Abstract
It has been postulated that ionizing radiation produces a unique form of cellular DNA damage called “clustered damages” or “multiply damaged sites”. Here, we show that clustered DNA damages are indeed formed in Escherichia coli by ionizing radiation and are converted to lethal double-strand breaks during attempted base-excision repair. In wild-type cells possessing the oxidative DNA glycosylases that cleave DNA at repairable single damages, double-strand breaks are formed at radiation-induced clusters during postirradiation incubation and also in a dose-dependent fashion. E. coli mutants lacking these enzymes do not form double-strand breaks postirradiation and are substantially more radioresistant than wild-type cells. Furthermore, overproduction of one of the oxidative DNA glycosylases in mutant cells confers a radiosensitive phenotype and an increase in the number of double-strand breaks. Thus, the effect of the oxidative DNA glycosylases in potentiating DNA damage must be considered when estimating radiation risk.
Approximately 70% of radiation-induced DNA damages are formed by reactive-free radicals produced by the radiolysis of water in the vicinity of DNA (1). These radiation-induced DNA damages are repaired by base-excision repair (2) and overlap substantially with those formed during normal oxidative metabolism (3), which are produced at significant rates in unirradiated cells (4). This overlap of damages has led to a controversy with respect to determining radiation risk. Proponents of a “threshold effect” claim that because endogenous oxidative damages are effectively repaired and adaptive responses have been demonstrated for certain radiation endpoints, then a threshold must exist for the carcinogenic and lethal consequences of ionizing radiation (5, 6). This issue is an important one because much of the projected radiation exposures associated with human activity over the next hundred years will come from low doses associated with medical tests, waste cleanup, and materials associated with nuclear weapons and nuclear power. Whether a threshold exists for the consequences of radiation damage will significantly influence risk estimates for low-dose exposures. Currently, risk estimates for individuals or groups exposed to low radiation doses are determined from epidemiological data obtained from populations exposed to high doses, and a “linear-no-threshold” estimation is used for assessment. Proponents of the linear-no-threshold model hold that the biologically important radiation damages in DNA, such as double-strand breaks, are substantially different from single, repairable oxidative lesions. It also has been predicted that ionizing radiation produces a unique form of DNA damage called “clustered damages” (1, 7, 8). If, indeed, clustered damages can be demonstrated in living cells and if, unlike the single lesions from which they are formed, they are poorly repaired, these facts would provide additional support for the linear-no-threshold model.
Modeling of radiation track structures (9, 10) has provided strong evidence for radiation-induced damage clusters. Energy from low linear energy transfer ionizing radiation (e.g., x-rays and γ-rays) is deposited in the water surrounding the DNA molecule such that between two and five radical pairs are generated within a radius of 1–4 nm. As a result, multiple single lesions, including oxidized purine or pyrimidine bases, apurinic, abasic (AP) sites, and single-strand breaks, can be formed in DNA from the same radiation energy deposition event. When two opposing single-strand breaks are formed within a cluster, a potentially lethal double-strand break results. In addition, a number of studies using irradiated DNA have shown that DNA double-strand breaks can be formed by postirradiation treatment with enzymes that recognize single DNA lesions (11–15), suggesting that these lesions may be closely opposed to each other or to single-strand breaks.
The single free-radical-induced DNA damages produced in clusters by ionizing radiation that overlap substantially with those produced during normal oxidative metabolism are repaired by base-excision repair (for reviews, see refs. 2 and 16–18). Oxidized purines and pyrimidines are recognized by DNA glycosylases that remove the damaged base and cleave the DNA backbone. The resulting strand break, as well as strand breaks that are produced directly by radiation, are processed by an apurinic endonuclease that can also directly incise an AP site. The gap then is filled in by a DNA polymerase and sealed by a DNA ligase. This process in its basic format has been conserved from bacteria to man.
