Abstract
Olfactory receptor (OR) genes represent ≈1% of genomic coding sequence in mammals, and these genes are clustered on multiple chromosomes in both the mouse and human genomes. We have taken a comparative genomics approach to identify features that may be involved in the dynamic evolution of this gene family and in the transcriptional control that results in a single OR gene expressed per olfactory neuron. We sequenced ≈350 kb of the murine P2 OR cluster and used synteny, gene linkage, and phylogenetic analysis to identify and sequence ≈111 kb of an orthologous cluster in the human genome. In total, 18 mouse and 8 human OR genes were identified, including 7 orthologs that appear to be functional in both species. Noncoding homology is evident between orthologs and generally is confined within the transcriptional unit. We find no evidence for common regulatory features shared among paralogs, and promoter regions generally do not contain strong promoter motifs. We discuss these observations, as well as OR clustering, in the context of evolutionary expansion and transcriptional regulation of OR repertoires.
Animals have evolved specialized sense organs that recognize olfactory information in the environment and transmit this information to the brain, where it then must be processed to create an internal representation of the external world. Humans, for example, are thought to recognize more than 10,000 discrete odors with exquisite discriminatory power such that subtle differences in chemical structure often can lead to profound differences in perceived odor quality.
Several divergent odorant receptor gene families, each encoding seven transmembrane domain proteins, have been identified in vertebrates and invertebrate species. In mammals, volatile odorants are detected by a family of as many as 1,000 receptors, each expressed in the main olfactory epithelium (1). Terrestrial vertebrates have a second anatomically and functionally distinct olfactory system, the vomeronasal organ, dedicated to the detection of pheromones (2, 3). Vomeronasal sensory neurons express at least two distinct families of receptors, each thought to contain 100–200 genes (4–9). In the invertebrate Caenorhabditis elegans, 900 chemosensory receptors are organized into four gene families (10). Two smaller gene families, each consisting of about 70 genes, encode the odorant receptors in Drosophila (11–13). Thus, chemosensory detection is accomplished by at least nine highly divergent gene families, each sharing little or no sequence similarity. The evolutionary requirement for odorant receptors therefore is met by the recruitment of novel gene families rather than exploiting preexisting odorant receptor families in ancestral genomes.
Odorant receptor genes are often highly divergent, and there are dramatic differences in the size of the gene family between species. During the relatively short period of terrestrial vertebrate evolution, for example, the olfactory receptor (OR) repertoire has expanded about 10-fold since the time of a common ancestor with aquatic fish. This striking diversification is likely to result from frequent recombination, gene conversion, duplication, and translocation (14–17). The rapid evolutionary change in OR repertoires may reflect the biological demands for adaptation to changing environments on time scales at least as frequent as speciation events.
Comparative genomics provides insight into the molecular events that generated these extraordinary gene families and may also facilitate the identification of regulatory elements governing the expression of olfactory receptor genes. An olfactory neuron expresses a given receptor from either the maternal or paternal allele, but never both (18). In addition, OR gene expression is spatially regulated such that a given receptor is expressed only in one of four topographic zones in the olfactory epithelium (19, 20). The transcriptional mechanisms that ensure that an individual olfactory neuron expresses only 1 of 2,000 OR alleles within a rapidly evolving genome remain unknown. We have performed a comparative genomic analysis of the orthologous mouse–human P2 cluster of OR genes to identify structural elements that may be involved in the dynamic evolution and transcriptional regulation of this gene family.
Materials and Methods
Clone Identification.
Using PCR primers designed from the murine P2 and I7 receptor sequences (1), we screened subpools of a 3-fold redundant mouse (strain 129 SVJ) embryonic stem cell-derived bacterial artificial chromosome (BAC) library (Genome Systems, St. Louis). Four positive clones were identified (BACs 22b5, 219o16, 59i3, and 139j24). Two m50 and three B5 clones were identified from screens from a mouse (strain 129 SVJ) genomic phage library (Stratagene). Mouse BAC RP23–388c2 (strain C57BL/6J) was identified by a BAC-End (http://www.tigr.org) database search, and human P1 artificial chromosome (PAC) 610i20 (AF065876 and AF065874), BAC RP11–560b16 (AC017103), BAC RP11–732a19 (AC027641), BAC RP11–413n10 (AC024729), cosmid Q25 (AAF00005), and cosmid Q27 (AAF00004) were all identified in GenBank by blast (http://www.ncbi.nlm.nih.gov). Before sequencing, clones were localized to metaphase chromosomes by using standard fluorescence in situ hybridization procedures (21).
