Abstract
Nuclear factors NF90 and NF45 form a complex involved in a variety of cellular processes and are thought to affect gene expression both at the transcriptional and translational level. In addition, this complex affects the replication of several viruses through direct interactions with viral RNA. NF90 and NF45 dimerize through their common ‘DZF’ domain (domain associated with zinc fingers). NF90 has additional double-stranded RNA-binding domains that likely mediate its association with target RNAs. We present the crystal structure of the NF90/NF45 dimerization complex at 1.9-Å resolution. The DZF domain shows structural similarity to the template-free nucleotidyltransferase family of RNA modifying enzymes. However, both NF90 and NF45 have lost critical catalytic residues during evolution and are therefore not functional enzymes. Residues on NF90 that make up its interface with NF45 are conserved in two related proteins, spermatid perinuclear RNA-binding protein (SPNR) and zinc-finger RNA-binding protein (Zfr). Using a co-immunoprecipitation assay and site-specific mutants, we demonstrate that NF45 is also able to recognize SPNR and Zfr through the same binding interface, revealing that NF45 is able to form a variety of cellular complexes with other DZF-domain proteins.
INTRODUCTION
Nuclear factors NF90 (also known as ILF3, DRBP76 or NFAR-1) and NF45 (also known as ILF2) form a protein complex that regulates gene expression at multiple levels (1,2). The complex was first identified as a transcription factor involved in IL-2 transcription in activated T-cells (3,4). Subsequently the NF90/NF45 complex has been identified in a number of different contexts. For example, NF90/NF45 has been implicated in DNA binding, transcription and repair, although it does not contain classical DNA-binding domains (3,5–8). On the other hand, NF90 contains classical double-stranded RNA-binding domains (dsRBDs) and is proposed to play a role in gene expression at the RNA level, either through direct association with mRNAs or through repression of miRNA metabolism (9–15).
In addition to its normal cellular roles, NF90/NF45 plays a critical role in the life cycle of several viruses, such as Hepatitis C virus (HCV), Bovine Viral Diarrhoea Virus (BVDV), Dengue virus and human immunodeficiency virus (HIV) that have a global impact on human and animal health (13,16–19). In the case of plus-stranded RNA viruses such as HCV and Dengue virus, NF90/NF45 is thought to associate with the viral genomic RNA through regulatory RNA structures in the 5′- and 3′-non-translated regions and has been observed to co-localize with viral replication foci in the cytoplasm (18,19). Plus-stranded RNA viral genomes resemble cellular mRNAs and it is possible that a normal cellular role of NF90/NF45 is co-opted by the viruses to further their own ends. Indeed, in the case of HIV, NF90 impacts on viral replication by modulating translation of Cyclin T1 through association with the 3′-untranslated region of Cyclin T1 mRNA (13). Knock-down of NF90 causes a shift of Cyclin T1 mRNA from polysomes to monosomes and the consequent reduction in Cyclin T1 protein reduces HIV proliferation.
NF90/NF45 has been shown to bind to several specific mRNAs other than Cyclin T1, including MKP-1, IL-2, acid β-glucosidase and to affect either translation or turnover of these mRNAs (9,11,15). Global RNA immunoprecipitation studies on the other hand, have shown that the complex can associate with a wide variety of mRNAs and non-coding RNAs (12,14,20). Together, these studies indicate a role for NF90/NF45 in post-transcriptional control of gene expression. However, what motifs are recognized by NF90/NF45 and what the molecular consequences are of NF90/NF45 association with mRNAs are not yet well understood. Clearly, a better understanding of the molecular role of this complex would provide valuable insights into its function both in viral infection and in normal cellular processes.
NF90 is found only in vertebrates while NF45 is found throughout the metazoa. In mammals, NF90 and NF45 show a broad tissue distribution but are particularly abundant in brain, spleen, testes and liver (14,21,22). Although NF90 and NF45 are nominally nuclear factors, the protein complex has been shown to shuttle between the nucleus and cytoplasm and certain splice variants of NF90 are preferentially located in the cytoplasm in normal tissue (14,15).
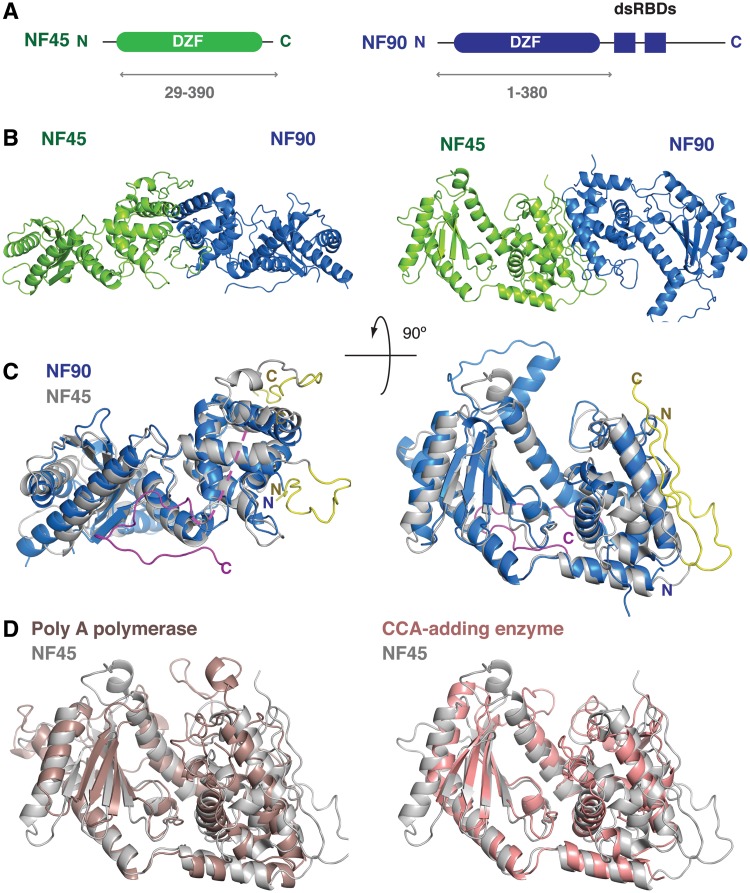
NF90 has a domain architecture that consists of an N-terminal domain with no known function, called a ‘domain associated with zinc fingers’ or DZF domain. This domain is followed by two dsRBDs and a low-complexity region at the C-terminus. The C-terminal region is the site of most sequence variations generated by alternative splicing and longer splice variants of NF90 show an enrichment of arginine, glycine and serine residues in this region (23). The DZF domain is found in three other proteins, namely NF45, spermatid perinuclear RNA-binding protein (SPNR, also known as STRBP) and Zinc-finger protein associated with RNA (Zfr). SPNR is a testes-specific paralogue of NF90 that has a similar domain organization while Zfr, an essential gene in mice and flies, has a different domain organization. In Zfr the DZF domain is found at the C-terminus and is preceded by several RNA-binding zinc fingers. NF45, the partner of NF90, is much smaller than the other proteins in the family. The DZF domain of NF45 is the only folded domain present in the protein sequence and is flanked by an arginine- and glycine-rich sequence at the N-terminus and a glutamate-rich sequence at the C-terminus (Figure 1A).
Figure 1.
