Abstract
We report a mass spectrometry-based comparative “bottom up” proteomics approach that combines d0/d4-succinic anhydride labeling with commercially available hydrazine (Hz)-functionalized beads (Affi-gel Hz beads) for detection, identification and relative quantification of site-specific oxylipid modifications in biological matrices. We evaluated and applied this robust and simple method for the quantitative analysis of oxylipid protein conjugates in cardiac mitochondrial proteome samples isolated from 3- and 24-month-old rat hearts. The use of d0/d4-succinic anhydride labeling, Hz-bead based affinity enrichment, nanoLC fractionation and MALDI-ToF/ToF tandem mass spectrometry yielded relative quantification of oxylipid conjugates with residue-specific modification information. Conjugation of acrolein (ACR), 4-hydroxy-2-hexenal (HHE), 4-hydroxy-2-nonenal (HNE) and 4-oxo-2-noneal (ONE) to cysteine, histidine and lysine residues were identified. HHE conjugates were the predominant subset of Michael-type adducts detected in this study. The HHE conjugates showed higher levels in mitochondrial preparations from young heart congruent with previous findings by others that the n-3/n-6 PUFA ratio is higher in young heart mitochondrial membranes. Although this study focuses on protein adducts of reactive oxylipids the method might be equally applicable to protein carbonyl modifications caused by metal catalyzed oxidation reactions.
Keywords: Protein carbonyls, oxidative stress, lipid peroxidation products, mitochondria, mass spectrometry, heart
INTRODUCTION
Mass spectrometry-based proteomics enables the identification and relative quantification of hundreds of proteins in complex biological matrices. Although progress has been made in the identification of post-translationally modified proteins, the site-specific assignment of distinct protein modifications remains a major analytical challenge. Successful strategies for site-specific mapping of PTMs combine protein chemistry, bioaffinity strategies and LC-MS/MS techniques [1–4]. Beside the classical key PTMs, phosphorylation and glycosylation, oxidative protein modifications have emerged as significant modulators of redox homeostasis and signaling, and are considered as emerging markers of oxidative stress insult in numerous chronic diseases and age-associated pathological conditions [5–8].
Protein carbonylation can be the result of insult by a wide variety of ROS and electrophilic lipids [9]. Electrophilic aldehydes, such as 2-alkenals and 4-hydroxy-2-alkenals, are derived from lipid peroxidation processes, [10, 11]. The best studied electrophilic keto-containing lipids are prostanoids with cyclopentenone structure [12]. These α,β-unsaturated carbonyl-containing compounds are highly reactive toward nucleophilic amino acid residues (cysteine, histidine, and lysine) and predominately form Michael-type rather than Schiff-base adducts [13–20]. The accumulation of oxidatively modified proteins in cells, tissue and bodily fluids has been discussed as a reflection of the severity of oxidative stress under normal and diseased conditions. Elevated levels of oxidatively modified proteins have been reported to be present in various tissues in the context of numerous age-associated diseases and aging [21–23]. We and others propose that oxidative protein modifications may play an important role in the molecular mechanisms that contribute to mitochondrial dysfunction and the development of age-related cardiovascular diseases [24–26].
Oxidative modifications of proteins are difficult to characterize and quantify in biological samples due to the complexity of protein extracts, the low abundance of the modified proteins, and absence of distinguishing UV or visible spectrophotometric absorbance/fluorescence properties. Instead, detection and quantification of carbonylated proteins require the use of specific chemical probes, which include 2,4-dinitrophenylhydrazine (DNPH) [27], tritiated sodium borohydride [27–29], and fluorescence probes [30, 31]. More recently, chemical proteomic approaches have emerged that combine chemoselective affinity labeling of protein carbonyls and mass spectrometry for the identification of protein targets of oxidative modifications. These approaches allow the assignment of the modification site in favorable cases. We and others have introduced strategies that employ biotin-containing probes that target the aldehyde/keto group specific to oxidatively modified proteins [32–34]. In addition, capture-release solid-phase-based approaches have been developed, in which hydrazidefunctionalized glass beads were synthesized and used for the enrichment of 4-HNE conjugated peptides [35–37]. Quantification or relative assessment of levels of oxidative protein modifications is achieved by combining chemical labeling with stable-isotope labeling approaches [36, 38].
An alternative to hydrazide-functionalized glass beads are commercially available Affi-Gel® hydrazide (Hz) -functionalized agarose beads. This hydrazide-functionalized resin has been used previously for the identification and quantification of N-linked glycoproteins in conjunction with isotope-coded succinylation and tandem mass spectrometry [39]. Here we describe the applicability of this strategy for the analysis of Michael-type protein adducts of α,β-unsaturated aldehydic lipids in complex mixtures. We applied this comparative profiling method to the analysis of protein-oxylipid conjugates present in mitochondria from young and old rat heart.
MATERIALS AND METHODS
Materials
4-Hydroxy-2-nonenal was purchased from Cayman Chemicals Inc., Ann Arbor, MI. Succinic-d0 anhydride and succinic-d4 anhydride were purchased from Sigma-Aldrich (St. Louis, MO) and ISOTEC (Miamisburg, OH), respectively. Handee™ Spin columns were obtained from Pierce Biotechnology (Rockford, IL). E.coli thioredoxin (TRX) and sequencing grade-modified trypsin were purchased from Promega Corporation, Madison, WI. α-Cyano-4-hydroxycinnamic acid was from Sigma Chemicals (St. Louis, MO). The Affi-Gel® Hz (Hydrazide) gel (product # 1536047) was obtained from BIO-RAD (Hercules, CA). According to the manufacture's specification the Affi-gel Hz gel loading is greater than or equal to 10 micromoles of Hz per ml of Affi-gel Hydrazide gel.