Sequence Generation.
Human PAC 610i20 (AF321237), mouse BACs 22b5 (AF321233), and the five mouse m50 (AF321236) and B5 (AF321235) phage clones were sequenced by the shotgun method to high redundancy (8×) and were finished by using PCR/primer walking. Contig assembly and finishing was accomplished by using PHRED/PHRAP (22, 23) and consed (24) software. Mouse BAC RP23–388c2 (AF321234) was sequenced to medium depth (5× redundancy), and the resulting 10 contigs that extend beyond BAC 22b5 were ordered by CAP3 assembly (25) and targeted PCR.
Isolation of 5′ OR Exons by Rapid Amplification of cDNA Ends (RACE).
The olfactory epithelium from B6CBAF1/J adult mice was dissected, and 1.3 μg of poly(A)+ mRNA was isolated by using oligo(dT) cellulose (Stratagene). Preparation of cDNA and RACE protocols were as described in the Marathon cDNA Amplification and SMART RACE PCR kits (CLONTECH).
Genomic Analysis Tools.
Repeat content was determined by the repeatmasker algorithm (http://ftp.genome.washington.edu; A. F. Smit and P. Green, unpublished results) and analyzed for homology by using blast (http://www.ncbi.nlm.nih.gov) and pipmaker algorithms (26). Genomic analysis tools available at the Baylor College of Medicine Search Launcher (http://www.hgsc.bcm.tmc.edu) were used, including TSSG/TSSW (V. V. Solovyev, A. A. Salamov, and C. B. Lawrence, unpublished data), nnpp (27), and matinspector (28).
Results and Discussion
Sequence of Orthologous P2 OR Clusters.
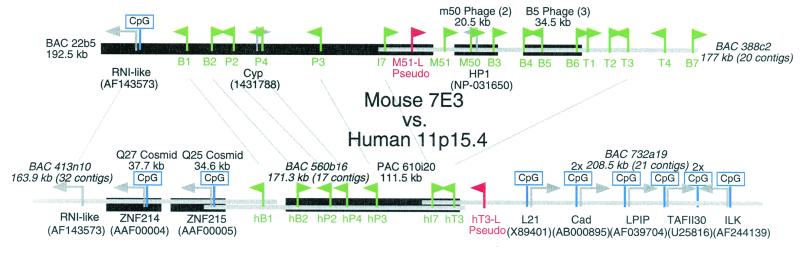
In an effort to relate the sequence and organization of the OR genes to their function, we have performed a comparative genomic analysis of the orthologous mouse and human P2 cluster of OR genes. The pattern of expression of these genes in the olfactory epithelium, the possible role of these genes in axon targeting, and ligand specificity of odorant receptors in the cluster have been examined extensively. Two BAC clones and five λ-phage clones containing OR genes from the mouse P2 locus were isolated. All clones map to a single location at mouse chromosome 7E3 by fluorescence in situ hybridization (data not shown). High-redundancy sequencing resulted in ≈350 kb of genomic sequence that contained 18 OR genes (including 1 pseudogene) and 3 non-OR genes (Fig. 1).
Figure 1.
Map of mouse and human orthologous OR clusters. Thick, black lines indicate finished or ordered sequence, encompassing mouse BAC 22b5, two mouse M50-phage contigs, three mouse B5-phage contigs, human Q25 cosmid, human Q27 cosmid, and human PAC 610i20. Thin, gray lines represent unfinished draft sequence (italicized BAC names, estimated sizes, and number of contigs indicated). OR genes are represented by green (ORF) and red (pseudogene) flags, with the orientation of the gene indicated by the flags. Non-OR genes are represented by gray arrows (accession numbers for highest blastx homologies are indicated in brackets below name). These include: mouse and human novel RNI-like genes, mouse cyclophilin-A processed pseudogene (Cyp) and HP1 chromobox-containing genes, two human zinc-finger genes (ZNF214, ZNF215), human L21 ribosomal, cadherin FIB1 (Cad), lysosomal pepstatin-insensitive protease (LPIP), TATA-binding factor II-30 (TAFII30), and integrin-linked kinase-1 (ILK) genes. Gene-linked CpG islands (CpG/GpC ≥ 0.75) are indicated by blue signs. Putative orthologous relationships are indicated by gray lines connecting genes from both clusters.