Recently, the potential consequences of base-excision repair processing of single lesions contained in clusters have been studied by using site-specifically opposed base damages or AP sites in oligonucleotide substrates together with the enzymes that initiate base-excision repair of these single damages (19–25). The overall conclusion is that cleavage of the first strand of a substrate containing two opposed base damages or AP sites is relatively unaffected by the position of the opposing lesion; however, once a strand break is introduced in one strand by an enzyme that recognizes the damage in that strand, cleavage at the damage site in the second strand depends on the position of the opposing strand break and the enzyme used. Incision of the second strand usually occurs if the opposing single-strand break is more than one base away. If incision of both strands does take place, a double-strand break is formed. The question remains, however, if all the base-excision repair enzymes necessary to repair the lesions on both strands were present, would sequential repair of each strand occur? With a reconstituted base-excision repair system using a substrate containing the oxidative DNA lesion, 7,8-dihydro-8-oxoguanine (8-oxoG), closely opposed to a single-strand break, we showed elsewhere that sequential repair of both strands only occurs if a double-strand break cannot be formed (26). In fact, whether or not a double-strand break is formed totally depends on whether the DNA glycosylase can incise the base damage opposite the strand break, which always occurs if the base damage and the single-strand break are opposed more than one base apart.
Because double-strand break formation at closely opposed damages depended solely on initial cleavage by the DNA glycosylase, the prediction was that cells devoid of the oxidative DNA glycosylases would be radioresistant because the abortive repair that would result in a potentially lethal double-strand break could not be initiated. In this study, we test this hypothesis and show that Escherichia coli cells devoid of all three oxidative DNA glycosylases that recognize oxidized purines and pyrimidines, as well as AP sites, are less sensitive to the lethal effects of ionizing radiation and form fewer double-strand breaks than wild-type cells. Moreover, the formation of double-strand breaks in wild-type cells increases during postirradiation incubation. Single mutant cells lacking formamidopyrimidine DNA glycosylase (Fpg) are also hyposensitive to ionizing radiation, and this phenotype can be complemented by overexpression of Fpg.
Materials and Methods
E. coli Cell Strains.
All strains were created by P1 transduction into BW35 (KL16) {Hfr KL16 PO-45:[lysA(61)-serA(62)]/thi relA spoT1}, which was generously provided by Bernard Weiss (Emory University School of Medicine, Department of Pathology, Atlanta) and have been described (27).
Construction of a Plasmid Expressing Fpg.
pCYSPC is an isopropyl β-d-thiogalactoside (IPTG)-inducible plasmid derived from the plasmid pCYB2 (New England BioLabs). A spectinomycin-resistance cassette from Enterococcus faecalis (28) was PCR-amplified with the phosphorylated primers 5′-PATCGATTTTCGTTCGTGAATAC-3′ and 5′-PGCAAGGGTTTATTGTTTTC-3′ and was cloned into the MscI restriction site of pCYB2. The resulting plasmid DNA then was PCR-amplified with the phosphorylated primers 5′-PGGATCTAGGTGAAGATCC-3′ and 5′-GAGTTTGTAGAAACGCAAAAAG-3′ and ligated, resulting in removal of the M13 and ampicillin-resistance regions. pCYSPC(Fpg) is the pCYSPC plasmid with the fpg gene (containing a stop codon) cloned into the multiple cloning site. Fpg enzyme activity of this construct was verified by using a DNA oligonucleotide substrate containing a single 8-oxoG lesion (data not shown). Each plasmid-harboring strain was grown in the presence of 100 μM IPTG.
X-Ray Sensitivity.
Cultures of exponentially growing cells were washed and starved in PBS (pH 7.4) at 37°C before irradiation with 0, 45, 90, 135, 180, 225, or 270 Gy with a Philips x-ray generator (Philips Electronic Instruments, Mt. Vernon, NY) at 50 kVp, 2 mA as described (29). The dose was determined with X8 thermoluminescent dosimeter chips (Landauer, Glenwood, IL). Cells were starved before irradiation to halt any initiated rounds of replication (30); this procedure enhances the reproducibility of the x-ray survival curves. When cells were incubated postirradiation, they were incubated in the same buffer without nutrients to continue to suppress initiation of new rounds of DNA replication. DNA synthesis was routinely monitored by fluorometric quantitation with an alkaline ethidium bromide solution (31); no DNA synthesis occurred under these conditions (data not shown).
