expression in the epidermis (skin explants), where proliferating cells (keratinocytes) are only found in the basal layer and within the bulge around the hair follicles. Indeed, FEN1-YFP expression was mainly observed in cells around the hair follicles and in cells in the basal layer (Figure 2C upper panel from the right). FEN1-YFP expressing cells were less abundant in the adult Fen1y/y brain than in higly proliferative organs (Skin, thymus and intestine in Figure 2C and spleen and testis in Supplementary Figure S1L), however we found some FEN1-YFP expressing cells in the Fen1y/y mouse brain, isolated or grouped (Figure 2C left panel and data not shown). These brain cells expressing FEN1-YFP were observed close to the lateral ventricles, the medial septal nucleus and in the cortex of coronal brain slices. Moreover, some of the grouped cells were observed next to blood vessels (data not shown), which could resemble proliferative aggregates of neuronal, glial and endothelial precursors (37).
FEN1 is highly mobile and rapidly accumulates at laser-induced DNA damage sites
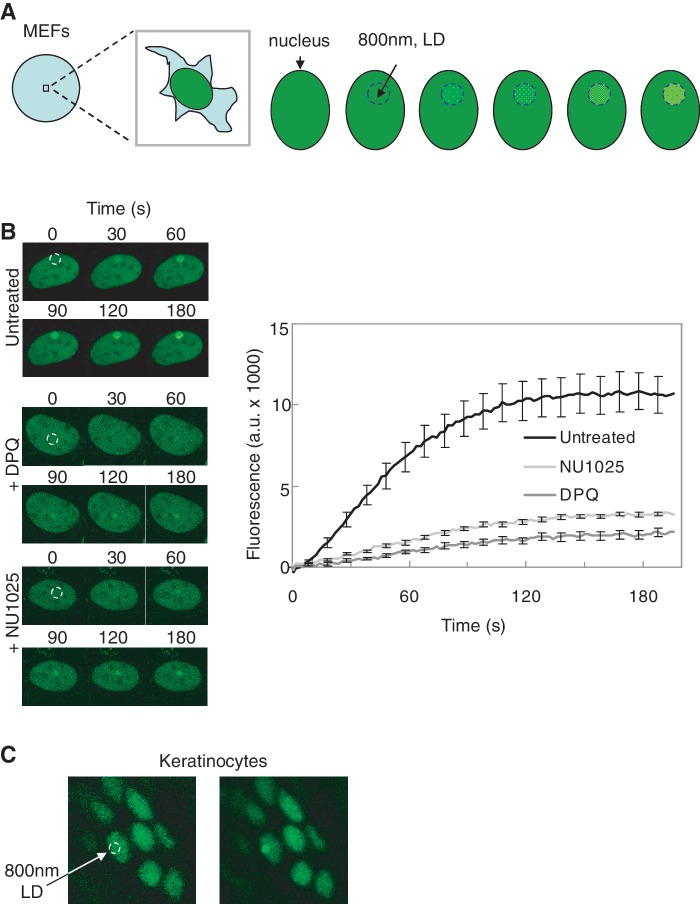
In order to study FEN1 mobility as well as its recruitment to sites of DNA damage, we induced local DNA damage by multiphoton laser treatment in the nucleus of targeted Fen1y/y MEF cells. Multiphoton laser micro-irradiation has been reported to induce a large spectrum of DNA lesions, including reactive oxygen species (ROS) induced DNA damage (38,39), base damage, cyclobutane pyrimidine dimers (40–42) and DNA strand breaks (41–43). We excluded from our local damage study cells showing a clear replicative focal pattern (Figure 6A and B) and avoided exposing nucleoli, to avoid possible involvement of FEN1 in DNA metabolic pathways (44) other than repair of the local DNA damage. Briefly, Fen1y/y MEFs received a short (34 ms) and very localized (∼2 μm) exposure to the output of a near infrared (800 nm) pulsed (200 fs) laser. Within ∼10 s after the multiphoton irradiation, the YFP fluorescence in the targeted ROI had increased to higher levels inside the ROI than outside the local damage spot (Figure 3A and B), reaching a plateau after ∼2 min. Fluorescence was also monitored in a limited number of Fen1y/y cells for a longer time period after laser-induced DNA damage (data not shown). Prolonged monitoring of YFP fluorescence on local DNA damage revealed a relatively early (∼4 min after local damage induction) and rapid decrease of the local FEN1 accumulation (∼75% drop of YFP fluorescence signal in about 15 min) at the irradiated sites (data not shown). Our results demonstrate that 15 min after DNA damage induction, less than 25% of FEN1 substrate remains, which is compatible with the known single strand break repair (SSBR) kinetics (45,46). We also performed local damage experiments on FEN1-YFP expressing skin cells, using the multiphoton 800 nm laser (Figure 3C). Like in MEFs, FEN1-YFP accumulated very fast to the site of the lesion in targeted keratinocytes (Figure 3C). For adult mouse brain, less proliferative cells would be expected than for skin, although proliferative zones and brain stem cells exist (37,47,48).