HNE-modification and succinic anhydride-labeling of E. coli thioredoxin followed by Affi-Gel Hz enrichment
Thioredoxin (1 mg/ml in 10 mM sodium phosphate, pH 7.4) was reacted with a 10-fold molar excess of HNE for 3 hr at 37°C. Excess reagent was removed by ultrafiltration (5 kDa MWCO, Amicon Ultrafree-MC, Millipore, Billerica, MA). The modified protein reaction mixture was subsequently digested with trypsin overnight (E:S = 1:50, 37°C). The digest was passed through an ultrafiltration unit and peptides below 5 kDa were collected. Succinic anhydride in acetonitrile was added to a final concentration of 5 mg/mL and the sample was incubated at 37°C for 2 hr.
Affi-Gel® Hz beads were washed in Handee™ Spin columns with100 mM sodium phosphate pH 4.5, and reacted with peptides for 2 hr at 37 °C with gentle shaking. For these experiments, the peptide: gel ratio ranged from 1–4 (by weight) and the total reaction volume was 2–4 times the gel volume. The mixture was subsequently rinsed 4 times each in 100 mM NH4HCO3, 30% acetonitrile, and H2O, using 2 times the gel bed volume per rinse. The mixture was subsequently incubated in 2% formic acid, 40% acetonitrile for 1 hr at 37°C to release hydrazone-linked peptides. Released peptides were collected and the elution step was repeated. These fractions were combined and lyophilized prior to MALDI-MS/MS analysis.
Detection and quantification of oxylipid conjugates in mitochondrial proteome samples
Rat heart mitochondria were isolated according to Suh et al. [40]. Cardiac mitochondrial samples from three 3-month and three 24-months old male Fischer 344 rats were mixed, and disrupted by several freeze-thaw cycles. Soluble protein fractions were obtained by centrifugation. Protein concentrations were determined by using Coomassie Plus™ protein assay reagent. Aliquots of the mitochondrial proteins (350 μg each) were digested with trypsin in 0.1 M sodium phosphate buffer (pH 8.0) at 37 °C for 16 hr prior to ultrafiltration (Amicon Ultrafree-MC centrifugal filter, 10 kDa MWCO, Millipore, Billerica, MA). The d0-and d4-succinic anhydride solution was added respectively to the aliquots of digested peptides in a final concentration of 5 mg/mL. The samples were incubated at 37°C for 2 hr, and then mixed prior to coupling with the Affi-Gel® Hz beads. The enrichment of d0/d4-succinic anhydride-labeled oxylipid-conjugated peptides was carried out according to the same protocol as outlined above. Comparison of cardiac mitochondrial samples from young rats vs. old rats was conducted in a similar way except that the aliquots of young and old rat mitochondrial proteins were digested with trypsin separately, and then treated with d0-and d4- succinic anhydride, respectively.
NanoLC MALDI-MS/MS analysis
An Ultimate LC Packing system (Dionex, Sunnyvale, CA) coupled to a MALDI target spotter (Probot™) was used. Peptide samples were loaded onto a 5 × 0.50 mm C18 trap cartridge at a flow rate of 30 μL/min. After 10 min the trap cartridge was automatically switched in-line with a 75 μm I.D. × 15 cm C18 PepMap 100 column. Peptides were eluted with a gradient from 20 % to 100 % B over 60 min using as solvent A, 5 % acetonitrile containing 0.1 % TFA, and as solvent B, 80 % acetonitrile containing 0.1 % TFA. The flow rate was 0.325 μL/min. The column effluent was mixed with α-cyano-4-hydroxycinnamic acid (2 mg/ml in 50% aqueous acetonitrile containing 0.1 % TFA) via a T-junction, and fractions (20 seconds each) were collected onto a stainless steel target plate.
MALDI-MS/MS analysis was performed on an ABI 4700 Proteomics Analyzer with TOF/TOF optics equipped with a Nd:YAG laser operating at a wavelength of 355 nm (Applied Biosystems, Inc. Framingham, MA) as previously described [33]. Briefly, MS spectra were acquired for each fraction on the target. Precursor ions with signal-to-noise greater than 40 were selected using a job-wide processing method. Precursor ions were selected by a time-gated window of approximately 3–10 Da width. Gas (air) pressure in the collision cell was set to 6 × 10−7 Torr, and the collision energy was 1 kV. The MALDI data was processed using the ABI Peaks-to-Mascot program to create Mascot-searchable files.
Mass spectral data processing and database searching
For protein identification, MS/MS data were searched against the SwissProt database (Taxonomy rodentia) using the Mascot search engine (Matrix Science, London, UK). Trypsin/P was selected as the digesting enzyme and one missed cleavage site was allowed. The following variable modifications were considered: oxidized Met (147.04 Da, monoisotopic mass); d0/d4- succinic anhydride-labeled N-terminal amine group (monoisotopic masses 100.02 and 104.05 Da); d0/d4-succinic anhydride-labeled lysine (monoisotopic masses 228.11 and 232.14 Da); acrolein-Cys, -His and -Lys conjugates (monoisotopic masses 159.04, 193.09 and 184.12 Da, respectively); HHE-Cys, -His and -Lys conjugates (monoisotopic masses 217.08, 251.13 and 242.16 Da, respectively); HNE-Cys, -His and -Lys conjugates (monoisotopic masses 259.12, 293.17 and 284.21 Da, respectively); ONE-Cys, -His and -Lys conjugates (monoisotopic masses 257.11, 291.16 and 282.19 Da, respectively). Searches were done with an initial mass tolerance of 0.2 Da in the MS mode and 0.2 Da in the MS/MS mode. In addition, tandem mass spectra of oxylipid-conjugated peptides were visually inspected for preferred fragmentation patterns, such as N-terminal cleavage to a Pro (P) and C-terminal cleavage to Asp (D), and the presence of ions indicating loss of the 2-enal from the peptide oxylipid adduct.
