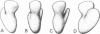Abstract
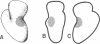
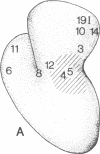
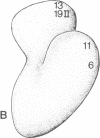
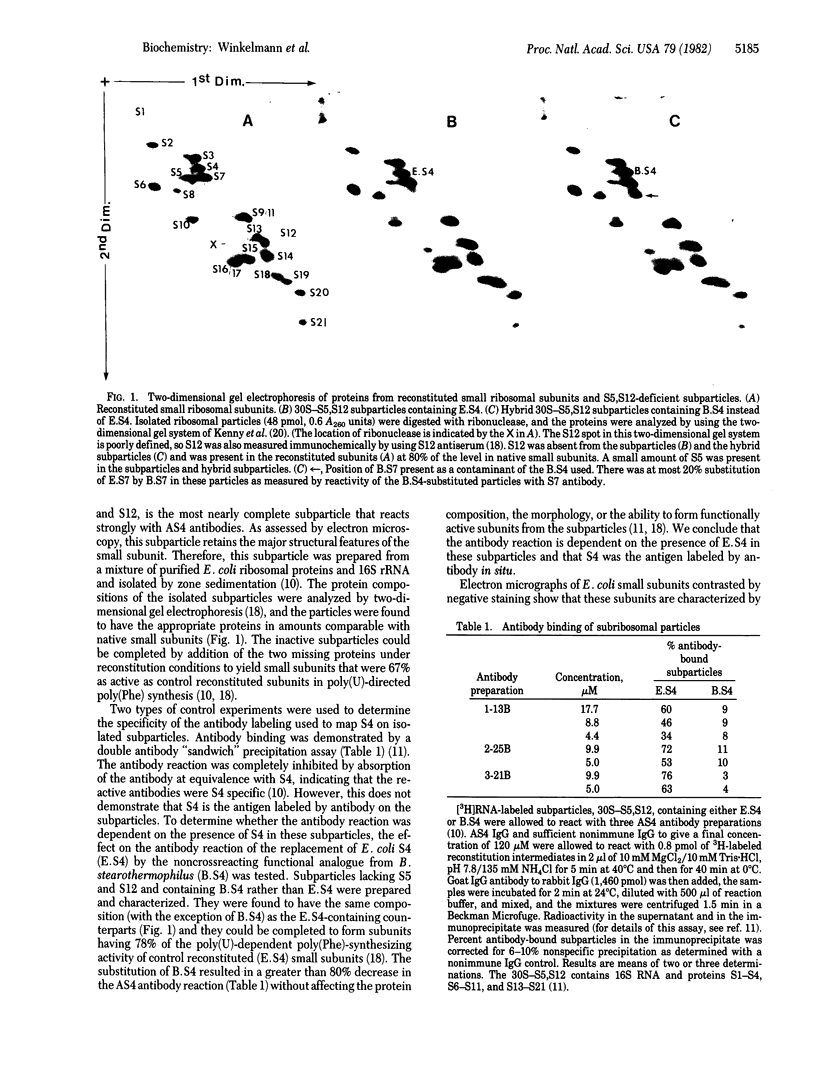
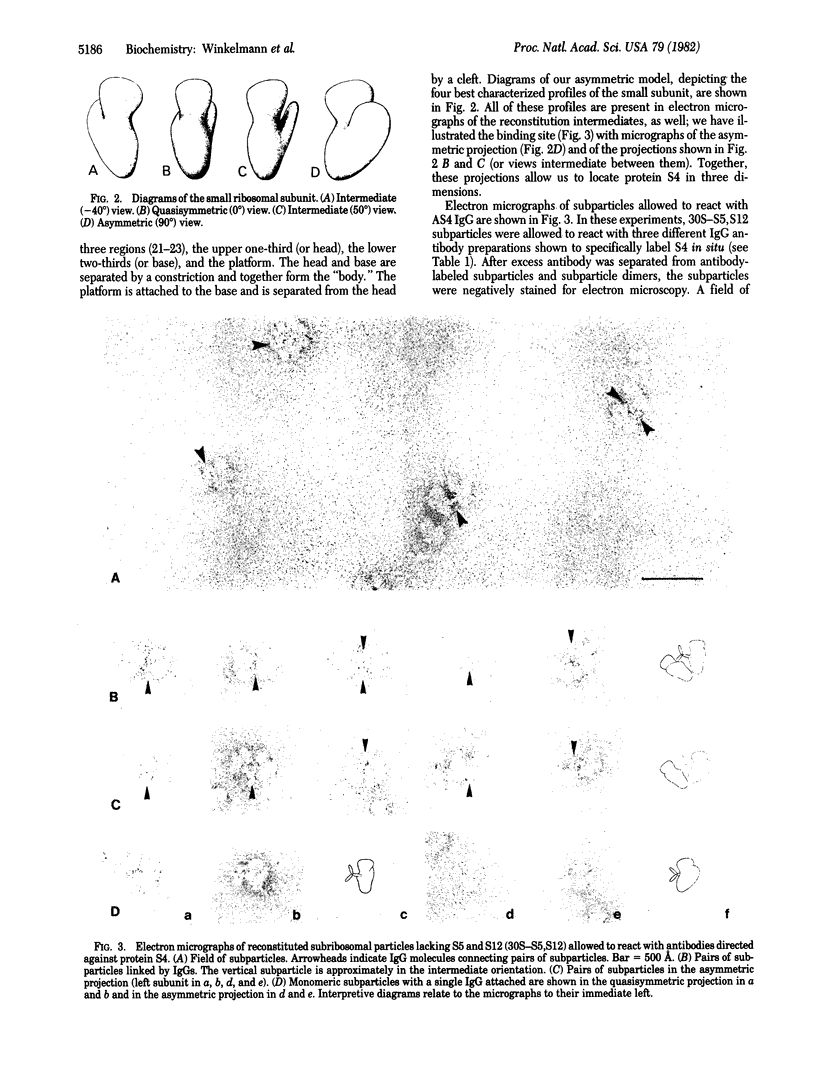
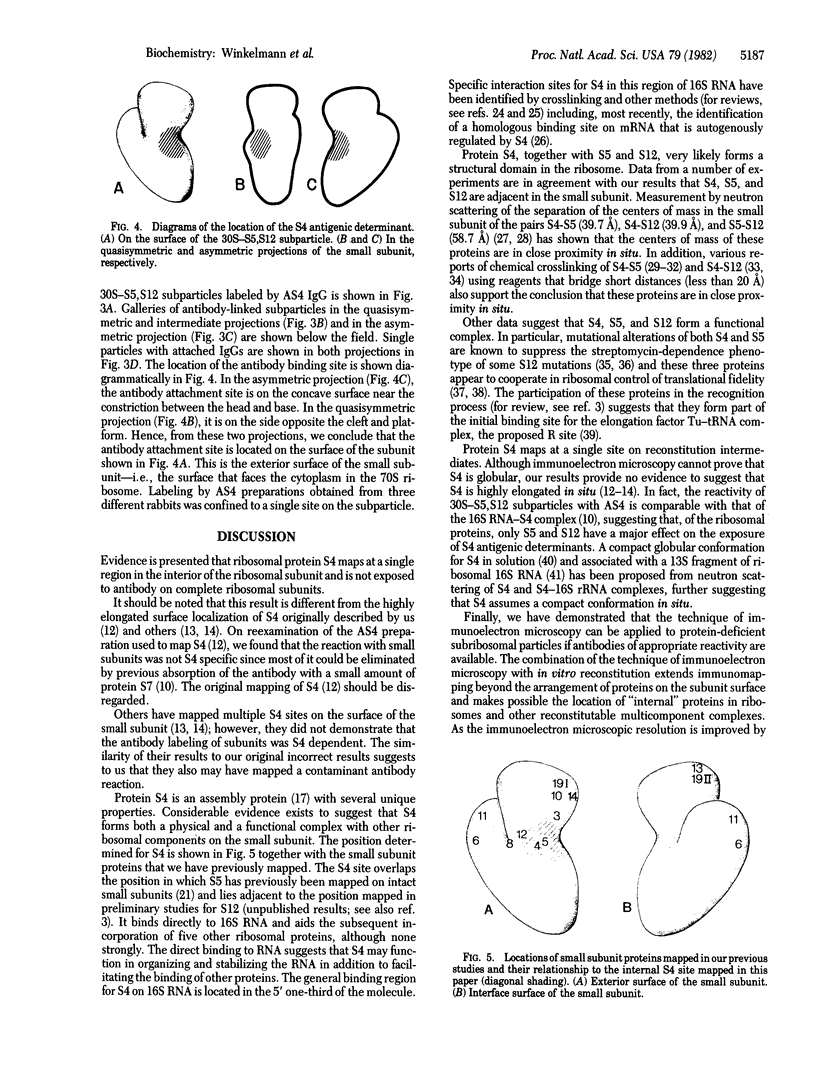
The location of protein S4 in the small ribosomal subunit has been identified by immunoelectron microscopy. Although intact small subunits are not reactive with antibodies directed against protein S4, subribosomal particles reconstituted without proteins S5 and S12 are reactive. By using these "incomplete" subparticles, we have mapped the position of S4. It is located at a single site on the exterior (cytoplasmic) side of the subunit, at the partition that separates the one-third, or head, from two-thirds, or base, of the subunit. In this location, protein S4 is "beneath" proteins S5 and S12. All three proteins are members of a complex on, or near, the external surface of the small ribosomal subunit that plays an important role in regulation of translational fidelity.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernabeu C., Lake J. A. Nascent polypeptide chains emerge from the exit domain of the large ribosomal subunit: immune mapping of the nascent chain. Proc Natl Acad Sci U S A. 1982 May;79(10):3111–3115. doi: 10.1073/pnas.79.10.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjare U., Gorini L. Drug dependence reversed by a ribosomal ambiguity mutation, ram, in Escherichia coli. J Mol Biol. 1971 May 14;57(3):423–435. doi: 10.1016/0022-2836(71)90101-x. [DOI] [PubMed] [Google Scholar]
- De Wilde M., Cabezón T., Villarroel R., Herzog A., Bollen A. Cooperative control of translation fidelity by ribosomal proteins in Escherichia coli. I. Properties of ribosomal mutants whose resistance to neamine is the cumulative effect of two distinct mutations. Mol Gen Genet. 1975 Dec 23;142(1):19–33. doi: 10.1007/BF00268752. [DOI] [PubMed] [Google Scholar]