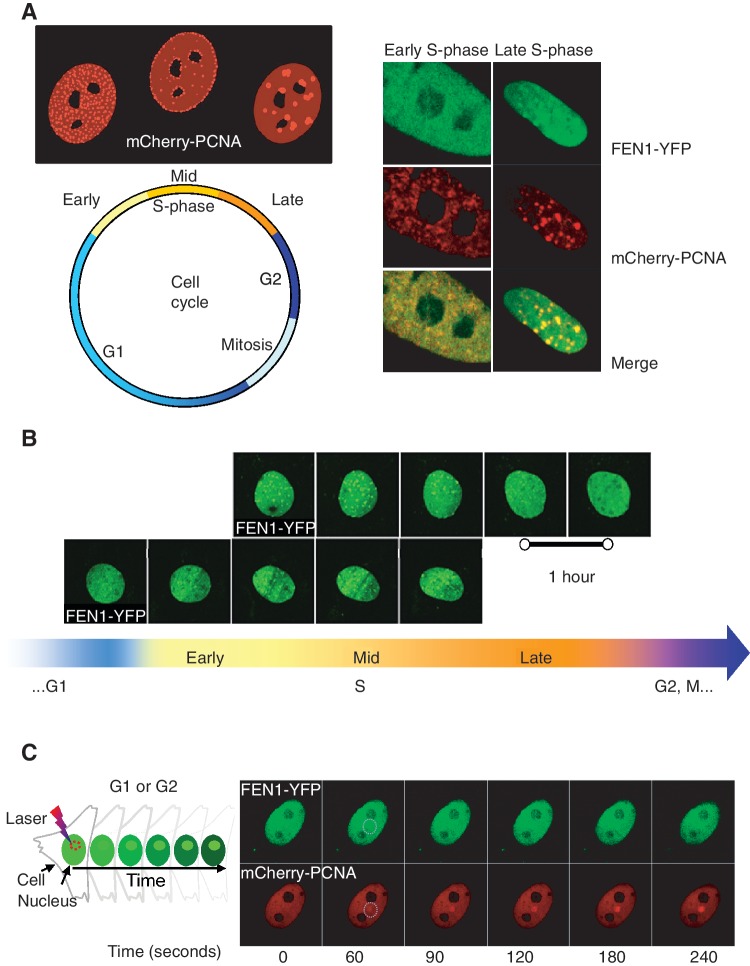
Figure 6.
FEN1-YFP dynamic behaviour throughout cell cycle and its colocalization with mCherry-PCNA. (A) Colocalization of FEN1-YFP and mCherry-PCNA in S-phase. Distinct PCNA replication structures are found in early, mid and late S-phase. In double colour MEFs expressing FEN1-YFP and mCherry-PCNA, PCNA appeared in very early S-phase and FEN1 foci could be detected soon thereafter (Multiphoton LSM 710 microscope). By late S-phase, perfect colocalization of FEN1 and PCNA foci was observed. (B) Live cell fluorescence imaging of FEN1 foci appearance and disappearance throughout S-phase. By mid S-phase distinct FEN1 foci were visualized, and the FEN1 foci were still present in late S-phase. By the end of late S-phase the FEN1 foci started to disappear (Multiphoton LSM 710 microscope). (C) Recruitment of FEN1 and PCNA to laser-induced DNA damage in G-phase cell. Non-S-phase MEFs expressing FEN1-YFP and mCherry-PCNA received irradiation with an 800 nm pulsed multiphoton laser (Multiphoton LSM 710 microscope). Fluorescence accumulation of FEN1-YFP and mCherry-PCNA to the DNA damage site was imaged over time. Early FEN1-YFP and mCherry-PCNA recruitment to the laser-induced DNA damage sites was found to be similar for both proteins.
Figure 3.
Mobility of FEN1-YFP in living cells. (A) Assay for local damage and fluorescence accumulation in MEFs. A circular area within the cell nucleus was irradiated with an 800 nm pulsed multiphoton laser to induce local DNA damage (dashed blue circle). Fluorescence in the locally damaged area was monitored over time. (B) FEN1-YFP accumulation after local damage. The PARP1-inhibitors DPQ and NU1025 was added to the indicated cells. FEN1-YFP showed a strong accumulation to the damaged site in untreated MEFs (n = 10), and a very weak accumulation to the region with DNA damage after addition of PARP1 inhibitors DPQ (n = 10) and NU1025 (n = 10) to the cells. The dashed white circle indicates the region in the cell nucleus where the DNA is damaged using a multiphoton laser at 800 nm. (C) Multiphoton laser at 800 nm was used to generate local DNA damage in keratinocytes of the Fen1y/y mouse skin, and FEN1-YFP was shown to accumulate in the regions with DNA damage. One representative image of skin cell nuclei before (left panel) and one image after (right panel) targeting with the multiphoton laser is shown (dashed white circle indicates damaged area of the targeted nucleus).