Extraction of labeling ratios for relative quantification of d0/d4-succinic anhydride labeled oxylipid peptide conjugates and statistical evaluation
Ion peaks separated by 4 or 8 Da, corresponding to the presence of one or two d0/d4 succinic anhydride modifications, were identified from the MS spectra of the entire LC-MALDI MS run. The ratio of these peaks was determined using the area under the isotope cluster, as identified by the ABI 4000 series explorer software. In the case that the isotope-labeled peptide ion pairs were detected in successive plat spots, we used the sum of the areas under isotopic clusters of peaks to determine the ratios. Identification of the d0/d4 (or d0/d8) pairs, summation of the cluster areas, and determination of the ratios was automated using a Visual Basic program implemented in Microsoft Excel. Ratios of significant peaks were confirmed by manual analysis of individual spectra.
P-values were calculated using the open source software R. Each d0/d4(8) peak area ratio determined for the oxylipid peptide conjugates was compared with the mean d0/d4(8) ratio of the non-oxylipid peptides using a confidence interval of 95% (Table 1). A two-sample t-test was applied to test the null hypothesis that there is no significant difference between the two samples. If the p-value was less than 0.05, the null hypothesis was rejected in favor of the alternative hypothesis that there is a significant difference between the two samples.
Table 1.
Differential proteomic analysis of oxylipid peptide conjugates in cardiac mitochondrial proteomes from 3- and 24-month old rats. Observed d0/d4,8 ratios of peptide oxylipid conjugates after post-digest labeling with isotope-coded succinic anhydride, Affi-gel Hz enrichment and nanoLC MALDI-MS/MS analysis.
| Protein Name | Sprot ID | Peptide sequence1 | d0/d4,8 | ||||
|---|---|---|---|---|---|---|---|
| sample 1 | sample 2 | sample 3 | Average ± SD | p-value | |||
| Complex I | |||||||
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 | NDUS3_MOUSE | ILTDYGFEGH*PFRb | 1.36 | 1.49 | 1.61 | 1.49 ± 0.13 | 0.023 |
| NADH-ubiquinone oxidoreductase 75 kDa subunit | NDUS1_RAT | VDSDNLC*TEEIFPTEGAGTDLRb | 2.33 | 1.89 | 2.17 | 2.13 ± 0.22 | 0.018 |
| NADH dehydrogenase 1 alpha subcomplex subunit 5 | NDUA5_RAT | TTGLVGLAVC*DTPHERa | 1.57 | 1.73 | 1.58 | 1.63 ± 0.09 | 0.005 |
| Complex II | |||||||
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit | DHSA_RAT | TYFSC*TSAHTSTGDGTAMVTRa | 1.32 | 1.25 | 1.47 | 1.35 ± 0.11 | 0.087 |
| TLNEADC*ATVPPAIRa | 1.38 | 1.30 | 1.47 | 1.38 ± 0.09 | 0.021 | ||
| TLNEADC*ATVPPAIRb | 1.27 | 1.33 | 1.08 | 1.23 ± 0.13 | 0.489 | ||
| Complex III | |||||||
| Ubiquinol-cytochrome-c reductase complex core protein I | UQCR1_MOUSE | NALISHLDGTTPVC*EDIGRa | 1.22 | 1.25 | 1.40 | 1.29 ± 0.10 | 0.149 |
| NALISHLDGTTPVC*EDIGRb | 1.54 | 1.83 | 1.63 | 1.67 ± 0.15 | 0.012 | ||
| YFYDQCPAVAGYGPIEQLSDYNRb | 1.76 | 1.81 | n.d. | 1.79 ± 0.04 | 0.001 | ||
| Ubiquinol-cytochrome-c reductase complex core protein II | UQCR2_RAT | NALANPLYC*PDYRa | 1.23 | 1.33 | 1.35 | 1.30 ± 0.06 | 0.049 |
| NALANPLYC*PDYRb | 1.76 | 1.78 | 1.72 | 1.75 ± 0.03 | 0.001 | ||
| Complex IV | |||||||
| Cytochrome c oxidase subunit Via isoform 2 | CX6A2_RAT | GDH*GGAGANTWRa | 1.01 | 1.25 | 1.25 | 1.17 ± 0.14 | 0.944 |
| GDH*GGAGANTWRb | 1.61 | 1.69 | 1.67 | 1.66 ± 0.04 | 0.001 | ||
| GDH*GGAGANTWRc | 1.28 | 1.61 | 1.68 | 1.52 ± 0.21 | 0.106 | ||
| Cytochrome c oxidase subunit VIb isoform 1 | CX6B1_RAT | GGDVSVC*EWYRb | 1.77 | 1.75 | 1.48 | 1.67 ± 0.16 | 0.015 |
| Cytochrome c oxidase subunit VIIa isoform 2 | CX7A2_RAT | LFQEDNGMPVH*LKb | 1.46 | 1.38 | 1.28 | 1.37 ± 0.09 | 0.043 |
| Cytochrome c oxidase subunit 4 isoform 1 | COX41_RAT | DYPLPDVAH*VKb | 0.95 | 1.10 | 1.30 | 1.12 ± 0.18 | 0.705 |
| Cytochrome c oxidase subunit 3 | COX3_RAT | EGTYQGHH*TPIVQKb | 1.68 | 1.93 | 2.10 | 1.90 ± 0.21 | 0.028 |
| Complex V | |||||||
| ATP synthase beta subunit | ATPβ_RAT | IMDPNIVGSEH*YDVARb | 1.14 | 1.18 | 1.11 | 1.14 ± 0.04 | 0.644 |
| EGNDLYH*EMIESGVINLKb | 1.38 | 1.27 | 1.23 | 1.29 ± 0.08 | 0.081 | ||
| ß-Oxidation | |||||||
| Long-chain specific acyl-CoA dehydrogenase | ACADL_RAT | C*IGAIAMTEPGAGSDLQGVRb | 0.98 | 1.05 | 1.14 | 1.06 ± 0.08 | 0.135 |
| AFVDSC*LQLHETKa | 0.90 | 1.10 | 1.18 | 1.06 ± 0.14 | 0.333 | ||
| Acetyl-CoA acetyltransferase | THIL_RAT | IHMGNC*AENTAKa | 1.27 | 1.38 | n.d. | 1.33 ± 0.08 | 0.128 |
| Other | |||||||
| ADP/ATP translocase 1 | ADT1_RAT | GADIMYTGTVDC*WRa | 1.64 | 1.65 | 1.48 | 1.59 ± 0.10 | 0.007 |
| GADIMYTGTVDC*WRb | 1.44 | 1.64 | 1.53 | 1.54 ± 0.10 | 0.011 | ||
| LLLQVQH*ASKb | 1.48 | 1.30 | 1.34 | 1.37 ± 0.09 | 0.046 | ||
| Creatine kinase, sarcomeric mitochondrial precursor | KCRS_RAT | ITH*GQFDERb | 1.86 | 1.59 | 1.94 | 1.80 ± 0.18 | 0.029 |
| Averaged d0/d4(8) ratio for non-oxylipid peptides | 1.13 | 1.13 | 1.23 | 1.16 ± 0.06 | |||
The site of oxylipid conjugation is marked with an asterisk (*). The chemical nature of the oxylipid modification is indicated by the following superscripts: a, acrolein, b, HHE, c, ONE, d, HNE.
