Abstract
Background
Human T-cell leukemia virus types 1 and 2 (HTLV-1 and HTLV-2) are delta retroviruses with similar genetic organization. Although both viruses immortalize T-cells in vitro, they exhibit distinct pathogenic potential in vivo. To search for possible differences in its expression strategy with respect to HTLV-1, we investigated the pattern of HTLV-2 expression in infected cell lines and peripheral blood mononuclear cells (PBMCs) from infected patients using splice site-specific quantitative RT-PCR.
Findings
A novel alternative splice acceptor site for exon 2 was identified; its usage in env transcripts was found to be subtype-specific. Time-course analysis revealed a two-phase expression kinetics in an infected cell line and in PBMCs of two of the three patients examined; this pattern was reminiscent of HTLV-1. In addition, the minus-strand APH2 transcript was mainly detected in the nucleus, a feature that was similar to its HTLV-1 orthologue HBZ. In contrast to HTLV-1, expression of the mRNA encoding the main regulatory proteins Tax and Rex and that of the mRNAs encoding the p28 and truncated Rex inhibitors is skewed towards p28/truncated Rex inhibitors in HTLV-2.
Conclusion
Our data suggest a general converging pattern of expression of HTLV-2 and HTLV-1 and highlight peculiar differences in the expression of regulatory proteins that might influence the pathobiology of these viruses.
Findings
Human T-cell leukemia virus types 1 and 2 (HTLV-1 and HTLV-2) are related deltaretroviruses [1] with similar genetic organization and expression strategies [2-4]. Both viruses immortalize T-cells in culture and establish a persistent infection in vivo [5]. However, unlike HTLV-1, which causes adult T-cell leukemia/lymphoma (ATLL) and tropical spastic paraparesis/HTLV-1-associated myelopathy (TSP/HAM), HTLV-2 has not been linked to lymphoproliferative diseases although an increase in lymphocytes counts was described in HTLV-2-infected patients [6] and coinfection with HTLV-2 plays an important role in the progression of HIV-infected patients to AIDS [7].
Like HTLV-1, HTLV-2 produces plus- and minus-strand alternatively spliced transcripts that code for virion components and non-structural proteins, including Tax and Rex that regulate viral expression at the transcriptional and post-transcriptional levels, respectively [8,9]. In contrast to HTLV-1, the kinetics of expression of the individual HTLV-2 transcripts has not been described so far. To address this point, we employed splice site-specific quantitative RT-PCR (qRT-PCR) to measure individual HTLV-2 mRNAs (Figure 1) and their expression kinetics in chronically infected cell lines and in peripheral blood mononuclear cells (PBMCs) obtained from HTLV-2-infected individuals. Results revealed interesting analogies and differences from HTLV-1 [10].
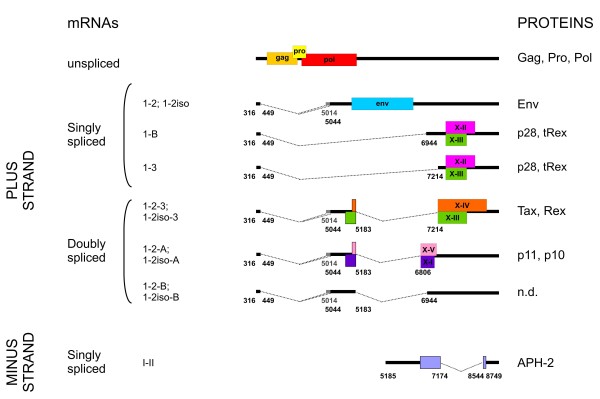
Figure 1.
Genetic organization, alternative splicing and coding potential of HTLV-2 mRNAs. Structure and coding potential of HTLV-2 alternatively spliced mRNAs. ORFs are indicated by boxes. Splice sites are indicated by numbers and correspond to the Mo-T isolate (GeneBank: M10060) in which + 1 is the first base of U3 in the 5’ LTR. Gray lines and numbers designate the 5’ end of exon 2iso identified in the present study. n.d.: the protein coding potential of this transcript has not yet been determined.