Three candidate human orthologs to OR genes that reside at the mouse P2 locus were identified from the GenBank database (HOR11–55A, HOR11–403A, and HOR11–610A) (29). These three genes reside in a single human PAC (610i20) that maps to human chromosome 11p15.4, a human region syntenic to mouse chromosome 7E3. We have generated high-redundancy genomic sequence of the ≈111-kb PAC 610i20. In addition, we have identified two cosmid and three BAC sequences that cover ≈400 kb at this human locus. Eight OR genes (including one pseudogene) and eight non-OR genes were identified within this 400-kb stretch of the human genome.
Both the mouse and human clusters are bounded on one side by novel genes with homology to RNase inhibitor (RNI) proteins. The mouse and human RNI-like genes have greater than 80% identity and have the same predicted intron–exon structures. Two multiexon zinc-finger genes, ZNF214 and ZNF215, reside between the RNI-like gene and the cluster of human OR genes. These zinc-finger genes are not present in the mouse P2 cluster, suggesting that there has been an insertion at the human locus or a deletion at the mouse locus. Both of these zinc-finger genes are associated with Beckwith–Wiedemann syndrome, a genetic disorder caused by abnormalities at an imprinted locus on human chromosome 11p15 (30). All nine of the non-OR ORFs identified in the regions flanking the mouse and human clusters have associated CpG islands except the human RNI-like gene, which differs in this regard from its mouse ortholog.
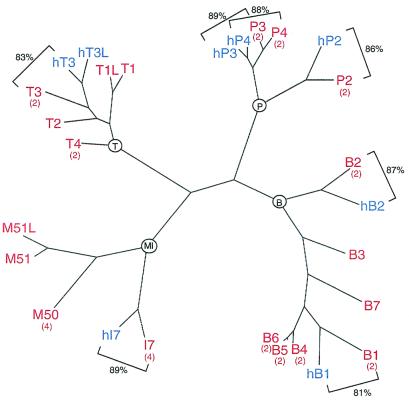
Seven putative OR orthologs were found upstream of the RNI-like gene at both the mouse and human locations. Each orthologous pair exhibits greater than 80% nucleotide sequence identity and resides in an identical position and orientation in the cluster. This high level of sequence identity among orthologs contrasts with an average value of about 58% identity among the human or mouse OR paralogs (Fig. 2). These data provide strong evidence that these two clusters are orthologous.
Figure 2.
Phylogenetic tree of OR genes identified at the mouse and human orthologous locations. All 26 OR genes identified from both mouse (red) and human (blue) OR clusters (plus one mouse T1-like gDNA fragment) are represented in a paup (Sinauer Associates, Sunderland, MA) parsimony tree. Zonal expression patterns in the mouse main olfactory epithelium (1 = dorsalmost to 4 = ventralmost zones) are indicated by bracketed red numbers beneath gene names. Percent identities for the seven pairs of putative mouse–human orthologs are indicated (square brackets). The four major clades in the tree are rooted by black circles (MI, P, T, and B). Average pairwise identities between OR genes from different major clades are 54%. Percent identities within the MI clade range between 74% (M51–M51L) and 60% (M50–I7), within the P clade range between 95% (P3–P4) and 74% (P2–P4), within the T clade range between 92% (hT1–hT1L) and 80% (T2–T4), and within the B clade range between 98% (B5–B6) and 65% (B2–B7).