Overview of the NF90/NF45 DZF dimerization domain structure. (A) Schematic representation of the domain organization of NF45 (green) and NF90 (blue). Double-headed arrows underneath indicate the sub-fragments of each protein that were used for crystallization. (B) Overview of the structure showing a ‘side’ view and a ‘top’ view rotated 90° around the horizontal axis. (C) Superposition of NF90 (blue) on NF45 (gray) using the same orientations of NF45 as shown in B. Extensions at the N- and C-termini of NF45 that are involved in dimerization interactions are shown in yellow. A C-terminal extension of NF90 is shown in magenta with a dotted line indicating a missing loop. (D) Superposition of NF45 (grey) on yeast Poly (A) polymerase (PDBid 2q66, brown) and CCA-adding enzyme from Archaeoglobus fulgidis (PDBid 1tfw, pink), shown from the ‘top’ view from (B).
While NF90/NF45 is thought to have a role in many aspects of gene expression and RNA metabolism, the molecular mechanisms responsible for these various processes are poorly understood. One possibility is that the dsRBDs of NF90 might direct the complex to its RNA targets, although the specificity of such an interaction remains undefined. Furthermore, the DZF domain is thought to mediate dimerization of NF90 with NF45 but any further role that this domain might have is not known. To gain insight into the possible function of NF90/NF45, we have solved the crystal structure of a truncated complex that contains the DZF dimerization domains. Our work demonstrates that the DZF domain has evolved from a family of RNA metabolizing enzymes. Based on the conservation of the NF90 dimer interface, we were able to identify other putative binding partners of NF45. We demonstrate, using a cell-based assay that NF45 can dimerize with other DZF-domain containing proteins through a structurally conserved interface. This finding suggests that NF45 may have a previously unappreciated role in other complexes that are involved in RNA metabolism.
MATERIALS AND METHODS
Purification of NF90/NF45 constructs
Mouse cDNAs for NF90 and NF45 were obtained from Imagene. Expression constructs of NF90 (residues 1–590 and residues 380–590) were cloned as N-terminal hexahistidine-tagged fusion proteins into pETMCN (24). Further C-terminal deletion constructs were made by site-directed mutagenesis to add stop codons to the NF90ΔC591 construct. NF45 (residues 29–390) was cloned into pGEX-6-p-1, with a N-terminal GST fusion (GE Healthcare). For crystallization, NF90ΔC381 and NF45ΔN28 were expressed separately in Escherichia coli BL-21 Gold pLysS cells and co-lysed in 20 mM Tris–HCl pH 7.5, 500 mM NaCl 1 mM DTT. The complex was purified in batch from cleared lysates using GSH resin and eluted by overnight digestion with tobacco etch virus (TEV) and 3C proteases. The protein was dialysed into a low salt buffer (20 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM DTT). Further purification was carried out using heparin sepharose with a salt gradient to 1000 mM NaCl, and size exclusion chromatography (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM DTT).
Crystallization
The NF90ΔC381/NF45ΔN28 complex was concentrated to 8 mg/ml and crystallized by vapour diffusion in a sitting drop with a reservoir solution of 100 mM MES pH 6.0, 0.22 M sodium malonate and 18% PEG 8000. Initial crystals diffracted poorly and showed multiple lattices. Crystals were improved using serial streak seeding. Highly diffracting crystals grew in space group I222. However many crystals were also found that grew in related C2 space groups where one of the symmetry axes of I222 had degenerated.
Data collection and structure solution
The crystals were cryoprotected in crystallization buffer supplemented with 10% glycerol and flash cooled to 100 K in a liquid nitrogen cryostream. For phasing experiments, the crystals were soaked for 1–2 h with K2AuCN4 prior to cryoprotection and cooling. Data were collected at the X06SA beamline at the Swiss Light Source (Villigen, Switzerland) using the PILATUS detector (25). The data were indexed and integrated using XDS (26). The heavy atom substructure was determined using SHELX (27) and phases were improved using SHARP (28). Approximately 70% of the structure was completed by autobuilding with BUCCANEER (29,30). Further building was carried out manually using the graphics software COOT (31) and the structure was further refined against a higher resolution native dataset with PHENIX (32). The final statistics for the geometry of the structure were calculated using MOLPROBITY (33). Figures were prepared using Pymol (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC).
Nucleotide triphosphate soaking/co-crystallization and data analysis
To overcome problems of dual occupancy of nucleotides and malonate in the putative actives site of NF90ΔN381/NF45ΔN28, the protein was rescreened for alternative crystallization conditions. Crystals were grown in 12–14% PEG3350, 200 mM MgCl2, 100 mM MES pH 6.5 and 5% glycerol and were either soaked in a cryoprotecant containing 10 mM of the nucleotide triphosphate (NTP) or co-crystallized in the presence of 10 mM NTP. The crystals were cryoprotected in the crystallization buffer supplemented with 20% glycerol and flash-cooled in liquid nitrogen to 100 K. Data were collected at beamlines I02 and I04-1 at Diamond Light Source (Didcot, UK). As with native crystals, data from NTP-bound crystals were indexed and integrated using XDS (26). Crystals grown in PEG3350 grew either in space group I222 with one molecule per asymmetric unit or P21 with two molecules per asymmetric unit. Molecular replacement using the native structure was carried out with Phaser (30,34) followed by refinement in PHENIX (32) and model building with COOT (31). In the absence of ligand, clear positive difference density was observed in the Fo–Fc maps for the nucleotides. However during refinement after fitting the ligands, negative Fo–Fc peaks appeared suggesting that the ligands were not present at full occupancy. The occupancy of the ligands was adjusted manually to model the best fit with the electron density map and not refined.
Co-immunoprecipitation
Sequences for human Zfr and SPNR were obtained as cDNAs from Imagene. Vectors containing human NF90b and NF45 cDNAs were a kind gift from S-E. Behrens (Martin Luther University, Halle). Zfr, SPNR, NF90b and NF45 were cloned into pEF-based mammalian expression vectors (a kind gift from Elena Conti, MPI Biochemistry, Martinsried) containing either an N-terminal HA or an N-terminal HA-FLAG epitope tag. Site-directed mutants were generated by PCR amplification of the vector by Pfu Ultra polymerase (Stratagene) with overlapping oligonucleotide primers that encoded the mutation. Vector DNA was transiently transfected into 293T cells in six-well plates (Iwaki) growing in DMEM with foetal bovine serum and penicillin/streptomycin (Gibco). An amount of 4 µg of vector DNA was transfected using 6 µl lipofectamine 2000 (Invitrogen) in a total volume of 200 µl OptiMEM (Gibco). The medium was replaced after 12 h and cells were harvested after 36 h. The cells were placed on ice and washed with 2 ml PBS and then lysed by incubation with 250 µl NET-G buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% NP40, 1 mM EDTA, 10% glycerol, protease inhibitor cocktail (Roche), 3U RNase A/T1 mix (Fermentas) and 0.6 U RQ1 DNase (Promega)) for 15 min at 4°C. The lysed cells were then resuspended from the plates and centrifuged at 16 000 g for 15 min at 4°C to remove insoluble debris. Ten percent of the soluble fraction was removed for sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis (input) while the remainder of the soluble fraction was incubated with 15 µl α-FLAG resin with rotation at 4°C for 1 h. The samples were then washed three times in NET-G buffer and excess liquid was removed with a gel-loading tip. Proteins were eluted from the α-FLAG resin by incubation with 3× FLAG peptide (0.2 mg/ml in NET-G) at 4°C for 15 min. Samples from input and eluted proteins were separated by SDS–PAGE, transferred onto a nitrocellulose membrane (Whatman) and probed with a mouse α-HA antibody (Covance) at a 1:2500 dilution in 5% milk in PBS-T. After washing in PBS-T, the blot was then probed with a secondary goat α-mouse antibody (Biorad) at a dilution of 1:10 000 in 5% milk/PBS-T. After washing off the secondary antibody, the blot was visualized with Super West pico ECL reagent (Pierce) and exposed to film.