Identification of a novel HTLV-2 splice acceptor-site
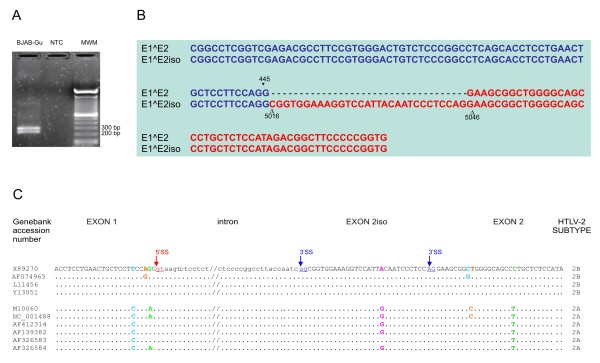
We first analyzed the pattern of viral mRNAs in the HTLV-2-infected cell lines Mo-T (infected with subtype A) and BJAB-Gu (infected with subtype B). Pilot experiments showed unexpectedly low (almost 3 orders of magnitude) levels of the env mRNA in BJAB-Gu cells compared to Mo-T. Further experiments in the BJAB-Gu cell line using a sense primer located at the 5’ end of exon 1 and an antisense primer located in exon 2 yielded the expected 275 bp band and a slightly longer product (Figure 2A). Sequence analysis of cloned PCR products indicated that the longer product was generated by usage of a novel splice acceptor site (termed "2iso") located 30 nucleotides upstream of exon 2 (Figure 2B). In silico analysis of different HTLV-2 isolates showed that the novel 2iso 3’splice-site (SS) defines a GU-AG type intron with a consensus “AGgtaagt” sequence at the exon 1-intron boundary that varied among the isolates (Figure 2).
Figure 2.
Identification of a novel HTLV-2 splice acceptor-site.A. Agarose gel electrophoresis of exon 1–2 and exon 1-2iso RT-PCR products. BJAB-Gu cDNA was amplified using the 6000-Gu forward primer (5’ GGCGGGCTCCTTCAACG 3’) located in the first exon and the 5183-Gu reverse primer (5’ CCATGGTGTTGGTGGTCT 3’) in the second exon. Amplification yielded the expected 275-bp band and a slightly longer product (lane 1). No bands were detected in the no template control (NTC, lane 2); lane 3 shows a 1-kbp molecular weight marker (MWM). Cell lines were cultured at 37°C, 5% CO2, in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM L-glutamine, penicillin (100 U/ml) and streptomycin (100 μg/ml). Cells were lysed in TRIzol® (Invitrogen) and total RNA was extracted and processed as detailed in Additional file 1: Table S1. B. Sequence alignment of cloned PCR products and identification of a novel splice acceptor site upstream of exon 2. Nucleotides in exon 1 are in blue and nucleotides in exon 2 are in red. Sequence analysis demonstrated that the 1-2iso junction is generated by splicing to a novel 3’splice-site (SS) located 30 nucleotides upstream of the exon 2 3'SS. Numbering of the sequence corresponds to that of the Gu isolate (GenBank: × 89270.1) in which + 1 is the first base of U3 in the 5’ LTR. ▼ indicates nucleotide position in exon 1; Δ indicates nucleotide position in exon 2 or 2iso. C. Sequence alignment of HTLV-2 isolates. DNA sequences containing exon 1 splice donor (5’SS) and exon 2iso or exon 2 splice-acceptor (3’SS) sites were compared between various HTLV-2 isolates retrieved from GenBank (accession numbers are indicated on the left, subtypes on the right). Exons and introns are indicated in upper and lower case, respectively. Underlined nucleotides indicate the 5’SS and 3’SS consensus sequences. The consensus sequence AGgtaagt at the exon 1-intron boundary varies among 2A and 2B subtypes, while the intronic gt 5’SS and ag 3’SS dinucleotides are invariable. The 3'SS defining exon 2iso is conserved among the different isolates examined.
