Abstract
Background
Interspecific hybrids between S. cerevisiae × S. kudriavzevii have frequently been detected in wine and beer fermentations. Significant physiological differences among parental and hybrid strains under different stress conditions have been evidenced. In this study, we used comparative genome hybridization analysis to evaluate the genome composition of different S. cerevisiae × S. kudriavzevii natural hybrids isolated from wine and beer fermentations to infer their evolutionary origins and to figure out the potential role of common S. kudriavzevii gene fraction present in these hybrids.
Results
Comparative genomic hybridization (CGH) and ploidy analyses carried out in this study confirmed the presence of individual and differential chromosomal composition patterns for most S. cerevisiae × S. kudriavzevii hybrids from beer and wine. All hybrids share a common set of depleted S. cerevisiae genes, which also are depleted or absent in the wine strains studied so far, and the presence a common set of S. kudriavzevii genes, which may be associated with their capability to grow at low temperatures. Finally, a maximum parsimony analysis of chromosomal rearrangement events, occurred in the hybrid genomes, indicated the presence of two main groups of wine hybrids and different divergent lineages of brewing strains.
Conclusion
Our data suggest that wine and beer S. cerevisiae × S. kudriavzevii hybrids have been originated by different rare-mating events involving a diploid wine S. cerevisiae and a haploid or diploid European S. kudriavzevii strains. Hybrids maintain several S. kudriavzevii genes involved in cold adaptation as well as those related to S. kudriavzevii mitochondrial functions.
Background
The development of molecular methods of yeast characterization has demonstrated that some wine and brewing Saccharomyces strains possess complex genomes composed by genetic elements from two or more species [1-7]. These strains are widely known as interspecific hybrids.
The best characterized industrial interspecific hybrid is the lager yeast S. pastorianus, originated from hybridization between S. cerevisiae and a S. bayanus-related yeast, which recently has been suggested to belong to the new species S. eubayanus[8]. The hybridization between S. cerevisiae and the cryotolerant S. eubayanus have been suggested as the result of selective pressures derived from brewing at low temperatures [8].
Other kind of natural Saccharomyces hybrids are those originated from hybridization between S. cerevisiae and S. kudriavzevii. These hybrids have mainly been isolated from wine and brewing environments [1-3].
The role of the S. kudriavzevii genome in these hybrids is unclear, since the known strains of this species have been found in decaying leaves from Japan and oak trees from Portugal and Spain [9,10], but not in fermentative industrial environments yet. The physiological evaluation of some of these S. kudriavzevii isolates showed that this species is characterized by a higher cryotolerance than S. cerevisiae, but a lower ethanol tolerance [11,12].
Albeit differences between S. cerevisiae × S. eubayanus and S. cerevisiae × S. kudriavzevii hybrids, the role of the S. eubayanus or S. kudriavzevii genomes in the hybrid seems to be similar, that is, the maintenance of good fermentative performance at low temperatures.
The characterization of a particular group of Swiss wine hybrids by PCR-RFLP, DNA arrays, ploidy analysis and gene dose determination by quantitative real-time PCR, evidenced the existence of a single common hybridization event to explain the origin of these hybrids followed by extensive chromosomal rearrangements including chromosome losses and the generation of chimerical chromosomes [13].
In this work, genome composition by array-CGH of a more diverse set of wine and brewing S. cerevisiae × S. kudriavzevii natural hybrids from diverse origins was evaluated to decipher their origins and evolution. The examination of gene losses and gains as well as the maintenance of specific metabolic pathways from the S. cerevisiae or S. kudriavzevii parental genomes was also analyzed with the aim of elucidating the role of each parental genome in the fermentative performance of the hybrid strains.
Methods
Yeast strains and culture media
The natural yeast hybrids S. cerevisiae × S. kudriavzevii used in this study have been isolated from wine and brewing fermentations in different locations (Table 1). The haploid strain S. cerevisiae S288c was used as control for microarray DNA hybridizations. Yeast strains were grown at 28°C in GPY medium (2% glucose, 0.5% peptone, 0.5% yeast extract).
Table 1.
List of hybrid strains used in this study
| Strain name | Isolation source |
|---|---|
| HA1841 |
wine, Perchtoldsdorf, Austria |
| HA1842 |
wine, Perchtoldsdorf, Austria |
| PB7 |
wine Pietro Picudo, León, Spain |
| Assmanhausen (AMH) |
wine, Geisenheim, Germany |
| Anchor VIN7 |
commercial strain, Anchor, South Africa |
| SOY3 |
wine, Daruvar, Croacia |
| CECT1388 |
ale beer, United Kingdom |
| CECT1990 |
beer, Göttinger Brauhaus AG, Germany |
| CECT11002 |
beer Chimay Trappist, Belgium |
| CECT11003 |
beer Orval Trappist, Belgium |
| CECT11004 |
beer, Westmalle Trappist, Belgium |
| CECT11011 | brewery, New Zealand |
Ploidy estimations by flow cytometry
Ploidy estimates are very important to interpret aCGH data from hybrids because hybridization signals are commonly normalized with respect to those of the reference haploid strain S288c.
The DNA content of both hybrid and control strains was assessed by flow cytometry by two different procedures. The first ploidy estimates were obtained in a FACScan cytometer (Becton Dickinson Inmunocytometry Systems, California, United States) by using the propidium iodide dye method described in Belloch et al. [13]. Due to discrepancies with the aCGH analysis, new estimates were later obtained in a Beckman Coulter FC 500 (Beckman Coulter Inc., California, USA) by using the SYTOX Green dye method described in Haase and Reed [14]. In both cases, ploidy levels were scored on the basis of the fluorescence intensity compared with the haploid (S288c) and diploid (FY1679) reference S. cerevisiae strains. Ploidy reported for each strain is the result of three independent measures. Results were tested by one way ANOVA and Tukey’s HSD tests.
DNA labeling and microarray competitive genome hybridization
Total DNA, extracted as described in Querol et al. [15], was resuspended in 50 μl of de-ionized water and digested with endonuclease Hinf I (Roche Applied Science, Germany), according to the manufacturer’s instructions, to fragments of an average length of 0.25 to 8 kbp. Each sample was purified using High Pure PCR Product Purification Kit (Roche Applied Science, Germany) and 2 μg was labelled using BioPrime Array CGH Genomic Labelling System (Invitrogen, California, USA). Unincorporated label was removed using MinElute PCR Purification Kit (Qiagen, Germany). Equal amounts of labelled DNA from the corresponding hybrid strains and the control S288c strain were used as probes for microarray hybridization.
Array competitive genomic hybridization (aCGH) was performed using a double-spotted array containing 6,240 ORFs of S. cerevisiae plus control spots totaling 6.4 K (Microarray Centre, University Health Network, Toronto, Canada). New microarrays were pre-treated for one hour at 65°C with pre-hybridization solution (7.5 ml 20× SSC, 0.5 ml 10% SDS, 0.5 ml 10 mg/ml bovine serum albumin in 50 ml final volume). Pre-hybridization solution was washed during 15 s in mili-Q H2O, 2 s in 2-propanol, 2 s in milli-Q H2O and dried by centrifugation at 1200 rpm, 10 min. Microarrays were treated with hybridization solution (15 μl SSC, 0.6 μl 10% SDS, 6 μl 1 mg/ml salmon DNA and DNA labelled in 60 μl final volume) at 95°C for 1 min and at room temperature for 5 min before DNA hybridization. Hybridization was performed for 18 h in chamber at 65°C, thus allowing hybridization of the S. cerevisiae part of the hybrid genome. A negative control of microarray hybridization was done by using DNA from S. kudriavzevii IFO 1802 strain vs. S288c. After hybridization microarrays were washed at 65°C for 5 min in 2× SSC, 0.1% SDS, at room temperature in 0.1× SSC− 0.1% SDS for 10 min and six times in 0.1× SSC 1 min and dried by centrifugation at 1200 rpm, 10 min.
Experiments were carried out in duplicates and Cy5-dCTP and Cy3-dCTP dye-swap assays were performed to reduce dye-specific bias. The aCGH was performed for all hybrid strains except for W27, W46, SPG16-91 and SPG441 previously analyzed by Belloch et al. [13].