PARP1 inhibition hinders FEN1 accumulation at DNA damage sites
PARP1 is rapidly activated by binding to nicks in DNA and catalyses the production of long PAR covalently linked to numerous substrate proteins, including itself, shortly after DNA break formation (49,50). PARylated PARP1 interacts with the C-terminal domain of the breast cancer susceptibility protein BRCA1 (BRCT) domain of X-ray repair cross complementing group 1 protein (XRCC1), and is responsible for a swift XRCC1 relocation to lesions (51,52). Due to the negative charge of the polymers, autoribosylated PARP1 dissociates from the nicked DNA indicating that the catalytic activity of PARP1 is needed for the dissociation of PARP1 from repair intermediates (53,54). In accordance, the addition of PARP1 inhibitor DPQ has resulted in PARP1 and PCNA persistent nuclear foci (55). Since PARP1 probably acts upstream of FEN1 in SSBR/BER, we wanted to test whether PARP1 inhibition would influence the binding kinetics of FEN1 to DNA lesions in living cells. To that aim we introduced local damage by pulsed 800 nm multiphoton laser irradiation in nuclei of Fen1y/y MEFs, either DPQ, NU1025 or untreated. Both the total amount and accumulation rate of FEN1-YFP in Fen1y/y MEFs was severely attenuated after inhibition of PARP1 (Figure 3B). PARP1 is not only implicated in SSBR/BER but also binds and responds to double strand breaks (DSBs) and stalled replication forks (56), the latter of which we circumvented by avoiding cells with replication foci in local damage experiments. To demonstrate that FEN1 accumulation was mainly to BER substrates, we compared the accumulation of FEN1 and non-homologous end-joining repair protein Ku80 to local damage sites (Supplementary Figure S1N). After micro-irradiation with our ‘low’ laser power sufficient to obtain fast and robust accumulation of FEN1, Ku80 barely accumulated (Supplementary Figure S1N). With a ‘high’ power laser setting defined in (41) to induce a clear recruitment of Ku80 to DSBs (approximately 1500 DSBs generated (41)), FEN1 accumulation was so high that it depleted the available nuclear pool of FEN1-YFP and completely saturated the detector of our confocal microscope setup (Supplementary Figure S1N). This indicates that, by far, DSBs are not the principle DNA break type induced when using our ‘low’ power setting.
Oxidized and alkylated base lesion trigger BER without affecting FEN1 mobility
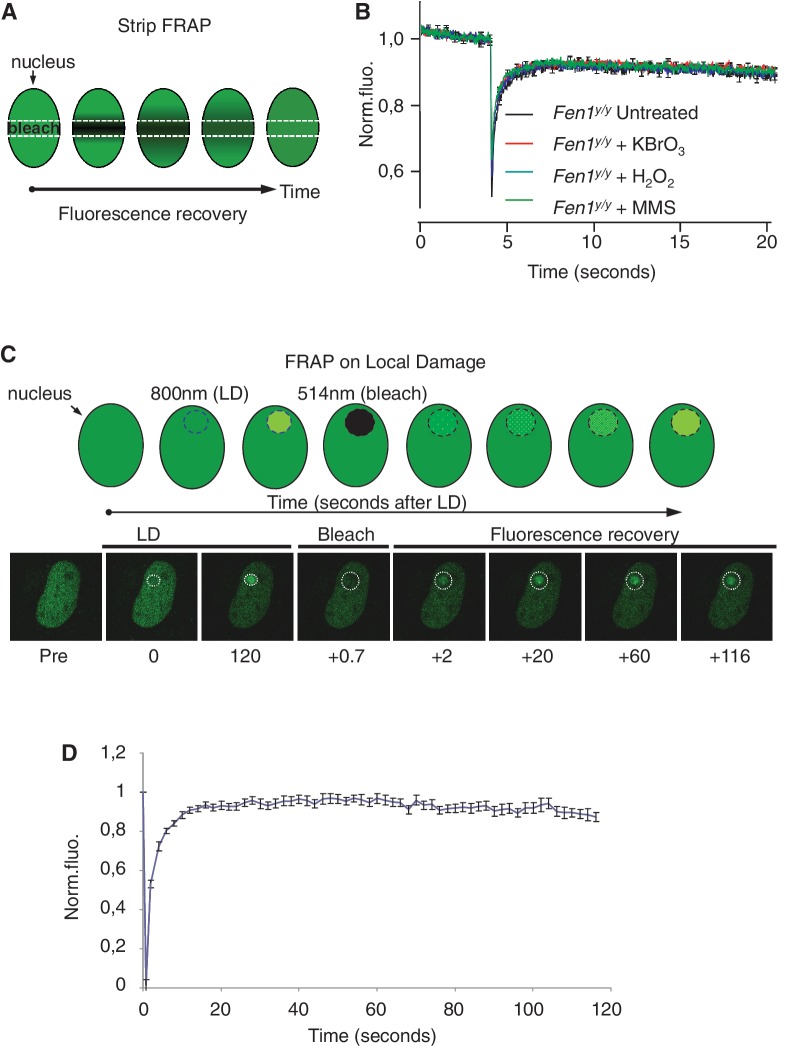
The accumulation of FEN1-YFP to local DNA damage suggests that this protein is transiently bound to lesions. To determine the fraction and time at which FEN1-YFP molecules are being bound to lesions or trapped in chromatin-associated BER complexes (substrate for FEN1) we performed FRAP in cells in the presence or absence of DNA damaging agents. FRAP was previously successfully applied to determine protein mobility of NER factors (57) and to deduce from these studies a kinetic framework of the repair reaction in living cells (58). We performed FRAP on FEN1-YFP expressing MEF’s treated with chemicals known to induce global oxidative or alkylation DNA damage. A narrow strip spanning the nucleus was photobleached and the fluorescence recovery was subsequently monitored in untreated and chemically damaged MEFs (Figure 4A). Surprisingly, treatment of Fen1y/y MEFs with the oxidizing chemicals KBrO3 and H2O2 and the alkylating agent MMS, each known to trigger BER, resulted in essentially unchanged FEN1-YFP mobility curves when compared to untreated cells (Figure 4B). Thus the global induction of oxidative and alkylation DNA damage in Fen1y/y MEFs did not produce a significant fraction of immobile FEN1-YFP bound to BER-intermediates in DNA. This indicates that either FEN1 is rapidly turned over from BER substrates (swift binding and dissociating from its flap DNA substrate) or only a limited amount of BER reactions are active at any moment, too low to be detected by FRAP. This could also indicate that continuous repair during drug incubation has substantially reduced the amount of DNA damage still present at the time of measurement, thus reducing the amount of immobilized FEN1.