n.d., not detected.
RESULTS AND DISCUSSION
Analytical strategy and use of isotope-coded succinic anhydride tagging in combination with Affi-Gel Hydrazide (Hz)-beads
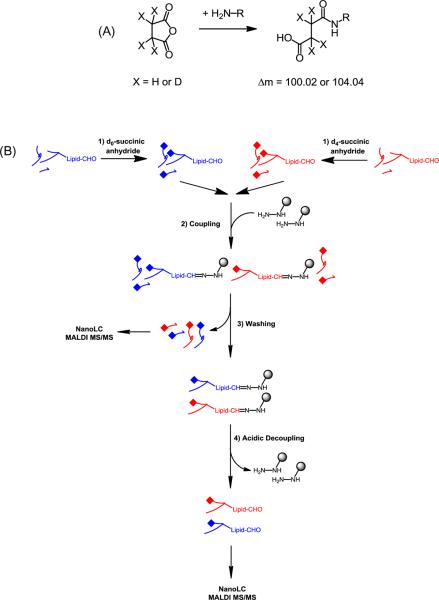
The overall strategy for labeling and isolating oxylipid-conjugated peptides is a three-step procedure (Figure 1). In the first step, all primary amine groups in the tryptic digest are converted to the succinate carboxylic acids and stable isotopes are introduced for quantitative analysis. In the second step, the isotopically labeled peptides are combined and subjected to an one-pot reaction in the presence of the Affi-Gel® Hydrazide (Hz) beads, in which hydrazide groups on the surface of the solid support form hydrazone bonds with aldehyde groups from the oxylipid-conjugated peptides. Finally, covalently bound oxylipid-conjugated peptides are detached from the gel through a mild acid hydrolysis of the hydrazone bonds, followed by nano-LC separation and MALDI-MS/MS analysis. Succinylation of tryptic peptides containing a C-terminal arginine residue results in a mass difference of 4 Da between the d0- and d4-labeled isotopomeric ion pair. Whereas peptides containing a C-terminal lysine are succinylated at both the N-terminal and the ε-amino group of the lysine residue, thus resulting in a mass difference of 8 Da between the isotopmeric ions. The use of d0/d4-succinic anhydride for determining relative levels of targeted tryptic peptides in paired samples has been described previously [39, 41–43].
Figure 1.
Analytical strategy used for the mass spectrometry-based site-specific identification and relative quantification of oxylipid peptide conjugates in complex biological mixtures. (A) Primary amine groups in peptides are chemically labeled with d0/d4 succinic anhydride. (B) For the selective capture of oxylipid peptide conjugates commercially available Affi gel-Hz beads are used. After removal of non-oxylipid-modified peptides, the hydrazone-linked oxylipid peptide conjugates are released and submitted to nanoLC fractionation and subsequent MALDI-MS/MS analysis. Peaks separated by 4 or 8 Da, corresponding to the presence of one or two d0/d4 modifications, were identified from the MS spectra and the d0/d4(8) (light/heavy) ratios were extracted by using the area under the isotope clusters (for details see experimental section).
We initially tested the method using E.coli thioredoxin (Trx) modified with HNE [33]. Two aliquots of the tryptic digests of HNE-modified Trx were treated with d0-and d4-succinic anhydride, respectively. The samples (10 nmol) were mixed and then bound to the Affi-Hz Gel. The experiment was monitored by MALDI-MS and the result is presented in the supplemental materials, Figure S1. Only the HNE-modified peptides, with m/z 1988.0 (d0), 1992.0 (d4), 2088.0 (d0) and 2096.0 (d8), were enriched with the expected 1:1 ratio as d0-and d4-labeled isotopomeric ion pairs, demonstrating the specificity of the method. The enriched peptides were subjected to tandem mass spectrometry to confirm the modification of the His residue at position 6 with HNE (supplemental materials, Figure S2). Succinylation converts basic to acidic sites which may affect ionization efficiencies and cause discrimination of succinylated peptides in the positive ionization mode. No corrections were made to adjust for possible differences in ionization efficiencies of succinylated versus unmodified peptides. The overall sample recovery efficiency was estimated at approximately 45% of the starting material by measuring the relative abundance of d0/d4(8)- succinic anhydride-labeled and HNE-modified peptides in the MALDI mass spectrum using the method described by Tao et al. [44] (data not shown).