Quantitative and temporal analysis of HTLV-2 mRNAs in infected cell lines
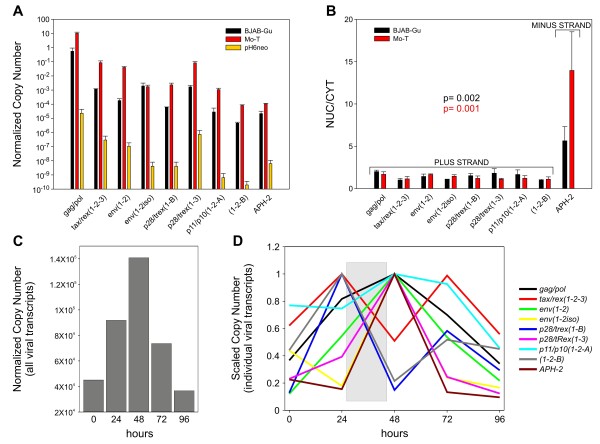
We next tested the expression levels of the different HTLV-2 mRNAs using splice site-specific primer sets and qRT-PCR (Additional file 1: Table S1), and we calculated the Normalized Copy Number (NCN) by dividing the absolute copy number of each transcript by the absolute copy number of the housekeeping gene (GAPDH or 18S rRNA, see Figures 3 and 4).
Figure 3.
Quantitative analysis, intracellular compartmentalization and expression kinetics of HTLV-2 mRNAs in infected cell lines. Normalized copy number (NCN) and Scaled Copy Numbers (SCN) were calculated as described in the text. Housekeeping genes used for normalization were GAPDH (panels A and B) and 18S rRNA (panels C and D) and in this latter case (panel C) the obtained NCN were multiplied for 107. A. NCN of viral mRNAs in the HTLV-2-infected cell lines BJAB-Gu (black bars) and Mo-T (red bars) and in cells transfected with the HTLV-2A molecular clone pH6neo (yellow bars). Cells were harvested at densities ranging from 100,000/ml (transfected cells) to 500,000/ml (infected cell lines). Shown are mean values of 3 independent experiments and standard error bars. B. Nucleo-cytoplasmic distribution (NUC/CYT) of plus- and minus-strand HTLV-2 mRNAs in infected cell lines Mo-T (red bars) and BJAB-Gu (black bars) was determined as detailed in Additional file 1: Table S1. Shown are mean values of 4 independent experiments, standard error bars and P values. The square brackets in the figure indicate a statistically significant difference between plus-and minus-strand transcripts calculated with a Mann–Whitney Rank Sum Test (p = 0.001 and p = 0.002 for the Mo-T and BJAB-Gu cell lines respectively). C,D. Temporal analysis of HTLV-2 expression. A confluent culture of BJAB-Gu cells was diluted to 25,000 cells/ml and aliquots were harvested every 24 h over a 96-h period. Shown are NCN of the sum of all (plus and minus strand) viral transcripts at each time point (C) and SCN of individual mRNAs (D). The grey box in the figure (panel D), highlights the switch in the pattern of expression from early to late transcripts.
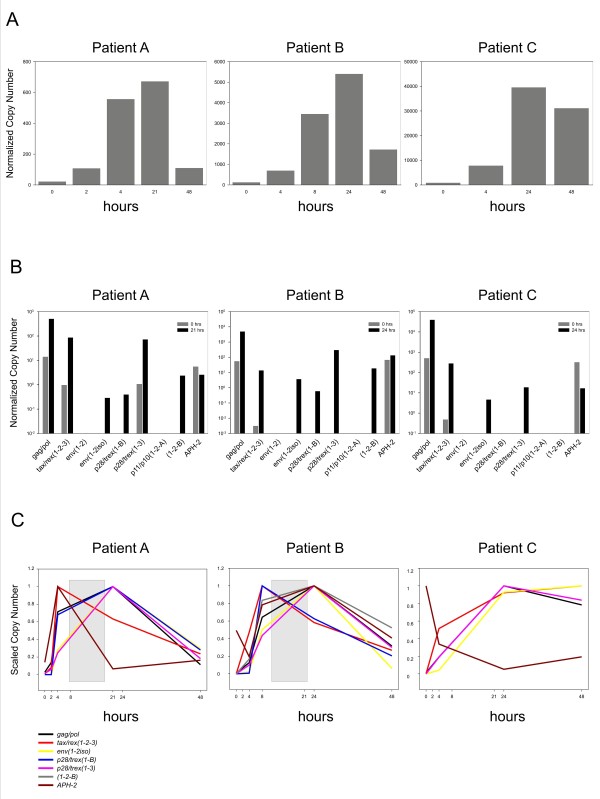
Figure 4.