RESULTS
Structural studies
Structural studies were carried out with mouse NF90 and NF45. Mouse and human NF90 show 95% sequence identity while mouse and human NF45 are identical at the protein level. Both NF90 and NF45 can be expressed recombinantly in E. coli as soluble proteins. However, attempts to purify the proteins separately were problematic as NF45 showed a tendency to aggregate and precipitate. Thermal denaturation studies of NF45 showed that it has a melting temperature (Tm) of 25°C while a complex of the DZF domains of NF90 and NF45 showed a Tm of 55°C indicating that the presence of NF90 greatly enhances the stability of NF45 (Supplementary Figure S1). A co-lysis and co-purification approach allowed a 1:1 complex of NF90/NF45 to be isolated that showed no aggregation. Indeed, the complex is stable even in high salt concentrations of 0.5 M.
NF90 contains two dsRBDs that have previously been shown to bind dsRNA (35,36). To assess the integrity of the purified proteins, electrophoretic mobility shift assays (EMSA) were carried out with both single-stranded (ss) and double-stranded (ds) RNA substrates. A long construct NF90ΔC591/NF45 (which includes the DZF and dsRBD domains of NF90 but not the C-terminal low complexity region) and a fragment containing only the dsRBDs of NF90 (residues 380–590) both showed dsRNA-binding activity as expected. A shorter construct containing only the DZF dimerization domains (NF90ΔC381/NF45ΔN28) showed no mobility shift with either ss or dsRNA indicating that this fragment is not necessary for RNA binding (Supplementary Figure S2). However, the long NF90ΔC591/NF45 complex shows qualitative differences compared with the dsRBD construct: the long complex shows apparently tighter binding and less heterogeneity than the dsRBDs alone. This suggests that the DZF domain, while not required for RNA binding, might nevertheless contribute to recognition of RNA substrates.
Initial attempts to crystallize the NF90ΔC591/NF45 construct were not successful, most likely because of the multiple domains present in NF90. A series of C-terminal truncations of NF90 were designed based on sequence analysis and limited proteolysis. Expression trials showed that NF90 constructs truncated beyond residue 380 were not soluble. Well-diffracting crystals were obtained with complexes that contained residues 1–380 of NF90 and 29–390 of NF45. An overview of the domain structure of NF90 and NF45 is shown in Figure 1A, with the constructs used for crystallization indicated underneath.
Phases for the structure were obtained using single wavelength anomalous dispersion (SAD) from gold-derivatized crystals that diffracted to 3.3 Å. The structure was refined against a native dataset that diffracted to 1.9 Å with a Rwork = 18.5% and Rfree = 21.0% and good stereochemistry (Table 1). The model includes 658 residues out of a total of 741 that were present in the crystal. Residues in the N- and C-termini and several loops (residues 1–5, 55–86, 341–353 and 375–380 of NF90 and 362–390 of NF45) could not be modelled due to poor electron density in these regions that made a clear interpretation of the map difficult.
Table 1.
Data collection, phasing and refinement statistics for NF90/NF45 native structure
| Dataset | Native | AuCN derivative |
|---|---|---|
| Beamline | Swiss light source PXII | |
| Wavelength (Å) | 1.0077 | 1.0077 |
| Spacegroup | I 2 2 2 | C2 |
| Unit cell (Å) | a = 82.9 | a = 229.9 α = 90.00 |
| b = 131.3 α = β = γ = 90 | b = 83.4 β = 127.1 | |
| c = 176.0 | c = 133.0 γ = 90.00 | |
| Resolution range (Å) | 79.0–1.9 (2.0–1.9)a | 81.0–3.3 (3.2–3.0) |
| Unique reflections | 74596 | 76165 |
| Multiplicity | 6.4 | 3.4 |
| Completeness (%) | 98.4 (95.4) | 99.0 (99.6) |
| I/σ(I) | 15.3 (3.4) | 16.4 (6.7) |
| Rsym (%) | 8.0 (60.6) | 6.7 (20.2) |
| Refinement | ||
| Rfree (%) | 21.0 | |
| Rwork (%) | 18.5 | |
| r.m.s.d. bonds (Å) | 0.007 | |
| r.m.s.d. angles (°) | 1.03 | |
| B-factors (Å2) | ||
| Overall | 30.8 | |
| Protein | 29.5 | |
| Solvent | 38.4 | |
| Ramachandran values | ||
| Most favoured (%) | 97.5 | |
| Allowed (%) | 2.5 | |
| Disallowed (%) | 0 | |
aValues in parentheses are for the highest resolution shell.
The DZF dimerization domains of NF90 and NF45 form an oblong structure with a flat face on one side and a curved face on the other (Figure 1B). Each protein shows a bipartite domain, characterized by an N-terminal mixed α–β region that contains a central anti-parallel β-sheet and a C-terminal α-helical region (Figures 1B and 2). NF90 and NF45 have 30% sequence identity over the DZF domain and superpose with a root mean square deviation (r.m.s.d.) of 2.0 Å over 261 residues out of 334 (Figure 1C). This gives the overall structure a pseudo 2-fold rotational symmetry. The central β-sheet of each domain forms the base of a cleft between the N- and C-terminal halves while dimerization of NF90 and NF45 is mediated by the α-helices at the C-terminus of each protein. NF45 has an extension to the DZF domain at the N-terminus, consisting of residues 29–47, that contributes to dimerization (see below). NF90 and NF45 also differ at their C-termini: residues 354–374 of NF90 pack in an extended conformation against the flat side of NF90. In contrast, residues 343–361 of NF45 follow a different path, forming a short β-hairpin and then binding in an extended conformation along the dimerization interface (Figure 1C).
Figure 2.
Conservation of NF90 and NF45 sequences. (A) Multiple sequence alignment of NF45 with sequences from mouse (M.m) zebrafish (D.r.) Drosphila (D.m.) and Ciona intestinalis (C.i.). A structure-based alignment of NF45 with yeast poly (A) polymerase is shown underneath. Human NF45 was not included as it has 100% identity with mouse NF45. Secondary structure elements are shown underneath as bars (α-helices) or arrows (β-strands). Blue dots indicate residues that interact with NF90, pink dots indicate residues that interact with ATP and black boxes show residues equivalent to those involved in catalysis in functional nucleotidyl transferases. Purple boxes indicate the two residues on helix 9 that were mutated to disrupt dimer formation. (B) Sequence alignment of NF90 with sequences from mouse (M.m.), human (H.s.), zebrafish (D.r.) and Xenopus (X.l.). NF90 is only found in vertebrates. Green dots indicate residues that are involved in dimerization interaction with NF45.
A structural similarity search using the DALI server showed that the DZF domains of NF90 and NF45 have significant structural similarity to members of the template-free nucleotidyltransferase family that includes poly (A) polymerases, TUTases, 2′–5′ oligoadenylate synthase and the CCA-adding enzymes of tRNAs (Figure 1D and Supplementary Figure S3). Template-free nucleotidyltransferases belong to the DNA polymerase β family of nucleotidyltransferases (37,38). None of the template-free nucleotidyltransferases for which structures are known encode dsRBDs for substrate recognition. However both poly (A) polymerase and the CCA-adding enzyme have RNA-binding domains in addition to their catalytic domains that aid with substrate recognition or processivity. The dsRBD domains of NF90 emanate from the flat side of the molecule, on the opposite face to that observed in other template-free nucleotidyltransferases (Supplementary Figure S3). However, the linker region between the end of the visible C-terminus and the first dsRBD is ∼23 residues, which would be sufficient to allow the dsRBDs to access the same face of the molecule as the auxiliary RNA-binding domains observed in other template-free nucleotidyltransferases.