Figure 4.
FRAP measurements of FEN1-YFP following global and local DNA damage. (A) Strip FRAP assay. Global alkylation or oxidative DNA damage was induced by chemical agents. A rectangular area within the nucleus was photobleached at 514 nm, followed by fluorescence recovery monitoring. (B) Strip FRAP graph in which the normalized fluorescence (Norm.fluo.) recovery after bleaching is plotted against time (seconds). No significant difference is observed in FEN1 after chemical induction of oxidative damage to trigger base excision/single strand break repair (BER/SSBR). The recovery of untreated live Fen1y/y MEF cells is shown in black, cells treated with 4 mM KBrO3 in red, cells treated with 150 μM H2O2 in blue and fluorescence recovery of 250 μM MMS treated cells is shown in green. Plotted data represents the average of at least 12 different strip-FRAP curves. (C) Assay for FRAP on local damage. DNA damage was induced in single nuclei of MEFs, by defining a circular area within the targeted nucleus to be scanned by a multiphoton laser at 800 nm. A slightly larger circular region covering the local DNA damage was subsequently photobleached at 514 nm, followed by fluorescence recovery monitoring. (D) FRAP on local damage. A fast and almost complete turnover of FEN1-YFP during BER was seen (n = 13) during FRAP on local damage (as described in C.).
Fast and almost complete turnover of FEN1 during SSBR/BER
In order to determine the binding or dwell time of FEN1 in active BER we performed FRAP on local laser-induced DNA damage, when binding is at equilibrium with dissociation in Fen1y/y MEFs nuclei (Figure 4C). We observed a fast and nearly complete turnover of FEN1-YFP on the local damage within less than 20 s (Figure 4D). The very small decrease in FEN1-YFP fluorescence seen after 1 min could be due to ongoing SSBR/BER that will reduce the amount of FEN1 substrate available. Thus, our FRAP on local damage data indicate, in agreement with the absence of a measurable immobilized fraction in our strip-FRAP experiments on global damage (Figure 4B), that FEN1 proteins rapidly bind to and dissociate from the DNA flaps formed as intermediates in LP-BER.
APE1, LIG1, PARP1 and PAR presence at laser-induced DNA damage
Local laser irradiation generates a wide spectrum of DNA lesions, including SSBs and DSBs. Both SSBs and DSBs attract PARP1. In order to determine whether the laser-induced DNA damage sites contained BER-substrates, immunofluorescent staining of BER-protein APE1 was performed in Fen1y/y primary MEFs (Figure 5). LIG1 functions in both DNA replication and BER (59,60). In local damage experiments we only laser irradiated non-S-phase cells. Staining of LIG1 in micro-irradiated Fen1y/y MEFs is thus expected to reflect LIG1 in BER. APE1 and LIG1 both accumulated at the site of DNA damage, together with FEN1-YFP, which could be imaged directly (Figure 5). Moreover, the laser irradiated strip regions in Fen1y/y primary MEFs were also stained against PARP1 and the PAR polymer, and their presence was confirmed (Figure 5).
FEN1 and PCNA localization during S-phase and at local DNA damage
PCNA is one of the proteins interacting with FEN1 both in DNA replication and DNA repair, and we wanted to compare FEN1 and PCNA’s dynamic behaviour in replication foci and recruitment kinetics to DNA damage sites. MEFs expressing FEN1-YFP and mCherry-PCNA were time-lapse imaged throughout the cell cycle (Figure 6). In (very) early S-phase, only PCNA foci were detected (Figure 6A). However, FEN1 foci appeared relatively soon after the PCNA foci became visible, and by mid S-phase distinct FEN1 foci were visualized (Figure 6B). In late S-phase, FEN1 and PCNA foci were both present, and the foci displayed perfect colocalization (Figure 6A). Early FEN1 and PCNA recruitment to laser-induced DNA damage was also monitored in non-S MEFs (Figure 6C) and was found to be similar for both factors.
DISCUSSION
DNA intermediates generated during DNA replication, recombination and repair need to be efficiently processed to avoid genomic instability. FEN1 has a key role in processing 5′-single-stranded DNA structures produced during Okazaki fragment maturation and in LP-BER (5,18,61,62). In order to visualize this process in living cells and tissues, we designed a knock-in mouse expressing FEN1-YFP fusion protein from the endogenous FEN1 promoter. Kinetics of FEN1-YFP in processing DNA repair intermediates were evaluated following induction of DNA damage by chemical agents or by multiphoton laser irradiation.