Microgel Electrophoresis.
Cultures of exponentially growing cells were starved and irradiated with 0, 45, or 90 Gy as described and then incubated in PBS for either 0, 4, or 8 min at 37°C. After incubation, cultures immediately were cooled to 4°C, and 5 μl of a dilution from each culture was embedded in 50 μl of low-melting-temperature agarose and transferred to an MGE microscope slide (Erie Scientific, Portsmouth, NH) as described (32, 33). The slides were electrophoresed in neutral conditions for 1 h at 12 volts (≈100 mA) in a modified electrophoretic unit (32) with buffer recirculated at ≈100 ml/min. The slides were dried as described (33) and stained with YOYO-1 (Molecular Probes). Observations were made on a Nikon E400 fluorescence microscope with a 40×/f 1.3 numerical aperture oil-immersion Fluor lens and an XF100 filter set (exciter, 475 nm; emitter, 535 nm). The number of double-strand breaks per cell was calculated by visually counting 1,000 chromosomes per experiment. All experiments were performed in triplicate.
Results
Attempted Repair of DNA in X-Irradiated E. coli Leads to both an Increase in Lethality and in the Number of DNA Double-Strand Breaks.
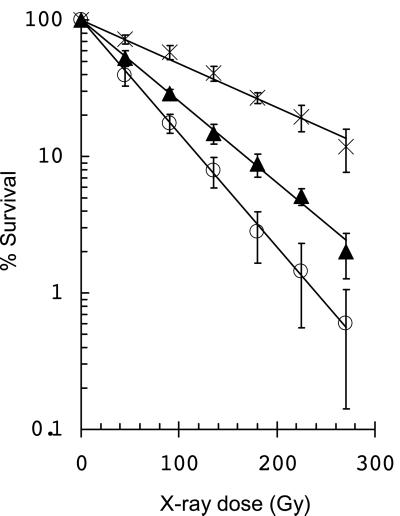
There are three DNA glycosylases in E. coli that recognize and remove the oxidative DNA base damages that are produced by ionizing radiation and cleave the DNA backbone (for reviews, see refs. 2 and 16). These include endonuclease III, the product of the nth gene and endonuclease VIII, the product of the nei gene, whose substrates are primarily oxidized pyrimidines. Fpg, the product of the fpg (mutM) gene, removes oxidized purines but has a fairly broad substrate specificity and also can remove oxidized pyrimidines. All three DNA glycosylases efficiently cleave an AP site, which is their best substrate. To test the idea that these enzymes actually produce lethal double-strand breaks from clustered damages in the DNA of irradiated cells, thus leading to increased cell killing, we determined the x-ray sensitivity of wild-type cells and compared it to a triple mutant lacking all three enzyme activities. Fig. 1 shows that, as predicted, the triple mutant cells, nth nei fpg (27), are substantially more radioresistant than wild-type cells—about 2-fold when comparing survival slopes. There was a 6-fold difference in survival at the highest dose tested. Double mutants, nth nei, unable to repair potentially lethal pyrimidine damages (2), are about 1.5-fold more sensitive than wild-type cells (29).
Figure 1.
X-ray inactivation of E. coli mutants lacking the oxidative DNA glycosylases compared with wild-type cells. ▴, wild type; ○, nei nth double mutant; and ×, nei nth fpg triple mutant cells. (Bars = standard deviations for the average of three experiments.)