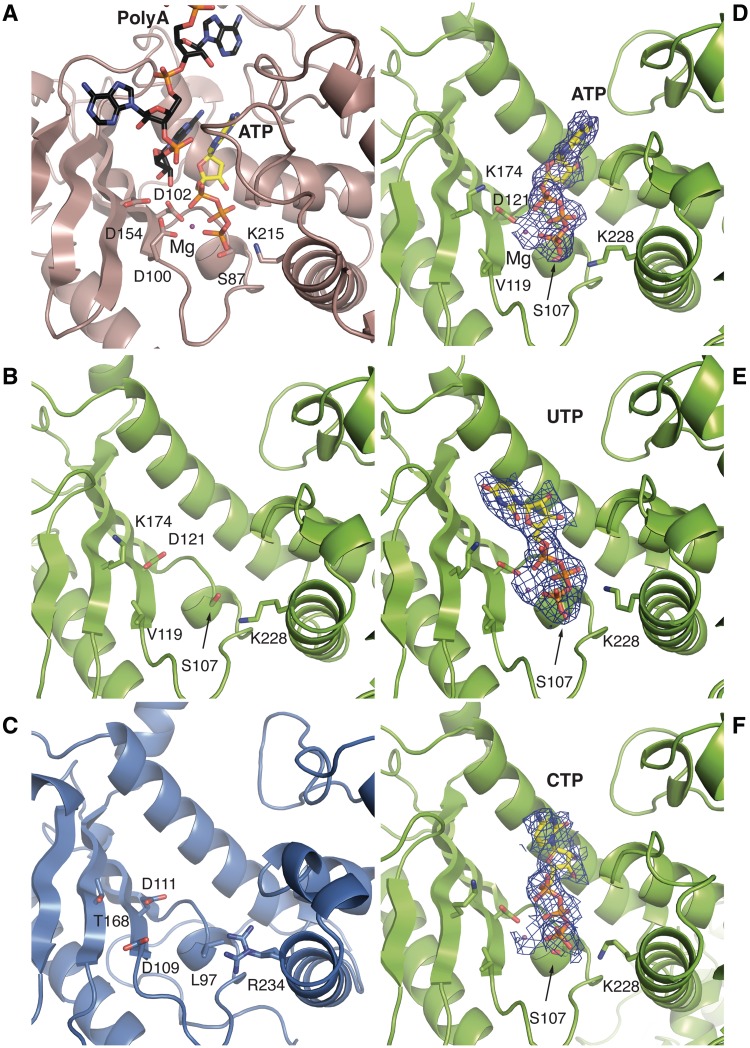
This family of enzymes catalyses the transfer of one NTP to the end of an existing oligonucleotide substrate. These enzymes have a catalytic signature sequence that includes three negatively charged residues (in yeast poly (A) polymerase, Pap1, these are residues Asp100, Asp102 and Asp154) at the base of the catalytic cleft, on the exposed surface of the β-sheet (Figure 3A). These negatively charged residues are required to position two divalent metal ions (typically two Mg2+ ions or Mg2+ and Mn2+) that co-ordinate the close approach of the phosphate groups of an incoming NTP to the substrate RNA primer (Figure 3A). In addition to this catalytic triad, a serine residue (Ser89 in Pap1) and a lysine residue (Lys215) are conserved in the template free nucleotidyltransferases and help to orient the γ-phosphate of the NTP.
Figure 3.
A close-up view of the active site of poly (A) polymerase and equivalent regions in NF90 and NF45. (A) The catalytic cleft of yeast poly (A) polymerase (derived from PDBids 2q66 and 3c66) showing the catalytic residues and the bound ATP and a poly (A) substrate. (B) and (C) top view of NF45 and NF90 oriented as in Figure 1C, looking into the cleft between the two domains. The residues shown are those in equivalent positions to catalytic and nucleotide-binding residues found in related nucleotidyl transferases. (D) ATP bound to the cleft of NF45. A 2Fo–Fc map from the final refined structure is shown, contoured at 1σ. (E) and (F) A similar view as in (D) showing complexes with UTP and CTP, respectively.
Neither NF90 nor NF45 has a full complement of these critical residues in their putative active sites. In NF45, two of the characteristic negatively charged residues, equivalent to Asp100 and Asp154, have been replaced with a valine residue (Val119) and a lysine residue (Lys174), respectively (Figure 3B). In NF90, the residue equivalent to Asp154 is replaced by a threonine residue (Thr168, Figure 3C). In addition, the serine and lysine residues at the base of the nucleotide-binding site are replaced by a leucine (Leu97) and an arginine residue (Arg234), respectively (Figure 3C). The structural analysis strongly indicates that the DZF domains of NF90 and NF45 are unlikely to possess nucleotidyltransferase activity. We further carried out an in vitro nucleotidyltransferase assay using α-labelled ATP or UTP. Although activity could be shown for a known poly (A) polymerase, no activity was detectable for NF90/NF45 in the conditions we used for this assay (Supplementary Figure S4)
Nucleotide binding to NF90/NF45
Although it is unlikely that NF90/NF45 could actually catalyse nucleotidyl transfer, prominent cleft regions of both NF90 and NF45 are conserved in both proteins (Supplementary Figure S5) suggesting that this region of the protein might nevertheless be functionally important. Furthermore, a malonate ion from the crystallization solution, which is negatively charged, was found in the cleft of both NF90 and NF45 in approximately the position where the triphosphate moiety of nucleotides is bound in other template-free nucleotidyltransferases. This suggested that NF90/NF45 might bind nucleotide ligands.
To identify potential nucleotide ligands, NF90ΔC381/NF45ΔN28 crystals were soaked with ATP, UTP, CTP and GTP. Initial data from soaking experiments showed extra density in the cleft of NF45 that was overlapping but larger than the electron density of the malonate ion. This suggested that nucleotides were competing with malonate ions for binding. An alternative crystallization form was identified that grew with PEG3350 as a precipitant and did not require sodium malonate. Crystals grown in PEG3350 belong either to I 222 or to P 21 space groups and diffract to lower resolution than crystals grown in sodium malonate. Several datasets of these crystals that were either grown or soaked with different nucleotides were collected and analysed (Table 2). Datasets from crystals co-crystallized with ATP or soaked with UTP (at 2.7 and 2.8 Å resolution, respectively) showed clear electron density for the ligand in the cleft of NF45. A dataset where CTP was co-crystallized with the complex diffracted to 3.1 Å resolution and also showed positive difference density at the same site but the density for the nucleotide was less well defined. This could be because CTP binds less well to this site and so does not saturate the nucleotide-binding sites in the crystal. Although a number of datasets were collected from crystals grown in the presence of GTP, none showed any evidence for nucleotide binding. Thermal denaturation assays with NF45 and the various nucleotides showed a similar trend: greater stabilization of the protein was seen in the presence of ATP and UTP (6–7°C) while CTP and GTP showed a lesser degree of stabilization (5°C and 3°C, respectively, Supplementary Figure S6A).
Table 2.
