Summary
Clathrin, a protein best known for its role in membrane trafficking, has been recognised for many years to localise to the spindle apparatus during mitosis but its function at the spindle remained unclear. Recent work has better defined the role of clathrin in the function of the mitotic spindle and proposed that it crosslinks microtubules (MTs) of the kinetochore fibres (K-fibres) in the mitotic spindle. This mitotic function is unrelated to clathrin’s role in membrane trafficking and occurs in partnership with two other spindle proteins: transforming acidic coiled-coil protein 3 (TACC3) and colonic hepatic tumour overexpressed gene (ch-TOG). This review summarises the role of clathrin in mitotic spindle organisation with an emphasis on the recent discovery of the TACC3/ch-TOG/clathrin complex.
Keywords: clathrin, TACC3, ch-TOG, mitotic spindle, microtubule
Introduction
Membrane trafficking and mitosis are two major areas of cell biological research. Investigators in the first area are interested in questions such as how do cells internalise nutrients from the extracellular medium, how are cell surface receptors downregulated and how are lysosomes formed? Researchers in the second area would like to determine how the mitotic spindle shares chromosomes equally between the two daughter cells. At first glance these two areas appear distinct and unrelated. However, if we look more closely, we find that some proteins are involved in both processes and arguably the best understood example of such a protein is clathrin (Royle, 2011; Scita and Di Fiore, 2010).
Clathrin is best known for its role in membrane trafficking but in recent years it has emerged that clathrin has another function that occurs during mitosis (Royle et al., 2005). In non-dividing cells, clathrin forms a coat around membranes that are to be moved from one part of the cell to another (Brodsky et al., 2001). During mitosis, a proportion of clathrin becomes localised to the mitotic spindle where it is proposed to crosslink microtubules (MTs) of the kinetochore fibres (K-fibres) (Booth et al., 2011; Royle et al., 2005). This function is unrelated to its role in membrane trafficking. Recently, it has become clear that clathrin carries out its mitotic function in a complex together with transforming acidic coiled-coil protein 3 (TACC3) and colonic hepatic tumour overexpressed gene (ch-TOG) (Booth et al., 2011; Fu et al., 2010; Hubner et al., 2010; Lin et al., 2010). TACC3 and ch-TOG are two major spindle proteins with important roles in mitosis (Al-Bassam and Chang, 2011; Peset and Vernos, 2008).
The aims of this Commentary are to describe our current understanding of the role of clathrin in mitotic spindle organisation; to discuss future directions and key questions for future work; and to present the work in a way that can be understood by investigators interested in membrane trafficking or mitosis but might be unfamiliar with some concepts from the other side of the fence.
A quick guide to clathrin
Clathrin is best known for its role in membrane trafficking. Clathrin-coated vesicles (CCVs) are formed at the plasma membrane and the trans-Golgi network (TGN) where they are involved in endocytosis and transport of secretory cargo, respectively (Brodsky et al., 2001). CCVs were first described in the 1960s in electron micrographs of several cellular systems; including, most famously, mosquito oocytes that uptake significant amounts of yolk protein (reviewed in Hirst and Robinson, 1998). Later, CCVs were purified from brain and their main protein constituent was identified and termed clathrin after the lattice-like (or clathrate) structures that it forms (Pearse, 1975) (Fig. 1A and B).
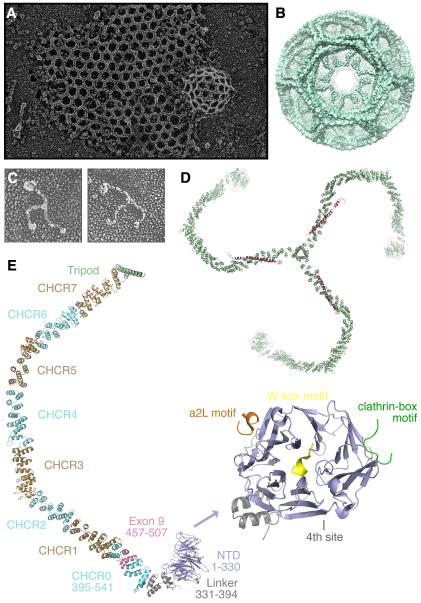
Fig. 1. The structure of clathrin.
(A) A clathrin lattice and budding vesicle at the plasma membrane as seen by deep-etch electron microscopy. © 1987 Rockefeller University Press. Originally published in J. Cell Biol. 105: 1999-2009 (Heuser et al., 1987). (B) Cryo-EM map of a clathrin-coated vesicle. Figure generated from EMD-5119 using UCSF Chimera. (C) Two examples of isolated clathrin triskelia viewed by electron microscopy. © 1988 Rockefeller University Press. Originally published in J. Cell Biol. 107: 877-86 (Heuser and Keen, 1988). (D) Molecular model of a clathrin triskelion. Clathrin heavy chains are green, the central portion of the clathrin light chain is shown in orange. Figure generated from PDB files 1XI4 and 3LVG using Pymol (Fotin et al., 2004; Wilbur et al., 2010). (E) Structural features of a single CHC. N-terminal domain (NTD 1-330, blue), linker (grey), CHCR0-7 (alternating blue and brown), hairpin (grey) and tripod helix (green). The regions implicated in mitotic function are annotated. Note the region encoded by exon 9 of CHC (residues 457-507, pink) is a subregion of the TACC3 binding region. Enlarged structure: view of the ‘foot’ region of CHC (1-363). The approximate positions of the various interacting motifs are shown. Clathrin-box motifs L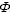 X
X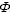 [DE] (yellow), W-box motifs PWXXW (green), arrestin-2L site [LI][LI]GXL (orange). The position of the fourth interaction site, for which the motif is unknown, is also indicated. Figure generated from PDB files 1UTC, 1C9I and 3GD using Pymol.
[DE] (yellow), W-box motifs PWXXW (green), arrestin-2L site [LI][LI]GXL (orange). The position of the fourth interaction site, for which the motif is unknown, is also indicated. Figure generated from PDB files 1UTC, 1C9I and 3GD using Pymol.
The assembly unit of the clathrin coat is a triskelion: a three-legged assembly of three clathrin heavy chains (CHCs), each with an associated light chain (Kirchhausen and Harrison, 1981; Ungewickell and Branton, 1981) (Fig. 1C and D). The three CHCs are noncovalently trimerised at their C-termini (Nathke et al., 1992) and the light chains are bound very tightly (Winkler and Stanley, 1983). In cells, triskelia are formed upon synthesis and monomers of clathrin heavy chain or light chain-free heavy chains are not readily observed (Brodsky, 1985; Hoffmann et al., 2010).
In humans there are four clathrin genes that encode two light (~25 kDa) and two heavy (~190 kDa) chains. The clathrin light chains a and b are encoded by the genes CLTA and CLTB on chromosome 9p13 and 5q35, respectively. The ubiquitously expressed clathrin heavy chain, referred to as CHC17 or CHC, is encoded by the gene CLTC, located at 17q11-qter. There is a CHC paralogue, referred to as CHC22, which is muscle-specific, that is encoded by the gene CLTCL1 at 22q11.21. However in most cells in the body, clathrin triskelia are trimers of CHC17 with a random distribution of light chain a and b. Clathrin is highly expressed in all cells with an estimated 500,000 triskelia per cell (Doxsey et al., 1987; Goud et al., 1985). Clathrin arose early in the evolution of eukaryotic life and is well conserved. For example, human CHC shows ~99% identity with CHCs from mammals and birds, ~70% homology with worms and >50% identity with CHC from yeast. In higher organisms, clathrin is an essential gene (Royle, 2006).