The array of human OR genes is bounded on its telomeric side by T3 and a T3-like pseudogene. At present, it is difficult to unequivocally assign a mouse T3 ortholog. The mouse and human T3 genes are 84% identical and are nearest one another in the phylogenetic tree. Moreover, they reside in the same relative orientation and share noncoding homology, suggesting that they are orthologs. However, four similar T genes (T1–T4) all share about 80% identity with hT3. Moreover, we have identified an additional mouse T-like gene that is not contained within the sequenced portion of the cluster and may reside distal to the mouse B7 gene. If the true mouse T3 ortholog is at the very telomeric end of the mouse cluster, as it is in the human array, a single human deletion event between hI7 and hT3 could account for the gene organization in both species.
All seven pairs of putative orthologs encode ORFs in both species. One possible exception is the hP4 gene, whose ORF, when compared with the mouse ortholog, begins with an ATA instead of the expected ATG. We confirmed this codon sequence in four individuals (data not shown). It is possible that either an upstream or downstream in-frame start codon is used. The G-to-A mutation in hP4 creates a strong acceptor splice site at bp 3–4, and the gene-structure-predicting program genie predicts a two-exon coding sequence with a novel hydrophobic leader peptide encoded by a 33-bp exon starting 1,228 bp upstream of the main ORF. Alternatively, an in-frame ATG, which could represent the translation initiation site, is found in both mP4 and hP4 42 nt downstream of the presumed start site of mP4.
The Size of the Human Cluster and the Human OR Repertoire.
The nature of the odorant receptor genes in a species and their number will reflect the repertoire of odorants to which a given organism is responsive. A total of seven orthologous OR pairs were identified between mouse and human that maintain greater than 80% sequence identity. These genes may recognize odorous ligands required by both species. The murine cluster, however, is approximately twice the size of the human cluster and contains 11 intact OR sequences that are absent from the human cluster. Efforts to identify orthologs to the murine M50 and B5 genes by library screens and database searches thus far have been unsuccessful, suggesting that this array of genes is deleted rather than translocated elsewhere in the human genome. These genes therefore are likely to reflect ORs that afford a selective advantage or adaptation to narrow ecological niches important to rodents but not primates.
Can we attach significance to the diminished size of the P2 cluster in the human chromosome? It has been suggested that humans are “microsmatic” and exhibit a decline in the breadth and discriminatory power of their olfactory system, reflected by a diminished number of receptor genes and a high frequency of OR pseudogenes in the human genome. The actual number of functional human OR genes, however, awaits the complete annotation of the human genome. Our studies at this locus suggest that if there is indeed a diminution of functional human OR sequences, this may be ascribed to block deletion rather than the accumulation of pseudogenes. Moreover, analysis of the T cell antigen receptor α/δ-linked OR cluster (R.P.L., J. Roach, I. Y. Lee, C. Boysen, A. F. Smit, B.J.R., and L.H., unpublished results), the OR genes surrounding the β-globin cluster (31), and the chromosome 17 OR cluster (32) reveals far lower frequencies of human OR pseudogenes than previously reported from PCR analysis. Of the 50 ORs identified at these loci, only 12 are apparent pseudogenes. The human OR genes appear to have undergone far more frequent duplication and translocation, such that clusters of OR genes are fewer in number in mouse than in humans (ref. 16; J.Y., unpublished data). It is possible, therefore, that estimates of the frequency of human pseudogenes, largely derived from PCR-amplified OR sequences, may reflect a large number of redundant OR copies residing at duplicated human loci. For example, the human 11p15 region contains two OR clusters that are duplicated on multiple chromosomes (16, 33) in addition to the single-copy P2 and β-globin OR clusters (34). Duplications and divergence also could lead to ORs with new specificities and at least transiently increase the effective copy number of specific ORs. The genomic OR repertoires of different species thus must reflect that balance between stability of OR genes recognizing ligands required over long periods of evolutionary time vs. duplication and divergence that afford rapid adaptability to environmental change.
Gene Clustering and Gene Twinning.
The P2 locus in mouse contains at least 18 odorant receptor genes. The organization of OR genes into large, linked arrays is characteristic of the OR gene family in mammals. We presume that the linkage of multiple homologous genes affords an evolutionary adaptability because this organization facilitates rapid crossing over and gene-conversion events that may serve to expand and diversify the gene family. The organization of genes into clusters also may have regulatory significance, as it does for the HOX (reviewed in ref. 35) and globin gene clusters (reviewed in ref. 36), but this has not been demonstrated for the OR genes.