Kinetics of FEN1 at endogenous DNA damage
In vivo imaging and kinetic studies revealed a highly mobile FEN1-YFP fusion protein, accumulating at sites of DNA lesion within 2 min with a high turnover rate. As specified above, PARP1 (and also PCNA) have roles in DSB repair. In the following we discuss kinetics of BER and assume that the amount of DSBs is too low to have an impact on FEN1 accumulation. FEN1-YFP accumulation at DNA damage sites presents a sigmoid shape. This might reflect that a step upstream of FEN1, such as the DNA glycosylase excision that initiates BER, must be completed prior to FEN1 recruitment. For nucleotide excision repair (NER), this sigmoid shape is not observed, even for late acting factors such as the single strand DNA binding protein RPA and the endonuclease ERCC1/XPF (58,63). The fluorescence intensity of recruited FEN1 reached a plateau ∼2–3 min after multiphoton induced local damage. In accordance with this, BER proteins, including the OGG1 DNA glycosylase, the scaffold protein XRCC1 and the DNA Ligase III, are known to accumulate at DNA repair sites, and also reach their maximum within 2 min (64–66). The replication associated BER proteins, DNA Ligase I and PCNA, reach a maximum after 5 min (59). The sliding clamp and processivity factor PCNA resembles DNA Ligase I recruitment kinetics, with slow and constant accumulation to DNA damage sites (64). However, DNA Ligase I has been reported to accumulate at irradiated sites with a delay of 30–60 s (64). The authors suggested that the DNA ligases are selectively recruited to DNA repair sites (64). DNA Ligase I is recruited by PCNA, via its PCNA-binding domain (PBD), to damaged DNA, while DNA Ligase III is recruited by XRCC1 (64). In a recent paper, methylation of FEN1 has been shown to suppress nearby phosphorylation and facilitate PCNA-binding (67). Taken together, the kinetic and biochemical data discussed here support similar recruitment mechanisms in LP-BER and DNA replication, as already suggested by Mortusewicz and colleagues (64). The processive association of PCNA with DNA favours continuous synthesis of long DNA stretches during DNA replication and LP-BER. The PCNA trimer is proposed to interact with DNA polymerases and methylated FEN1 simultaneously, and allows rapid exchange between DNA polymerase and the active site of FEN1 for flap cleavage (67). Phosphorylated (and demethylated) FEN1 with decreased affinity for PCNA allows DNA Ligase I to interact with PCNA and seal the nick (67). This is well in accordance with our time-lapse studies showing mCherry-PCNA appearing in early S-phase foci, followed by FEN1-YFP foci colocalizing with PCNA foci soon thereafter. FEN1 foci disappear by the end of late S-phase.
BER complex(ity) and stepwise repair
FEN1 binding to BER substrates was observed in local damage experiments. FRAP on LD data indicates that FEN1 binding to BER intermediates is short-lived. In accordance, FEN1 mobility was not significantly different in globally damaged versus untreated cells, as measured by recovery of FEN1-YFP fluorescence in a photobleached strip of the nucleus; strip-FRAP analysis. This suggests that DNA damage bound FEN1 is rapidly released, i.e. in a similar time range to FEN1’s typical diffusion time or faster. This indicates a highly mobile FEN1, rapidly binding and dissociating from its DNA flap substrate, without necessarily being part of a repair complex that strongly binds the DNA but rather transiently interacting with the sliding clamp PCNA, BER proteins and DNA flap substrate. PARP1 inhibition (DPQ and NU1025) interrupted FEN1 accumulation at sites of DNA damage, indicating that FEN1 is dependent upon active PARP1 for recruitment to DNA damage intermediates in BER. This is in agreement with previous findings where PARP1 was found to be required for the assembly of the scaffold protein XRCC1 at sites of oxidative DNA damage (55,68). Moreover, PCNA’s access to DNA damage sites is affected in a contradictory manner by inactive PARP1. Decreased relocalization of PCNA to DNA lesions after PARP1 inhibition has been reported by several groups (69–71). Recruitment of PCNA to DNA damage was not hindered in cells lacking PARP1 (71). On the contrary, increased PCNA accumulation at DNA damage sites following PARP1 inhibition has also been reported (55,72). Hanssen-Bauer et al. (72) observed increased recruitment of FEN1 and PCNA to sites of DNA damage following 4-amino-1,8-naphtalimide (4-AN) treatment, whereas another PARP-inhibitor, N-5,6-dihydro-6-oxo-2-phenanthridinyl)-2-acetamide hydrochloride (PJ34), did not affect FEN1 and PCNA recruitment (72). Recruitment of aprataxin, a DNA-binding protein implicated in removal of DNA adenylates arising from abortive DNA ligase reactions in BER, is hindered by PARP1 inhibition by PJ34 and 3-aminobenzamide (3AB) (73). It emerges that treatment with some PARP-inhibitors, including DPQ, DIQ and NU1025, result in reduced recruitment of BER-protein(s) to DNA lesions following microirradiation, including FEN1 (this study and data not shown for DIQ), PCNA, XRCC1, polymerase β and Ligase IIIα (69–71). On the other hand, 4-AN PARP inhibitor causes increased accumulation of FEN1 and PCNA (55,72). Furthermore, recruitment of PCNA and FEN1 require high doses of irradiation, and recruitment is enhanced by XRCC1 and PARP1 at the site of DNA damage (72). In the experimental setup by Hanssen-Bauer and colleagues XRCC1/polynucleotide kinase/polymerase β was recruited to sites with low levels of induced DNA damage independent of PARP1 accumulation and poly (ADP-ribosyl)ation (PARylation) (72). Thus, the type and dose of DNA damaging agent and the PARP-inhibitor used will affect the DNA damage formed and the SSBR/BER factor recruitment and organization. The actual function of PARylation in SSBR/BER is not completely understood, however, several cellular regulatory mechanisms are likely to be directed by PARylation (72).