We have a good structural knowledge of clathrin as summarised in Fig. 1. Several domains of clathrin have been studied by X-ray crystallography and an atomic model of a complete clathrin lattice has been built using this structural information together with electron microscopy of purified CCVs (Fotin et al., 2004). The globular N-terminal domain (NTD) of CHC comprises a seven-bladed beta propeller (ter Haar et al., 1998). Four interaction sites on the NTD that are important for endocytic function have been described (Kang et al., 2009; Miele et al., 2004; ter Haar et al., 2000; Willox and Royle, 2011) and are important for interactions with adaptor proteins. Next to the NTD, moving towards the C-terminus there is a short linker followed by eight CHC repeats (CHCRs) numbered CHCR0-7. Finally, there is a short linker followed by the trimerisation domain, whereas final C-terminal residues are unresolved (Fotin et al., 2004). The triskelion is often described as a leg, with the NTD being the foot. Interactions between CHCRs on adjacent clathrin triskelia regulate lattice formation. The light chain binds to the heavy chain at the region most proximal to the trimerisation domain (Fotin et al., 2004) and recent evidence indicates that the light chains could influence triskelion structure (Wilbur et al., 2010).
A key concept in the function of clathrin in membrane trafficking is its inability to bind membranes or cargo directly (Unanue et al., 1981). Instead, clathrin binds to adaptor proteins, which in turn can bind to membranes or to proteins destined for trafficking (Reider and Wendland, 2011). In interphase cells, assembled clathrin can be visualised in coated pits and vesicles at the plasma membrane, the TGN and free in the cytoplasm (Fig. 2 A). Some of these clathrin spots co-localise with adaptors such as AP-2 that are involved in endocytosis at the plasma membrane, whereas another subset of clathrin co-localises with AP-1 at the TGN (Robinson, 2004). In addition, there are many other adaptors and accessory proteins involved in clathrin-mediated membrane traffic that form an extensive network of interactions, with clathrin as a hub, interacting with many of these proteins (Traub, 2011). It is worth noting that accessory proteins for membrane trafficking are sometimes referred to as CLASPs (clathrin associated sorting proteins) (Reider and Wendland, 2011), whereas in cell division research, CLASP refers to a family of microtubule-binding proteins (cytoplasmic linker protein (CLIP)-associated proteins) (Al-Bassam and Chang, 2011).
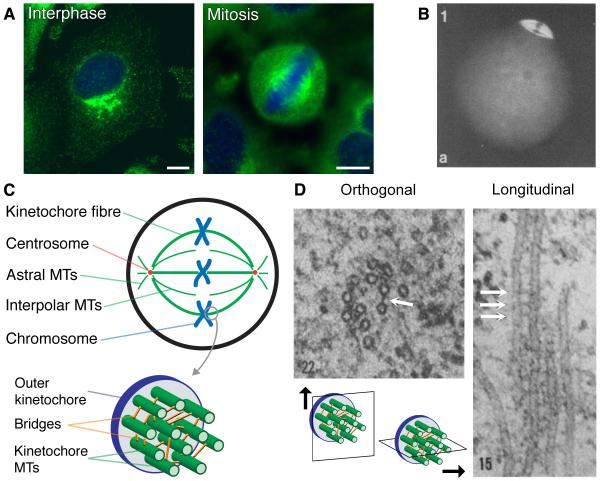
Figure 2. Clathrin localisation during mitosis.
(A) The subcellular distribution of clathrin in HeLa cells in interphase and mitosis. Clathrin was detected using monoclonal antibody X22 (green), DNA was stained with DAPI (blue). Scale bar, 10 μm. (B) Micrograph of clathrin as detected by indirect immunoflourescence on the second metaphase spindle in an unfertilised mouse egg. Reproduced with permission © 1985 Company of Biologists. Originally published in (Maro et al., 1985). (C) Schematic diagram of the microtubule (MT) organisation of the mitotic spindle at metaphase. The kinetochore (K)-fibre is a bundle of parallel MTs that extend from the spindle pole and terminate with their plus end at the kinetochore. Only 11 MTs are shown for clarity, bridges are represented as orange sticks. (D) Electron micrographs of inter-microtubule bridges in the mitotic spindle. Two views are seen: on the left, a fibre is sectioned orthogonally so that microtubules appear as circles; and on the right, a fibre is sectioned in parallel with the spindle axis so that microtubules are long stripes. Bridges are the electron-dense connections between microtubules marked by white arrows. A schematic diagram (inset) shows the sectioning orientation. © 1970 Rockefeller University Press. Originally published in J. Cell Biol. 54: 438-44 (Hepler et al., 1970).
The mitotic function of clathrin: background
The first clues to a mitotic function for clathrin can be traced back to studies more than 30 years ago that examined the subcellular distribution of clathrin using immunofluorescence. Maro et al. found that clathrin is located on the second metaphase spindle in unfertilised mouse eggs (Maro et al., 1985) (Fig. 2B). Much later, the subcellular localisation of clathrin during mitosis was studied using several different antibodies and various cell lines (Okamoto et al., 2000). In addition, clathrin was identified as a spindle protein by two untargeted methods. First, a gene trap intended to find nuclear proteins revealed clathrin to be localised to spindles (Sutherland et al., 2001). Second, clathrin was identified by mass spectrometry of purified mitotic spindles (Mack and Compton, 2001). For technical reasons, this study only found a subset of the total spindle proteome (Sauer et al., 2005), but, interestingly, clathrin and the factors that recruit it to the spindle were present in this subset (see below). With hindsight, we can see clearly that clathrin is a bona fide spindle protein; however, at the time there were several reasons why these results were not pursued further. Uncertainty over antibody specificity was a worry with the original study (Maro et al., 1985). Also, the presence of clathrin at the spindle could have been a contaminant – especially as it is a large protein that frequently appears erroneously in mass spectrometry studies (Trinkle-Mulcahy et al., 2008).
In addition, with no known role at the mitotic spindle, the simplest conclusion was that clathrin’s presence at the spindle reflected a large store of coated vesicles, a pervasive idea in the field. Seminal studies of mitotic cell morphology did report coated vesicles amongst microtubules of the spindle, but did not quantify whether they were enriched here (Robbins and Jentzsch, 1969). Attempts to label membranes in the spindle generally show that, while membranes might be present, they are not enriched at the spindle (Waterman-Storer et al., 1993), although this is somewhat dependent on the cell line. EM studies have confirmed a relative lack of endoplasmic reticulum/nuclear envelope (Puhka et al., 2007), Golgi-derived membranes (Lucocq et al., 1989), and coated vesicles (Tooze and Hollinshead, 1992) within the spindle itself. The spindle apparatus is surrounded by membranes and it has been proposed that these membranes are important for spindle function (Zheng, 2010). If a large store of CCVs was present, adaptors should be found together with clathrin, but they are not (Royle et al., 2005). Finally, immunogold labelling of clathrin shows that the label is not associated with membranes, but with MTs (Booth et al., 2011; Royle et al., 2005). It is therefore unlikely that the clathrin that is located at the spindle is associated with membranes.
The subcellular localisation of clathrin in mitosis that was first observed with antibodies was revisited recently using GFP-tagged clathrins in parallel with antibody staining (Royle et al., 2005). This study confirmed that clathrin is colocalised with MTs of the spindle apparatus early in mitosis. It is localised to the kinetochore fibres of the spindle and there is no obvious localisation to astral, interpolar or midzone MTs as the cell goes through mitosis. The association with MTs becomes less obvious in telophase and cytokinesis (Royle et al., 2005). Evidence for a functional role of clathrin at the spindle came from RNA interference (RNAi) of CHC, which caused a delay in mitosis as a result of defects in chromosome congression at the metaphase plate (Royle et al., 2005). The congression defects stem from a destabilisation of the kinetochore fibres of the mitotic spindle (Royle et al., 2005). In the next section, the mechanism by which clathrin stabilises kinetochore fibres will be considered.