Evidence for duplication and divergence is apparent in the phylogenetic tree of the 26 OR genes in the mouse and human P2 clusters. These genes fall into four distinct clades. Within a clade, human and mouse sequences share greater than 80% identity, but the shared identity between clades is less than 60%. The most similar genes most often reside next to one another, although there are exceptions (e.g., B7 is physically close to T4, yet it is most homologous to the physically distant B3 gene).
In general, OR genes most similar to one another tend to be expressed in the same zone of the olfactory epithelium. With the exception of the M50-I7 miniclade, the phylogenetically diverse genes at this locus are all expressed in zone 2, the second, dorsalmost zone (Fig. 2). In no case do we find OR genes of the same clade in the phylogenetic tree expressed in different zones. These data would imply that genes that share maximal sequence identity through their coding regions also have retained those regulatory sequences presumably governing zonal expression.
One particularly interesting example of extreme conservation is observed for the mouse and human P3–P4 pairs and the mouse B4-B5-B6 trio. In both mouse and human, the P3 and P4 genes are more similar to each other (>90%) than they are to their respective orthologs. Two lines of evidence suggest that this unexpectedly high sequence conservation between neighboring paralogs in both mouse and human is not due to a recent block duplication. First, the high level of paralogous sequence identity ends abruptly at the coding-sequence boundaries. Second, mouse and human P3 orthologs, as well as the mouse and human P4 orthologs, share sequence homology in 5′ upstream DNA, consistent with the suggestion that both orthologous pairs existed in an ancestor common to mouse and human.
Recent gene conversions could be responsible for the P3–P4 similarity. These conversion events, however, must be nonrandom because they occur between the same pairs of genes in both mouse and human and must be coincidentally timed such that the twins are diverged to similar extents in both species despite different rates of evolution (37). It is more likely that these highly homologous sets of ORs afford a selective advantage not as a single OR gene, but as a pair. The presence of a pair of extremely similar but nonidentical receptors, for example, might afford the opportunity for fine discrimination among structurally similar odorants.
Gene Structure and Transcription.
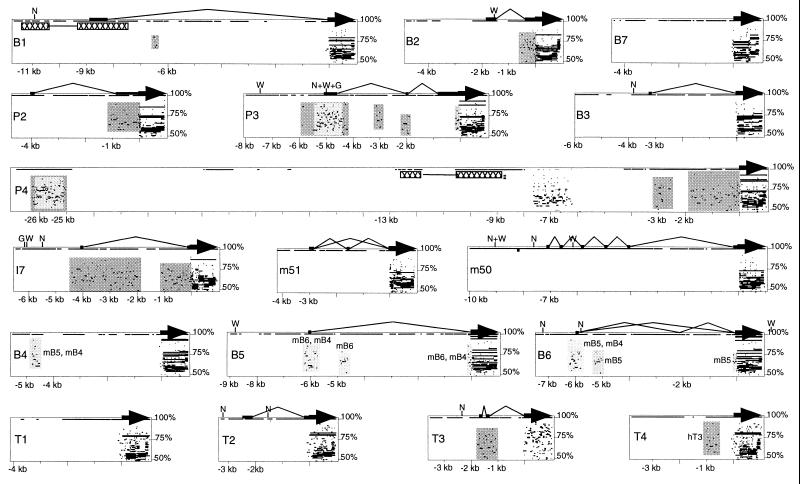
We next analyzed the RNA transcripts from 12 individual OR genes within the mouse P2 cluster. The availability of the complete nucleotide sequence of the cluster, along with transcriptional analysis, allows us to determine the precise intron–exon structure of these 12 OR genes. We generated RACE-PCR products for 12 mouse OR genes to define the transcription start site (TSS) and identify putative upstream promoter regions (Fig. 3). Transcription initiates at variable distances from the initiator ATG, with the most distant start site 9 kb (B1) and the closest start site 1.7 kb (T3) from the initiator. All of the 5′ untranslated regions have at least one upstream intron. In no case do we observe introns that span exons of other genes. Most OR genes in the cluster contain a single 5′ intron, with M50 as an unusual exception; it has five introns within its untranslated region (UTR). Two of the genes, M51 and B6, reveal alternative splicing within the 5′ UTR, but this does not alter the reading frame of the receptor. We cannot, at present, attach functional significance to alternatively spliced forms. Differential splicing in untranslated regions may affect the stability, the translational efficiency, or localization of RNA within the neuron. However, analysis of UTR sequences between paralogs reveals little sequence identity and, therefore, does not allow us to identify conserved elements in RNA of potential functional significance.