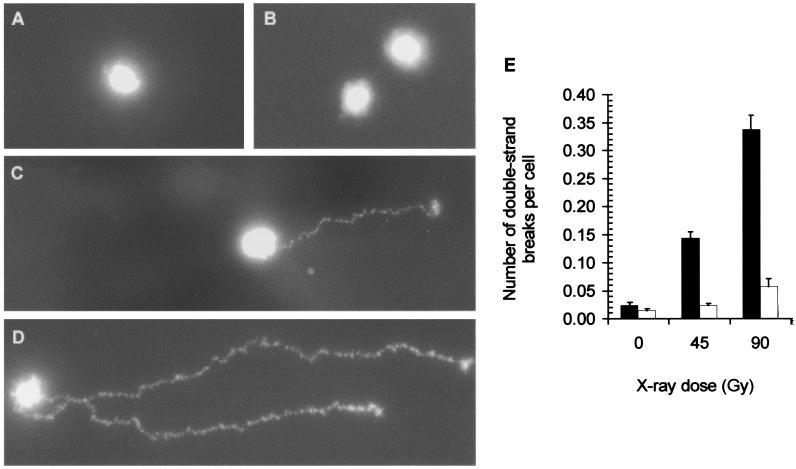
To determine whether the hypersensitivity observed in wild-type cells when compared with the triple mutant lacking the oxidative DNA glycosylases was, in fact, caused by double-strand breaks resulting from attempted repair of clustered lesions, we used neutral microgel electrophoresis to visualize the double-strand breaks (33). Here, the irradiated cells were gently lysed on a microscope slide, subjected to electrophoresis to “electrostretch” the DNA, and the number of double-strand breaks visualized by an intense fluorescent dye, YOYO-1, were counted. Fig. 2 shows the results of such an experiment for wild-type E. coli cells irradiated with 45 Gy, a dose at which 60% of the wild-type cells survive. Fig. 2 A and B show chromosome molecules with no apparent double-strand breaks. In Fig. 2C, the DNA migration indicates the presence of one double-strand break, whereas in Fig. 2D, the migration indicates the presence of two double-strand breaks. Fig. 2E shows the dose–response for the number of double-strand breaks present after an 8-min incubation after irradiation of wild-type and nth nei fpg triple mutant cells. In wild-type cells, the number of double-strand breaks was significantly greater (about 6-fold) compared with the triple mutants lacking the oxidative DNA glycosylases.
Figure 2.
Formation of DNA double-strand breaks in x-irradiated E. coli cells. (A–D) Representative results from the experiment with wild-type cells shown in E in which 1,000 chromosomes were observed in each of three experiments and the number of double-strand breaks followed a Poisson distribution. (A and B) Photomicrographs of E. coli chromosomes showing no apparent migration representing zero double-strand breaks. (C) DNA migration representing one double-strand break. (D) DNA migration representing two double-strand breaks. (E) The number of double-strand breaks as determined by microgel electrophoresis for wild-type (black bars) and nei nth fpg triple mutant (white bars) cells after an 8-min postirradiation incubation at 37°C in PBS. (Bars = standard deviations for the average of three experiments.)
Single E. coli Mutants Lacking Only Fpg DNA Glycosylase Are Also Radioresistant.
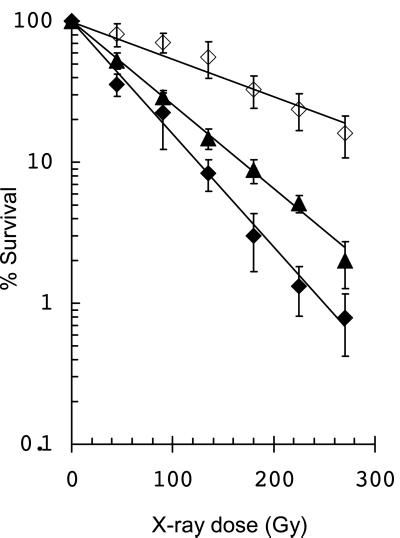
When we examined the radiosensitivity of other combinations of mutants, we observed that single mutants lacking only Fpg protein, which repairs 8-oxoG and recognizes other oxidized purines and pyrimidines, as well as AP sites, were as radioresistant as the triple mutant (Fig. 3). Fig. 3 also shows that the radioresistance of fpg mutants could be complemented by overexpressing the Fpg protein in a plasmid construct. In the experiment shown here, the overexpressing cells contained ≈13-fold more Fpg than wild-type cells and were about 25% more sensitive. Expression was determined by Fpg activity on an oligonucleotide substrate containing 8-oxoG (data not shown). In the Fpg-overexpressing cells, which contain active endonucleases III and VIII, presumably all of the potentially lethal base damages recognized by these proteins are being repaired, while at the same time double-strand breaks at clusters are being introduced by Fpg, endonuclease III, and endonuclease VIII.