Evaluation of the comparative d0 and d4-succinic anhydride labeling strategy for the analysis of mitochondrial proteome samples
We first tested the enrichment performance of Affi-Gel® Hz beads for profiling of oxylipid-modified peptides in a trypic digest of the soluble protein fraction of mitochondria isolated from rat heart (Table S1, Figures S4–1 through S4–34). Next, we combined the Affi-Gel® Hz bead enrichment strategy for oxylipid peptide conjugates with succinic anhydride labeling (Table S2). The compiled MALDI tandem mass spectral data sets (Table S1 and S2) show that the majority of oxylipid-conjugates were HHE conjugates (28 characterized modifications, 52 %), followed by acrolein derivatives (22 modifications, 41%). The longer chain oxylipids, HNE and ONE, were only found on 4 peptides (7 %). Multiple oxylipid modifications were identified on 7 sites: Succinate dehydrogenase [ubiquinone] flavoprotein subunit (DHSA_RAT) Cys-258 and Cys-646; Ubiquinol-cytochrome-c reductase complex core protein I (UQCR1_MOUSE) Cys-410; Ubiquinol-cytochrome-c reductase complex core protein II (UQCR2_RAT) Cys-191; Cytochrome c oxidase subunit VIa isoform 2 (CX6A2_RAT) His-20; ATP synthase β subunit (ATPβ_RAT) His-417 and ADP/ATP translocase 1 (ADT_RAT) Cys-256. As in previous work, Michael-type adduction of reactive oxylipids was found on constituents of the major mitochondrial energy pathways including oxidative phosphorylation, the TCA cycle and β-oxidation [26].
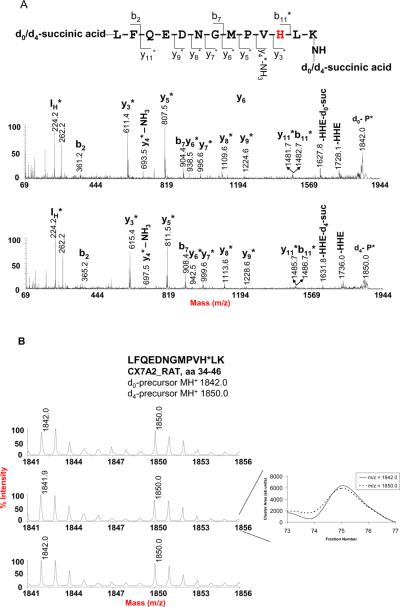
Aliquots of the digest from the same mitochondrial protein sample were used to evaluate the precision and accuracy of this method for quantification of endogenous oxylipid-protein conjugates using d0- and d4- succinylation coupled with Affi-Gel® Hz. The two aliquots were reacted separately with d0-and d4- succinic anhydride and combined prior to Affi-Gel® Hz enrichment. The enriched fraction was further processed as described above. For determining the isotopomeric ratios of d0- and d4(8)-SA-tagged and oxylipid-conjugated peptide pairs, ion traces observed in survey scans were evaluated to calculate the d0/d4(8)-ratios using the sum of the areas under isotopic clusters. In Figure 2, the isotope cluster observed for the peptide LFQEDNGMPVH*LK from Cytochrome c oxidase subunit VIIa isoform 2 (CX7A2_RAT) is shown as an example. The peak area ratio of the isotopomeric pair was determined with a variability of 9.4% RSD and an average ratio of 0.96 ± 0.09 (mean ± SD) from three independently prepared samples. The peak area ratios for 19 d0- and d4(8)-SA-tagged oxylipid-peptide conjugates are listed in Table S3 (in supplemental materials). The average ratios of these d0/d4(8)-SA tagged peptides range from 0.89 ± 0.07 for the peptide from NADH dehydrogenase 1 alpha subcomplex subunit 5 (NDUA5_RAT) to 1.17 ± 0.27 for the peptide derived from the long-chain specific Cytochrome c oxidase subunit 3 (COX3_RAT). The overall average ratio was 1.03 ± 0.07. The overall variability in peptide quantification for these peptides was 6.8% in accord to previous findings that relative quantification protocols based on stable isotope coding show a variability of 6 – 8 % [45]. The variability of the current isotope-coded succinylation approach is also comparable to a recently reported method for the differential quantification of HNE-modification sites in proteins which employs reductive dimethylation with isotope-coded formaldehyde [46].
Figure 2.
Reproducibility of relative peak area measurements illustrated for the d0/d4-succinic anhydride-labeled peptide LFQEDNGMPVH*LK (aa 34–46) of cytochrome c oxidase polypeptide VIIa-liver/heart (CX7A2_RAT) with endogenous HHE conjugate on His-44. (A) MALDI mass spectra of the d0 and d4-SA-tagged HHE peptide conjugate. The bn and yn fragment ions indicate that the N-terminal Leu residue and C-terminal Lys residue were both tagged by the d0/d4- succinic anhydride, which is consistent with a mass difference of 8 Da from the paired set of mass peaks of the precursor ions as shown in B. The fragment ions at m/z 1728.1 and 1736.0 indicate loss of HHE from the d0 and d8-tagged precursor, respectively. IH m/z 224.2, immonium ion of HHE-His. (B) MALDI survey mass spectra from three samples prepared from rat heart mitochondria. The reconstructed ion chromatograms were generated using Peak Explorer™ software version 3.0, in which each fraction number corresponds to 20 seconds eluting time. The peak area ratio of the d0/d8 pair was determined by summation of the peak cluster areas of each fraction across the LC-MALDI peak. The peak cluster areas were determined by ABI 4000 Data Explorer® software version 4.3, and a custom Visual Basic program was used to automatically sum the peak areas.