Temporal analysis of HTLV-2 expression in PBMCs from infected patients. Normalized copy number (NCN) and Scaled Copy Numbers (SCN) were calculated as described in the text. The housekeeping gene used for the normalization was 18S rRNA and the resulted NCN was multiplied for 107. A. NCN of the sum of all viral mRNAs (plus- and minus- strands) viral mRNAs in PBMCs from infected patients in a 48-h time course of culture in vitro. PBMCs were cultured and harvested for total RNA at the indicated time points. B. NCN of the individual mRNAs in HTLV-2-infected patients at time 0 (grey bars) and after 21 (patient A) or 24 (patients B and C) hours of culture (black bars). C. SCN of individual mRNAs are plotted over a 48-h time period. The grey box in the figure, highlights the switch in the pattern of expression from early to late transcripts.
Using two splice site-specific primer pairs, we quantified the levels of the alternative env transcripts [env(1–2) and env(1-2iso)] from the Mo-T and BJAB-Gu cell lines and in cells transfected with the HTLV-2A molecular clone pH6neo (kindly provided by Dr. P. L. Green). Results showed that the env(1-2iso) mRNA was expressed at similar levels in both cell lines, while the canonic env(1–2) transcript was about 100-fold less abundant in BJAB-Gu cells (Figure 3A). Interestingly, the HTLV-2A molecular clone pH6neo also expressed mainly env(1–2), suggesting a subtype-specific usage of the 2 vs. 2iso 3’SS (Figure 3A). While the sequence of the acceptor and donor dinucleotides is conserved between the two isolates, the adjacent nucleotides are different (Figure 2C), suggesting that this could result in a different splice site usage in the different isolates. The 1-2iso splice acceptor site was also used in the context of a doubly spliced tax/rex(1-2iso-3) mRNA which was readily detected in both cell lines and in the molecular clone pH6neo (data not shown). Selection of 1-2iso 3'SS as opposed to the 1–2 3'SS is not predicted to affect the coding potential of the env(1–2; 1-2iso), tax/rex(1-2-3; 1-2iso-3) or 1-2-B and 1-2iso-B mRNAs, as both 3'SS are located in the non-coding portion of the transcripts. Interestingly, the 1-2iso splice site is not unique to only the BJAB-Gu cell line, as it is efficiently used in the patients, which are infected with subtype 2B (see next paragraph).
In the chronically infected cell lines Mo-T and BJAB-Gu as well as in cells transfected with the pH6neo molecular clone the most abundant viral mRNA was gag/pol followed by tax/rex(1-2-3), env and p28/trex(13) (Figure 3A); the latter codes for p28 and the truncated isoforms of Rex (p22/20 and p19/p18). We proposed to term these truncated isoforms of Rex as “tRex” [11]. Both p28 and tRex may act as latency factors by inhibiting the tax/rex mRNA [12] and the Rex protein, respectively [13]. Interestingly, in HTLV-2 tax/rex was detected at levels comparable to p28/trextaxrex/trex(1–3) ratio = 0.5 in pH6neo-transfections; 1.04 in Mo-T; 0.69 in BJAB-Gu]. This is in contrast with HTLV-1, where tax/rex is much more abundant than the mRNAs coding for p30Tof and p21Rex, the orthologs of p28 and tRex, respectively [10]. These data suggest that the ratio of expression of the tax/rex mRNA vs. the mRNAs encoding potential inhibitors is higher in HTLV-1[10] compared to HTLV-2. The p28 and tRex proteins are also coded by the less abundant p28/trex(1-B) mRNA. Transcript 1-2-B, whose protein product has not yet been identified and is also expressed as 1-2iso-B; the p11/p10(1-2-A) transcript and the minus-strand APH-2 transcript were all expressed at lower levels. These data are consistent with a previous study by Li et al. [14].