Figure 3.
X-ray inactivation of single E. coli fpg mutants and fpg mutants overexpressing Fpg protein. ▴, wild type; ⋄, fpg single mutant + pCYSPC; and ♦, fpg single mutant + pCYSPC(Fpg). (Bars = standard deviations for the average of three experiments.)
Double-Strand Breaks Are Produced in the DNA of X-Irradiated Wild-Type Cells as a Function of Time After Irradiation.
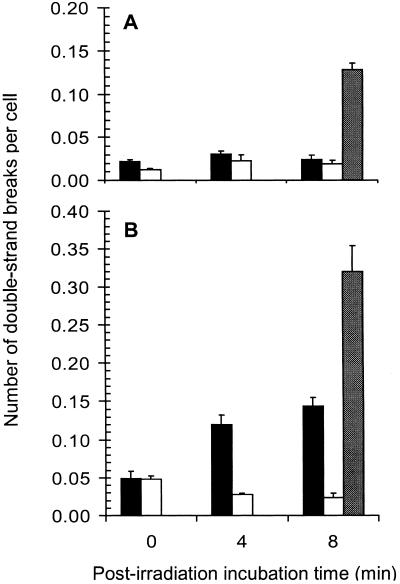
If, in fact, double- strand breaks are being produced in cellular DNA because of attempted repair of clustered DNA lesions, as suggested by the correlation between double-strand breaks and survival of wild-type and mutant cells, then we would expect that the number of these breaks would increase in wild-type cells as a function of postirradiation incubation time. In Fig. 4, it can be clearly seen that at a dose of 45 Gy, the number of double-strand breaks increases about 3-fold between 0 min and 8 min after irradiation in wild-type cells under conditions where no replication occurs. No further increase was seen after 8 min (data not shown). In fpg mutant cells, the number of double-strand breaks did not increase with time after irradiation as would be expected if no incisions at clustered lesions were formed or if repair kept up with their production. In fact, by 8 min, the number of double-strand breaks returned to background levels in the fpg mutant, suggesting they had been repaired. The level of frank double-strand breaks (those present immediately after irradiation) in both wild-type cells and fpg mutants was approximately the same after 45-Gy irradiation, about three times background levels. Because the number of double-strand breaks returned to background levels after 8 min in fpg mutant cells, a double-strand break may not be the ultimate lethal event in these cells. Fig. 4 also shows that the number of double-strand breaks formed in irradiated cells overexpressing Fpg is increased some 6.5-fold over the number of initial double-strand breaks observed in irradiated wild-type cells and fpg mutants. Interestingly, the number of double-strand breaks observed in unirradiated cells containing the overexpressing plasmid was significantly higher (about 6.5-fold) than in unirradiated wild-type cells without the plasmid, suggesting that some clustered damages are present in unirradiated cells. Taken together, these data show that the formation of double-strand breaks at sites of clustered DNA damage by attempted repair in wild-type cells contributes significantly to the lethal effects of radiation in these cells, either by placing an increased burden on the recombination system that repairs double-strand breaks or by forming products that are poor substrates for recombination repair.
Figure 4.
DNA double-strand breaks formed during postirradiation incubation. Wild-type (black bars), fpg single mutant (white bars), and fpg single mutant + pCYSPC(Fpg) (gray bars) cells were irradiated with 0 Gy (A) or 45 Gy (B) as described and then incubated in PBS for either 0, 4, or 8 min. (Bars = standard deviations for the average of three experiments.)