The mitotic function of clathrin is apparently distinct from its role in membrane trafficking. This may seem obvious given that the clathrin at the spindle is not associated with membranes and that the mitotic function of clathrin occurs when clathrin-mediated endocytosis is inhibited (Warren, 1993). However, the function of clathrin in membrane trafficking is so well established that this point needed to be thoroughly investigated. The best evidence for the distinction between membrane trafficking and mitotic function of clathrin came from experiments that separated the two clathrin functions using CHC mutants that are capable of fulfilling only one function but not the other (Blixt and Royle, 2011; Hood and Royle, 2009; Royle and Lagnado, 2006).
Mitotic spindle structure: role for non-motor proteins in fibre stability
In mammalian cells, the spindle apparatus is composed of three classes of MT: astral, interpolar and kinetochore (Fig. 2C). Astral MTs radiate from the spindle pole to the cell cortex, whereas interpolar MTs run along most of the way from the spindle pole to the opposing pole (Mastronarde et al., 1993; McDonald et al., 1992). Kinetochore MTs connect the spindle pole with the kinetochore, and are responsible for chromosome movement (McDonald et al., 1992; Rieder, 2005). Numerous kinetochore MTs are bundled together to form a K-fibre. The number of MTs in such a fibre depends on the size of the kinetochore and not necessarily on the size of the chromosome it has to move (Rieder, 1982), and range from 20-40 MTs in humans to only a few in yeast (Ding et al., 1993).
In many cellular systems, bundles of MTs are cross-linked by electron-dense inter-MT bridges (Stephens and Edds, 1976) that are composed of motor proteins or non-motor microtubule associated proteins (MAPs). A good example is in dendrites, in which the parallel MT bundles are cross-linked by single MAP2 molecules conferring a spacing of ~62 nm and those in the axon by tau resulting in a shorter inter-MT distance of ~20 nm (Chen et al., 1992; Kim et al., 1979). In the mitotic spindle, MTs of K-fibres are similarly cross-linked by bridges (Hepler et al., 1970; Mastronarde et al., 1993; McDonald et al., 1992; Witt et al., 1981), which likely are also formed by non-motor structural proteins (Manning and Compton, 2008; Peterman and Scholey, 2009) (see Fig. 2D for examples). However, unlike the inter-MT bridges in neuronal cells, which are formed by single MAPs, the bridges in K-fibres appear to be of varying lengths suggesting that a number of different proteins mediate the cross-linking (Booth et al., 2011).
The “bridge hypothesis” for clathrin function
As described above, depletion of clathrin, a protein found on K-fibres, results in their destabilisation (Royle et al., 2005). So what is the mechanism by which clathrin stabilises K-fibres? There are two regions of clathrin that are important for its localisation at the spindle. The first is in the NTD (Royle et al., 2005; Royle and Lagnado, 2006) and the second is a stretch of 50 amino acids in the CHCR0 or ‘ankle’ region (Hood and Royle, 2009) (Fig. 1E). As these regions, which are closely located on one CHC, are spaced far away from each other in the clathrin triskelion, it was proposed that clathrin could stabilise kinetochore fibres by physically cross-bracing adjacent MTs (Royle et al., 2005). This idea, the ‘bridge hypothesis’, was supported by experiments with CHC mutants in the context of cells depleted of endogenous CHC (Royle and Lagnado, 2006) as CHC mutants that are unable to either trimerise or to bind to the spindle (i.e. unable to act as a bridge) do not rescue the mitotic defects caused by CHC RNAi. Moreover, trimeric or dimeric CHCs that contain the spindle interaction sites on the NTD and the ankle but lack most of the leg region were capable of functional rescue (Blixt and Royle, 2011; Royle and Lagnado, 2006). While these experiments provide support for the bridge hypothesis, they are not a direct test.
Clearly, if clathrin acts as an inter-MT bridge in K-fibres, upon its depletion, the bridges should disappear. Using EM, it was found that ~40% of inter-MT bridges are missing from clathrin-depleted K-fibres compared to a control RNAi condition (Booth et al., 2011). Importantly, it was found that inter-MT bridges in K-fibres exist in several populations of different lengths. The ~40% decrease in bridges in clathrin-depleted K-fibres could be almost entirely explained by loss of the shortest inter-MT bridges (Fig. 2E). A second test of whether clathrin is indeed an inter-MT bridge is their labelling with anti-clathrin antibodies. Booth et al. observed anti-clathrin immuno-gold labelling of inter-MT bridges which could be clearly distinguished from tubulin labelling (Booth et al., 2011) providing strong evidence that clathrin indeed acts as an inter-MT bridge.
The simplest model for the function of inter-MT bridges, such as clathrin, is that they stabilise K-fibres by physically cross-bracing them (Rieder, 1982). However, clathrin-depleted K-fibres also exhibit MT loss and as a result the fibres are thinner and there is more space between MTs (Booth et al., 2011). Therefore, destabilisation of K-fibres could be the result of the loss of bridges and/or of MTs. It was suggested that bridge loss precedes MT loss and that bridges may protect from MT catastrophe (Booth et al., 2011). Further work is needed to determine the relative contribution of physical cross-linking and prevention of MT catastrophe to K-fibre stabilisation.
Clathrin is different from the other non-motor MAPs that have been shown to cross-link MTs. Besides clathrin’s trimeric structure rather than the bipolar MAP structure, most MAPs, such as protein regulator of cytokinesis 1 (PRC1) are able to bind to MTs directly (Peterman and Scholey, 2009). Clathrin has no known microtubule-binding domains and it does not bind to MTs that have been assembled in vitro unless other mitotic proteins are added (Booth et al., 2011), suggesting that other factor(s) are required to recruit clathrin to the mitotic spindle. This is a familiar concept of clathrin function. As mentioned above, during interphase, clathrin requires adaptor proteins to mediate membrane or cargo interactions for membrane trafficking and it appears that during mitosis, clathrin also needs adaptor protein(s) to be able to bind to kinetochore MTs.
Interaction of clathrin with TACC3 and ch-TOG
The question of which protein(s) clathrin associates with at the spindle was answered independently by four groups (Booth et al., 2011; Fu et al., 2010; Hubner et al., 2010; Lin et al., 2010). Using a variety of approaches and systems, these groups found that clathrin is in a complex with TACC3 and ch-TOG at the mitotic spindle. These two proteins are well-characterised spindle components with established roles in MT growth and stabilisation (Barr and Gergely, 2007). Schematic diagrams of TACC3 and ch-TOG are shown in Fig. 3A, and molecular details of their homologues presented in Table 1. Previously, ch-TOG has been described to be important for MT outgrowth from the centrosome and TACC proteins were thought to be important for loading ch-TOG onto spindle MTs (Barr and Gergely, 2008; Gergely et al., 2003; Kinoshita et al., 2005; Lee et al., 2001). Below, I will discuss that although there is consensus regarding the composition of the TACC3/ch-TOG/clathrin complex and the importance of TACC3 phosphorylation by the kinase Aurora-A in regulation of the complex, there is some disagreement over the mechanism by which the complex binds to MTs and the role phosphorylation may have in this event (Fu et al., 2011; Hood and Royle, 2011).
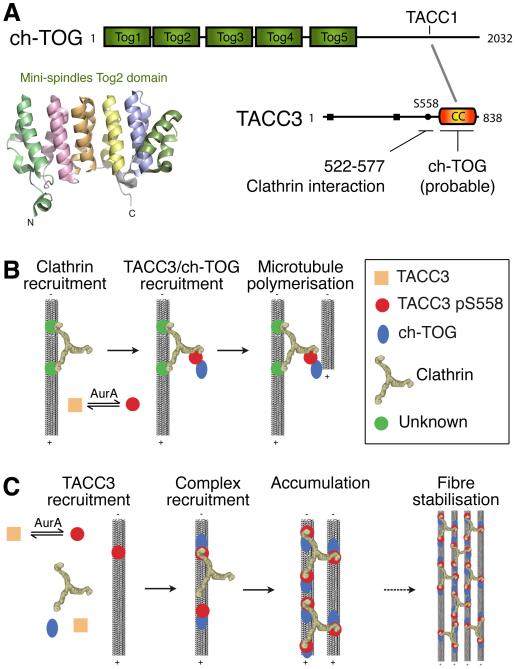
Figure 3. Models for the interaction of clathrin with TACC3 and ch-TOG at the spindle.