Whether FEN1 is part of a LP-BER complex and whether FEN1 is taking part in a coordinated handoff of DNA repair intermediates has been and is still being investigated by several research groups. Working models suggest both pre-assembled multi-protein complexes and short-lived transient complexes assembled at the damage site, where DNA repair substrates are sequestered in a coordinated manner from one protein to the next in the BER pathway, the latter being proposed as ‘passing the baton’ or ‘substrate channelling’ (74–77). For SP-BER, a complex or ‘repairosome’ proficient in SP-BER of uracil from DNA was found after examination of a bovine testis nuclear extract, supporting the first mechanism (78). Moreover, it has recently been shown in vitro by Prasad and co-workers that SP-BER can proceed from an AP-site to a ligated product through channelling (79). However, in the case of LP-BER, the intermediate after DNA polymerase β gap filling was not channelled to FEN1 in their in vitro assay (79). In support of LP-BER sequential enzyme actions, a recent experiment utilizing purified human proteins concludes that the LP-BER proteins mechanistically function in a defined sequential manner (80). Balakrishnan and co-authors also present a structural model for LP-BER, where the proteins are part of a multi-enzyme complex (80). In the context of a LP-BER repairosome, they suggest a model where FEN1 binds and cleaves its flap substrate after DNA polymerase β has dissociated from the DNA substrate, and is stimulated and counter-stimulates other LP-BER proteins in the complex (80). The recently resolved crystal structures of human FEN1 with DNA are also consistent with the ‘passing the baton’ concept (81). The authors propose a model where FEN1 binds dsDNA exposed in the DNA polymerase β complex and displaces DNA polymerase β to access the 5′ and 3′ flaps, resulting in a direct handoff of dangerous DNA intermediates (81). The way FEN1 was observed to bind DNA, with dsDNA directing FEN1 and 5′ nuclease specificity, and their model of DNA polymerase β and DNA Ligase 1 buried interfaces, is well in accordance with a model of coordinated stepwise BER (81).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figure 1, Supplementary Methods and Supplementary References [82–84].
FUNDING
South-Eastern Norway Regional Health Authority, Norway (to L.K. and A.K.); the Norwegian Cancer Society (to A.K., in part); La Ligue Nationale Contre le Cancer (to C.G.); the Association for International Cancer Research [AICR: 07-0129 to C.G. and P.O.M.]; ATIP InCa/CNRS [contract n°039438 to P.O.M. and G.G.M.]; the Association pour la Recherche sur le Cancer [ARC contract N 8505 to acquire the Zeiss LSM710 microscope of the Toulouse microscopy platform]. Funding for open access charge: South-Eastern Norway Regional Health Authority.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks to the Norwegian transgenic centre, Oslo, and to the animal facilities at Rikshospitalet, Oslo, Erasmus MC, Rotterdam and CNRS, Toulouse. We thank Rajikala Suganthan for dissecting out p7.5 mice cerebellum and Cristina Ceballos Iglesias for the production of the mCherry-PCNA stable cell line. This work also benefited from the TRI RIO Optical Imaging Platform at IPBS (Genotoul, Toulouse, France) supported by grants from the Région Midi-Pyrénées (CPER), the Grand Toulouse community, the ARC (ARC Equipement N°8505), the CNRS and the EU through the FEDER program.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol. Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bambara RA, Murante RS, Henricksen LA. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 5.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedrich-Heineken E, Hubscher U. The Fen1 extrahelical 3′-flap pocket is conserved from archaea to human and regulates DNA substrate specificity. Nucleic Acids Res. 2004;32:2520–2528. doi: 10.1093/nar/gkh576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosfield DJ, Frank G, Weng Y, Tainer JA, Shen B. Newly discovered archaebacterial flap endonucleases show a structure-specific mechanism for DNA substrate binding and catalysis resembling human flap endonuclease-1. J. Biol. Chem. 1998;273:27154–27161. doi: 10.1074/jbc.273.42.27154. [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi K, Taniguchi Y, Hatanaka A, Sonoda E, Hochegger H, Adachi N, Matsuzaki Y, Koyama H, van G, Jasin M, et al. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol. Cell Biol. 2005;25:6948–6955. doi: 10.1128/MCB.25.16.6948-6955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negritto MC, Qiu J, Ratay DO, Shen B, Bailis AM. Novel function of Rad27 (FEN-1) in restricting short-sequence recombination. Mol. Cell Biol. 2001;21:2349–2358. doi: 10.1128/MCB.21.7.2349-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parrish JZ, Yang C, Shen B, Xue D. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J. 2003;22:3451–3460. doi: 10.1093/emboj/cdg320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Wilson TE, Lieber MR. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl Acad. Sci. USA. 1999;96:1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordenin DA, Kunkel TA, Resnick MA. Repeat expansion–all in a flap? Nat. Genet. 1997;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- 13.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 14.Parenteau J, Wellinger RJ. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell Biol. 1999;19:4143–4152. doi: 10.1128/mcb.19.6.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parenteau J, Wellinger RJ. Differential processing of leading- and lagging-strand ends at Saccharomyces cerevisiae telomeres revealed by the absence of Rad27p nuclease. Genetics. 2002;162:1583–1594. doi: 10.1093/genetics/162.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, Stewart SA. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallur AC, Maizels N. Distinct activities of exonuclease 1 and flap endonuclease 1 at telomeric g4 DNA. PLoS One. 2010;5:e8908. doi: 10.1371/journal.pone.0008908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD, Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 19.Kim IS. Down-regulation of human FEN-1 gene expression during differentiation of promyelocytic leukemia cells. Exp. Mol. Med. 1998;30:252–256. doi: 10.1038/emm.1998.37. [DOI] [PubMed] [Google Scholar]