- Expert-Bezançon A., Barritault D., Milet M., Guérin M. F., Hayes D. H. Identification of neighbouring proteins in Escherichia coli 30 S ribosome subunits. J Mol Biol. 1977 Jun 5;112(4):603–629. doi: 10.1016/s0022-2836(77)80166-6. [DOI] [PubMed] [Google Scholar]
- Gorini L. Ribosomal discrimination of tRNAs. Nat New Biol. 1971 Dec 29;234(52):261–264. doi: 10.1038/newbio234261a0. [DOI] [PubMed] [Google Scholar]
- Hasenbank R., Guthrie C., Stöffler G., Wittmann H. G., Rosen L., Apirion D. Electrophoretic and immunological studies on ribosomal proteins of 100 Escherichia coli revertants from streptomycin dependence. Mol Gen Genet. 1973 Dec 14;127(1):1–18. doi: 10.1007/BF00267778. [DOI] [PubMed] [Google Scholar]
- Held W. A., Mizushima S., Nomura M. Reconstitution of Escherichia coli 30 S ribosomal subunits from purified molecular components. J Biol Chem. 1973 Aug 25;248(16):5720–5730. [PubMed] [Google Scholar]
- Held W. A., Nomura M. Rate determining step in the reconstitution of Escherichia coli 30S ribosomal subunits. Biochemistry. 1973 Aug 14;12(17):3273–3281. doi: 10.1021/bi00741a020. [DOI] [PubMed] [Google Scholar]
- Higo K., Held W., Kahan L., Nomura M. Functional correspondence between 30S ribosomal proteins of Escherichia coli and Bacillus stearothermophilus. Proc Natl Acad Sci U S A. 1973 Mar;70(3):944–948. doi: 10.1073/pnas.70.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahan L., Winkelmann D. A., Lake J. A. Ribosomal proteins S3, S6, S8 and S10 of Escherichia coli localized on the external surface of the small subunit by immune electron microscopy. J Mol Biol. 1981 Jan 5;145(1):193–214. doi: 10.1016/0022-2836(81)90340-5. [DOI] [PubMed] [Google Scholar]