Microarray scanning and data normalization
Microarray scanning was done by using a GenePix Personal 4100A scanner (Axon Instruments/Molecular Devices Corp., California, USA). Microarray images and raw data were produced with the GenePix Pro 6.1 software (Axon Instruments/Molecular Devices Corp., California, USA) and background was subtracted by applying the local feature background median option. M-A plots (M = Log2 ratios; A = log2 of the product of the intensities) were represented to evaluate if ratio data were intensity-dependent. The normalization process and filtering were done using Acuity 4.0 (Axon Instruments/Molecular Devices Corp., California, USA). Raw hybridization signals from hybrids were normalized with respect to those of the reference haploid strain S228c by using the ratio-based option, in which average hybridization ratios are adjusted to 1 (and hence, the corresponding log2 values to 0).
Normalized data were filtered by regression correlations 635/532 > 0.6, signal intensity in both channels more than 350 units, and signal to noise (SNR) > 2.5. Features with artifacts or flagged as bad were removed from the analysis. Replicates were averaged after filtering. It is worth to remark that strong normalization factors were applied to the negative control signal in each channel (2 to the red and 0.46 to the green one). Raw data and normalized microarray data are available in ArrayExpress [16], under the ref. E-MEXP-3114.
Chromosome structure and recombination sites in the chimerical chromosomes
The log2 of normalized Cy5/Cy3 signal ratio obtained for each ORF was represented with respect to its corresponding chromosomal location using the completely sequenced reference S. cerevisiae strain S288c. These plots, called caryoscopes, were generated using ChARM v.1.1 [17]. Highly stringent hybridization conditions (65°C) were used to avoid the cross hybridization of S. kudriavzevii DNA present in the hybrids. The caryoscope of the negative control experiment showed that most S. kudriavzevii genes did not hybridize under these conditions and in the case of cross hybridization (red signal) this was due to the very strong normalization factors applied in these control, which increased the red signal and reduced the green one by factors not applied in the case of the experiments performed with DNA from hybrids (see Additional file 1: Figure S1). Accordingly, differences in the log2 ratio values observed in the caryoscopes revealed variations in the relative copy number of S. cerevisiae genes present in the hybrid strains.
The identification of over- and underrepresented regions was confirmed due to the normalization procedure, the hybridization ratios derived from aCGH analysis show the relative proportions of each gene with respect to the average number of copies in the hybrid, allowing the identification of over- and underrepresented regions in the hybrid genome by a one-way ANOVA test to determine the different levels of hybridization observed in the aCGH analysis. The approximate locations of the recombination points in the mosaic chromosomes were determined from the up and down jump locations in the ORFs mapping by microarray analysis of the hybrid yeast genomes.
Finally, by considering the collinearity of S. kudriavzevii and S. cerevisiae genomes [18], the S. kudriavzevii gene content in the hybrid genomes can be deduced from the presence/absence of the chromosome regions coming from each parental species, obtained in a previous PCR-RFLP analysis of these hybrids [19].
Gene Ontology (GO) analysis of S. kudriavzevii genes
GenMAPP v2.1 software [20] was used to perform gene ontology analysis of the S. kudriavzevii fraction in the hybrid genomes. Four different GO analyses were carried out using S. kudriavzevii genes present in all hybrid strains, including those previously characterized [13], these analyses corresponded to: i) the complete set of wine and brewing hybrids, except strain AMH, showing the lowest S. kudriavzevii gene content, ii) only wine hybrids, except AMH, iii) only brewing hybrids and iv) only AMH. In all cases, statistically significant GO term enrichments were shown by computing a p-value using the hypergeometric distribution (the background set of genes was 6241, the number of ORFs measured in microarray experiments). GO terms showing significant values (z-score >2 and p-value <0.05) were sorted according to their corresponding GO category.
Maximum parsimony tree
A list of minimal number of chromosomal rearrangements, chromosomal losses and restriction site changes were used to reconstruct the maximum parsimony tree. Data obtained from a previous study [13] were again included in this analysis. A binary matrix was constructed to codify each particular event (Additional file 2: Table S1). Parsimony trees were constructed by PHYLIP 3.66 package using the Mix program [21], taking chromosomal rearrangements and gain/losses as irreversible events (Camin-Sokal model) and the RFLP changes as reversible events (Wagner model). The consensus tree was obtained with Consense program using the Majority rule.
Results
Hybrid genome structures
Caryoscopes, representing log2 hybridization ratios for each gene mapped onto its corresponding chromosome position, of six hybrid strains from wine and 6 hybrids from brewing were obtained by array comparative genomic hybridization (aCGH) (Additional file 3: Figure S2). Due to the normalization procedure, the hybridization ratios derived from aCGH analysis show the relative proportions of each gene with respect to the average number of copies in the hybrid, allowing the identification of over- and underrepresented regions in the hybrid genome. However, aCGH analysis in combination with ploidy estimates and with information on the presence/absence of the chromosome regions coming from each parental species, obtained in a previous PCR-RFLP analysis of these hybrids [2,19], allowed us to decipher the genome composition of hybrids.
This way, ploidy estimates for these hybrids were obtained by flow cytometry. The initial estimates with the propidium iodide method suggested that most hybrids were diploids or close to diploidy (relative C-values of 2.0 to 2.6). However, these ploidy values were not congruent with the caryoscope and PCR-RFLP data. The ratio-based normalization of hybridization signals adjusts the average signal ratios (problem strain/reference strain) to 1, and hence the log2 values to 0. In the analysis of hybrids, ploidy estimates were 2n-2.6n, corresponding on average to a subgenome coming from each parental species, i.e. for each gene there are on average a copy coming from S. cerevisiae and another from S. kudriavzevii. Due to the high astringent hybridization conditions used in the aCGH analysis of hybrids, only the S. cerevisiae subgenome is hybridizing, as confirmed by the negative control performed with S. kudriavzevii DNA. Therefore, in the normalization of hybridization signals, these ratios correspond to the adjustment of average signals coming from 1 S. cerevisiae gene copy from the hybrid to 1 gene copy form the reference haploid S. cerevisiae strain. In the case of an increase of copy numbers in specific genes or chromosomal regions, log2 values should be higher than 0 (1, 2, etc. depending on the number of copies), but in the case of loss of S. cerevisiae gene copies in the hybrid, a ratio of 0 (log2 of – ∞) should be observed. However, 3–4 levels of log2 values, including negative but not infinite, are observed for some hybrids (Additional file 3: Figure S2), which made difficult the interpretation of the aCGH results and suggested that ploidy estimates with propidium iodide were wrong.
Therefore, new ploidy estimates of hybrids were obtained by using SYTOX Green as the DNA-binding dye, because Haase and Reed [14] demonstrated that improves linearity between DNA content and fluorescence, and decreases peak drift associated with changes in dye concentration, growth conditions or cell size. In this new ploidy analysis, Swiss wine hybrids analyzed in our previous study [13] were also included.
The statistical analysis of the new estimates showed two significantly different groups of hybrids according to ploidy levels: most hybrids, including the Swiss wine strains, appear as allotriploids and hybrids AMH and PB7 as allotetraploid yeasts (Table 2). The new ploidy estimates are in agreement with the different levels of hybridization observed in the aCGH analyses and also with the previous PCR-RFLP analysis of hybrids [19].
Table 2.
DNA contents of natural hybrids, estimated by flow cytometry using the SYTOX green method with respect to the reference haploid and diploid S. cerevisiae strains, S288c and FY 1679, respectively
| Strain | DNA content relative to haploid strain S288c |
|---|---|
| FY1679 |
2.00a ± 0.00 |
| HA 1841 |
3.01b ± 0.08 |
| HA 1842 |
3.07b ± 0.07 |
| VIN7 |
3.04b ± 0.08 |
| SOY3 |
2.89b ± 0.09 |
| CECT 1388 |
3.25b ± 0.09 |
| CECT 1990 |
2.86b ± 0.07 |
| CECT 11002 |
3.02b ± 0.14 |
| CECT 11003 |
3.21b ± 0.09 |
| CECT 11004 |
3.13b ± 0.07 |
| CECT 11011 |
2.99b ± 0.05 |
| W27 |
3.18b ± 0.08 |
| W46 |
3.20b ± 0.07 |
| 441 |
3.10b ± 0.09 |
| SPG16-91 |
3.14b ± 0.08 |
| PB7 |
3.96c ± 0.08 |
| AMH | 3.85c ± 0.18 |
Results are the mean value of three replicates. Means with the same letters do not differ significantly by one way ANOVA and Tukey’s HSD tests (p < 0.05).