(A) Schematic diagrams of ch-TOG and TACC3. Human ch-TOG has five TOG domains (green) that bind soluble tubulin dimers. The structure of the second TOG domain from mini-spindles (msps), the Drosophila ch-TOG homologue, is shown (PDB code 2QK2). TACC3 has several Aurora-A phosphorylation sites (shown in black) and a C-terminal coiled-coil domain (CC, orange). The interaction between TACC3 and ch-TOG probably occurs between the CC domain of TACC3 and ch-TOG C-terminal domain, the site of TACC1 interaction (Peset and Vernos, 2008). (B) The ‘clathrin recruits TACC3’ model. In this model, clathrin is initially recruited to microtubules by an unknown protein. Phosphorylated TACC3 binds clathrin and is therefore recruited to the spindle. TACC3 also binds ch-TOG and the microtubule polymerisation activity of ch-TOG enhances spindle stability (Fu et al., 2010; Hubner et al., 2010; Lin et al., 2010). (C) The inter-microtubule bridge model. In this model, Aurora A-phosphorylated TACC3 is the initial recruitment factor. Clathrin can bind to TACC3/ch-TOG complexes, which might be located on adjacent microtubules allowing clathrin to cross-link microtubules. The resulting TACC3/ch-TOG/clathrin complex is more stable owing to multiple interactions and therefore accumulates on the spindle (Booth et al., 2011). Microtubules, triskelia and the distances between them are shown to scale based on electron microscopy.
Table 1. Details of TACC3/ch-TOG/clathrin complex members.
| Membe r |
Compositio n |
Humans | Homologues | Notes | Referenc e |
|---|---|---|---|---|---|
| TACC3 | 2 × 90 kDa (monomer runs at 150 kDa by SDS PAGE). |
TACC3/ERIC 1 (838) |
Tacc3/AINT (mm, 630), TACC3/Maskin (xl, 931), dTACC (dm, 1226), C7G031 (dd, 616), TAC-1/2P40 (ce, 260), ALP7/Mia1 (sp, 474). |
TACC1 and TACC2 expressed in mammals. No clear homologue in plants or S. cerevisiae. Best sc candidates are SLK19 (sc, 821) with weak homology or Spc72 (sc, 622) due to interaction with STU2. TACC domain is best conserved and N- terminal region is variable. |
(Still et al., 2004) |
| Ch- TOG |
1 × 225 kDa | CKAP5/ch- TOG (2032) |
Ckap5 (mm, 2032), XMAP215 (xl, 2065), Msps (dm, 2050), MOR1 (at, 1978), CP224 (dd, 2013), Zyg-9 (ce, 1415), Alp14/Dis1 (sp, 809), STU2 (sc, 888). |
Higher eukaryotes have 5 TOG domains. Zyg-9 has 3 TOG domains and yeast homologues have 2 TOG domains. |
(Al-Bassam and Chang, 2011) |
| Clathrin | 3 × (190 + 27 kDa) |
CLTC/CHC (1675) CLTCL1/CHC 22 (1640) |
Cltc(mm, 1675), cltc (xl, 1675), Chc (dm, 1678), AT3G11130 (at, 1705), chcA (dd, 1694), chc-1 (ce, 1681), chc1 (sp, 1666), CHC1 (sc, 1653). |
Domain organisation of CHC is completely conserved down to yeast. Homologues of CHC22 are present in mammals (pseudogene in mice), birds, amphibians and fish. |
(Wakeha m et al., 2005) |
| CLTA/LCa (248) CLTB/LCb (229) |
Clta (mm, 216), Clta (xl, 203), clc(dd, 194), F4J5M9 (at, 258), Clc (dm, 219), clic-1 (ce, 226), clc1 (sp, 229), CLC1 (sc, 233) |
Homologues of both LCa and LCb in mammals, birds, amphibians and fish. Only LCa homologues are shown here. Single light chains in flies, worms, slime mold and fungi. Three light chain genes in Arabidopsis. |
(Wakeha m et al., 2005) |
Numbers in parentheses refer to the number of amino acids of the longest isoform. Abbreviations: mm, mouse; xl, frog; dm, fruitfly; at, plant; dd, slime mould; ce, nematode; sp, fission yeast; sc, budding yeast.
The TACC3/ch-TOG/clathrin complex
Using quantitative proteomics, clathrin was identified as a binding protein for TACC3 (Hubner et al., 2010) and ch-TOG was found to be associated with clathrin and TACC3 (Hubner et al., 2010). Lin et al. showed that clathrin can interact with a minimal 50-residue fragment of TACC3 (Lin et al., 2010). The corresponding region on clathrin for the interaction with TACC3 in vitro was found to be residues 331-542 (Fig. 1E), a region that contains the previously identified site required for recruitment of clathrin to the spindle (Hood and Royle, 2009). The interaction between clathrin and the Xenopus TACC3 homologue, maskin, demonstrates that this functional complex is conserved in frogs (Fu et al., 2010). This study also showed that the complex is under the regulation of Importin β and RanGTP. Booth et al. isolated the native clathrin complex from purified mitotic spindles from human cells and found it contains a complex of TACC3, ch-TOG and clathrin (Booth et al., 2011).
Depletion of TACC3 results in strikingly similar mitotic defects (Gergely et al., 2003) to those observed for clathrin depletion (Royle et al., 2005). Lin et al. nicely demonstrated that the mitotic phenotypes observed with either clathrin or TACC3 RNAi are equivalent to the depletion of both proteins (Lin et al., 2010). Using Xenopus egg extracts, Fu et al. showed that depletion of either TACC3 or clathrin results in defective spindle assembly (Fu et al., 2010). These results suggest that both proteins work together in the same pathway and that independent mitotic functions for TACC3 and clathrin are unlikely (Hood and Royle, 2011). This is underscored by the observation that TACC3-depleted K-fibres have fewer inter-MT bridges and that the extent of bridge loss is very similar to that seen in CHC-depleted K-fibres (Booth et al., 2011). Depletion of ch-TOG leads to mitotic defects that are much more severe compared to the effects of TACC3 or clathrin RNAi (Booth et al., 2011; Gergely et al., 2003), suggesting that, although ch-TOG is part of the TACC3/ch-TOG/clathrin complex in spindles, it has additional independent functions.
Regulation by Aurora-A kinase
TACC3 is a well-characterised substrate for Aurora-A kinase (Barr and Gergely, 2007). Several serine and threonine residues outside of its coiled-coil region are phosphorylated during mitosis, but of these, only S25, T30, S402 and S558 are sensitive to inhibition of Aurora-A kinase (Kettenbach et al., 2011). The phosphorylation of TACC3 on residue S558 by Aurora-A in particular is thought to be critical for spindle localisation (Booth et al., 2011; Kinoshita et al., 2005; LeRoy et al., 2007; Lin et al., 2010). Mutation of S558 to alanine results in a TACC3 variant that is unable to localise properly to spindles. Similarly, treatment of cells with the selective Aurora-A kinase inhibitors MLN8054 or MLN8237 effectively removes TACC3 from spindles (Booth et al., 2011; Cheeseman et al., 2011; Hubner et al., 2010; LeRoy et al., 2007).