Data collection and refinement statistics for NF90ΔC381/NF45ΔN28 crystals with bound NTPs
| Dataset (ligand) | ATP | UTP | CTP |
|---|---|---|---|
| Beamline | Diamond I04-1 | Diamond I02 | Diamond I04-1 |
| Wavelength (Å) | 0.9173 | 0.9795 | 0.9173 |
| Spacegroup | P 1 21 1 | I 2 2 2 | P 1 21 1 |
| Unit cell (Å) | a = 70.9 α = 90.00 | a = 83.9 α = 90.00 | a = 71.16 α = 90.00 |
| b = 84.1 β = 93.34 | b = 133.5 β = 90.00 | b = 83.97 β = 93.57 | |
| c = 159.6 γ = 90.00 | c = 178.0 γ = 90.00 | c = 159.68 γ = 90.00 | |
| Resolution range (Å) | 66.11–2.69 (2.84–2.69)a | 66.76–2.80 (2.95–2.80) | 66.43–3.10 (3.27–3.10) |
| Unique reflections | 50 589 | 23 950 | 33 184 |
| Multiplicity | 2.9 | 4.2 | 2.8 |
| Completeness (%) | 97.1 (93.7)a | 96.7 (95.4) | 96.8 (98.7) |
| I/σ(I) | 8.0 (2.9) | 10.1 (2.4) | 7.6 (2.8) |
| Rsym (%) | 10.1 (37.6) | 13.1 (62.7) | 12.5 (36.5) |
| Refinement | |||
| Rfree (%) | 26.6 | 26.3 | 25.3 |
| Rwork (%) | 24.0 | 23.6 | 21.5 |
| r.m.s.d. bonds (Å) | 0.004 | 0.011 | 0.012 |
| r.m.s.d. angles (°) | 0.68 | 1.06 | 0.87 |
| B-factors (Å2) | |||
| Overall | 37.0 | 49.1 | 39.8 |
| Ligand | 42.1 | 31.5 | 30.79 |
| Ramachandran values | |||
| Most favoured (%) | 95.5 | 95.5 | 94.7 |
| Allowed (%) | 4.2 | 4.3 | 4.6 |
| Disallowed (%) | 0.3 | 0.2 | 0.7 |
aValues in parentheses are for the highest resolution shell.
In all of the datasets analysed, the electron density was clearly interpretable for the triphosphate moiety of the NTPs and a magnesium ion but less clear for the bases. A similar effect has been noted in previous studies with mitochondrial editosome-like complex associated TUTase1 (MEAT1) and RNA editing terminal uridylyltransferase 2 (RET2) where clear density for the triphosphate moiety could be seen with non-cognate NTPs but the bases remained poorly defined (39,40). The γ phosphate of each of the NTPs makes a close approach to Ser107 at the base of the cleft and is further stabilized by Lys228 (Figure 3D–F). The Mg2+ ion is co-ordinated by an oxygen atom of Asp121 and three oxygen atoms from the α, β and γ phosphates of the nucleotide. The metal is also likely to be co-ordinated by water molecules that were not visible at this resolution. None of the datasets analysed showed any evidence for nucleotide binding to the NF90 cleft. This is unlikely to be because of inaccessibility of the cleft. This region of NF90 shows a similar exposure to solvent channels as the equivalent region of NF45. The structural analysis points to a crucial difference in the pocket between NF90 and NF45, namely the presence of a leucine residue, Leu97, at the base of the binding pocket. This residue is found in the equivalent position to the conserved serine, Ser107, which is present in NF45 and in functional template-free nucleotidyltransferases. The proximity of Leu97 to Arg234, which is normally a lysine residue in other nucleotidyltransferases, may form a steric block that prevents the triphosphate moiety from binding.
Surprisingly, when the binding mode of ATP, UTP and CTP are compared in the three structures, the orientation of the triphosphate moiety is retained while the positions of the bases vary. The orientation of ATP in NF45 shows remarkable similarity to ATP binding in Pap1. The adenine base contacts the edges of His207 on helix 4, which in turn is oriented by a pi-stacking interaction with Trp243. The adenine moiety makes further van der Waals interactions with the side chains of Thr241 and Pro242 at the beginning of helix 6. The base of UTP flips over to make an alternative set of van der Waals contacts with Leu176 on strand 5 and Ser200 and Ala201 on helix 4. The cytosine base is placed in an intermediate position and has van der Waals interactions with His207, Ala203 and Ala204 of helix 4.
The binding of ATP and UTP with NF45 is something of a puzzle. The binding site for these ligands is clearly conserved and the position and orientation of the triphosphate moiety is similar to that observed in catalytically active template-free nucleotidyltransferases. However, the protein does not seem able to use the nucleotides to catalyse nucleotidyl transfer and also shows little structural perturbation on nucleotide binding (r.m.s.d. values between native and nucleotide-bound structures range from 0.4 to 0.8 Å). One possibility is that nucleotide binding is an evolutionary relic. This would be analogous to many pseudokinases of the serine/threonine protein kinase family. In several cases, structural studies on non-functional protein kinases have revealed that some of these proteins retain the ability to bind ATP (41,42). A more intriguing possibility is that nucleotide binding might represent an as yet undefined ligand such as a peptide partner, a small molecule or an RNA-associated chemical moiety.
The NF90/NF45 dimerization interface
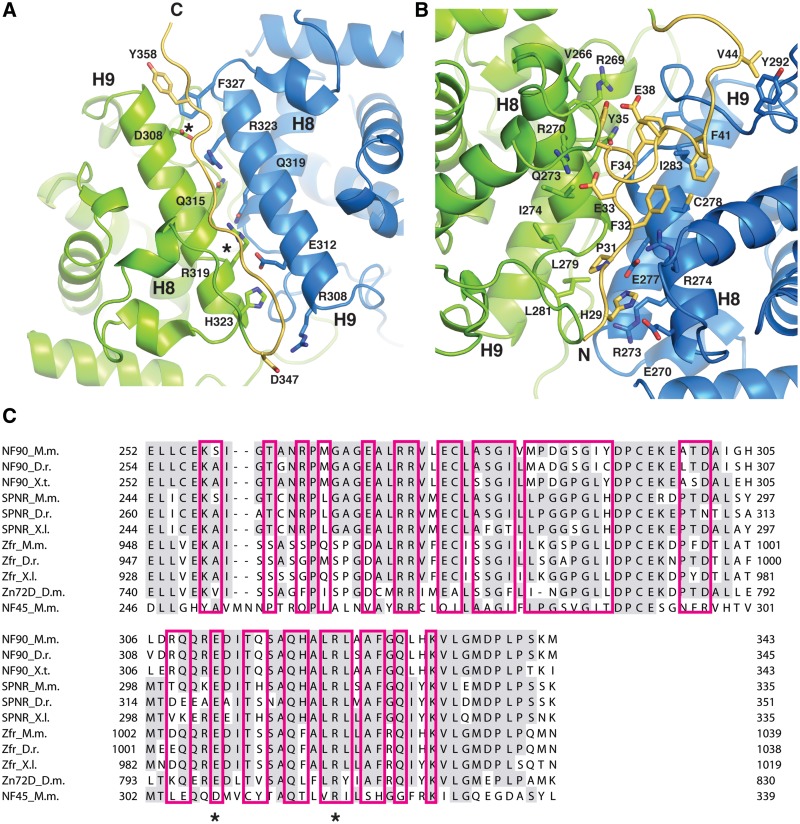
The dimerization of NF90 and NF45 is mediated through a close anti-parallel association of helix 9 and the interaction of the preceding loop region and C-terminal end of helix 8 in each DZF domain. This interaction buries a total area of 1698 Å2, which is sufficient for a biologically relevant interaction (43). The helix 9 interface shows a remarkable symmetry at its core with three conserved residues on NF90 (Glu312, Gln319, Arg323) interacting with three similar conserved residues on NF45 (Asp308, Gln315, Arg319) (Figure 4A). The glutamine residues make a polar contact at the centre of the interaction while the Arg323 from NF90 forms a salt bridge with Asp308 of NF45, as does Arg319 of NF45 with Glu312 on NF90. From this interaction one can imagine how this heterodimerization interface could have evolved from a homodimeric precursor. The symmetry of the interaction breaks down at either end of helix 9, with Phe327 at the C-terminal end of helix 9 in NF90 contributing a hydrophobic interaction while His323 at the C-terminal end of NF45 helix 9 and Arg308 and Gln309 at the N-terminal end of NF90 helix 9 contribute to a more hydrophilic interface (Figure 4A). Packed against the helix 9–helix 9 interface is an extended region of the C-terminus of NF45, which makes primarily main chain interactions with residues on both NF45 and NF90 and contributes a tyrosine side chain (Tyr358) to the hydrophobic pocket at the N-terminal end of NF45 helix 9.