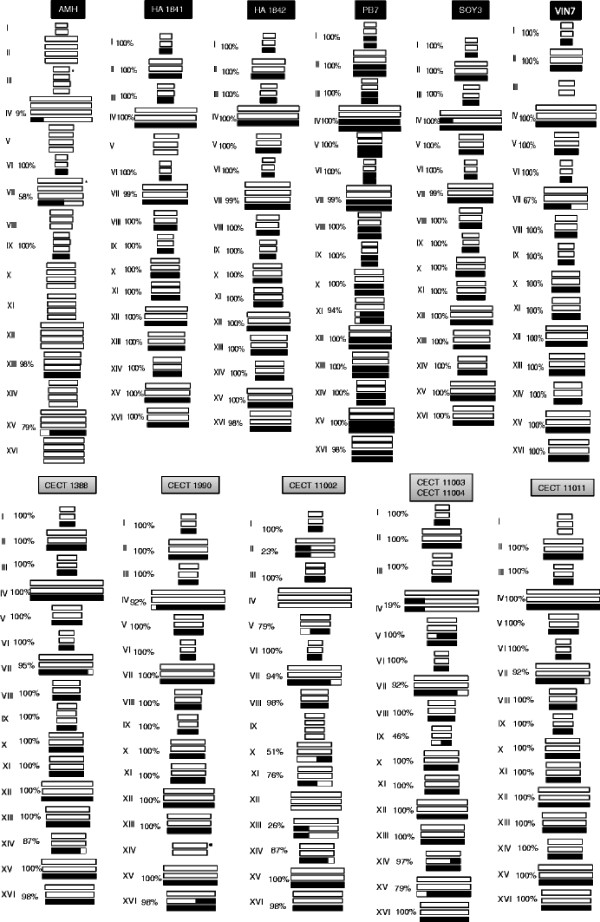
Final genome compositions were inferred for all hybrids as depicted in Figure 1, taking into consideration flow cytometry data, aCGH and PCR-RFLP data. For example, in the case of the partial allotetraploid hybrid AMH, its caryoscope shows four different hybridization levels, which correspond to 2 copies of S. cerevisiae genes located in chromosomes (chr.) I and VI; 3 copies of S. cerevisiae genes located in chr. VIII, IX and XIII, short chr. IV left region, chr. VII left arm, and chr. XV right arm; 4 copies of S. cerevisiae genes located in chr. II, III left, IV right, V, VII right, X, XI, XII, XIV, XV left, and XVI; and 5 copies of S. cerevisiae genes located in chr. III left region and in a segment of chr. VII (Figure 1; Additional file 3: Figure S2).
Figure 1.
Genome composition of hybrids deduced from aCGH analysis, ploidy estimates and a previous analysis of absence/presence of parental genes by RFLP analysis [[2],[19]]. White and black bars are used to represent the S. cerevisiae and S. kudriavzevii genome fractions, respectively. Chromosomes showing black and white regions correspond to chimerical chromosomes. The percentages of S. kudriavzevii genes maintained in each chromosome are shown for each chormosome. Strains names are depicted on a black or a gray background corresponding to wine or brewing strains, respectively. Asterisks in AMH Chr. III and VII indicate regions where non-reciprocal translocations or segmental duplications can be present.
According to this combined analysis, 11 different patterns were differentiated in the 12 hybrids under analysis. As a general rule, different degrees of loss of S. kudriavzevii gene content in most hybrids were observed. Only the allotetraploid hybrid PB7 maintains a complete diploid set of chromosomes from each parental species, with the exception of a small segment located in the left arm of chromosome XI of the S. kudriavzevii subgenome. On the contrary, the largest reduction of the S. kudriavzevii gene content is observed in the partial allotetraploid hybrid AMH, which lost 72% of the S. kudriavzevii genes. The rest of hybrids, all of them allotriploid, showed intermediate situations derived from ancestors containing a diploid set of S. cerevisiae chromosomes and a haploid set of S. kudriavzevii chromosomes.
These combined analyses also allowed us to detect different types of chromosome rearrangements present in hybrids: i) the complete loss of a S. kudriavzevii parental chromosome compensated by an extra copy of the S. cerevisiae chromosome (chr. II,,III, V, X, XI, XII, XIV and XVI in AMH; chr. V in HA1841; chr. IV, IX and XII in CECT 11002; chr. I in CECT 11011); ii) aneuploidies (chr. I, VI and VIII in AMH; chr. IX in CECT 1388; chr. XIV in CECT 1990; chr. IX in CECT 11002; chr. III and V in CECT 11003 and CECT 11004; chr. III in VIN7), and iii) the presence of chimerical chromosomes (chr. IV, VII and XV in AMH; chr. XI in PB7; chr. IV in SOY3; chr. VII in VIN7; chr. VII and XIV in CECT 1388; chr. IV and XVI CECT 1990; chr. II, V, VII, X, XI, XIII and XIV in CECT 11002; chr. IV, V, VII, IX, XIV and XV in CECT 11003 and CECT 11004; and chr. VII in CECT 11011); (see Figure 1).
These chimerical chromosomes are characterized by over- and underrepresented regions evidenced as up and down jumps in the log2 ratio in the caryoscopes, which are indicative of probable non-reciprocal recombination events between homeologous chromosomes (homologous from different species) (Table 3). The recombination sites in the chimerical chromosomes were mapped according to the genome browser from Saccharomyces genome database (SGD). Using a windows size of 15–20 Kb (four genes in the left and right of the most plausible recombination point) we found Ty elements, ARS sequences, clusters of homologous regions (CHRs) and tRNA elements that may have facilitated the recombination of the two homologous parental chromosomes (Table 3). In several cases, a common recombination site was observed in chromosomes belonging to two or more hybrids, indicative of common ancestry. This is the case of chromosomes IV, V, IX, XIV and XV in brewing hybrids CECT 11003 and 11004; chromosome XIV in CECT 1388 and 11002; chromosome XV in CECT 11003, 11004 and AMH and chromosome VII in hybrids CECT 11003, CECT 11004, CECT 11002, CECT 11011 and CECT 1388 (Table 3 and Additional file 3: Figure S2).
Table 3.
List of chimerical chromosome (CC) types found in the different S. cerevisiae × S. kudriavzevii hybrids
| Chr. | CC type | Strains | Breakpoint mapping interval | Putative recombining sequences |
|---|---|---|---|---|
| II |
type 1 |
CECT 11002 |
YBL018C-YBL011W |
Ty1 LTR, Ty3 LTR, tRNA-Ile, tRNA-Gly, ARS |
| IV |
type 1 |
W27, W46, 441, SPG16-91, CECT 11003, CECT 11004 |
YDL095W |
PMT1 (ref. [13]) |
| |
type 2 |
AMH, SOY3 |
YDL185W-YDL179W |
CHR 12 |
| |
type 3 |
CECT 1990 |
YDL185W-YDL179W |
CHR 12 |
| V |
type 1 |
W27, W46, 441, SPG16-91, CECT 11003, CECT 11004 |
YER006W |
NUG1 (ref. [13]) |
| |
type 2 |
CECT 11002 |
YEL018C-YEL011W |
Ty1 LTR, Ty4 LTR, tRNA-Gln |
| VII |
type 1 |
W46,CECT 11003, CECT 11004, CECT 11002, CECT 11011, CECT 1388 |
YGR249W-YGR244C |
ARS, CHR 29 |
| |
type 2 |
AMH |
YGR062C-YGR058W |
CHR 30 |
| |
type 3 |
VIN7 |
YGR106C-YGR112C |
tRNA-Leu, tRNA-Lys, Ty1 LTR, tRNA-Cys, Ty3 LTR, ARS |
| IX |
type 1 |
W27, W46, 441, SPG16-91, CECT 11003, CECT 11004 |
YIL053W |
RHR2-RPL34B (ref. [13]) |
| X |
type 1 |
CECT 11002 |
YJL039C-YJL036C |
tRNA-Asp, tRNA-Arg, Ty1 LTR, ARS, tRNA-Val |
| XI |
type 1 |
CECT 11002 |
YKR025C-YKR028W |
Ty1 LTR |
| |
type 2 |
PB7 |
YKL203C-YKL204W |
ARS |
| XIII |
type 1 |
CECT 11002 |
YML012C-YML009W-B |
CEN13, ARS |
| XIV |
type 1 |
W27, W46, 441, SPG16-91, CECT 11003, CECT 11004 |
YNR001C |
CEN14 (ref. [13]) |
| |
type 2 |
CECT 1388, CECT 11002 |
YNR029C-YNR032W |
ARS |
| XV |
type 1 |
W27, W46, 441, SPG16-91, CECT 11003, CECT 11004,AMH |
YOL053W |
THI20-PSH1 (ref. [13]) |
| XVI | type 1 | CECT 1990 | YPR007C-YPR011C | Ty1LTR, tRNA-Gly, tRNA-Lys, |
Chr., chromosome number; CHR, cluster of homology region. Strain names in italics correspond to wine hybrids and in bold to brewing hybrids. Some recombination sites were described elsewhere [13], as indicated.