Several lines of evidence indicate that the phosphorylation of TACC3, probably at S558, is required for the binding of clathrin by TACC3. Firstly, no interaction with clathrin was found with a TACC3 mutant with alanines replacing S558 and two other serine residues (Hubner et al., 2010). Secondly, phosphorylation of the 50-residue TACC3 fragment containing S558 by Aurora-A is necessary to be able to detect an interaction with clathrin (Lin et al., 2010). Finally, the interaction between clathrin and maskin is enhanced by Aurora-A kinase activity with the most N-terminal Aurora-A phosphorylation site not required for the interaction (Fu et al., 2010).
Two models for binding of TACC3/ch-TOG/clathrin to MTs
Depletion of clathrin by RNAi reduces the amount of TACC3 on the spindle (Booth et al., 2011; Fu et al., 2010; Hubner et al., 2010; Lin et al., 2010), and this observation together with the phosphorylation data suggests a model, in which clathrin recruits TACC3 to the mitotic spindle (Fig. 3B). Here, TACC3 phosphorylation is needed for TACC3 to be able to bind to clathrin. Spindle stability is then achieved by TACC3 recruiting ch-TOG to promote MT assembly and therefore stability.
The ‘clathrin recruits TACC3’ model explained some of the results, but not all. Firstly, the majority (>80%) of clathrin is not at the spindle, whereas TACC3 is localised almost exclusively at the spindle. Thus, as a targeting protein, clathrin would not be very efficient and in fact, on the basis of localisation, it would make more sense if TACC3 recruited clathrin to the spindle. Secondly, trimerisation-deficient CHCs cannot function in mitosis (Royle and Lagnado, 2006), but if clathrin simply recruited TACC3 to achieve spindle stability, we would not expect any mitotic defects with these mutants. It is also difficult to reconcile this model with the finding that TACC3/ch-TOG/clathrin complexes form inter-MT bridges. Thirdly, as described above, clathrin cannot bind MTs, but TACC3 (or maskin) and ch-TOG are able to do so (Charrasse et al., 1998; O’Brien et al., 2005; Peset et al., 2005). Therefore the question arises why these proteins would require clathrin for their recruitment to MTs, and if this was the case, which protein is responsible for recruiting clathrin to the spindle?
The native clathrin complex found on spindles only contains TACC3, ch-TOG and clathrin (Booth et al., 2011). Therefore, unless an unknown protein dissociated from the complex during the purification, the factor recruiting the complex to MTs is one of these three. Although depletion of clathrin reduces the amount of TACC3 and ch-TOG at the spindle (Booth et al., 2011; Lin et al., 2010), depletion of TACC3 (and to a lesser extent ch-TOG) also decreases the recruitment of the other two complex members (Booth et al., 2011). If the converse experiment is performed, i.e. TACC3 is over-expressed in mitotic cells, increased amounts of ch-TOG and clathrin are recruited to the spindle (Booth et al., 2011; Fu et al., 2010). However, over-expression of ch-TOG or clathrin has no effect on the distribution of the other complex members, and, furthermore, over-expression of clathrin quickly saturates its binding sites on the spindle (Booth et al., 2011). Thus, TACC3 appears to be the limiting factor in the recruitment of the complex to MTs, suggesting that TACC3 recruits clathrin to the spindle, and not the other way around.
However, as depletion of any member of the TACC3/ch-TOG/clathrin complex affects the localisation of the other two, a simple ‘single-step’ serial recruitment model, whereby either clathrin recruits TACC3 or TACC3 recruits clathrin (Fu et al., 2011) is not the full story. Booth et al. therefore proposed a two-step model, in which the complex is recruited to MTs, where, after initial recruitment, further complexes accumulate (Fig. 3C). Such an accumulation is made possible by the triskelion structure of clathrin, which allows it to bind multiple TACC3 and ch-TOG molecules. If subcomplexes of TACC3 and ch-TOG are bound by different MTs, clathrin will be able to bridge between those. This model consolidates the known structures of the proteins involved, the ultrastructural data showing TACC3/ch-TOG/clathrin complexes as inter-MT bridges and the observations from RNAi and overexpression studies. The model is not without its problems however, and it requires further refinement.
It is proposed that phosphorylation of TACC3 by Aurora-A kinase is required for TACC3 to bind to MTs prior to the recruitment of ch-TOG and clathrin (Fig. 3C). There are several observations that suggest this is indeed the case. Firstly, inhibition of Aurora-A kinase activity using Alisertib (MLN8237) after the spindle has assembled results in loss of TACC3 and clathrin from spindles with similar kinetics (Booth et al., 2011), and also initiates a loss of inter-MT bridges from K-fibres, which causes their subsequent destabilisation (Cheeseman et al., 2011). Secondly, non-phosphorylatable TACC3 mutants do not localise to spindles and, in these cells, clathrin is also not found on the spindle (Booth et al., 2011). An alternative view is that phosphorylation of TACC3 is only required for binding to clathrin and not for interaction with MTs. For example, TACC3 phosphorylated at S558 was originally thought to be restricted to the centrosome (Kinoshita et al., 2005). However, recent work shows that the phosphorylated form of TACC3 is found along the length of the spindle MTs and that the original antibody erroneously detected an unrelated protein on centrosomes (Lin et al., 2010). It was also reported that chronic inhibition of Aurora-A kinase (16 hrs, MLN8054) displaces TACC3 but not clathrin from the spindle (Hubner et al., 2010). However, the normal spindle morphology and intact clathrin-TACC3 binding under these conditions point to an incomplete inhibition of the kinase. There are some additional concerns about the phospho-dependent recruitment of TACC3 to MTs that remain valid. Firstly, there is evidence that maskin can bind to MTs independently of its phosphorylation (Fu et al., 2010; Kinoshita et al., 2005). Secondly, the interaction between clathrin and TACC3 requires, or at least is enhanced by, phosphorylation of TACC3 at S558 (Fu et al., 2010; Hubner et al., 2010; Lin et al., 2010). S558 cannot simultaneously act as both a MT binding site and a clathrin binding site. One possible solution to this apparent paradox is that the interaction between phosphorylated S558 of TACC3 and clathrin creates the MT binding site.
Despite the many pieces of evidence described here, more work is needed to determine the precise conformation of the TACC3/ch-TOG/clathrin complex on MTs. The binding sites that govern the interactions between these proteins are currently poorly defined and many questions remain. For example, does the NTD of CHC bind to TACC3 or ch-TOG? Further insight into TACC3/ch-TOG/clathrin function will come from detailing the interactions between complex members at high resolution. Furthermore, understanding the role of phosphorylation of TACC3 at S558 in the interactions with complex members and MTs is crucial to refining the current models.
Other proteins involved in mitotic function of clathrin
Clathrin-interacting proteins
Prior to the identification of the TACC3/ch-TOG/clathrin complex, two other proteins had been implicated in the role of clathrin in mitosis. Firstly, the transcription factor B-myb was found to be in a complex with clathrin and filamin in mitotic cells and depletion of B-myb resulted in a loss of clathrin at the spindle (Yamauchi et al., 2008). However, neither B-myb nor filamin are enriched on the mitotic spindle, so they are not involved directly in recruiting clathrin to the spindle. Interestingly, B-Myb is important for the transcription of a number of cell cycle-regulated genes including Aurora-A kinase (Knight et al., 2009) and it is possible that the observed effect on clathrin was an indirect result of the reduced expression of one of these target genes. The second protein implicated in clathrin function is cyclin G-associated kinase (GAK), a protein involved in uncoating CCVs (Greener et al., 2000). Depletion of GAK results in an accumulation of cells at prometaphase indicating a problem in spindle function (Shimizu et al., 2009; Tanenbaum et al., 2010). GAK RNAi also reduces the amount of clathrin on the mitotic spindle. Again, like B-myb, GAK is not enriched at the spindle and therefore may only have an indirect effect on clathrin localisation. For example, a reduction in the amount of free clathrin that is able to bind the spindle may be the result of the inhibition of vesicle uncoating in GAK-depleted cells (Hirst et al., 2008; Tanenbaum et al., 2010). The mitotic defects in GAK-depleted cells are more severe than those observed in cells depleted of clathrin or TACC3, which suggests that GAK has a mitotic function that is independent of clathrin.