Discussion
The presence of the oxidative DNA glycosylases that recognize the free radical damage produced in DNA by ionizing radiation are a detriment to the survival of irradiated cells (Figs. 1 and 3). This effect is caused by the attempted but abortive repair of single lesions that are produced in clusters by ionizing radiation, as demonstrated by the correlation between the survival of wild-type and mutant strains and the presence of double-strand breaks in the DNA (Fig. 2). Further substantiation for this conclusion comes from the observation that the number of double- strand breaks increases in wild-type cells containing the oxidative DNA glycosylases during postirradiation incubation but not in mutant cells (Fig. 4) and that the radioresistance observed in mutant cells can be complemented by overexpressing Fpg DNA glycosylase with a concomitant increase in double-strand breaks (Figs. 3 and 4). In this study, the proposed deleterious consequences of clustered lesions produced by ionizing radiation have been demonstrated in living cells, and it validates the physical modeling studies that predicted these effects. The data clearly show that not only are such clusters formed in living cells by radiation, but their abortive processing contributes significantly to the number of lethal damages. The results presented in this paper also explain a previous observation that the number of double-strand breaks continues to increase after the exposure of bacterial cells to ionizing radiation (34).
Does base-excision repair processing also protect cells from the lethal effects of ionizing radiation? E. coli exonuclease III (Xth) and endonuclease IV (Nfo) are required for processing all of the single lesions produced by ionizing radiation including base damages, AP sites, and single-strand breaks (for a review, see ref. 2). Thus, xth nfo double mutants are completely devoid of base-excision repair and are also almost 4.5-fold more sensitive to ionizing radiation than wild-type cells (35). Double mutants defective in endonucleases III (nth) and VIII (nei) are also about 1.5-fold more hypersensitive to ionizing radiation (ref. 29, Fig. 1). Thus, it can be concluded that single lesions contribute significantly to the lethal effects of ionizing radiation in bacteria. Interestingly, the addition of the nth mutation to the xth nfo double mutant increases its resistance to ionizing radiation (35). This result was unexpected at the time of the experiments because nth is epistatic to xth and nfo, but the results undoubtedly represent a sparing effect because endonuclease III is no longer present to create additional lethal double-strand breaks at clustered damages. In keeping with the observations presented here, it also would be predicted that the addition of the fpg mutation to xth nfo mutants would result also in a sparing effect on radiation sensitivity.
The sensitivity of the fpg single mutant is the same as the nth nei fpg triple mutant; the nth nei double mutant is radiosensitive, whereas nth and nei single mutants exhibit wild-type sensitivity (ref. 29 and Fig. 1). If a Poisson distribution of damage is assumed, it seems that the combination of endonucleases III and VIII repair lethal lesions, such as thymine glycol (2), and induce lethal lesions, presumably because of the production of double-strand breaks at clustered single lesions, and that these effects compensate for one another. Despite the survival data, and the fact that addition of an nth mutation to a mutant defective in the second step of base-excision repair confers some radioresistance (35), no increase in double-strand breaks was observed after irradiation of fpg mutant cells that contain endonucleases III and VIII (Fig. 4). This observation suggests that the latter enzymes may not be major contributors to the production of double-strand breaks formed by attempted repair at clusters, or they are repaired as readily as they are formed in the single-mutant background.
Fpg may play a more important role than endonucleases III and VIII in producing double-strand breaks at clusters because Fpg has a broader substrate specificity than endonuclease III. Not only does it recognize purine lesions produced by ionizing radiation such as 8-oxoG and formamidopyrimidines, but it also recognizes the oxidized pyrimidines with catalytic efficiencies only about an order of magnitude less than those of endonucleases III and VIII (36, 37). In addition, Fpg recognizes and incises AP sites of the C4′ type produced by ionizing radiation (38) about 3-fold more efficiently than endonuclease III (39). Finally, guanine is readily oxidized at position 8, and although the levels of endogenous 8-oxoG remain controversial (40, 41), they seem to be high in E. coli because in the absence of repair, the spontaneous mutation frequency caused by 8-oxoG is about 30-fold higher than that caused by oxidized pyrimidines (27). Therefore, endogenous 8-oxoG also may contribute to the formation of clusters in irradiated cells. High endogenous levels of 8-oxoG and spontaneous depurinations also may contribute to the formation of clustered damages and to the double-strand breaks observed in unirradiated cells overexpressing Fpg (Fig. 4). This formation of double-strand breaks may be exacerbated not only by the broad substrate specificity of Fpg but also by the increased ratio of Fpg to the subsequent enzymes in the repair pathway, because recent evidence suggests that the base-excision repair reactions for free radical damage are sequentially coordinated (42).