S. cerevisiae gene depletions in hybrids
Although hybrids maintain in their genomes at least a complete set of S. cerevisiae chromosomes, aCGH data from all hybrids analyzed in this work, as well as from those previously analyzed [13], can be used to determine the common fraction of S. cerevisiae genes showing gene copy variations in hybrids compared to the reference strain S288c. A common set of genes showing the same copy number variations in hybrids may be indicative of common origins.
The analysis of the S. cerevisiae gene content from all hybrids revealed the presence of less copies of a common set of genes. Among them, the most interesting were CUP1, ASP3, and ENA gene families, as well as Ty elements and 13 ORFs of unknown function (Additional file 4: Table S2). In general, copy variations in the S. cerevisiae genome fraction of the hybrids were found in genes located in subtelomeric regions (Additional file 3: Figure S2), although in some cases involve genes located in intrachromosomal regions, such as CUP1.
Short segment amplifications were also detected in the aCGH analysis. This was the case of hybrid AMH that showed three short region amplifications in chr. III, VII and XIII. The higher hybridization signals of genes located in the two first regions could be postulated as indicative of the presence of chimerical chromosomes, however according to the previous PCR-RFLP analysis S. kudriavzevii genes were absent. Other amplifications of S. cerevisiae segments located in chromosome XVI are observed in hybrids CECT 1388 (between genes YPL159C and YPL126W) and CECT 11002 (between YPL141C and YPL126W). Finally, a deleted region was found in one of the two copies of S. cerevisiae chromosome XIV from strain CECT 1990 (between loci YNR013C and YNR031C) (Additional file 3: Figure S2).
S. kudriavzevii gene content and Gene Ontology (GO) analyses
Data obtained from all hybrids analyzed in this work as well as from those previously analyzed [13] were also used to evaluate the presence of common S. kudriavzevii genes (Additional file 5: Table S3). These common set of genes could be interesting to unveil potentially genes of adaptive value in hybrids.
As a general rule, most hybrids maintained around 90% of the S. kudriavzevii genome, with the exception of the brewing strain CECT 11002 and the wine strain AMH which only maintain 56.9% and 30.5% respectively.
To determine if a group of S. kudriavzevii genes associated with particular cellular components, molecular functions or biological processes may have been maintained in all hybrids due to potential adaptive value, four different gene ontology (GO) term enrichment analyses were performed (Additional file 6: Table S4). The first analysis included all wine and brewing hybrids. Due to the low representation of the S. kudriavzevii genome fraction in AMH, this strain was removed from this first analysis. Gene ontology analysis was also separately performed according to the source of isolation of hybrids, wine and brewing fermentations. GO terms showing significant values were sorted according to their corresponding GO categories (Additional file 6: Table S4). Table 4 shows only those significantly represented GO terms of putative importance for wine or brewing fermentations.
Table 4.
Summary of the most relevant metabolic pathways and biological processes obtained after Gene Ontology analysis using the S. kudriavzevii genes retained in each group of hybrids
| Group of hybrids | GO ID | GO Term | Npresent/Nmeasured | % | p-value |
|---|---|---|---|---|---|
|
WINE |
6487 |
Protein amino acid N-linked glycosylation |
36/42 |
85.7 |
0.013 |
| |
6839 |
Mitochondrial transport |
10/10 |
100 |
0.033 |
| |
|
Ergosterol Biosynthesis |
17/19 |
89.5 |
0.049 |
|
BREWING |
6487 |
Protein amino acid N-linked glycosylation |
28/42 |
66.7 |
0.017 |
| |
|
Fatty acid elongation saturated |
4/4 |
100 |
0.039 |
| |
|
Glycine serine and threonine metabolism |
27/42 |
64.3 |
0.03 |
| |
|
Arginine_and_proline_metabolism |
16/23 |
69.6 |
0.049 |
| |
|
Sulfur_Degradation |
4/4 |
100 |
0.048 |
|
ALL |
6487 |
Protein amino acid N-linked glycosylation |
25/42 |
59.5 |
0.003 |
| |
15908 |
Fatty acid transport |
4/4 |
100 |
0.025 |
| |
|
Glutamate metabolism |
15/27 |
55.6 |
0.046 |
| |
|
Sulfur metabolism |
8/11 |
72.7 |
0.021 |
| |
|
NAD salvage pathway |
5/6 |
83.3 |
0.027 |
| |
|
Sulfate assimilation pathway II |
5/6 |
83.3 |
0.019 |
|
AMH |
6972 |
Hyperosmotic response |
5/7 |
71.4 |
0.036 |
| |
9331 |
Glycerol 3 phosphate dehydrogenase complex |
3/3 |
100 |
0.033 |
| |
|
Histidine biosynthesis |
5/7 |
71.4 |
0.039 |
| Fatty acid metabolism | 11/17 | 64.7 | 0.010 |
Due to the massive S. kudriavzevii gene losses in AMH, this strain was not included in any grouping, and hence, analyzed alone.
Significantly represented GO terms common to both wine and brewing hybrids mainly corresponded to genes related to fatty acid metabolism (particularly transport), sulfur metabolism and the NAD+ salvage pathway. Genes associated with amino acid metabolism (N-linked glycosylation and glutamate metabolism) were also represented (Table 4).
GO terms related to amino acid N-linked glycosilation were also significantly present in hybrids from wine and brewing analyzed independently. Moreover, GO terms associated with ergosterol biosynthesis and mitochondrial transport were also significantly detected in wine hybrids; while those related to metabolism of amino acids such as glycine, threonine, arginine and proline, sulfur metabolism, as well as fatty acid elongation were significant present in brewing strains (Table 4). Finally, an independent analysis of significant GO terms for AMH hybrid revealed the presence of genes involved in hyperosmotic response, glycerol-3-phosphate dehydrogenase complex, histidine biosynthesis and fatty acid metabolism (Table 4).
Phylogenetic relationships among hybrids
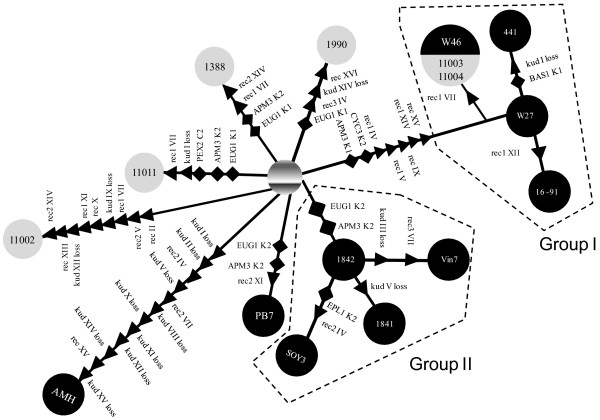
A maximum parsimony tree was constructed based in presence/absence of chromosomes and chromosome regions data obtained for each particular genetic event in all analyzed hybrids. The tree topology revealed the presence of two main groups containing most allotriploid hybrids, particularly those from wine (Figure 2).
Figure 2.