Differential proteomics analysis of mitochondrial proteomes from 3 and 20-month old hearts
Finally, we used our new method for a comparative analysis of oxylipid-conjugates present in mitochondrial proteomes obtained from hearts of 3-months and 20-month old rats. In this report, we selected 26 most frequently and fully identified isotopomeric ion pairs from a single experiment to demonstrate the feasibility of this method for the relative quantification of oxylipid-conjugated peptides in a biological matrix. The analysis of the d0/d4(8) ratio implies that in average the content of the oxylipid conjugates was higher in the proteome samples obtained from the 3-months old hearts. The current solid phase approach allows the quantitative analysis of d0/d4(8)-tagged non-oxylipid conjugated peptides from the non-bound fraction that is obtained after step 3 of the method (Figure 1B). We used this information to confirm that the protein concentrations of the d0 and d4(8) preparations, which were mixed for the comparative analyses, were indeed close to uniformity (Table 1).
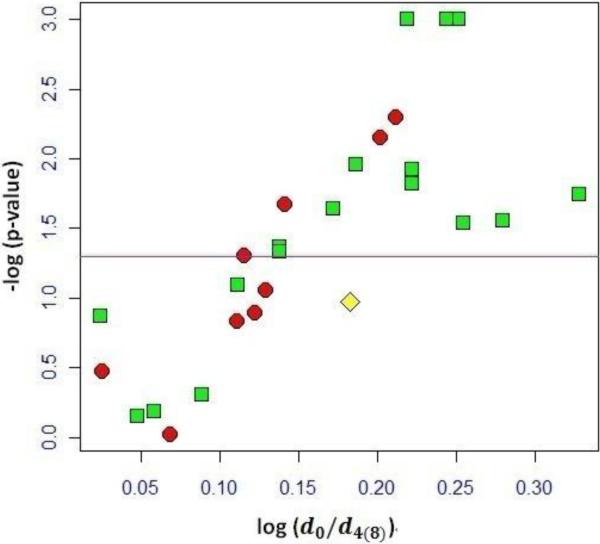
We used a Volcano plot to evaluate the statistical significance of the d0/d4(8) ratios of distinct oxylipid peptide conjugates. Figure 3 shows a volcano plot for the data compiled in table 1. Each labeling ratio was compared to the normalization factor by using a confidence interval of 95%. The two-sample t-test was applied to test the null hypothesis of whether or not the d0/d4(8) ratio of the oxylipid peptide conjugates was different from the average d0/d4(8)-SA labeled non-oxylipid conjugated peptides. The ordinate shows the −log10 p-values; the abscissa shows the as log10 values of the mean of the labeling ratio of the oxylipid peptides from our three paired samples. Oxylipid peptides conjugates that show −log(p-values) ≥ 1.30 are considered as being significant different. The majority of the labeling ratios that had p-values <0.05 were HHE conjugates (11 out 15) and within this subset 6 conjugates were on His residues and 5 on Cys residues. Overall, the current dataset implied that HHE conjugation was more prominently observable in the mitochondrial proteome samples from the 3-month hearts compared to the preparation that originated from the 20-month old animals. This finding is in agreement with previous reports that oxidation of n-3 polyunsaturated fatty acids yields HHE and that mitochondrial membranes from young hearts have higher content of n-3 polyunsaturated fatty acids [47].
Figure 3.
Volcano plot compiling the d0/d4(8)-SA labeling data for the differential profiling analysis of endogenous oxylipid conjugates in proteome samples from 3-month and 20-month old rat heart mitochondria. Each data point represents the d0/d4(8) ratio observed for a distinct oxylipid peptide conjugate. Red circles represent peptide conjugates with acrolein, green squares indicate HHE peptide adducts, and the data point representing the peptide adduct with ONE is shown as the yellow diamond. The horizontal line at -log10 (p-value) = 1.30 indicates where p = 0.05; data above the line have p < 0.05 and are considered as significant different. The plot shows that 15 d0/d4-SA labeled ratios observed for oxylipid peptides were significantly different from the mean ratio extracted for the non-oxylipid peptides. Of the d0/d4-SA labeled oxylipid peptide conjugates with p < 0.05, the majority was related to HHE adducts (12 out of 15).
In the current datasets the majority of the oxylipid conjugates, that were identified and subsequently quantitatively assessed, were acrolein and HHE adducts. However, ONE and HNE conjugates may seem to be underrepresented. There are numerous factors that may contribute to this bias and some of these may be related to sample storage, preparation, discrimination during separation and mass spectrometry analysis, and the automated peptide/protein identification including the statistical techniques used for scoring of uninterpreted tandem mass spectral data [48].
An important experimental detail relates to the pH value of the peptide solution during the capture step which needs to be slightly acidic (pH 4.5) to allow conditions that promote presence of the “open” reactive aldehydic form of the peptide oxylipdid conjugate; 4-hydroxy alkenal peptide conjugates readily form half-acetals that would escape the capture reactions.
In the current study, post-digestion succinylation was performed in phosphate buffer at pH 7.4, i.e. under conditions at which succinic anhydride modifies the N-terminal α-NH2 group of peptides and the ε-amino group of lysine residues. Therefore, peptides were observed that contain one or two SA tags depending on the C-terminal residue of the tryptic peptide (Arg or Lys). Selective succinylation of α-amino groups in sodium acetate buffer at pH 7.6 was described by Koehler et al. [49].
CONCLUSIONS
We report a mass spectrometry-based comparative “bottom up” proteomics approach that combines d0/d4-succinic anhydride labeling in combination with hydrazine-functionalized beads (Affi-Hz gel) for detection, identification and relatively quantification of distinct site-specific oxylipid modifications in biological matrices. We evaluated and applied this method for the analysis of oxylipid protein conjugates in proteome samples from 3- and 24-month-old rat heart mitochondria. The described method may be useful for a wide range of comparative profiling studies with a focus on protein carbonyls when used with nanoLC coupled to robotic sample spotting and MALDI tandem mass spectrometry. However, the applicability of the method may be limited when online nanoLC-ESI-MS is used due to the chromatographic isotope shift associated with deuterium-containing tags [43, 50]. Although the current study focused on Michael adducts of reactive oxylipids the method might be equally applicable to aldehydic protein modifications caused by metal catalyzed oxidation reactions [51].