- Kenny J. W., Lambert J. M., Traut R. R. Cross-linking of ribosomes using 2-iminothiolane (methyl 4-mercaptobutyrimidate) and identification of cross-linked proteins by diagonal polyacrylamide/sodium dodecyl sulfate gel electrophoresis. Methods Enzymol. 1979;59:534–550. doi: 10.1016/0076-6879(79)59112-5. [DOI] [PubMed] [Google Scholar]
- Keren-Zur M., Boublik M., Ofengand J. Localization of the decoding region on the 30S Escherichia coli ribosomal subunit by affinity immunoelectron microscopy. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1054–1058. doi: 10.1073/pnas.76.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. Aminoacyl-tRNA binding at the recognition site is the first step of the elongation cycle of protein synthesis. Proc Natl Acad Sci U S A. 1977 May;74(5):1903–1907. doi: 10.1073/pnas.74.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A., Kahan L. Ribosomal proteins S5, S11, S13 and S19 localized by electron microscopy of antibody-labeled subunits. J Mol Biol. 1975 Dec 25;99(4):631–644. doi: 10.1016/s0022-2836(75)80177-x. [DOI] [PubMed] [Google Scholar]
- Lake J. A., Pendergast M., Kahan L., Nomura M. Localization of Escherichia coli ribosomal proteins S4 and S14 by electron microscopy of antibody-labeled subunits. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4688–4692. doi: 10.1073/pnas.71.12.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. Practical aspects of immune electron microscopy. Methods Enzymol. 1979;61:250–257. doi: 10.1016/0076-6879(79)61014-5. [DOI] [PubMed] [Google Scholar]
- Lake J. A. Ribosome structure determined by electron microscopy of Escherichia coli small subunits, large subunits and monomeric ribosomes. J Mol Biol. 1976 Jul 25;105(1):131–139. doi: 10.1016/0022-2836(76)90200-x. [DOI] [PubMed] [Google Scholar]
- Langer J. A., Engelman D. M., Moore P. B. Neutron-scattering studies of the ribosome of Escherichia coli: a provisional map of the locations of proteins S3, S4, S5, S7, S8 and S9 in the 30 S subunit. J Mol Biol. 1978 Mar 15;119(4):463–485. doi: 10.1016/0022-2836(78)90197-3. [DOI] [PubMed] [Google Scholar]