- 20.Warbrick E, Coates PJ, Hall PA. Fen1 expression: a novel marker for cell proliferation. J. Pathol. 1998;186:319–324. doi: 10.1002/(SICI)1096-9896(1998110)186:3<319::AID-PATH184>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Otto CJ, Almqvist E, Hayden MR, Andrew SE. The "flap" endonuclease gene FEN1 is excluded as a candidate gene implicated in the CAG repeat expansion underlying Huntington disease. Clin. Genet. 2001;59:122–127. doi: 10.1034/j.1399-0004.2001.590210.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim IS, Lee MY, Lee IH, Shin SL, Lee SY. Gene expression of flap endonuclease-1 during cell proliferation and differentiation. Biochim. Biophys. Acta. 2000;1496:333–340. doi: 10.1016/s0167-4889(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 23.Kucherlapati M, Yang K, Kuraguchi M, Zhao J, Lia M, Heyer J, Kane MF, Fan K, Russell R, Brown AM, et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl Acad. Sci. USA. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen E, Kleppa L, Meza TJ, Meza-Zepeda LA, Rada C, Castellanos CG, Lien GF, Nesse GJ, Neuberger MS, Laerdahl JK, et al. Early-onset lymphoma and extensive embryonic apoptosis in two domain-specific Fen1 mice mutants. Cancer Res. 2008;68:4571–4579. doi: 10.1158/0008-5472.CAN-08-0168. [DOI] [PubMed] [Google Scholar]
- 26.Zheng L, Dai H, Qiu J, Huang Q, Shen B. Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol. Cell Biol. 2007;27:3176–3186. doi: 10.1128/MCB.01652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng L, Dai H, Zhou M, Li M, Singh P, Qiu J, Tsark W, Huang Q, Kernstine K, Zhang X, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat. Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- 28.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dianov G, Price A, Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell Biol. 1992;12:1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokhansanj BA, Rodrigue GR, Fitch JP, Wilson DM., III A quantitative model of human DNA base excision repair. I. Mechanistic insights. Nucleic Acids Res. 2002;30:1817–1825. doi: 10.1093/nar/30.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 33.Petermann E, Ziegler M, Oei SL. ATP-dependent selection between single nucleotide and long patch base excision repair. DNA Repair (Amst) 2003;2:1101–1114. doi: 10.1016/s1568-7864(03)00117-4. [DOI] [PubMed] [Google Scholar]
- 34.Giglia-Mari G, Theil AF, Mari PO, Mourgues S, Nonnekens J, Andrieux LO, de WJ, Miquel C, Wijgers N, Maas A, et al. Differentiation driven changes in the dynamic organization of Basal transcription initiation. PLoS. Biol. 2009;7:e1000220. doi: 10.1371/journal.pbio.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinant C, de JM, Essers J, van Cappellen WA, Kanaar R, Houtsmuller AB, Vermeulen W. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 2007;120:2731–2740. doi: 10.1242/jcs.004523. [DOI] [PubMed] [Google Scholar]
- 36.Houtsmuller AB, Vermeulen W. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem. Cell Biol. 2001;115:13–21. doi: 10.1007/s004180000234. [DOI] [PubMed] [Google Scholar]
- 37.Cleaver O, Melton DA. Endothelial signaling during development. Nat. Med. 2003;9:661–668. doi: 10.1038/nm0603-661. [DOI] [PubMed] [Google Scholar]
- 38.Tirlapur UK, Konig K, Peuckert C, Krieg R, Halbhuber KJ. Femtosecond near-infrared laser pulses elicit generation of reactive oxygen species in mammalian cells leading to apoptosis-like death. Exp. Cell Res. 2001;263:88–97. doi: 10.1006/excr.2000.5082. [DOI] [PubMed] [Google Scholar]
- 39.King BA, Oh DH. Spatial control of reactive oxygen species formation in fibroblasts using two-photon excitation. Photochem. Photobiol. 2004;80:1–6. doi: 10.1562/2004-03-01-RA-093.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meldrum RA, Botchway SW, Wharton CW, Hirst GJ. Nanoscale spatial induction of ultraviolet photoproducts in cellular DNA by three-photon near-infrared absorption. EMBO Rep. 2003;4:1144–1149. doi: 10.1038/sj.embor.7400028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong X, Mohanty SK, Stephens J, Heale JT, Gomez-Godinez V, Shi LZ, Kim JS, Yokomori K, Berns MW. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009;37:e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harper JV, Reynolds P, Leatherbarrow EL, Botchway SW, Parker AW, O'Neill P. Induction of persistent double strand breaks following multiphoton irradiation of cycling and G1-arrested mammalian cells-replication-induced double strand breaks. Photochem. Photobiol. 2008;84:1506–1514. doi: 10.1111/j.1751-1097.2008.00378.x. [DOI] [PubMed] [Google Scholar]