Supplementary Material
HIGHLIGHTS
Site-specific information is reported for oxylipid modifications of rat heart mitochondrial proteins.
The new method combines isotope-coded succinylation and hydrazide-based capture/release.
Relative changes are reported for oxylipid protein modifications in mitochondrial preparations of 3- and 24-month-old rat heart.
Acknowledgements
This study was supported by NIH/NIA grant R01AG025372. We acknowledge the use of Oregon State University's Mass Spectrometry Facility and Core supported in part by NIH/NIEHS grant P30ES00210. B.H. acknowledges financial support from “the Fundamental Research Funds for the Central Universities of China (2010QNA4014)”, and “NSF from ZheJiang Province, China (Y2100044)”.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Leonard SE, Carroll KS. Chemical “omics” approaches for understanding protein cysteine oxidation in biology. Current Opinion in Chemical Biology. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Chouchani ET, James AM, Fearnley IM, Lilley KS, Murphy MP. Proteomic approaches to the characterization of protein thiol modification. Current Opinion in Chemical Biology. 2011;15:120–128. doi: 10.1016/j.cbpa.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heal WP, Tate EW. Getting a chemical handle on protein post-translational modification. Org Biomol Chem. 2010;8:731–738. doi: 10.1039/b917894e. [DOI] [PubMed] [Google Scholar]
- 4.Maier CS, Chavez J, Wang J, Wu J. Protein adducts of aldehydic lipid peroxidation products identification and characterization of protein adducts using an aldehyde/keto-reactive probe in combination with mass spectrometry. Methods Enzymol. 2010;473:305–330. doi: 10.1016/S0076-6879(10)73016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schopfer FJ, Cipollina C, Freeman BA. Formation and signaling actions of electrophilic lipids. Chem Rev. 2011;111:5997–6021. doi: 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritz KS, Petersen DR. Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem Res Toxicol. 2011;24:1411–1419. doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marnett LJ, Riggins JN, West JD. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J Clin Invest. 2003;111:583–593. doi: 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan LJ, Forster MJ. Chemical probes for analysis of carbonylated proteins: a review. J Chromatogr B Analyt Technol Biomed Life Sci. 879:1308–1315. doi: 10.1016/j.jchromb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchida K. Current status of acrolein as a lipid peroxidation product. Trends Cardiovasc Med. 1999;9:109–113. doi: 10.1016/s1050-1738(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 11.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 12.Garzón B, Oeste CL, Díez-Dacal B, Pérez-Sala D. Proteomic studies on protein modification by cyclopentenone prostaglandins: Expanding our view on electrophile actions. Journal of Proteomics. 2011;74:2243–2263. doi: 10.1016/j.jprot.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 13.Carini M, Aldini G, Facino RM. Mass spectrometry for detection of 4-hydroxy-trans-2-nonenal (HNE) adducts with peptides and proteins. Mass Spectrom Rev. 2004;23:281–305. doi: 10.1002/mas.10076. [DOI] [PubMed] [Google Scholar]
- 14.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 15.Grasse LD, Lame MW, Segall HJ. In vivo covalent binding of trans-4-hydroxy-2-hexenal to rat liver macromolecules. Toxicol Lett. 1985;29:43–49. doi: 10.1016/0378-4274(85)90198-5. [DOI] [PubMed] [Google Scholar]
- 16.Grune T, Davies KJ. The proteasomal system and HNE-modified proteins. Mol Aspects Med. 2003;24:195–204. doi: 10.1016/s0098-2997(03)00014-1. [DOI] [PubMed] [Google Scholar]
- 17.Lee JY, Je JH, Kim DH, Chung SW, Zou Y, Kim ND, Ae Yoo M, Suck Baik H, Yu BP, Chung HY. Induction of endothelial apoptosis by 4-hydroxyhexenal. Eur J Biochem. 2004;271:1339–1347. doi: 10.1111/j.1432-1033.2004.04042.x. [DOI] [PubMed] [Google Scholar]
- 18.Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol Aspects Med. 2003;24:149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 19.Shibata N, Yamada S, Uchida K, Hirano A, Sakoda S, Fujimura H, Sasaki S, Iwata M, Toi S, Kawaguchi M, Yamamoto T, Kobayashi M. Accumulation of protein-bound 4-hydroxy-2-hexenal in spinal cords from patients with sporadic amyotrophic lateral sclerosis. Brain Res. 2004;1019:170–177. doi: 10.1016/j.brainres.2004.05.110. [DOI] [PubMed] [Google Scholar]
- 20.Zarkovic K. 4-hydroxynonenal and neurodegenerative diseases. Mol Aspects Med. 2003;24:293–303. doi: 10.1016/s0098-2997(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 21.Salmon AB, Richardson A, Perez VI. Update on the oxidative stress theory of aging: does oxidative stress play a role in aging or healthy aging? Free Radic Biol Med. 2010;48:642–655. doi: 10.1016/j.freeradbiomed.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toda T, Nakamura M, Morisawa H, Hirota M, Nishigaki R, Yoshimi Y. Proteomic approaches to oxidative protein modifications implicated in the mechanism of aging. Geriatrics & Gerontology International. 2010;10:S25–S31. doi: 10.1111/j.1447-0594.2010.00606.x. [DOI] [PubMed] [Google Scholar]
- 23.Beal MF. Oxidatively modified proteins in aging and disease. Free Radical Biology and Medicine. 2002;32:797–803. doi: 10.1016/s0891-5849(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 24.Lesnefsky EJ, Hoppel CL. Oxidative phosphorylation and aging. Ageing Res Rev. 2006;5:402–433. doi: 10.1016/j.arr.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Lesnefsky EJ, Hoppel CL. Ischemia-reperfusion injury in the aged heart: role of mitochondria. Arch Biochem Biophys. 2003;420:287–297. doi: 10.1016/j.abb.2003.09.046. [DOI] [PubMed] [Google Scholar]