Maximum parsimony tree indicating the minimum number of chromosomal rearrangements and restriction site changes (presence/absence matrix is given in Additional file2: Table S1) necessary to connect the different genotypes exhibited by the S. cerevisiae × S. kudriavzevii hybrids to a putative hybrid ancestor. This putative ancestor is not necessarily the same for all lineages, it just corresponds to an ancestral state containing the complete S. cerevisiae and S. kudriavzevii genomes, but it could be generated several times from different parental strains, as discussed in the main text. Genotypes are represented by white and gray circles for wine and brewing hybrids, respectively. Rearrangements are indicated by arrows giving the direction of the irreversible change and were treated under the Camin-Sokal criterion. Rearrangements were assumed to be caused by nonreciprocal recombination (rec) among homoeologous chromosomes (roman numbers) and whole chromosome losses (loss) of one of the parental chromosomes (kud, S. kudriavzevii). Restriction site changes can be reversible (gains/losses represented by diamonds) and were treated under the Wagner criterion. The gene region and the restriction patterns involved are also indicated (for a description see references [2] and [19]).
Group I was constituted by Swiss wine strains W46, 441, W27 and SPG 16-91 as well as the brewing strains CECT 11003 and CECT 11004. This group is supported by the presence of five shared chimerical chromosomes as well as the CYC3 K2 allele [2].
Group II includes the remaining allotriploid wine hybrids HA1841, HA 1842, VIN7 and SOY3. This group is only supported by the common presence of S. kudriavzevii K2 alleles for genes EUG1 and APM3[19], and the possession of a higher fraction of S. kudriavzevii genome.
The rest of the allotriploid hybrids, isolated from brewing, and the wine allotetraploid PB7 and AMH strains, appeared in separated branches with strain-specific chromosomal rearrangements. The only exception is the shared loss of S. kudriavzevii chr. XII between the partial allotetraploid AMH and the allotriploid CECT 11002, which can be considered a convergent event. PB7 also shared similar restriction alleles with Group II but this strain is also allotetraploid (Table 2).
This most parsimonious tree shows several convergent events, such as chromosomal losses, chromosomal rearrangements and restriction site changes (evidencing different allelic variants). S. kudriavzevii chr. I seems to have been lost independently in hybrids SPG 441, CECT 11011, and AMH. In a similar way, the lack of chr. V in hybrids HA 1841 and AMH, and chromosome XIV in CECT 1990 and AMH seem to be independent events according to this parsimony analysis.
Convergent events involving recombinant chromosomes were also found. This is the case of the type 2 recombination in chr. IV (shared by AMH and SOY3), type 1 recombination in chr. VII (shared by CECT 11002, CECT 11011, 1388, W46, CECT 11003 and CECT 11004), type 2 recombination in chr. XIV (CECT 1388 and CECT 11002) and the recombinant chr. XV (AMH and Group I hybrids). This could be indicative of the presence of recombination hotspots in the Saccharomyces genomes.
Discussion
The genome diversity in S. cerevisiae × S. kudriavzevii hybrids
The genome composition of 11 new wine and brewing S. cerevisiae x S. kudriavzevii hybrid strains was described in this work by means of aCGH analysis. Additionally, a comparison between them and other four wine hybrids already described by [13] was also performed. Individual and differential chromosomal composition patterns were found for each particular strain, except for brewing strains CECT 11003 and CECT 11004 which appear closely related to the previously described Swiss wine hybrids [13]. The close relationships between wine hybrid strains from Switzerland and the brewing strains CECT 11003 and 11004 was already observed in a previous study based on PCR-RFLP analysis of hybrids as well as in the phylogenetic reconstruction based on COX2 sequences [19]. In that work, a recombination in chromosome XV was proposed as the unique difference between strains 11003 and 11004; however, aCGH analysis carried out in this study demonstrated that this recombination is present in both strains (Figure 2). These Swiss wine hybrids were previously described as diploids [13] on the basis of ploidy estimations with propidium iodide. However, in the reanalysis of ploidy with SYTOX Green, they also resulted to be allotriploids as CECT 11003 and CECT 11004.
Flow cytometry results with SYTOX Green were in accordance with genome structure deduced from aCGH analysis carried out in this work and with the presence/absence of parental genes deduced from a previous PCR-RFLP analysis of hybrids [19]. Most S. cerevisiae × S. kudriavzevii hybrid strains were allotriploids, with the exception of AMH and PB7 which were allotetraploids. Some aneuploidies were also found in several hybrids. Aneuploidies seem to be common in Saccharomyces hybrids since this phenomenon have also been observed in S. cerevisiae × S. bayanus hybrids [22,23]. The role of aneuploidies in the hybrid genomes is not clear, but their presence in S. cerevisiae affected both the transcriptome and proteome, generating significant phenotypic variation and bringing fitness gains under diverse conditions [24].
Recently, the hybrid genome of VIN7, one the hybrids analyzed in the present study, has completely been sequenced [25], concluding that this strain is an almost perfect allotriploid hybrid that contains a heterozygous diploid S. cerevisiae genome and a haploid S. kudriavzevii genome. The genome constitution of VIN7 deduced from the sequencing analysis is basically similar to the one inferred by aCGH in the present study, but there are some differences. The genome sequence analysis detected a homeologous recombination generating a chimerical chromosome VII, a genomic substitution of a region of 15 kb, of S. kudriavzevii genomic DNA from chromosome IV by the orthologous sequences from S. cerevisiae and a genomic substitution of a 13 kb region of S. cerevisiae genomic DNA from chromosome IV by S. kudriavzevii sequences combined with homeologous recombination between the S. kudriavzevii and S. cerevisiae alleles. The first rearrangement involving a chimerical chromosome VII was clearly detected in the aCGH analysis, but not the two genomic substitutions. Both genomic substitution involve short segmental replacements of a few genes (7 and 8), and the second an almost reciprocal recombination between homeologous chromosomes that cannot be observed by aCGH analysis. However, the presence/absence analysis of parental genes in hybrids [19] detected the loss of S. kudriavzevii chromosome III in our VIN7. As an ongoing project, our group is also sequencing the whole genome of several S. cerevisae × S. kudriavzevii hybrids, including the commercial VIN7 yeast. We checked in the preliminary sequencing of our VIN7 strain for the presence of S. kudriavzevii chromosome III sequences and the result was negative, confirming our aCGH results and indicating that our VIN7 strain is different. These differences may be due to the fact that our VIN7 strain was isolated from a commercial dry yeast sample provided by Anchor Yeast but Borneman et al. [25] sequenced the original mother culture of VIN7, as they mention in their acknowledgements. Therefore, the continuous propagation of this yeast in molasses under aerobic conditions to obtain commercial dry yeasts may have promoted a new chromosomal rearrangement, the loss of the S. kudriavzevii chromosome III.
Taking into consideration the ploidy data as well the fact that most hybrids possess either trisomic (2 S. cerevisiae chromosomes: 1 S. kudriavzevii chromosome) or tetrasomic chromosomes (2 S. cerevisiae chromosomes: 2 S.kudriavzevii chromosomes), two scenarios on the hybridization process are plausible. In the case of allotriploid hybrids, the simplest explanations for their origins are hybridization events by rare-mating between a diploid cell of S. cerevisiae and a haploid cell or spore of S. kudriavzevii. This is also supported by the genome sequencing of VIN7, one of the allotriploid strains, which resulted to contain heterozygous diploid genome from S. cerevisiae and a haploid genome from S. kudriavzevii[25].
On the other hand, diploid and diploid cell rare-mating between S. cerevisiae and S. kudriavzevii should be invoked to explain the origin of allotetraploid hybrids. In the case of PB7 it was observed high spore viability (95%) due to the presence of the two chromosomes copies of each parental strain.
Rare-mating between diploid cells was already proposed as a probable mechanism for hybrids generation [13,26]. However, haploid cell or spore mating between S. cerevisiae and S. kudriavzevii, followed by whole genome duplications due to endoreplication or chromosome duplications due to non-disjunction, and subsequent chromosomal rearrangements, although less plausible, cannot totally be discarded.