- 44.Guo Z, Qian L, Liu R, Dai H, Zhou M, Zheng L, Shen B. Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol. Cell Biol. 2008;28:4310–4319. doi: 10.1128/MCB.00200-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frankenberg-Schwager M. Review of repair kinetics for DNA damage induced in eukaryotic cells in vitro by ionizing radiation. Radiother. Oncol. 1989;14:307–320. doi: 10.1016/0167-8140(89)90143-6. [DOI] [PubMed] [Google Scholar]
- 46.Frankenberg-Schwager M. Induction, repair and biological relevance of radiation-induced DNA lesions in eukaryotic cells. Radiat. Environ. Biophys. 1990;29:273–292. doi: 10.1007/BF01210408. [DOI] [PubMed] [Google Scholar]
- 47.Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 48.Migaud M, Batailler M, Segura S, Duittoz A, Franceschini I, Pillon D. Emerging new sites for adult neurogenesis in the mammalian brain: a comparative study between the hypothalamus and the classical neurogenic zones. Eur. J. Neurosci. 2010;32:2042–2052. doi: 10.1111/j.1460-9568.2010.07521.x. [DOI] [PubMed] [Google Scholar]
- 49.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 50.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de MJ, de MG. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferro AM, Olivera BM. Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. J. Biol. Chem. 1982;257:7808–7813. [PubMed] [Google Scholar]
- 54.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Godon C, Cordelieres FP, Biard D, Giocanti N, Megnin-Chanet F, Hall J, Favaudon V. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De VM, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: Current state of the art. Biochem. Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 57.Vermeulen W. Dynamics of mammalian NER proteins. DNA Repair (Amst) 2011;10:760–771. doi: 10.1016/j.dnarep.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 58.Luijsterburg MS, von BG, Gourdin AM, Politi AZ, Mone MJ, Warmerdam DO, Goedhart J, Vermeulen W, van DR, Hofer T. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J. Cell Biol. 2010;189:445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soderhall S, Lindahl T. Mammalian DNA ligases. Serological evidence for two separate enzymes. J. Biol. Chem. 1975;250:8438–8444. [PubMed] [Google Scholar]
- 60.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 61.Harrington JJ, Lieber MR. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Politi A, Mone MJ, Houtsmuller AB, Hoogstraten D, Vermeulen W, Heinrich R, van DR. Mathematical modeling of nucleotide excision repair reveals efficiency of sequential assembly strategies. Mol. Cell. 2005;19:679–690. doi: 10.1016/j.molcel.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 64.Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34:3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zielinska A, Davies OT, Meldrum RA, Hodges NJ. Direct visualization of repair of oxidative damage by OGG1 in the nuclei of live cells. J. Biochem. Mol. Toxicol. 2011;25:1–7. doi: 10.1002/jbt.20346. [DOI] [PubMed] [Google Scholar]
- 66.Berquist BR, Singh DK, Fan J, Kim D, Gillenwater E, Kulkarni A, Bohr VA, Ackerman EJ, Tomkinson AE, Wilson DM., III Functional capacity of XRCC1 protein variants identified in DNA repair-deficient Chinese hamster ovary cell lines and the human population. Nucleic Acids Res. 2010;38:5023–5035. doi: 10.1093/nar/gkq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Z, Zheng L, Xu H, Dai H, Zhou M, Pascua MR, Chen QM, Shen B. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat. Chem. Biol. 2010;6:766–773. doi: 10.1038/nchembio.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mortusewicz O, Fouquerel E, Ame JC, Leonhardt H, Schreiber V. PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 2011;39:5045–5056. doi: 10.1093/nar/gkr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanssen-Bauer A, Solvang-Garten K, Sundheim O, Pena-Diaz J, Andersen S, Slupphaug G, Krokan HE, Wilson DM, III, Akbari M, Otterlei M. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ. Mol. Mutagen. 2011;52:623–635. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harris JL, Jakob B, Taucher-Scholz G, Dianov GL, Becherel OJ, Lavin MF. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum. Mol. Genet. 2009;18:4102–4117. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- 74.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat. Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 75.Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Curr. Opin. Struct. Biol. 1999;9:37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 76.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 77.Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- 78.Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE, Wilson SH. Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 79.Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. J. Biol. Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balakrishnan L, Brandt PD, Lindsey-Boltz LA, Sancar A, Bambara RA. Long patch base excision repair proceeds via coordinated stimulation of the multienzyme DNA repair complex. J. Biol. Chem. 2009;284:15158–15172. doi: 10.1074/jbc.M109.000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holen T, Moe SE, Sorbo JG, Meza TJ, Ottersen OP, Klungland A. Tolerated wobble mutations in siRNAs decrease specificity, but can enhance activity in vivo. Nucleic Acids Res. 2005;33:4704–4710. doi: 10.1093/nar/gki785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl Acad. Sci. USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spiro C, McMurray CT. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol. Cell Biol. 2003;23:6063–6074. doi: 10.1128/MCB.23.17.6063-6074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.