In summary, the presence of the oxidative DNA glycosylases that recognize repairable single free radical-induced DNA lesions is a detriment to the survival of cells exposed to ionizing radiation. This effect is caused by attempted but abortive repair of single lesions that are produced in clusters. Because base-excision repair is conserved from bacteria to man, and extracts from mammalian cells behave similarly to the prokaryotic enzymes on substrates containing closely opposed damages (24), the observations presented here have critical implications on how we think about risk assessment. The oxidative DNA glycosylases perform important cellular housekeeping activities in human cells and even may be induced further by ionizing radiation (43–45); thus, it will be critical to take into consideration when estimating risk at low radiation doses that the major impact of the oxidative DNA glycosylases in irradiated cells may be to potentiate the formation of double-strand breaks known to give rise to radiation-induced cellular lethality and mutations.
Acknowledgments
We thank Drs. John Ward, Michael Laspia, Douglas Johnson, and Nicklas Heintz for critically reading the manuscript and Dr. Jeffrey Bond for the Poisson analysis of the survival curves. This work was supported by National Institutes of Health Grant R37 CA33657, awarded by the National Cancer Institute.
Abbreviations
- AP
apurinic, abasic
- 8-oxoG
oxidative DNA lesion 7,8-dihydro-8-oxoguanine
- Fpg
formamidopyrimidine DNA glycosylase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ward J F. Radiat Res. 1985;104:S103–S111. [PubMed] [Google Scholar]
- 2.Wallace S S. Radiat Res. 1998;150:S60–S79. [PubMed] [Google Scholar]
- 3.Henle E S, Linn S. J Biol Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 4.Beckman K B, Ames B N. J Biol Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 5.Feinendegen L E, Loken M K, Booz J, Muhlensiepen H, Sondhaus C A, Bond V P. Stem Cells (Dayton) 1995;13, Suppl. 1:7–20. [PubMed] [Google Scholar]
- 6.Feinendegen L E, Bond V P, Sondhaus C A, Muehlensiepen H. Mutat Res. 1996;358:199–205. doi: 10.1016/s0027-5107(96)00121-2. [DOI] [PubMed] [Google Scholar]
- 7.Ward J F. Radiat Res. 1981;86:185–195. [PubMed] [Google Scholar]
- 8.Ward J F. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 9.Brenner D J, Ward J F. Int J Radiat Biol. 1992;61:737–748. doi: 10.1080/09553009214551591. [DOI] [PubMed] [Google Scholar]
- 10.Goodhead D T. Int J Radiat Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 11.Prise K M, Pullar C H, Michael B D. Carcinogenesis. 1999;20:905–909. doi: 10.1093/carcin/20.5.905. [DOI] [PubMed] [Google Scholar]
- 12.Milligan J R, Aguilera J A, Nguyen T T, Paglinawan R A, Ward J F. Int J Radiat Biol. 2000;76:1475–1483. doi: 10.1080/09553000050176234. [DOI] [PubMed] [Google Scholar]
- 13.Rydberg B. Radiat Res. 2000;153:805–812. doi: 10.1667/0033-7587(2000)153[0805:rihlst]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Sutherland B M, Bennett P V, Sidorkina O, Laval J. Proc Natl Acad Sci USA. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutherland B M, Bennett P V, Sidorkina O, Laval J. Biochemistry. 2000;39:8026–8031. doi: 10.1021/bi9927989. [DOI] [PubMed] [Google Scholar]
- 16.Wallace S S. In: Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Scandalios J, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 49–90. [Google Scholar]
- 17.David S S, Williams S D. Chem Rev (Washington, D.C.) 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl T, Wood R D. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi M, Lillis R, Demple B, Takeshita M. J Biol Chem. 1994;269:21907–21914. [PubMed] [Google Scholar]