Characterization of the S. kudriavzevii subgenome from hybrids
According to Sipiczki [26], genomes from each parental species interact in the new hybrid genome. This interaction can be observed in the loss of large parts of one or both genomes as well as in the presence of chimerical chromosomes that make the hybrid genome as stable as possible to future genetic modifications. Additionally, adaptive evolution of these hybrid genomes under fermentative environmental conditions could make hybrid genome to conserve the chromosomes, or part of them, which grant a selective advantage [27]. According to the results obtained in this work as well as in our previous studies [2,13,19], S. cerevisiae × S. kudriavzevii hybrids seem to have the common trend to lose the S. kudriavevii parental chromosomes maintaining the S. cerevisiae ones. The reduction of the non-S. cerevisiae genome observed in both wine and brewing S. cerevisiae × S. kudriavzevii hybrids was already reported for artificial S. cerevisiae × S. uvarum hybrids genetically stabilized by successive sporulation steps [28]. In contrast, S. pastorianus (S. cerevisiae × S. eubayanus hybrids) Group 1 strains obtained from different brewing processes and studied by aCGH analysis, showed a trend to lose the S. cerevisiae genome fraction [22]. The cause of the predominance of one or the other parental genome in the hybrids remains unclear yet. However, selective pressures acting under harsh environmental conditions and cytonuclear interactions have been suggested as the main factors affecting the genome conformation of hybrids. In S. cerevisiae × S. eubayanus lager strains, supposed to be naturally selected after years of use in brewing, the predominance of a S. eubayanus-like genome has been related to the maintenance of the S. eubayanus mitochondria [22,29]. However, artificial hybrids constructed from the same two parental species, but without selective pressures, inherited their mitochondrial genome from either one or the other parental species randomly [29,30]. The conservation of the mitochondrial genome from the parental species most represented in the nuclear genome was also observed in the stable artificial S. cerevisiae × S. uvarum hybrids, which maintained the mitochondrial genome of the S. cerevisiae parental strain [28]. All S. cerevisiae × S. kudriavzevii natural hybrids analyzed in this work, except for AMH, maintained a S. kudriavzevii mitochondrial genome [2,19]. However, S. cerevisiae × S. kudriavzevii artificial hybrids, randomly inherited the S. cerevisiae or the S. kudriavzevii mitochondrial DNA (Pérez-Través et al. personal communication). This discrepancy between the mtDNA inheritance in artificial vs. natural hybrids has been associated with the result of an unwitting human-driven selection of naturally generated hybrid strains for fermentations at low temperature [29]. A common origin for all hybrids could be another possible explanation, but the present analysis of the genome constitutions in hybrids suggests diverse origins.
Interestingly, the hybrid AMH, which maintained the S. cerevisiae mitochondria, has lost a 69% of the nuclear genes of S. kudriavzevii coding for proteins with functions associated to the mitochondria; while the rest of the analyzed hybrids with S. kudriavzevii mitochondria have lost only 0.67%–42.48% of the S. kudriavzevii genes related to mitochondrial functions. Due to the fact that a number of mitochondrial proteins encoded in the nuclear genome play an important role in the mtDNA replication and transmission, both the type of mitochondrial DNAs and the functions of the mitochondria in a hybrid strain are clearly under the control of the nuclear genome [31]. One of the most interesting evidence about nuclear-mitochondrial genome interactions were described by Lee et al. [32], who demonstrated that the presence of the S. bayanus nuclear gene AEP2 together with the S. cerevisiae mitochondrial gene OLI1 cause a cytonuclear incompatibility. More recently, Chou et al. [33] identified other two genes, MRS1 and AIM22, associated with cytonuclear incompatibility among S. cerevisiae, S. paradoxus and S. bayanus. A similar behavior involving the same or other different genes in S. cerevisiae × S. kudriavzevii hybrids was not yet demonstrated.
aCGH and GO analysis carried out with those S. kudriavzevii genes conserved in all S. cerevisiae × S. kudriavzevii hybrids with S. kudriavzevii mitochondria (excluding AMH) evidenced a significant enrichment in nuclear genes related to mitochondrial function (a total of 328 genes) supporting the hypothesis of a necessary interaction between the S. kudriavzevii nuclear-encoded proteins and the mitochondrial genomes or their products. Taking into consideration that a total of 751 proteins encoded by the nuclear genome are associated with the mitochondrial function in S. cerevisiae[34], and considering a similar number in S. kudriavzevii, we can assume that the remaining genes up to 751 might be non-essential for the maintenance of the S. kudriavzevii mitochondria in hybrids. In particular the S. kudriavzevii gene AEP2 reported by Lee et al. [32] was not common to all analyzed hybrids, indicating that different incompatible nuclear-mitochondrial pair of genes could be associated with each particular pair of Saccharomyces parental species involved in hybrid generation.
GO analysis was also very informative with regards to the conservation in hybrids of particular groups of genes, inherited from each parental species, that may be potentially related to adaptive advantage for fermentation at low temperatures. A significant overrepresentation of S. kudriavzevii genes associated with the physiological adaptation of yeasts to grow at low temperatures, such as fatty acid transport and N-glycosilation of proteins in all hybrids, and ergosterol biosynthesis in the case of wine hybrids [35-37] was observed (Table 4). Changes in membrane fluidity are the primary signal triggering the cold shock response [35]. This response involves certain groups of genes: members of the DAN/TIR family of cell-wall mannoproteins, genes coding for temperature inducible protein (TIP1) and seripauperins (PAU), genes related to ergosterol and phospholipid synthesis (ERG, INO1 and OPI3) and the gene coding for the only known desaturase in S. cerevisiae (OLE1), among others [35]. These sets of genes are present in the S. kudriavzevii subgenome of all hybrids analyzed in this work, with some exceptions mainly involving AMH (Table 4 and Additional file 6: Table S4).
Our results are in agreement with results about stress olerance, including adaptation to low temperatures, previously obtained in our laboratory using some of the S. cerevisiae × S. kudriavzevii hybrids analyzed in this work [11,38]. Physiological implications of possessing S. kudriavzevii genes in those particular functions or metabolic pathways must be elucidated in future studies involving both transcriptomic and metabolomic analyses.
Wine yeast signatures in the S. cerevisiae subgenome from hybrids
An interesting result obtained from aCGH analysis was the detection of a common set of S. cerevisiae genes that are in lower copies in the genome of all S. cerevisiae × S. kudriavzevii hybrids (Additional file 4: Table S2). This finding might indicate that the S. cerevisiae parental strains involved in the different hybridization events shared a similar genetic background and were closely related yeasts.
Using a similar methodology, a trend to loss some particular set of genes in S. cerevisiae wine strains, with regards to strains belonging to the same species but isolated from different sources, was previously demonstrated [39,40]. Dunn et al. [39] proposed the term “commercial wine yeast signature” to refer to this set of genes. Most of these genes that are frequently depleted in wine strains are also depleted in the S. cerevisiae fraction of the hybrid genomes of all hybrids. This finding supports the hypothesis that these hybrids have likely been generated from wine S. cerevisiae parental strains.
On the origin of hybrids
The maximum parsimony analysis of the relationships between the wine and beer hybrids are congruent with diverse origins for the strains according to chromosomal rearrangement differences, mainly due to the presence of chimerical chromosomes, and S. kudriavzevii chromosome losses, in some cases compensated by the presence of an extra copy of the homeologous S. cerevisiae chromosome (Figure 2).
While the brewing strains seem to represent different and divergent lines (except strains CECT11003 and 11004), most wine hybrids clustered in two main groups of strains sharing common events, with the exception of AMH and PB7 that were independently originated. Brewing strains CECT 11003 and 11004 shared the same genome than wine hybrid W46 probably evidencing that either an original strain with this common genome structure was introduced in both fermentative processes, or colonize one fermentative process from the other. The parsimony tree obtained in this study is congruent with previous phylogenetic reconstructions of hybrids based on COX2 sequences [19].