Figure 4.
The NF90/NF45 dimerization interface. (A) Close up view of the dimerization interface in the same orientation as the top view in Figure 1B. The C-terminal extension of NF45 (yellow) lies in an extended conformation over helix 9 (H9) of NF90 (blue) and NF45 (green). Helix 9 shows symmetrical binding interactions at the core. Mutated residues in NF45 are marked with an asterisk. (B) A close up view of the interface rotated 180° around the horizontal axis. The N-terminal extension of NF45 is shown in yellow. Conserved residues involved in the dimerization interface and in stabilizing the N-terminal extension are shown. (C) A sequence alignment of the dimerization interface of mouse NF90 with related DZF domain-containing proteins, SPNR and Zfr. Sequences from mouse (M.m.), zebrafish (D.r.) Xenopus species (X.l. and X.t.) and salmon (S.s.) are shown.
NF45 has an N-terminal extension in addition to the C-terminal extension that also contributes to the dimerization interface (Figures 1C and 4B). The N-terminal extension of NF45 packs against the opposite face of the molecule, on helix 8 and the loop region that connects it to helix 9 (Figure 4B). Like the C-terminal extension, the N-terminal extension of NF45 interacts with both NF90 and NF45 at the dimerization interface. However, unlike the C-terminal extension, these interactions are primarily mediated by side chain interactions that are contributed by His29, Pro31, Phe32, Asp33, Phe34, Tyr35 and Phe41. Consequently, the N-terminal extension of NF45 buries a large hydrophobic surface area that presumably contributes to stabilization of the complex. Additional polar interactions are contributed by Asp33, which forms a salt bridge with Arg270 on NF45, and His29, which hydrogen bonds with Glu270 and Glu277 on NF90.
The contribution of the N- and C-terminal extensions of NF45 to its interaction with NF90 lends asymmetry to the interaction interface. It is unlikely that NF45 would be able to homodimerize because the extension sequences would likely cause a steric clash. Similarly, it also seems unlikely that NF90 could homodimerize as the hydrophobic surface that is protected by the N-terminus of NF45 would be exposed. Interestingly, when NF90 is compared with other DZF domain containing proteins such as SPNR and Zfr, the residues involved in NF45 interactions are relatively well conserved (Figure 3B). Based on these observations, two predictions can be made: first; that NF90 and NF45 are unable to form homodimers and second; that NF45 might be a binding partner for other DZF domain-containing proteins, namely SPNR and Zfr. Indeed, a TAP-tagging study of the binding partners of the Drosophila homologue of Zfr, Zn72D, showed that it co-purifies with Drosophila NF45 (44).
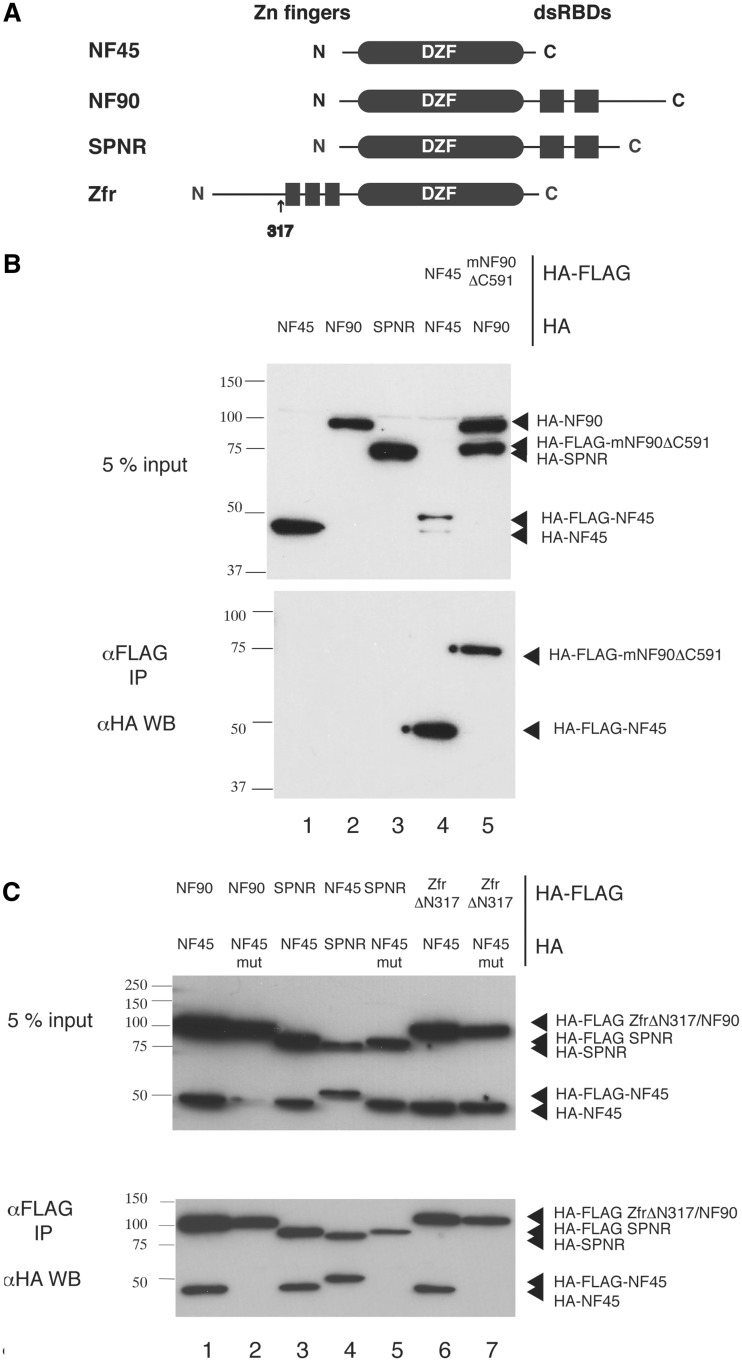
To test whether NF90 and NF45 can homodimerize and whether NF45 is a binding partner of other DZF-domain containing proteins, a co-immunoprecipitation assay using tagged human proteins in mammalian cells was set up. For this assay isoform 6 of NF90, NF90b, was used. Each protein was cloned as an N-terminally HA-FLAG or HA tagged protein and pairs of differentially tagged constructs were co-transfected into 293 T cells. Tagged proteins were immunoprecipitated with anti-FLAG resin and the presence of binding partners was examined by anti-HA western blot (Figure 5). All experiments were carried out in the presence of nucleases to remove RNA. Control experiments showed that HA-tagged proteins do not bind non-specifically to the anti-FLAG resin (Figure 5B, lanes 1–3). Co-transfection of HA-NF45 with HA-FLAG NF45 did not show a complex form on co-immunoprecipitation (Figure 5B, lane 4), confirming previous observations with in vitro pull down experiments (45). Similarly HA-FLAG NF90 is unable to co-precipitate HA-NF90. In this case, a C-terminally truncated HA-FLAG-tagged mouse NF90ΔC591 was co-transfected with full-length NF90b to differentiate the two constructs by size. This confirms that neither NF45 nor NF90 are able to homodimerize.