Figure 3.
pipmaker and analysis of the 5′ UTRs and promoter regions of mouse OR genes. Gene structures for 12 mouse OR genes are plotted as determined by 5′ RACE-PCR. Coding sequences are shown as thick arrows, upstream exons are shown as thin lines, and introns are shown as bent lines. The B6 and M51 genes are alternatively spliced as shown. Relative positions to translation start sites are shown in kilobases (kb). The positions of putative promoters (excluding those found within repeats) between 3 kb upstream and 1 kb downstream of transcription start sites as predicted by three algorithms (N = NNPP, G = TSSG, W = TSSW) are indicated. Only NNPP scores >0.90 (0.33% false-positive rate) and TSSG/TSSW LDF scores >4.0 are shown. The sequence of each gene was compared with all other mouse and human OR genes at the P2 loci. Detected sequence homology (>50% identity) is plotted according to position along the test gene and level of sequence similarity. Dark-gray-shaded boxes surround regions of homology detected between the mouse OR gene and its human ortholog. Light-gray-shaded boxes indicate paralogy detected between P3 and P4 and among B4, B5, and B6. A region at the putative promoter of B1 has high sequence identity to an inverted sequence upstream of P4 (hatched rectangles). All other plots in noncoding regions represent sequence similarity of the test sequence to itself at some other position.
The determination of TSS for 12 OR genes allows us to identify putative upstream promoter regions in orthologous mouse and human genes. Sequence comparison between other orthologous gene pairs has been used to delineate regulatory regions controlling specific gene expression (e.g., refs. 38 and 39). We therefore have searched for conserved motifs in regions within 3 kb of the experimentally determined TSS for the OR genes. It is noteworthy that promoter-predicting algorithms, which search for combinations of TATA boxes, associated motifs, and canonical initiator sequences, fail to identify upstream promoters in the vicinity of TSSs for any of the 12 genes examined. The highest-scoring promoter is positioned just within the 5′ UTR of P3, 120 bp downstream of the start site, and was the only instance in which all three search algorithms used were in agreement.
pipmaker analysis detects upstream homology within orthologous UTRs, but rarely does this homology encompass putative promoter regions upstream of the start site. We detect only a small number of putative regulatory regions shared among the genes in the cluster and no conserved regions shared by more than two genes. The P3–P4 and B4–B5–B6 genes are the only exceptions to the general observation that promoter regions of paralogs lack sequence similarity. A search against the transfac database reveals only six conserved transcription factor-binding motifs within the homologous mouse and human P3 and P4 promoter regions. Three of these are Ikaros motifs, along with two S8 homeodomain motifs and a methylation-sensitive vMYB motif. The general lack of paralogous homology and strong promoter motifs is also evident from analyses of the mouse–human β-globin (31) and T cell receptor (R.P.L., J. Roach, I. Y. Lee, C. Boysen, A. F. Smit, B.J.R., and L.H., unpublished results) OR clusters. Although we find few clues to regulatory function in the putative promoter regions, orthology and promoter motifs detected downstream of the TSSs suggests that these internal regions may have regulatory importance. Evolution of compact gene structures could have facilitated the genomic expansion of the OR family.
An individual olfactory sensory neuron expresses one receptor gene from a repertoire of 1,000 genes. Moreover, an OR gene is expressed from only one of the two alleles in a given cell, but the products of the paternal and maternal alleles are represented equally in a population of cells (18). A second level of spatial regulation restricts each OR gene expression to one of four zones of the olfactory epithelium (19, 20). Finally, and most interestingly, within a spatial zone a neuron expresses only 1 of the ≈250 genes permitted within that zone.