A recent study on the intracellular distribution of HTLV-1 mRNAs indicated that the plus-strand transcripts showed similar partition in the nucleus and cytoplasm, while the minus-strand mRNAs were over 10-fold more abundant in the nucleus, suggesting that they may play a role as a non-coding RNAs [10]. To test whether this is the case for HTLV-2, we analyzed the nucleo-cytoplasmic distribution of plus- and minus-strand HTLV-2 mRNAs. Results obtained in the Mo-T and BJAB-Gu infected cell lines showed that all the plus-strand mRNAs were equally distributed in the nucleus and cytoplasm, while the minus-strand APH-2 mRNA [15] was about 10-fold or 4-fold higher (Mo-T and BJAB-Gu cell lines, respectively) in the nucleus (Figure 3B; Mann–Whitney Rank Sum Test; p = 0.001 and p = 0.002 for the Mo-T and BJAB-Gu cell lines respectively). This finding is reminiscent of cell lines chronically infected with HTLV-1 [10].
We next investigated whether, in analogy to HTLV-1 [11], HTLV-2 is characterized by an early/late temporal pattern of expression. To test this possibility in an in vitro model, we induced HTLV-2 expression in BJAB-Gu cells by diluting to 25,000 cells/ml starting from a confluent culture and measured transcript levels over 96 h by qRT-PCR. As indicated in Figure 3C, this treatment results in an overall upregulation of viral expression. To ensure that the pattern of viral expression was not affected by cell death, we tested cell viability by trypan blue exclusion at each time point and found that cell death did not exceed 4% for up to 96 h of culture. The amount of p19 released by the BJAB-Gu cells was below the threshold of detection by ELISA at 0 and 24 h and rose to 62 pg/ml at 48 h, 121 pg/ml at 72 h, and 169 pg/ml at 96 h.
To better visualize the timing of expression of individual mRNAs, we calculated the scaled copy number (SCN) by dividing the normalized copy number (NCN) of each transcript at each time point by the maximum NCN measured for that mRNA during the experiment. Results showed 2-phase kinetics with the expression of tax/rex(1-2-3), p28/tRex(1-B) and 1-2-B preceding that of gag/pol, env and p28/trex(1–3) mRNAs (Figure 3D), a finding that is consistent with the key role of Tax and Rex as master regulatory proteins driving expression of the other viral genes.
Temporal analysis of HTLV-2 expression in PBMCs from infected patients
We next used qRT-PCR to investigate HTLV-2 expression kinetics in PBMCs obtained from 3 HTLV-2B-infected patients; these patients had similar proviral loads and were HTLV-1- and HIV-1-negative (Additional file 2: Table S2). All of the recruited patients gave informed consent, according to the Italian laws and the Declaration of Helsinki. The study was approved by the institutional ethics committees of all participating institutions. As described for HTLV-1 [10], a sharp upregulation of HTLV-2 transcripts was observed upon culture of the PBMCs in all the examined patients (Figure 4A). Before in vitro culture only gag/pol, APH2 and tax/rex(1-2-3) were detected in all patients; p28/trex(13) was detected only in patient A (Figure 4B, grey bars). Upon in vitro culture the expression of all transcripts was sharply upregulated (Figure 4B, black bars); however, patient C exhibited a marked downregulation of APH-2. Following reactivation, gag/pol was the most abundant transcript. Tax/rex(1-2-3), p28/trex(13), APH-2, and in patients A and B, 1-2-B were also readily detected, while the other transcripts were expressed at much lower levels. Consistent with the fact that the patients were infected with HTLV-2 subtype B, env(1-2iso) was efficiently detected, while env(1–2) was undetectable. Patient C did not express p28/trex(1-B) or (1-2-B). p11/p10(1-2-A) was below the threshold of detection in all patients. The tax/rex over p28/trex(1–3) ratio ranged between 0.9 and 7.6 (depending on the time point) in patient A, and was even more skewed in favor of p28/trex(1–3) in patient B (ratios between 0.04 and 0.37, depending on the time point).