Figure 5.
Co-immunoprecipitation studies of NF45 with other DZF domain-containing proteins. (A) Overview of domain organization of DZF-domain containing proteins. (B) Co-immunoprecipitation studies carried out with HA-tagged proteins (1. HA-NF45, 2. HA-NF90, 3. HA-STRBP) and with differentially tagged pairs of NF45 (lane 4) and NF90 (lane 5). (C) Co-immunoprecipitation studies of NF45 wild-type (lanes 1, 3, 4 and 6) or NF45 mutant (lanes 2, 5 and 7) co-transfected with NF90 (lanes 1 and 2), SPNR (lanes 3,4 and 5) and Zfr (lanes 6 and 7).
NF45 forms alternative complexes with SPNR and Zfr
NF90 and NF45 show a robust co-precipitation in this assay (Figure 5C, lane 1). To disrupt dimerization, a double mutant of NF45 was created where the charges were reversed on two of the residues forming the core interaction on helix 9 i.e. Asp308 was mutated to arginine (D308R) and Arg319 was mutated to glutamate (R319E). Although the mutated protein is expressed in the transfected cells, there is no association with NF90 in the co-precipitation assay, showing that the mutant does indeed disrupt binding to NF90 (Figure 5C, lane 2).
Similar co-immunoprecipitation reactions showed a robust interaction between SPNR and NF45 (Figure 5C, lanes 3 and 4) and between Zfr and NF45 (lane 6). However, when either SPNR or Zfr were co-transfected with the mutant NF45, no interaction could be seen (lanes 5 and 7). This demonstrates that NF45 can form complexes with both Zfr and SPNR. The lack of interaction with the mutant shows that NF45 uses the same interface to interact with Zfr and SPNR as it uses to interact with NF90. The absence of an interaction with the mutant is unlikely to be caused by a disruption to the structural integrity of the mutant. Thermal denaturation assays carried out with a purified NF45 mutant showed that it has a similar melting profile to the wild-type protein and is also stabilized in the presence of UTP and ATP (Supplementary Figure S6B).
Interestingly, in each case where NF90, SPNR and Zfr are co-expressed with the mutant NF45, the overall amount of protein expressed is lower than when functional pairs are expressed together. This effect is seen consistently over multiple experiments and so is unlikely to be caused by variable expression levels that are normally seen with transient transfections. It has previously been shown that NF90 protein levels are correlated with NF45 levels (46). Given the difference in stability of NF45 on its own versus in a complex (Tm = 25°C versus 55°C) this difference in expression is likely to come from the mutual stabilization effect of the two partner proteins.
DISCUSSION
The structural studies on the dimerization domain of NF90 and NF45 show that these proteins belong to the family of template-free nucleotidyltransferase enzymes. However, the lack of conserved catalytic residues suggests that the DZF domain encodes a ‘pseudotransferase’ that is no longer able to catalyse transfer of nucleotides. Ligand-binding studies indicate that NF45, but not NF90, retains the ability to bind NTPs in a conserved binding pocket at the degenerated active site. Whether NTPs are the true ligands of this site remains to be established.
The co-precipitation studies confirm two predictions based on the structure of the NF90/NF45 complex. We have demonstrated that NF45 is not able to homodimerize in a cellular context. This is most likely because of the intrinsic asymmetry of the dimerization interaction with NF90, as well as the potential steric clashes with the N- and C-terminal extensions of NF45. The second prediction was based on the observation that residues in NF90 that are involved in binding are conserved in SPNR and Zfr. The prediction that NF45 should interact with both of these related proteins is borne out by the co-precipitation studies and shows that the interaction between Zfr and NF45 is present in mammals as well as in flies. Furthermore, this interaction is disrupted when these proteins are co-transfected with NF45 that is mutated at critical residues on helix 9, indicating that NF90, SPNR and Zfr use a conserved interface for their interaction with NF45.
These interactions help to explain the evolutionary relationship between NF90 and NF45. The DZF domains of NF90 and NF45 share ∼30% sequence identity and a high degree of structural similarity. The complex that the two proteins form is very intimate and, based on our thermal denaturation assays and previous in vivo observations (46), NF45 is unstable in the absence of a binding partner both in vitro and in vivo. Both NF45 and Zfr are found in worms and insects and a Zfr-like protein that has only one annotated zinc finger is found, along with NF45, in early metazoans such as Trichoplax adherans and Ciona intestinalis. NF90 and SPNR, on the other hand, are only found in vertebrates. We would predict that NF45 is always found in a complex with NF90, SPNR or Zfr/Zfr-like proteins. It is also possible that some early metazoa might have a symmetrical NF45 homodimer. Indeed, the symmetrical interactions at the core of the dimerization interface suggest that all of these complexes might have arisen from a homodimeric precursor.
It is interesting that NF45, which retains a conserved ligand-binding site, is also the component that is able to participate in other protein complexes. That NF45 can participate in a variety of different complexes also has important implications for functional studies of all of the DZF proteins. Both NF90 and Zfr are widely expressed in a variety of tissues and so NF90/NF45 and Zfr/NF45 complexes could co-exist in the same cells. SPNR shows a more restricted tissue distribution and is found primarily in brain and testes. However both NF90 and Zfr are also present in neuronal cells, suggesting that in this system there is an even richer variety of this family of complexes present. Since levels of NF90 and NF45 are known to be co-regulated, caution should be exercised when overexpressing or knocking down individual proteins as this could potentially lead to a redistribution of other DZF domain complexes.
Further cell-based, biochemical and structural studies are required to determine the functional role of the DZF dimerization domain in its various different guises. The dimer presents an extended protein surface that is highly conserved (Supplementary Figure S5). This extended surface could potentially provide a docking platform for protein–protein interactions. Furthermore, although the DZF domain is not required for dsRNA binding, it might nevertheless contribute to RNA recognition, either through the conserved ligand-binding site or through sequence-specific interactions. Future studies will elucidate the role of this platform in the various biological roles proposed for NF90 and NF45.
ACCESSION NUMBERS
Data have been deposited in the PDB with the following accession codes: 4AT7, 4AT8, 4AT9 and 4ATB.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6 and Supplementary Methods.
FUNDING
Medical Research Council [G1000520 to A.G.C.]; Wellcome Trust [077707, 092076] and the early part, Max Planck Gesellschaft. Funding for open access charge: Wellcome Trust [092076].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Elena Conti (MPI for Biochemistry, Martinsried) for supporting the early part of this work. The authors thank Doris Lindner for cloning of NF90 constructs and Katherina Nagel (Conti Department) for supplying Gld2/Gld3 complex. The authors thank Fabien Bonneau for constructive discussions and help with setting up EMSA, nucleotidyltransferase and coIP assays. The authors thank the crystallization facility at the MPI for Biochemistry in Martinsried for initial crystallization screening and beam-line scientists at the Swiss Light Source and Diamond Light Source for support during data collection. The authors thank the staff of the Edinburgh Protein Production Facility and Biophysical Core Facility for support with protein purification and characterization. The authors thank Elena Conti, Fulvia Bono and Esben Lorentzen for critical reading of the manuscript.