What regulatory mechanisms might be responsible for this hierarchy of transcriptional control? It is reasonable to assume that zonal control results from spatial information imparted on OR genes and therefore might require common motifs flanking all genes expressed in a given zone. We and others (40) have defined the boundary for such putative motifs. Correct zonal expression, for example, is observed with P2 transgenes containing as little as 2.0 kb of upstream DNA flanking the TSS and 1.3 kb of DNA downstream of the poly(A) addition site (R.A., unpublished observations). Comparison of the DNA flanking the eight mouse genes expressed in zone II (including P2 itself) at 11-bp resolution reveals no shared elements above noise, and transfac searches fail to identify smaller, conserved transcription factor-binding sites. Thus, if common regulatory elements direct OR gene expression in a given zone, they are likely to involve novel, scattered and/or small motifs not detected in our analysis.
Perhaps most enigmatic are the control mechanisms ensuring that only a single OR gene is expressed in an individual sensory neuron. In one extreme model, each of the 1,000 genes might retain different regulatory motifs that interact with a unique combination of trans-acting factors. Such a model would be consistent with our inability to detect conserved sequence motifs surrounding paralogs. However, recent data demonstrating that OR transgenes are faithfully expressed in olfactory neurons, but are never coexpressed in the same cell as the endogenous homolog, argue against models that invoke 1,000 different combinations of activators (41). A second mechanism consistent with our data invokes recombination of OR genes to a single expression site. Regulatory information, for example, may not reside adjacent to the OR transcription unit, and OR transcription may require recombination into a single, active locus. Evidence for such a model awaits analysis of the DNA from homogeneous populations of sensory neurons expressing the same receptor gene. Finally, we might invoke a novel mechanism of control in which only a single OR transcription complex resides within olfactory neurons that can stably accommodate only a single OR gene. Whatever the mechanism, analysis of DNA in several OR paralogs has failed to define a conserved regulatory motif, suggesting that either further refined analysis is required or novel mechanisms are operative in regulating OR gene expression.
Acknowledgments
We acknowledge Jim Edmondson for the initial isolation of numerous OR genes at this locus, Jill Buettner for the initial identification of human OR genes on PAC 610i20, Michelle Kim for providing RACE template cDNA from mouse olfactory epithelium, Lena Linardopoulou for help in constructing mouse and human OR alignments from GenBank entries, Steve Swartzell for his help in the assembly of BAC 388c2, and Joe Osborne for his help in the early aspects of this project. We thank Webb Miller for generous support on the use of pipmaker. We also thank numerous individuals in the Department of Molecular Biotechnology Core Sequencing Facility. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health Grants R01-DC04209 and HG00202-09.
Abbreviations
- OR
olfactory receptor
- BAC
bacterial artificial chromosome
- RACE
rapid amplification of cDNA ends
- PAC
P1 artificial chromosome
- RNI
RNase inhibitor
- TSS
transcription start site
- UTR
untranslated region
Footnotes
References
- 1.Buck L, Axel R. Cell. 1991;65:165–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 2.Keverne E B. Science. 1999;286:716–720. doi: 10.1126/science.286.5440.716. [DOI] [PubMed] [Google Scholar]
- 3.Leinders-Zufall T, Lane A P, Puche A C, Ma W, Novotny M V, Shipley M T, Zufall F. Nature (London) 2000;405:792–796. doi: 10.1038/35015572. [DOI] [PubMed] [Google Scholar]
- 4.Dulac C, Axel R. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 5.Herrada G, Dulac C. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- 6.Ryba N J, Tirindelli R. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- 7.Matsunami H, Buck L B. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- 8.Pantages E, Dulac C. Neuron. 2000;28:835–845. doi: 10.1016/s0896-6273(00)00157-4. [DOI] [PubMed] [Google Scholar]