SCN calculations (Figure 4C) indicated a two-phase kinetics of HTLV-2 gene expression in patients A and B, with an early sharp rise in tax/rex(1-2-3) expression followed by expression of all other genes. Patient B also expressed p28/trex(1-B) as an early transcript, although it its expression levels were lower by about 3 orders of magnitude compared to the p28/tRex(1–3) (Figure 4B).
The expression pattern of patients A and B is similar to that of the BJAB-Gu cell line (Figure 3D) and suggests an early/late switch in the pattern of viral expression. While plus-strand transcripts showed an overall common pattern of expression in two patients and in the BJAB-Gu cell line, the temporal expression of APH-2 appeared to be highly variable, at least in the restricted number of samples examined. Further studies carried out on larger cohorts of samples should be performed, to test the possible correlation of different APH-2 expression patterns with virus load and patient status. In this regard, a recent study showed that both Tax and APH-2 mRNA levels correlate with proviral load; interestingly, no correlation was found between APH-2 levels and the number of lymphocytes in the patients, suggesting that, in contrast to HBZ, APH-2 does not promote cell proliferation [16].
Although the temporal pattern of HTLV-2 expression was reminiscent of that of HTLV-1, it is noteworthy that the most abundant HTLV-2 mRNA encoding p28 and tRex (i.e. mRNA1-3) was expressed from a “late” transcript, while HTLV-1 p21Rex was expressed as an early transcript in most patients examined in a previous study [10].
Taken together with previous analyses of HTLV-1 expression, the present study suggests a converging expression strategy of the two viruses characterized by 2-phase expression kinetics of viral mRNAs and by marked nuclear localization of minus-strand transcripts. Our results also highlight the following differences between the 2 viruses: (i) the relative expression of the tax/rex mRNA compared to the mRNAs coding for potential inhibitors of Tax (p30Tof and p28) and Rex [(p21Rex and p28/tRex(1–3)] is skewed toward tax/rex in HTLV-1 and towards the inhibitors in HTLV-2; (ii) the p28/trex mRNA was expressed as a “late” transcript in HTLV-2 while mRNAs for HTLV-1 orthologs p30/Tof and p21Rex are expressed early in most patients examined, suggesting interesting differences in the fine-tuning of Rex function between the two viruses. Nevertheless, it is appropriate to be cautious in the interpretation of these data, since only three patients were examined. This limitation was due to the fact that the kinetics analysis required (i) large numbers of PBMC, (ii) a high proviral load, and (iii) no coinfection with HIV or HTLV-1, which severely limited the number of samples suitable for this analysis.
Future studies should be aimed at further investigating the functions of these regulatory proteins in HTLV-1 and HTLV-2 and at understanding how they may contribute to the different pathobiology of the two viruses.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CB identified the novel splice acceptor site and carried out kinetics of expression in cell lines and patients. FR carried out nucleo-cytoplasmic fractionations and real-time RT-PCR assays; AC and PR set real-time RT-PCR and measured mRNA expression in cell lines; PR carried out cell lines and patients cultures; IC carried out cell culture and transfection; CC provided patient samples and supervised cell cultures; VC and UB designed the experiments and prepared the manuscript; and all authors contributed to the analysis and interpretation of the data. All authors read and approved the final manuscript.
Supplementary Material
Table S1. List of primer sets and probes used in qPCR detection of HTLV-2 proviral load or HTLV-2 mRNAs expression.
Table S2. Clinical and immunological features of HTLV-2 infected patients.
Contributor Information
Cecilia Bender, Email: ce_bender@yahoo.com.ar.
Francesca Rende, Email: francesca.rende@unipd.it.
Alessia Cotena, Email: alessiacotena@hotmail.com.
Paola Righi, Email: paola.righi@gmail.com.
Paola Ronzi, Email: paolaronzi@alice.it.
Ilaria Cavallari, Email: ilaria.cavallari@unipd.it.