Finally, the quantitative proteomics study that identified the TACC3-clathrin interaction also reported that G2 and S phase-expressed protein 1 (GTSE1) strongly interacts with TACC3 and clathrin (Hubner et al., 2010). GTSE1 is a cell cycle-regulated protein that is strongly phosphorylated during mitosis. It localises to the mitotic spindle, which suggests a role in spindle function. Curiously, the interaction profile of GTSE1 was reported to be identical to that of clathrin (Hubner et al., 2010), including binding of coated vesicle components. However, it needs to be clarified whether or not GTSE1 is truly a co-factor of clathrin. Although it is unclear whether any other proteins are directly involved in the function of the clathrin/TACC3/ch-TOG complex, it is apparent that this complex is not exclusively responsible for all inter-MT bridges in K-fibres and other factors might help to stabilise K-fibres.
Inter-MT bridges in K-fibres
As mentioned above, there are four apparent populations of inter-MT bridges in K-fibres with lengths that range from 15 to 53 nm with the shortest bridge type corresponding to the TACC3/ch-TOG/clathrin complex, but the identity of the remaining bridges is unclear (Booth et al., 2011). Several proteins have been proposed to act as MT crosslinkers in spindles (Manning and Compton, 2008; Peterman and Scholey, 2009), but only a few of these are found on K-fibres. Good candidates for the longer inter-MT bridges are HSET (also known as KIFC1) and hepatoma upregulated protein (HURP). HSET is a kinesin-related protein that preferentially localises between parallel MTs in K-fibres (Mountain et al., 1999) and overexpression of HSET causes bundling of spindle MTs (Cai et al., 2009). HURP is a MT-associated protein that localises to the mitotic spindle in a gradient with the highest concentration nearest to the chromosomes. Depletion of HURP causes problems in K-fibre attachment (Wong and Fang, 2006). Ultrastructural studies will be important to confirm whether or not these proteins are indeed involved in inter-MT bridges.
For many years, research into spindle function has focused on microtubule motors and only recently non-motor proteins have begun to receive attention (Manning and Compton, 2008). As an example, a recent study into the biophysics of the spindle highlighted the importance of proteins that cross-link spindle MTs (Shimamoto et al., 2011). Identifying and characterising these proteins is definitely an area that should be prioritised. Once we have a good understanding of the proteins that can cross-link parallel MTs in K-fibres, it will be interesting to determine how these bridge complexes differ from those formed by proteins known to cross-link other MT arrays, e.g. MAP2, nuclear mitotic apparatus protein 1 (NuMA) and PRC1. Another outstanding question is how K-fibre bridge complexes select parallel rather than anti-parallel MTs. Analysis of MT dynamics in the mitotic spindle shows that the MTs in K-fibres are turned over more slowly (McIntosh et al., 2002). This is likely to be due to the stabilisation conferred by attachment to the kinetochore and also by cross-linking MTs along their length. It will be important to understand the contribution of inter-MT bridges to K-fibre stabilisation and to determine whether all inter-MT bridge complexes stabilise MTs in similar ways.
Perspectives
Over the last couple of years our understanding of the role of clathrin in mitosis has accelerated considerably. We have progressed from the curious observation of clathrin on the spindle, through validation of clathrin as a genuine spindle protein with a role in K-fibre stabilisation, to recent work showing that clathrin works in concert with two core spindle proteins. Despite this progress, many questions remain and some of these are discussed below.
How conserved is the function of clathrin in mitosis? Equivalent observations to those in human and rat cells (Royle et al., 2005) have been made in mouse (Han et al., 2010; Yamauchi et al., 2008), pig (Holzenspies et al., 2010), frog (Fu et al., 2010) and plants (Tahara et al., 2007). Removal of clathrin is lethal in chicken DT40 cells, but an apoptosis-resistant strain could be isolated (Wettey et al., 2002), which however only exhibits mild mitotic defects after removal of CHC (Borlido et al., 2008). It remains to be seen whether the function of clathrin in mitosis is not conserved in birds or if these results were specific for the strain used in the study. Similarly, it is unclear whether or not the mitotic function of clathrin is conserved in lower organisms. As discussed above, the conservation of the clathrin protein is excellent in eukaryotes, but TACC3 is less well conserved with no clear homologue in S. cerevisiae (Table 1). S. pombe has the TACC homologue Alp7 (also known as Mia1p) that works together with the ch-TOG homologue Alp14 in spindle assembly (Sato and Toda, 2007). Interestingly, S. cerevisiae have a single MT acting as a ‘K-fibre’, obviously not requiring inter-MT bridges; whereas S. pombe has four MTs that have inter-MT bridges (Ding et al., 1993). Alp7 has been shown to be able to crosslink MTs in S. pombe pointing to a minimalistic version of inter-MT bridges (Thadani et al., 2009).
A related question is why clathrin needs three legs to fulfil its bridging function in mitosis as bipolar MT cross-linkers are probably the most logical configuration (Peterman and Scholey, 2009). A trimeric cross-linker would allow for bridging between three MTs simultaneously and, in theory, would result in less parallel movement during the cross-linking of two MTs compared to a bipolar linker. However, the answer could also be that it may not be optimal to have a tripolar MT cross-linker, but that clathrin acquired this function later in evolution. The fact that clathrin is a triskelion is owing to its role in membrane trafficking, a process that arose earlier in evolution than either open mitosis or K-fibres that contain more than one MT and thus require cross-linking. As the resolution of clathrin inter-MT bridges is lagging behind that of proteins such as PRC1 that cross-link anti-parallel MTs (Subramanian et al., 2010), a full answer to this question will therefore require analysis of the fine structure of these bridges.
Finally, it is also uncertain whether clathrin is multimerised into a lattice at the spindle. The shape of electron-dense inter-MT bridges observed in single EM sections suggests that clathrin is present as individual triskelia. However, CHC mutants that are unlikely to be able to form lattices are still able to localise and function at the spindle, arguing that lattices are not necessary for mitotic function (Blixt and Royle, 2011; Royle and Lagnado, 2006). To answer this question will also require a more detailed visualisation of the clathrin inter-MT bridges.
We are just beginning to understand at the molecular level how K-fibres are stabilised by inter-MT bridges. The fibres must be strong enough to perform their function but not overstabilised to the point that MT dynamics are affected. The expression of many non-motor spindle proteins, such as TACC3 and ch-TOG, is altered in many cancers (Manning and Compton, 2008). Determining how K-fibre stabilisation is optimised may therefore be important for uncovering new targets for anti-cancer therapeutics in the future.
Acknowledgements
I would like to thank Ian Prior and members of my lab for useful discussion. In particular, Fiona Hood who helped with Fig. 3 and also commented on the review. I am very grateful to the authors and publishers for allowing their work to be reproduced in this review. As always, not all important work could be discussed for space reasons. The work in my lab on clathrin’s mitotic function is supported by a Career Establishment Award from Cancer Research UK (C25425/A8722).