- Lutter L. C., Zeichhardt H., Kurland C. G. Ribosomal protein neighborhoods. I. S18 and S21 as well as S5 and S8 are neighbors. Mol Gen Genet. 1972;119(4):357–366. [PubMed] [Google Scholar]
- Nomura M., Yates J. L., Dean D., Post L. E. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein MRNA. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7084–7088. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz H., Towbin H., Elson D. The use of a cleavable crosslinking reagent to identify neighboring proteins in the 30-S ribosomal subunit of Escherichia coli. Eur J Biochem. 1976 Mar 16;63(1):83–92. doi: 10.1111/j.1432-1033.1976.tb10210.x. [DOI] [PubMed] [Google Scholar]
- Politz S. M., Glitz D. G. Ribosome structure: localization of N6,N6-dimethyladenosine by electron microscopy of a ribosome-antibody complex. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1468–1472. doi: 10.1073/pnas.74.4.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. R., Yabuki S., Sillers I. Y., Schindler D. G., Engelman D. M., Moore P. B. Positions of proteins S6, S11 and S15 in the 30 S ribosomal subunit of Escherichia coli. J Mol Biol. 1981 Dec 15;153(3):739–760. doi: 10.1016/0022-2836(81)90416-2. [DOI] [PubMed] [Google Scholar]
- Serdyuk I. N., Gogia Z. V., Venyaminov S. Y., Khechinashvili N. N., Bushuev V. N., Spirin A. S. Compact globular conformation of protein S4 from Escherichia coli ribosomes. J Mol Biol. 1980 Feb 15;137(1):93–107. doi: 10.1016/0022-2836(80)90159-x. [DOI] [PubMed] [Google Scholar]
- Serdyuk I. N., Shpungin J. L., Zaccai G. Neutron scattering study of the 13 S fragment of 16 S RNA and its complex with ribosomal protein S4. J Mol Biol. 1980 Feb 15;137(1):109–121. doi: 10.1016/0022-2836(80)90160-6. [DOI] [PubMed] [Google Scholar]