REFERENCES
- 1.Marcoulatos P, Koussidis G, Mamuris Z, Velissariou V, Vamvakopoulos NC. Mapping interleukin enhancer binding factor 2 gene (ILF2) to human chromosome 1 (1q11-qter and 1p11-p12) by polymerase chain reaction amplification of human-rodent somatic cell hybrid DNA templates. J. Interferon Cytokine Res. 1996;16:1035–1038. doi: 10.1089/jir.1996.16.1035. [DOI] [PubMed] [Google Scholar]
- 2.Barber GN. The NFAR's (nuclear factors associated with dsRNA): evolutionarily conserved members of the dsRNA binding protein family. RNA Biol. 2009;6:35–39. doi: 10.4161/rna.6.1.7565. [DOI] [PubMed] [Google Scholar]
- 3.Corthesy B, Kao PN. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J. Biol. Chem. 1994;269:20682–20690. [PubMed] [Google Scholar]
- 4.Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, Corthesy B. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem. 1994;269:20691–20699. [PubMed] [Google Scholar]
- 5.Orford RL, Robinson C, Haydon JM, Patient RK, Guille MJ. The maternal CCAAT box transcription factor which controls GATA-2 expression is novel and developmentally regulated and contains a double-stranded-RNA-binding subunit. Mol. Cell Biol. 1998;18:5557–5566. doi: 10.1128/mcb.18.9.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamanna RA, Hoque M, Lewis-Antes A, Azzam EI, Lagunoff D, Pe'ery T, Mathews MB. The NF90/NF45 complex participates in DNA break repair via nonhomologous end joining. Mol. Cell Biol. 2011;31:4832–4843. doi: 10.1128/MCB.05849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi L, Qiu D, Zhao G, Corthesy B, Lees-Miller S, Reeves WH, Kao PN. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 2007;35:2302–2310. doi: 10.1093/nar/gkm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichman TW, Parrott AM, Fierro-Monti I, Caron DJ, Kao PN, Lee CG, Li H, Mathews MB. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J. Mol. Biol. 2003;332:85–98. doi: 10.1016/s0022-2836(03)00885-4. [DOI] [PubMed] [Google Scholar]
- 9.Xu YH, Grabowski GA. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab. 1999;68:441–454. doi: 10.1006/mgme.1999.2934. [DOI] [PubMed] [Google Scholar]
- 10.Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T, Agata Y. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuwano Y, Pullmann R, Jr, Marasa BS, Abdelmohsen K, Lee EK, Yang X, Martindale JL, Zhan M, Gorospe M. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010;38:225–238. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoque M, Shamanna RA, Guan D, Pe'ery T, Mathews MB. HIV-1 replication and latency are regulated by translational control of cyclin T1. J. Mol. Biol. 2011;410:917–932. doi: 10.1016/j.jmb.2011.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neplioueva V, Dobrikova EY, Mukherjee N, Keene JD, Gromeier M. Tissue type-specific expression of the dsRNA-binding protein 76 and genome-wide elucidation of its target mRNAs. PLoS One. 2010;5:e11710. doi: 10.1371/journal.pone.0011710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shim J, Lim H, R.Yates J, Karin M. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell. 2002;10:1331–1344. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- 16.Isken O, Grassmann CW, Sarisky RT, Kann M, Zhang S, Grosse F, Kao PN, Behrens SE. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 2003;22:5655–5665. doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isken O, Grassmann CW, Yu H, Behrens SE. Complex signals in the genomic 3' nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA. 2004;10:1637–1652. doi: 10.1261/rna.7290904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isken O, Baroth M, Grassmann CW, Weinlich S, Ostareck DH, Ostareck-Lederer A, Behrens SE. Nuclear factors are involved in hepatitis C virus RNA replication. RNA. 2007;13:1675–1692. doi: 10.1261/rna.594207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomila RC, Martin GW, Gehrke L. NF90 binds the dengue virus RNA 3' terminus and is a positive regulator of dengue virus replication. PLoS One. 2011;6:e16687. doi: 10.1371/journal.pone.0016687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parrott AM, Mathews MB. Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue-restricted distribution. Nucleic Acids Res. 2007;35:6249–6258. doi: 10.1093/nar/gkm668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3'-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J. Biol. Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 22.Zhao G, Shi L, Qiu D, Hu H, Kao PN. NF45/ILF2 tissue expression, promoter analysis, and interleukin-2 transactivating function. Exp. Cell Res. 2005;305:312–323. doi: 10.1016/j.yexcr.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 23.Duchange N, Pidoux J, Camus E, Sauvaget D. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene. 2000;261:345–353. doi: 10.1016/s0378-1119(00)00495-9. [DOI] [PubMed] [Google Scholar]
- 24.Diebold ML, Fribourg S, Koch M, Metzger T, Romier C. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J. Struct. Biol. 2011;175:178–188. doi: 10.1016/j.jsb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Kraft P, Bergamaschi A, Broennimann C, Dinapoli R, Eikenberry EF, Henrich B, Johnson I, Mozzanica A, Schleputz CM, Willmott PR, et al. Performance of single-photon-counting PILATUS detector modules. J. Synchrotron. Radiat. 2009;16:368–375. doi: 10.1107/S0909049509009911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 28.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 29.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative computing project, n. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scarlett GP, Elgar SJ, Cary PD, Noble AM, Orford RL, Kneale GG, Guille MJ. Intact RNA-binding domains are necessary for structure-specific DNA binding and transcription control by CBTF122 during Xenopus development. J. Biol. Chem. 2004;279:52447–52455. doi: 10.1074/jbc.M406107200. [DOI] [PubMed] [Google Scholar]
- 36.Liao HJ, Kobayashi R, Mathews MB. Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl Acad. Sci. USA. 1998;95:8514–8519. doi: 10.1073/pnas.95.15.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L, Sander C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 38.Yue D, Maizels N, Weiner AM. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA. 1996;2:895–908. [PMC free article] [PubMed] [Google Scholar]
- 39.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stagno J, Aphasizheva I, Bruystens J, Luecke H, Aphasizhev R. Structure of the mitochondrial editosome-like complex associated TUTase 1 reveals divergent mechanisms of UTP selection and domain organization. J. Mol. Biol. 2010;399:464–475. doi: 10.1016/j.jmb.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda K, Gupta S, Chen K, Wu C, Qin J. The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions. Mol. Cell. 2009;36:819–830. doi: 10.1016/j.molcel.2009.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeqiraj E, Filippi BM, Goldie S, Navratilova I, Boudeau J, Deak M, Alessi DR, van Aalten DM. ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 2009;7:e1000126. doi: 10.1371/journal.pbio.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 44.Worringer KA, Chu F, Panning B. The zinc finger protein Zn72D and DEAD box helicase Belle interact and control maleless mRNA and protein levels. BMC Mol. Biol. 2009;10:33. doi: 10.1186/1471-2199-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker LM, Fierro-Monti I, Reichman TW, Gunnery S, Mathews MB. Double-stranded RNA-binding proteins and the control of protein synthesis and cell growth. Cold Spring Harb. Symp. Quant. Biol. 2001;66:485–497. doi: 10.1101/sqb.2001.66.485. [DOI] [PubMed] [Google Scholar]
- 46.Guan D, Altan-Bonnet N, Parrott AM, Arrigo CJ, Li Q, Khaleduzzaman M, Li H, Lee CG, Pe'ery T, Mathews MB. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol. Cell Biol. 2008;28:4629–4641. doi: 10.1128/MCB.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.