- 9.Del Punta K, Rothman A, Rodriguez I, Mombaerts P. Genome Res. 2000;10:1958–1967. doi: 10.1101/gr.10.12.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Troemel E R. BioEssays. 1999;21:1011–1020. doi: 10.1002/(SICI)1521-1878(199912)22:1<1011::AID-BIES5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 11.Vosshall L B, Amrein H, Morozov P S, Rzhetsky A, Axel R. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 12.Clyne P J, Warr C G, Freeman M R, Lessing D, Kim J, Carlson J R. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 13.Scott K, Brady R, Jr, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Arie N, Lancet D, Taylor C, Khen M, Walker N, Ledbetter D H, Carrozzo R, Patel K, Sheer D, Lehrach H, et al. Hum Mol Genet. 1993;3:229–235. doi: 10.1093/hmg/3.2.229. [DOI] [PubMed] [Google Scholar]
- 15.Glusman G, Clifton S, Roe B, Lancet D. Genomics. 1996;37:147–160. doi: 10.1006/geno.1996.0536. [DOI] [PubMed] [Google Scholar]
- 16.Trask B J, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, et al. Hum Mol Genet. 1998;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- 17.Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D. Genomics. 1999;63:227–245. doi: 10.1006/geno.1999.5900. [DOI] [PubMed] [Google Scholar]
- 18.Chess A, Simon I, Cedar H, Axel R. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 19.Ressler K J, Sullivan S L, Buck L B. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 20.Vassar R, Ngai J, Axel R. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 21.Trask B J. In: Genome Analysis, A Laboratory Manual. Birren B, Green E, Heiter P, Klapholz S, Myers R, Reithman H, Roskams J, editors. Vol. 4. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 303–413. [Google Scholar]
- 22.Ewing B, Hillier L, Wendl M C, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 23.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 24.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 25.Huang X, Madan A. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz S, Zhang Z, Frazer K A, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese M G, Harris N L, Eeckman F H. In: Biocomputing, Proceedings of the 1996 Pacific Symposium. Hunter L, Klein T E, editors. Singapore: World Scientific Publishing; 1996. pp. 2–7. [Google Scholar]
- 28.Quandt K, Frech K, Karas H, Wingender E, Werner T. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buettner J A, Glusman G, Ben-Arie N, Ramos P, Lancet D, Evans G A. Genomics. 1998;53:56–68. doi: 10.1006/geno.1998.5422. [DOI] [PubMed] [Google Scholar]
- 30.Alders M, Ryan A, Hodges M, Bliek J, Feinberg A P, Privitera O, Westerveld A, Little P F, Mannens M. Am J Hum Genet. 2000;66:1473–1484. doi: 10.1086/302892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulger M, Bender M A, van Doorninck J H, Wertman B, Farrell C M, Felsenfeld G, Groudine M, Hardison R. Proc Natl Acad Sci USA. 2000;97:14560–14565. doi: 10.1073/pnas.97.26.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glusman G, Sosinsky A, Ben-Asher E, Avidan N, Sonkin D, Bahar A, Rosenthal A, Clifton S, Roe B, Ferraz C, et al. Genomics. 2000;63:227–245. doi: 10.1006/geno.1999.6030. [DOI] [PubMed] [Google Scholar]
- 33.Trask B J, Massa H, Brand-Arpon V, Chan K, Friedman C, Nguyen O T, Eichler E, van den Engh G, Rouquier S, Shizuya H, et al. Hum Mol Genet. 1998;7:2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- 34.Bulger M, van Doorninck H J, Saitoh N, Telling A, Farrell C, Bender M A, Felsenfeld G, Axel R, Groudine M. Proc Natl Acad Sci USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duboule D. Curr Opin Genet Dev. 1998;8:514–518. doi: 10.1016/s0959-437x(98)80004-x. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Harju S, Peterson K R. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 37.Li W H, Ellsworth D L, Krushkal J, Chang B H, Hewett-Emmett D. Mol Phylogenet Evol. 1996;5:182–187. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
- 38.Williams S C, Altmann C R, Chow R L, Hemmati-Brivanlou A, Lang R A. Mech Dev. 1998;73:225–229. doi: 10.1016/s0925-4773(98)00057-4. [DOI] [PubMed] [Google Scholar]
- 39.Stojanovic N, Florea L, Riemer C, Gumicio D, Slighton J, Goodman M, Miller W, Hardison R. Nucleic Acids Res. 1999;27:3899–3910. doi: 10.1093/nar/27.19.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qasba P, Reed R R. J Neurosci. 1998;18:227–236. doi: 10.1523/JNEUROSCI.18-01-00227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, et al. Nat Neurosci. 2000;3:687–693. doi: 10.1038/76641. [DOI] [PubMed] [Google Scholar]