Claudio Casoli, Email: claudio.casoli@gemiblab.com.
Vincenzo Ciminale, Email: v.ciminale@unipd.it.
Umberto Bertazzoni, Email: umberto.bertazzoni@univr.it.
Acknowledgement
We thank Donna D'Agostino, Luigi Chieco-Bianchi, Elisabetta Pilotti and Maria Grazia Romanelli for discussions. This work was supported by grants from the European Union (“The role of chronic infections in the development of cancer,” contract no. 2005–018704), the Associazione Italiana per la Ricerca sul Cancro (AIRC), the AIRC 2008-Cariverona Regional grant (UB and VC), the Ministero per l’Università e la Ricerca Scientifica e Tecnologica, and the Universities of Padova and Verona.
References
- Lairmore M, Franchini G. In: Fields Virology. Fifth. Knipe D, Howley P, editor. Philadelpia: Lippincott Williams and Wilkins; 2007. pp. 2071–2106. vol. 2. [Google Scholar]
- Ciminale V, Pavlakis GN, Derse D, Cunningham CP, Felber BK. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I. J Virol. 1992;66:1737. doi: 10.1128/jvi.66.3.1737-1745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralnik IJ. et al. Protein isoforms encoded by the pX region of human T-cell leukemia/lymphotropic virus type I. Proc Natl Acad Sci U S A. 1992;89:8813. doi: 10.1073/pnas.89.18.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciminale V. et al. Expression and characterization of proteins produced by mRNAs spliced into the X region of the human T-cell leukemia/lymphotropic virus type II. Virology. 1995;209:445. doi: 10.1006/viro.1995.1277. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- Bartman MT. et al. Long-term increases in lymphocytes and platelets in human T-lymphotropic virus type II infection. Blood. 2008;112:3995. doi: 10.1182/blood-2008-05-155960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casoli C, Pilotti E, Bertazzoni U. Molecular and cellular interactions of HIV-1/HTLV coinfection and impact on AIDS progression. AIDS Rev. 2007;9:140. [PubMed] [Google Scholar]
- Feuer G, Green PL. Comparative biology of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2. Oncogene. 2005;24:5996. doi: 10.1038/sj.onc.1208971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis I, Green PL. The human T-cell leukemia virus Rex protein. Front Biosci. 2005;10:431. doi: 10.2741/1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rende F. et al. Kinetics and intracellular compartmentalization of HTLV-1 gene expression: nuclear retention of HBZ mRNA. Blood. 2011;117(18):4855–4859. doi: 10.1182/blood-2010-11-316463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rende F, Comparison of the genetic organization, expression strategies, and oncogenic potential of HTLV-1 and HTLV-2. Leukemia Research and Treatment. 2012. Volume 2012 (2012), Article ID 876153, 14 pages. [DOI] [PMC free article] [PubMed]
- Nicot C. et al. HTLV-1-encoded p30(II) is a post-transcriptional negative regulator of viral replication. Nat Med. 2004;10(2):197–201. doi: 10.1038/nm984. [DOI] [PubMed] [Google Scholar]
- Ciminale V, Zotti L, D'Agostino DM, Chieco-Bianchi L. Inhibition of human T-cell leukemia virus type 2 Rex function by truncated forms of Rex encoded in alternatively spliced mRNAs. J Virol. 1997;71:2810. doi: 10.1128/jvi.71.4.2810-2818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Green PL. Detection and quantitation of HTLV-1 and HTLV-2 mRNA species by real-time RT-PCR. J Virol Methods. 2007;142:159. doi: 10.1016/j.jviromet.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halin M. et al. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood. 2009;114:2427. doi: 10.1182/blood-2008-09-179879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douceron E. et al. HTLV-2 APH-2 expression is correlated with proviral load but APH-2 does not promote lymphocytosis. J Infect Dis. 2012;205:82. doi: 10.1093/infdis/jir708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of primer sets and probes used in qPCR detection of HTLV-2 proviral load or HTLV-2 mRNAs expression.
Table S2. Clinical and immunological features of HTLV-2 infected patients.