- 26.Choksi KB, Papaconstantinou J. Age-related alterations in oxidatively damaged proteins of mouse heart mitochondrial electron transport chain complexes. Free Radic Biol Med. 2008;44:1795–1805. doi: 10.1016/j.freeradbiomed.2008.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, Ahn BW, Shaltiel S, Stadtman ER. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 28.Yan LJ, Orr WC, Sohal RS. Identification of oxidized proteins based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunochemical detection, isoelectric focusing, and microsequencing. Anal Biochem. 1998;263:67–71. doi: 10.1006/abio.1998.2799. [DOI] [PubMed] [Google Scholar]
- 29.Lenz AG, Costabel U, Shaltiel S, Levine RL. Determination of carbonyl groups in oxidatively modified proteins by reduction with tritiated sodium borohydride. Anal Biochem. 1989;177:419–425. doi: 10.1016/0003-2697(89)90077-8. [DOI] [PubMed] [Google Scholar]
- 30.Chaudhuri AR, de Waal EM, Pierce A, Van Remmen H, Ward WF, Richardson A. Detection of protein carbonyls in aging liver tissue: A fluorescence-based proteomic approach. Mech Ageing Dev. 2006;127:849–861. doi: 10.1016/j.mad.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Vaishnav RA, Getchell ML, Poon HF, Barnett KR, Hunter SA, Pierce WM, Klein JB, Butterfield DA, Getchell TV. Oxidative stress in the aging murine olfactory bulb: redox proteomics and cellular localization. J Neurosci Res. 2007;85:373–385. doi: 10.1002/jnr.21130. [DOI] [PubMed] [Google Scholar]
- 32.Yoo BS, Regnier FE. Proteomic analysis of carbonylated proteins in two-dimensional gel electrophoresis using avidin-fluorescein affinity staining. Electrophoresis. 2004;25:1334–1341. doi: 10.1002/elps.200405890. [DOI] [PubMed] [Google Scholar]
- 33.Chavez J, Wu J, Han B, Chung WG, Maier CS. New role for an old probe: affinity labeling of oxylipid protein conjugates by N'-aminooxymethylcarbonylhydrazino D-biotin. Anal Chem. 2006;78:6847–6854. doi: 10.1021/ac0607257. [DOI] [PubMed] [Google Scholar]
- 34.Han B, Stevens JF, Maier CS. Design, synthesis, and application of a hydrazide-functionalized isotope-coded affinity tag for the quantification of oxylipid-protein conjugates. Anal Chem. 2007;79:3342–3354. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 35.Rauniyar N, Stevens SM, Prokai-Tatrai K, Prokai L. Characterization of 4-Hydroxy-2-nonenal-Modified Peptides by Liquid Chromatography-Tandem Mass Spectrometry Using Data-Dependent Acquisition: Neutral Loss-Driven MS3 versus Neutral Loss-Driven Electron Capture Dissociation. Anal Chem. 2008;81:782–789. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauniyar N, Prokai L. Isotope-coded dimethyl tagging for differential quantification of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. J Mass Spectrom. 2011;46:976–985. doi: 10.1002/jms.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roe MR, Xie H, Bandhakavi S, Griffin TJ. Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Anal Chem. 2007;79:3747–3756. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 38.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 40.Suh JH, Heath SH, Hagen TM. Two subpopulations of mitochondria in the aging rat heart display heterogenous levels of oxidative stress. Free Radic Biol Med. 2003;35:1064–1072. doi: 10.1016/s0891-5849(03)00468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R, Sioma CS, Wang S, Regnier FE. Fractionation of isotopically labeled peptides in quantitative proteomics. Anal Chem. 2001;73:5142–5149. doi: 10.1021/ac010583a. [DOI] [PubMed] [Google Scholar]
- 42.Che FY, Fricker LD. Quantitative peptidomics of mouse pituitary: comparison of different stable isotopic tags. J Mass Spectrom. 2005;40:238–249. doi: 10.1002/jms.743. [DOI] [PubMed] [Google Scholar]
- 43.Zhang R, Sioma CS, Thompson RA, Xiong L, Regnier FE. Controlling deuterium isotope effects in comparative proteomics. Anal Chem. 2002;74:3662–3669. doi: 10.1021/ac025614w. [DOI] [PubMed] [Google Scholar]
- 44.Tao WA, Wollscheid B, O'Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 45.Madian AG, Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res. 9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rauniyar N, Prokai L. Isotope-coded dimethyl tagging for differential quantification of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. J Mass Spectrom. 46:976–985. doi: 10.1002/jms.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pepe S. Effect of dietary polyunsaturated fatty acids on age-related changes in cardiac mitochondrial membranes. Exp Gerontol. 2005;40:369–376. doi: 10.1016/j.exger.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Guo J, Prokai L. To tag or not to tag: a comparative evaluation of immunoaffinity-labeling and tandem mass spectrometry for the identification and localization of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. J Proteomics. 74:2360–2369. doi: 10.1016/j.jprot.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koehler CJ, Arntzen MO, Strozynski M, Treumann A, Thiede B. Isobaric peptide termini labeling utilizing site-specific N-terminal succinylation. Anal Chem. 2011;83:4775–4781. doi: 10.1021/ac200229w. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Wang Y, Li S. Deuterium isobaric amine-reactive tags for quantitative proteomics. Anal Chem. 82:7588–7595. doi: 10.1021/ac101306x. [DOI] [PubMed] [Google Scholar]
- 51.Chavez JD, Bisson WH, Maier CS. A targeted mass spectrometry-based approach for the identification and characterization of proteins containing alpha-aminoadipic and gamma-glutamic semialdehyde residues. Anal Bioanal Chem. 398:2905–2914. doi: 10.1007/s00216-010-4289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.