- Shatsky I. N., Mochalova L. V., Kojouharova M. S., Bogdanov A. A., Vasiliev V. D. Localization of the 3' end of Escherichia coli 16 S RNA by electron microscopy of antibody-labelled subunits. J Mol Biol. 1979 Oct 9;133(4):501–515. doi: 10.1016/0022-2836(79)90404-2. [DOI] [PubMed] [Google Scholar]
- Sommer A., Traut R. R. Identification by diagonal gel electrophoresis of nine neighboring protein pairs in the Escherichia coli 30 S ribosome crosslinked with methyl-4-mercaptobutyrimidate. J Mol Biol. 1975 Oct 5;97(4):471–481. doi: 10.1016/s0022-2836(75)80054-4. [DOI] [PubMed] [Google Scholar]
- Sommer A., Traut R. R. Identification of neighboring protein pairs in the Escherichia coli 30 S ribosomal subunit by crosslinking with methyl-4-mercaptobutyrimidate. J Mol Biol. 1976 Oct 5;106(4):995–1015. doi: 10.1016/0022-2836(76)90348-x. [DOI] [PubMed] [Google Scholar]
- Strycharz W. A., Nomura M., Lake J. A. Ribosomal proteins L7/L12 localized at a single region of the large subunit by immune electron microscopy. J Mol Biol. 1978 Dec 5;126(2):123–140. doi: 10.1016/0022-2836(78)90355-8. [DOI] [PubMed] [Google Scholar]
- Tischendorf G. W., Stöffler G. Localization of Escherichia coli ribosomal protein S4 on the surface of the 30S ribosomal subunit by immuno electron microscopy. I. Distribution of antibody-binding sites as obtained with immunoglobulins and monovalent antibody fragments from various S4-specific antisera. Mol Gen Genet. 1975 Dec 30;142(3):193–208. doi: 10.1007/BF00425645. [DOI] [PubMed] [Google Scholar]
- Vasiliev V. D., Koteliansky V. E., Rezapkin G. V. The complex of 16 S RNA with proteins S4, S7, S8, S15 retains the main morphological features of the 30 S ribosomal subparticle. FEBS Lett. 1977 Jul 1;79(1):170–174. doi: 10.1016/0014-5793(77)80376-1. [DOI] [PubMed] [Google Scholar]
- Winkelmann D., Kahan L. The accessibility of antigenic determinants of ribosomal protein S4 in situ. J Supramol Struct. 1979;10(4):443–455. doi: 10.1002/jss.400100407. [DOI] [PubMed] [Google Scholar]