References
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–14. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J Cell Sci. 2007;120:2987–96. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- Barr AR, Gergely F. MCAK-independent functions of ch-Tog/XMAP215 in microtubule plus-end dynamics. Mol Cell Biol. 2008;28:7199–211. doi: 10.1128/MCB.01040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt MKE, Royle SJ. Clathrin heavy chain gene fusions expressed in human cancers: analysis of cellular functions. Traffic. 2011;12:754–61. doi: 10.1111/j.1600-0854.2011.01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DG, Hood FE, Prior IA, Royle SJ. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 2011;30:906–19. doi: 10.1038/emboj.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlido J, Veltri G, Jackson AP, Mills IG. Clathrin is spindle-associated but not essential for mitosis. PLoS ONE. 2008;3:e3115. doi: 10.1371/journal.pone.0003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM. Clathrin structure characterized with monoclonal antibodies. II. Identification of in vivo forms of clathrin. J Cell Biol. 1985;101:2055–62. doi: 10.1083/jcb.101.6.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–68. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Cai S, Weaver LN, Ems-McClung SC, Walczak CE. Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol Biol Cell. 2009;20:1348–59. doi: 10.1091/mbc.E08-09-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S, Schroeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, Larroque C. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J Cell Sci. 1998;111(Pt 10):1371–83. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- Cheeseman LP, Booth DG, Hood FE, Prior IA, Royle SJ. Aurora A kinase activity is required for localization of TACC3/ch-TOG/clathrin inter-microtubule bridges. Commun Integr Biol. 2011;4:409–412. doi: 10.4161/cib.4.4.15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kanai Y, Cowan NJ, Hirokawa N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature. 1992;360:674–7. doi: 10.1038/360674a0. [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–51. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Brodsky FM, Blank GS, Helenius A. Inhibition of endocytosis by anti-clathrin antibodies. Cell. 1987;50:453–63. doi: 10.1016/0092-8674(87)90499-5. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–9. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Fu W, Jiang Q, Zhang C. Novel functions of endocytic player clathrin in mitosis. Cell Res. 2011 doi: 10.1038/cr.2011.106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Tao W, Zheng P, Fu J, Bian M, Jiang Q, Clarke PR, Zhang C. Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J Cell Sci. 2010;123:3645–51. doi: 10.1242/jcs.075911. [DOI] [PubMed] [Google Scholar]
- Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–41. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goud B, Huet C, Louvard D. Assembled and unassembled pools of clathrin: a quantitative study using an enzyme immunoassay. J Cell Biol. 1985;100:521–7. doi: 10.1083/jcb.100.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greener T, Zhao X, Nojima H, Eisenberg E, Greene LE. Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from nonneuronal cells. J Biol Chem. 2000;275:1365–70. doi: 10.1074/jbc.275.2.1365. [DOI] [PubMed] [Google Scholar]
- Han Z, Liang CG, Cheng Y, Duan X, Zhong Z, Potireddy S, Moncada C, Merali S, Latham KE. Oocyte spindle proteomics analysis leading to rescue of chromosome congression defects in cloned embryos. J Proteome Res. 2010;9:6025–32. doi: 10.1021/pr100827j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, McIntosh JR, Cleland S. Intermicrotubule bridges in mitotic spindle apparatus. J Cell Biol. 1970;45:438–44. doi: 10.1083/jcb.45.2.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE, Keen J. Deep-etch visualization of proteins involved in clathrin assembly. J Cell Biol. 1988;107:877–86. doi: 10.1083/jcb.107.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE, Keen JH, Amende LM, Lippoldt RE, Prasad K. Deep-etch visualization of 27S clathrin: a tetrahedral tetramer. J Cell Biol. 1987;105:1999–2009. doi: 10.1083/jcb.105.5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Robinson MS. Clathrin and adaptors. Biochim Biophys Acta. 1998;1404:173–93. doi: 10.1016/s0167-4889(98)00056-1. [DOI] [PubMed] [Google Scholar]
- Hirst J, Sahlender DA, Li S, Lubben NB, Borner GH, Robinson MS. Auxilin depletion causes self-assembly of clathrin into membraneless cages in vivo. Traffic. 2008;9:1354–71. doi: 10.1111/j.1600-0854.2008.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Dannhauser PN, Groos S, Hinrichsen L, Curth U, Ungewickell EJ. A comparison of GFP-tagged clathrin light chains with fluorochromated light chains in vivo and in vitro. Traffic. 2010;11:1129–40. doi: 10.1111/j.1600-0854.2010.01084.x. [DOI] [PubMed] [Google Scholar]
- Holzenspies JJ, Roelen BA, Colenbrander B, Romijn RA, Hemrika W, Stoorvogel W, van Haeften T. Clathrin is essential for meiotic spindle function in oocytes. Reproduction. 2010;140:223–33. doi: 10.1530/REP-10-0045. [DOI] [PubMed] [Google Scholar]
- Hood FE, Royle SJ. Functional equivalence of the clathrin heavy chains CHC17 and CHC22 in endocytosis and mitosis. J Cell Sci. 2009;122:2185–90. doi: 10.1242/jcs.046177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood FE, Royle SJ. Pulling it together: The mitotic function of TACC3. BioArchitecture. 2011;1:105–9. doi: 10.4161/bioa.1.3.16518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–54. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DS, Kern RC, Puthenveedu MA, von Zastrow M, Williams JC, Benovic JL. Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking. J Biol Chem. 2009;284:29860–72. doi: 10.1074/jbc.M109.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of aurora and polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Binder LI, Rosenbaum JL. The periodic association of MAP2 with brain microtubules in vitro. J Cell Biol. 1979;80:266–76. doi: 10.1083/jcb.80.2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW, Hyman AA. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J Cell Biol. 2005;170:1047–55. doi: 10.1083/jcb.200503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC. Protein organization in clathrin trimers. Cell. 1981;23:755–61. doi: 10.1016/0092-8674(81)90439-6. [DOI] [PubMed] [Google Scholar]
- Knight AS, Notaridou M, Watson RJ. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28:1737–47. doi: 10.1038/onc.2009.22. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol. 2001;3:643–9. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- LeRoy PJ, Hunter JJ, Hoar KM, Burke KE, Shinde V, Ruan J, Bowman D, Galvin K, Ecsedy JA. Localization of human TACC3 to mitotic spindles is mediated by phosphorylation on Ser558 by Aurora A: a novel pharmacodynamic method for measuring Aurora A activity. Cancer Res. 2007;67:5362–70. doi: 10.1158/0008-5472.CAN-07-0122. [DOI] [PubMed] [Google Scholar]
- Lin CH, Hu CK, Shih HM. Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J Cell Biol. 2010;189:1097–105. doi: 10.1083/jcb.200911120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq JM, Berger EG, Warren G. Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J Cell Biol. 1989;109:463–74. doi: 10.1083/jcb.109.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack GJ, Compton DA. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc Natl Acad Sci USA. 2001;98:14434–9. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Compton DA. Structural and regulatory roles of nonmotor spindle proteins. Curr Opin Cell Biol. 2008;20:101–6. doi: 10.1016/j.ceb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro B, Johnson MH, Pickering SJ, Louvard D. Changes in the distribution of membranous organelles during mouse early development. J Embryol Exp Morphol. 1985;90:287–309. [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–89. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR. Kinetochore microtubules in PTK cells. J Cell Biol. 1992;118:369–83. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, West RR. Chromosomemicrotubule interactions during mitosis. Annu Rev Cell Dev Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- Miele AE, Watson PJ, Evans PR, Traub LM, Owen DJ. Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Nat Struct Mol Biol. 2004;11:242–8. doi: 10.1038/nsmb736. [DOI] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–66. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathke IS, Heuser J, Lupas A, Stock J, Turck CW, Brodsky FM. Folding and trimerization of clathrin subunits at the triskelion hub. Cell. 1992;68:899–910. doi: 10.1016/0092-8674(92)90033-9. [DOI] [PubMed] [Google Scholar]