The occurrence of several chimerical chromosomes sharing similar—if not the same—recombination points, common to some S. cerevisiae × S. kudriavzevii hybrids located in different branches of the parsimony tree, indicates the presence of recombination hot spots. Recombination between homeologous chromosomes are probably mediated by highly recombining regions located in the recombination sites, such as ARS sequences [41], Ty elements [42], Y’ elements, rRNA regions and conserved coding genes [13,43]. When recombination is initiated in a region with high homology, the mismatch repair system (MMR) stimulates the loss of one partner of the recombination event in the hybrids and the fixation of the other, thus generating a chimerical recombinant chromosome. With the exception of the almost perfect allotetraploid PB7, hybrids have low spore viability (<1%) indicating that they are maintained by mitotic budding. Therefore, mitotic homeologous recombination, although much less frequent than meiotic, may also explain the generation of chimerical chromosomes.
The genome composition of hybrids reveals that the ancestral hybrid strains were allotriploid or allotetraploid, resulting from rare mating between diploid S. cerevisiae and haploid or diploid S. kudriavzevii[4,25]. The presence of triple hybrids also supports this hypothesis [1,19]. Finally, the presence of S. kudriavzevii alleles shared between most hybrids and the European S. kudriavzevii population [10], as well as the presence of the gene GAL4 from S. kudriavzevii[2,19], which is a functional gene in the European populations of S. kudriavzevii but a pseudogene in the Japanese strains [44], indicate that these hybrids were originated from a European S. kudriavzevii parental strain.
Conclusions
Hybridization between S. cerevisiae and S. kudriavzevii have occurred several times by rare-mating between different wine S. cerevisiae diploid and European S. kudriavzevii haploid or diploid progenitors. After hybridization, the hybrid genome suffered random genomic rearrangements mediated by crossing-over between homologous chromosomes and non-disjunction, promoting the loss of variable fractions of the parental subgenomes. Both the restrictions imposed byinteractions between both parental genomes as well as between nuclear and mitochondrial genomes, together with the selective environmental conditions prevailing during fermentation modulated the final composition of the hybrid genomes, characterized by the maintaining of the S. cerevisiae genome and the progressive reduction of the S. kudriavzevii contribution.
Competing interests
The author(s) declare that they have no competing interests.
Authors’ contributions
This study is the result of the collaboration between AQ and EB laboratories. CB, AQ and EB conceived and supervised this study. DP, CB, AQ and EB designed the experiments. DP and CL performed the experimental work and data analyses. DP and CB wrote the first version of the manuscript. CL, AQ and EB participated in the final manuscript revision. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Caryoscope representation of microarray data of S.kudriavzevii IFO1802. Array CGH data are shown in numerical order with chromosome I at the top and chromosome XVI at the bottom. Red signal indicates hybridization signal but it’s important to note the high normalization factor applied to the red signal (2) and the correction applied to the green one (0.49). This figure indicate that no cross hybridization has occurred between S. kudriavzevii genes and S. cerevisiae genes. (PPTX 106 kb)
able S1. Binary table showing the presence/absence of a particular event for each hybrid strain (coded as 1/0 respectively). kud, S. kudriavzevii; loss, chromosome loss; rec, recombination generating a chimerical chromosome. Roman numerals indicate chromosome numbers and Arabic numerals the types of chimerical chromosomes or the restriction sites in the analysis of RFLP patterns in 34 genes. Parsimony tree was computed using a mixture model in which chimerical chromosomes, S. kudriavzevii chromosome losses and double S. cerevisiae chromosome were considered under the Camin-Sokal criterion and restriction site gains/losses under the Wagner criterion. (XLS 32 kb)
Figure S2. Caryoscope representation of microarray data of 11 S. cerevisiae × S.kudriavzevii hybrids. Array CGH data are shown in numerical order with chromosome I at the top and chromosome XVI at the bottom for each strain. Regions with higher red signal correspond to S. cerevisiae genome regions that are overrepresented in the hybrid genome and those with higher green signal to those regions that are underrepresented. aCGH of wine and brewery hybrids are depicted on black and gray backgrounds, respectively. Arrows indicate potential non-reciprocal recombination events between homeologous chromosomes involved in the generation of chimerical chromosomes. (PPT 607 kb)
Table S2. List of S. cerevisiae genes depleted in all S. cerevisiae × S. kudriavzevii hybrids under analysis. (XLS 29 kb)
Table S3. S. kudriavzevii gene composition for each hybrid. A cross indicates the putative presence of that gene by considering colinearity between S. kudriavzevii and S. cerevisiae genomes. R indicates interspecies recombinant genes. Wine and brewing hybrids are indicated in black and gray lettering, respectively. (XLS 2001 kb)
Table S4. Metabolic pathways and biological processes obtained from a Gene Ontology analysis using the S. kudriavzevii genes retained in each hybrid grouping. (XLS 38 kb)
Contributor Information
David Peris, Email: david.peris-navarro@uv.es.
Christian A Lopes, Email: clopes@conicet.gov.ar.
Carmela Belloch, Email: belloch@iata.csic.es.
Amparo Querol, Email: aquerol@iata.csic.es.
Eladio Barrio, Email: eladio.barrio@uv.es.
Acknowledgements
This work was supported by Spanish Governments projects AGL2009-12673-CO2-01 and AGL2009-12673-CO2-02 to AQ and EB respectively, and by grant PROMETEO/2009/019 from Generalitat Valenciana to AQ, EB and CB. DP and CL acknowledges to the Spanish Government for a FPI fellowship and a postdoctoral contract, respectively. We also thank Ksenija Lopandić, José Manuel Álvarez-Pérez, Sandi Orlić, Lallemand Bio and Anchor Wine Yeasts for providing yeast strains.
References
- González SS, Barrio E, Gafner J, Querol A. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 2006;6:1221–1234. doi: 10.1111/j.1567-1364.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- González SS, Barrio E, Querol A. Molecular characterization of new natural hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii from brewing. Appl Environ Microbiol. 2008;74:2314–2320. doi: 10.1128/AEM.01867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopandić K, Gangl H, Wallner E, Tscheik G, Leitner G, Querol A, Borth N, Breitenbach M, Prillinger H, Tiefenbrunner W. Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 2007;7:953–965. doi: 10.1111/j.1567-1364.2007.00240.x. [DOI] [PubMed] [Google Scholar]
- de Barros Lopes M, Bellon JR, Shirley NJ, Ganter PF. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 2002;1:323–331. doi: 10.1111/j.1567-1364.2002.tb00051.x. [DOI] [PubMed] [Google Scholar]
- Groth C, Hansen J, Piškur J. A natural chimeric yeast containing genetic material from three species. Int J Syst Bacteriol. 1999;49:1933–1938. doi: 10.1099/00207713-49-4-1933. [DOI] [PubMed] [Google Scholar]
- Liti G, Peruffo A, James SA, Roberts IN, Louis EJ. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast. 2005;22:177–192. doi: 10.1002/yea.1200. [DOI] [PubMed] [Google Scholar]
- Masneuf I, Hansen J, Groth C, Piškur J, Dubourdieu D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl Environ Microbiol. 1998;64:3887–3892. doi: 10.1128/aem.64.10.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libkind D, Hittinger CT, Valério E, Gonçalves C, Dover J, Johnston M, Gonçalves P, Sampaio JP. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci. 2011;108:14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio JP, Gonçalves P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and sympatric with S. cerevisiae and S. paradoxus. Appl Environ Microbiol. 2008;74:2144–2152. doi: 10.1128/AEM.02396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes CA, Barrio E, Querol A. Natural hybrids of Saccharomyces cerevisiae × Saccharomyces kudriavzevii share alleles with European wild populations of S. kudriavzevii. FEMS Yeast Res. 2010;10:412–421. doi: 10.1111/j.1567-1364.2010.00614.x. [DOI] [PubMed] [Google Scholar]
- Arroyo-López FN, Orlić S, Querol A, Barrio E. Effects of temperature, pH and sugar concentration on the growth parameters of Saccharomyces cerevisiae, S. kudriavzevii and their interspecific hybrids. Int J Food Microbiol. 2009;131:120–127. doi: 10.1016/j.ijfoodmicro.2009.01.035. [DOI] [PubMed] [Google Scholar]