- 20.Wilson D M, III, Takeshita M, Grollman A P, Demple B. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhry M A, Weinfeld M. J Mol Biol. 1995;249:914–922. doi: 10.1006/jmbi.1995.0348. [DOI] [PubMed] [Google Scholar]
- 22.Chaudhry M A, Weinfeld M. J Biol Chem. 1997;272:15650–15655. doi: 10.1074/jbc.272.25.15650. [DOI] [PubMed] [Google Scholar]
- 23.Harrison L, Hatahet Z, Purmal A A, Wallace S S. Nucleic Acids Res. 1998;26:932–941. doi: 10.1093/nar/26.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David-Cordonnier M H, Laval J, O'Neill P. J Biol Chem. 2000;275:11865–11873. doi: 10.1074/jbc.275.16.11865. [DOI] [PubMed] [Google Scholar]
- 25.David-Cordonnier M H, Boiteux S, O'Neill P. Nucleic Acids Res. 2001;29:1107–1113. doi: 10.1093/nar/29.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison L, Hatahet Z, Wallace S S. J Mol Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 27.Blaisdell J O, Hatahet Z, Wallace S S. J Bacteriol. 1999;181:6396–6402. doi: 10.1128/jb.181.20.6396-6402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LeBlanc D J, Lee L N, Inamine J M. Antimicrob Agents Chemother. 1991;35:1804–1810. doi: 10.1128/aac.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang D, Hatahet Z, Blaisdell J O, Melamede R J, Wallace S S. J Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Billen D, Bruns L. J Bacteriol. 1970;103:400–403. doi: 10.1128/jb.103.2.400-403.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaeffer W I, Melamede R. J Clin Microbiol. 1993;31:1303–1307. doi: 10.1128/jcm.31.5.1303-1307.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh N P, Stephens R E. Mutat Res. 1997;383:167–175. doi: 10.1016/s0921-8777(96)00056-0. [DOI] [PubMed] [Google Scholar]
- 33.Singh N P, Stephens R E, Singh H, Lai H. Mutat Res. 1999;429:159–168. doi: 10.1016/s0027-5107(99)00124-4. [DOI] [PubMed] [Google Scholar]
- 34.Bonura T, Smith K C, Kaplan H S. Proc Natl Acad Sci USA. 1975;72:4265–4269. doi: 10.1073/pnas.72.11.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham R P, Saporito S M, Spitzer S G, Weiss B. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatahet Z, Kow Y W, Purmal A A, Cunningham R P, Wallace S S. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 37.D'Ham C, Romieu A, Jaquinod M, Gasparutto D, Cadet J. Biochemistry. 1999;38:3335–3344. doi: 10.1021/bi981982b. [DOI] [PubMed] [Google Scholar]
- 38.von Sonntag C. The Chemical Basis of Radiation Biology. London: Taylor and Francis; 1987. [Google Scholar]
- 39.Häring M, Rudiger H, Demple B, Boiteux S, Epe B. Nucleic Acids Res. 1994;22:2010–2015. doi: 10.1093/nar/22.11.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cadet J, Douki T, Ravanat J-L. Environ Health Perspect. 1997;105:1034–1039. doi: 10.1289/ehp.105-1470384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cadet J, D'Ham C, Douki T, Pouget J-P, Ravant J-L, Sauvaigo S. Free Radical Res. 1998;29:541–550. doi: 10.1080/10715769800300581. [DOI] [PubMed] [Google Scholar]
- 42.Hill J W, Hazra T K, Izumi T, Mitra S. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grombacher T, Mitra S, Kaina B. Carcinogenesis. 1996;17:2329–2336. doi: 10.1093/carcin/17.11.2329. [DOI] [PubMed] [Google Scholar]
- 44.Le X C, Xing J Z, Lee J, Leadon S A, Weinfeld M. Science. 1998;280:1066–1069. doi: 10.1126/science.280.5366.1066. [DOI] [PubMed] [Google Scholar]
- 45.Ramana C V, Boldogh I, Izumi T, Mitra S. Proc Natl Acad Sci USA. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]