- O’Brien LL, Albee AJ, Liu L, Tao W, Dobrzyn P, Lizarraga SB, Wiese C. The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Mol Biol Cell. 2005;16:2836–47. doi: 10.1091/mbc.E04-10-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto CT, McKinney J, Jeng YY. Clathrin in mitotic spindles. Am J Physiol Cell Physiol. 2000;279:C369–74. doi: 10.1152/ajpcell.2000.279.2.C369. [DOI] [PubMed] [Google Scholar]
- Pearse BM. Coated vesicles from pig brain: purification and biochemical characterization. J Mol Biol. 1975;97:93–8. doi: 10.1016/s0022-2836(75)80024-6. [DOI] [PubMed] [Google Scholar]
- Peset I, Seiler J, Sardon T, Bejarano LA, Rybina S, Vernos I. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J Cell Biol. 2005;170:1057–66. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18:379–88. doi: 10.1016/j.tcb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Peterman EJ, Scholey JM. Mitotic microtubule crosslinkers: insights from mechanistic studies. Curr Biol. 2009;19:R1089–94. doi: 10.1016/j.cub.2009.10.047. [DOI] [PubMed] [Google Scholar]
- Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider A, Wendland B. Endocytic adaptors--social networking at the plasma membrane. J Cell Sci. 2011;124:1613–22. doi: 10.1242/jcs.073395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Rieder CL. Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to a draw. Chromosoma. 2005;114:310–8. doi: 10.1007/s00412-005-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E, Jentzsch G. Ultrastructural changes in the mitotic apparatus at the metaphase-to-anaphase transition. J Cell Biol. 1969;40:678–91. doi: 10.1083/jcb.40.3.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–74. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Royle SJ. The cellular functions of clathrin. Cell Mol Life Sci. 2006;63:1823–32. doi: 10.1007/s00018-005-5587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ. Mitotic moonlighting functions for membrane trafficking proteins. Traffic. 2011;12:791–8. doi: 10.1111/j.1600-0854.2011.01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ, Bright NA, Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434:1152–7. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ, Lagnado L. Trimerisation is important for the function of clathrin at the mitotic spindle. J Cell Sci. 2006;119:4071–8. doi: 10.1242/jcs.03192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–7. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH. Proteome analysis of the human mitotic spindle. Mol Cell Proteomics. 2005;4:35–43. doi: 10.1074/mcp.M400158-MCP200. [DOI] [PubMed] [Google Scholar]
- Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463:464–73. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- Shimamoto Y, Maeda YT, Ishiwata S, Libchaber AJ, Kapoor TM. Insights into the micromechanical properties of the metaphase spindle. Cell. 2011;145:1062–74. doi: 10.1016/j.cell.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Nagamori I, Yabuta N, Nojima H. GAK, a regulator of clathrin-mediated membrane traffic, also controls centrosome integrity and chromosome congression. J Cell Sci. 2009;122:3145–52. doi: 10.1242/jcs.052795. [DOI] [PubMed] [Google Scholar]
- Stephens RE, Edds KT. Microtubules: structure, chemistry, and function. Physiol Rev. 1976;56:709–77. doi: 10.1152/physrev.1976.56.4.709. [DOI] [PubMed] [Google Scholar]
- Still IH, Vettaikkorumakankauv AK, DiMatteo A, Liang P. Structure-function evolution of the transforming acidic coiled coil genes revealed by analysis of phylogenetically diverse organisms. BMC Evol Biol. 2004;4:16. doi: 10.1186/1471-2148-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142:433–43. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland HG, Mumford GK, Newton K, Ford LV, Farrall R, Dellaire G, Caceres JF, Bickmore WA. Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum Mol Genet. 2001;10:1995–2011. doi: 10.1093/hmg/10.18.1995. [DOI] [PubMed] [Google Scholar]
- Tahara H, Yokota E, Igarashi H, Orii H, Yao M, Sonobe S, Hashimoto T, Hussey PJ, Shimmen T. Clathrin is involved in organization of mitotic spindle and phragmoplast as well as in endocytosis in tobacco cell cultures. Protoplasma. 2007;230:1–11. doi: 10.1007/s00709-006-0226-7. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Vallenius T, Geers EF, Greene L, Makela TP, Medema RH. Cyclin G-associated kinase promotes microtubule outgrowth from chromosomes during spindle assembly. Chromosoma. 2010;119:415–24. doi: 10.1007/s00412-010-0267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci USA. 2000;97:1096–100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Musacchio A, Harrison SC, Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;95:563–73. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani R, Ling YC, Oliferenko S. The fission yeast TACC protein Mia1p stabilizes microtubule arrays by length-independent crosslinking. Curr Biol. 2009;19:1861–8. doi: 10.1016/j.cub.2009.09.063. [DOI] [PubMed] [Google Scholar]
- Tooze J, Hollinshead M. Evidence that globular Golgi clusters in mitotic HeLa cells are clustered tubular endosomes. Eur J Cell Biol. 1992;58:228–42. [PubMed] [Google Scholar]
- Traub LM. Regarding the amazing choreography of clathrin coats. PLoS biology. 2011;9:e1001037. doi: 10.1371/journal.pbio.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol. 2008;183:223–39. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unanue ER, Ungewickell E, Branton D. The binding of clathrin triskelions to membranes from coated vesicles. Cell. 1981;26:439–46. doi: 10.1016/0092-8674(81)90213-0. [DOI] [PubMed] [Google Scholar]
- Ungewickell E, Branton D. Assembly units of clathrin coats. Nature. 1981;289:420–2. doi: 10.1038/289420a0. [DOI] [PubMed] [Google Scholar]
- Wakeham DE, Abi-Rached L, Towler MC, Wilbur JD, Parham P, Brodsky FM. Clathrin heavy and light chain isoforms originated by independent mechanisms of gene duplication during chordate evolution. Proc Natl Acad Sci USA. 2005;102:7209–14. doi: 10.1073/pnas.0502058102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. Membrane partitioning during cell division. Annu Rev Biochem. 1993;62:323–48. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Sanger JW, Sanger JM. Dynamics of organelles in the mitotic spindles of living cells: membrane and microtubule interactions. Cell Motil Cytoskeleton. 1993;26:19–39. doi: 10.1002/cm.970260104. [DOI] [PubMed] [Google Scholar]
- Wettey FR, Hawkins SF, Stewart A, Luzio JP, Howard JC, Jackson AP. Controlled elimination of clathrin heavy-chain expression in DT40 lymphocytes. Science. 2002;297:1521–5. doi: 10.1126/science.1074222. [DOI] [PubMed] [Google Scholar]
- Wilbur JD, Hwang PK, Ybe JA, Lane M, Sellers BD, Jacobson MP, Fletterick RJ, Brodsky FM. Conformation switching of clathrin light chain regulates clathrin lattice assembly. Dev Cell. 2010;18:841–8. doi: 10.1016/j.devcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willox AK, Royle SJ. Functional analysis of interaction sites on the N-terminal domain of clathrin heavy chain. Traffic. 2011 doi: 10.1111/j.1600-0854.2011.01289.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler FK, Stanley KK. Clathrin heavy chain, light chain interactions. EMBO J. 1983;2:1393–400. doi: 10.1002/j.1460-2075.1983.tb01597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt PL, Ris H, Borisy GG. Structure of kinetochore fibers: microtubule continuity and inter-microtubule bridges. Chromosoma. 1981;83:523–40. doi: 10.1007/BF00328277. [DOI] [PubMed] [Google Scholar]
- Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006;173:879–91. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Ishidao T, Nomura T, Shinagawa T, Tanaka Y, Yonemura S, Ishii S. A B-Myb complex containing clathrin and filamin is required for mitotic spindle function. EMBO J. 2008;27:1852–62. doi: 10.1038/emboj.2008.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. A membranous spindle matrix orchestrates cell division. Nat Rev Mol Cell Biol. 2010;11:529–35. doi: 10.1038/nrm2919. [DOI] [PMC free article] [PubMed] [Google Scholar]