- Salvadó Z, Arroyo-López FN, Barrio E, Querol A, Guillamón JM. Quantifying the individual effects of ethanol and temperature on the fitness advantage of Saccharomyces cerevisiae. Food Microbiol. 2011;28:1155–1161. doi: 10.1016/j.fm.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Belloch C, Pérez-Torrado R, González SS, Pérez-Ortín JE, García-Martínez J, Querol A, Barrio E. Chimeric genomes of natural hybrids of Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Appl Environ Microbiol. 2009;75:2534–2544. doi: 10.1128/AEM.02282-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- Querol A, Barrio E, Ramón D. A comparative study of different methods of yeast-strain characterization. Syst Appl Microbiol. 1992;15:439–446. doi: 10.1016/S0723-2020(11)80219-5. [DOI] [Google Scholar]
- Brazma A. ArrayExpress, a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CL, Dunham MJ, Kung SY, Troyanskaya OG. Accurate detection of aneuploidies in array CGH and gene expression microarray data. Bioinformatics. 2004;20:3533–3543. doi: 10.1093/bioinformatics/bth440. [DOI] [PubMed] [Google Scholar]
- Cliften PF, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Peris D, Belloch C, Lopandić K, Álvarez-Pérez JM, Querol A, Barrio E. The molecular characterization of new types of S. cerevisiae × S. kudriavzevii hybrid yeasts unveils a high genetic diversity. Yeast. 2012;29:81–91. doi: 10.1002/yea.2891. [DOI] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: Phylogeny Inference Package. v. 369. University of Washington, Seattle; 2005. [Google Scholar]
- Dunn B, Sherlock G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 2008;18:1610–1623. doi: 10.1101/gr.076075.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao Y, Kanamori T, Itoh T, Kodama Y, Rainieri S, Nakamura N, Shimonaga T, Hattori M, Ashikari T. Genome sequencing of the lager brewing yeast, an interspecies hybrid. DNA Res. 2009;16:115–129. doi: 10.1093/dnares/dsp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ. The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS Yeast Res. 2012;12:88–96. doi: 10.1111/j.1567-1364.2011.00773.x. [DOI] [PubMed] [Google Scholar]
- Sipiczki M. Interspecies hybridisation and recombination in Saccharomyces wine yeasts. FEMS Yeast Res. 2008;8:996–1007. doi: 10.1111/j.1567-1364.2008.00369.x. [DOI] [PubMed] [Google Scholar]
- Barrio E, González SS, Arias A, Belloch C, Querol A. In: Yeasts in Food and Beverages. 1. Querol A, Fleet GH, editor. Springer, Berlin; 2006. Molecular mechanisms involved in the adaptive evolution of industrial yeasts; pp. 153–174. [Google Scholar]
- Antunovics Z, Nguyen HV, Gaillardin C, Sipiczki M. Gradual genome stabilisation by progressive reduction of the Saccharomyces uvarum genome in an interspecific hybrid with Saccharomyces cerevisiae. FEMS Yeast Res. 2005;5:1141–1150. doi: 10.1016/j.femsyr.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Rainieri S, Kodama Y, Nakao Y, Pulvirenti A, Giudici P. The inheritance of mitochondrial DNA in lager brewing strains. FEMS Yeast Res. 2008;8:586–596. doi: 10.1111/j.1567-1364.2008.00363.x. [DOI] [PubMed] [Google Scholar]
- Solieri L, Antúnez O, Pérez-Ortín JE, Barrio E, Giudici P. Mitochondrial inheritance and fermentative: oxidative balance in hybrids between Saccharomyces cerevisiae and Saccharomyces uvarum. Yeast. 2008;25:485–500. doi: 10.1002/yea.1600. [DOI] [PubMed] [Google Scholar]
- Vero LDE, Pulvirenti A, Gullo M, Bonatti P, Giudici P. Sorting of mitochondrial DNA and proteins in the progeny of Saccharomyces interspecific hybrids. Ann Microbiol. 2003;53:219–231. [Google Scholar]
- Lee HY, Chou JY, Cheong L, Chang NH, Yang SY, Leu JY. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 2008;135:1065–1073. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- Chou JY, Hung YS, Lin KH, Lee HY, Leu JY. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 2010;8:e1000432. doi: 10.1371/journal.pbio.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera J, Rández-Gil F, Prieto JA. Cold response in Saccharomyces cerevisiae: new functions for old mechanisms. FEMS Microbiol Rev. 2007;31:327–341. doi: 10.1111/j.1574-6976.2007.00066.x. [DOI] [PubMed] [Google Scholar]
- Beltrán G, Novo M, Leberre V, Sokol S, Labourdette D, Guillamón JM, Mas A, François J, Rozès N. Integration of transcriptomic and metabolic analyses for understanding the global responses of low-temperature winemaking fermentations. FEMS Yeast Res. 2006;6:1167–1183. doi: 10.1111/j.1567-1364.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- Higgins VJ, Beckhouse AG, Oliver AD, Rogers PJ, Dawes IW. Yeast genome-wide expression analysis identifies a strong ergosterol and oxidative stress response during the initial stages of an industrial lager fermentation. Appl Environ Microbiol. 2003;69:4777–4787. doi: 10.1128/AEM.69.8.4777-4787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloch C, Orlić S, Barrio E, Querol A. Fermentative stress adaptation of hybrids within the Saccharomyces sensu stricto complex. Int J Food Microbiol. 2008;122:188–195. doi: 10.1016/j.ijfoodmicro.2007.11.083. [DOI] [PubMed] [Google Scholar]
- Dunn B, Levine RP, Sherlock G. Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics. 2005;6:53. doi: 10.1186/1471-2164-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreto L, Eiriz M, Gomes A, Pereira P, Schuller D, Santos M. Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics. 2008;9:524. doi: 10.1186/1471-2164-9-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Collingwood D, Raghuraman MK, Brewer BJ. Fragile genomic sites are associated with origins of replication. Genome Biol Evol. 2009;1:350–363. doi: 10.1093/gbe/evp034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- Pérez-Ortín JE, Querol A, Puig S, Barrio E. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 2002;12:1533–1539. doi: 10.1101/gr.436602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Gonçalves P, Sampaio JP, Dover J, Johnston M, Rokas A. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature. 2010;464:54–58. doi: 10.1038/nature08791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Caryoscope representation of microarray data of S.kudriavzevii IFO1802. Array CGH data are shown in numerical order with chromosome I at the top and chromosome XVI at the bottom. Red signal indicates hybridization signal but it’s important to note the high normalization factor applied to the red signal (2) and the correction applied to the green one (0.49). This figure indicate that no cross hybridization has occurred between S. kudriavzevii genes and S. cerevisiae genes. (PPTX 106 kb)
able S1. Binary table showing the presence/absence of a particular event for each hybrid strain (coded as 1/0 respectively). kud, S. kudriavzevii; loss, chromosome loss; rec, recombination generating a chimerical chromosome. Roman numerals indicate chromosome numbers and Arabic numerals the types of chimerical chromosomes or the restriction sites in the analysis of RFLP patterns in 34 genes. Parsimony tree was computed using a mixture model in which chimerical chromosomes, S. kudriavzevii chromosome losses and double S. cerevisiae chromosome were considered under the Camin-Sokal criterion and restriction site gains/losses under the Wagner criterion. (XLS 32 kb)
Figure S2. Caryoscope representation of microarray data of 11 S. cerevisiae × S.kudriavzevii hybrids. Array CGH data are shown in numerical order with chromosome I at the top and chromosome XVI at the bottom for each strain. Regions with higher red signal correspond to S. cerevisiae genome regions that are overrepresented in the hybrid genome and those with higher green signal to those regions that are underrepresented. aCGH of wine and brewery hybrids are depicted on black and gray backgrounds, respectively. Arrows indicate potential non-reciprocal recombination events between homeologous chromosomes involved in the generation of chimerical chromosomes. (PPT 607 kb)
Table S2. List of S. cerevisiae genes depleted in all S. cerevisiae × S. kudriavzevii hybrids under analysis. (XLS 29 kb)
Table S3. S. kudriavzevii gene composition for each hybrid. A cross indicates the putative presence of that gene by considering colinearity between S. kudriavzevii and S. cerevisiae genomes. R indicates interspecies recombinant genes. Wine and brewing hybrids are indicated in black and gray lettering, respectively. (XLS 2001 kb)
Table S4. Metabolic pathways and biological processes obtained from a Gene Ontology analysis using the S. kudriavzevii genes retained in each hybrid grouping. (XLS 38 kb)