Summary
Streptococcus mutans regulates genetic competence through a complex network that receives inputs from a number of environmental stimuli, including two signaling peptides designated as CSP and XIP. The response of the downstream competence genes to these inputs shows evidence of stochasticity and bistability and has been difficult to interpret. We have used microfluidic, single-cell methods to study how combinations of extracellular signals shape the response of comX, an alternative sigma factor governing expression of the late competence genes. We find that the composition of the medium determines which extracellular signal (XIP or CSP) can elicit a response from comX and whether that response is unimodal or bimodal across a population of cells. In a chemically defined medium, exogenous CSP does not induce comX, whereas exogenous XIP elicits a comX response from all cells. In complex medium, exogenous XIP does not induce comX, whereas CSP elicits a bimodal comX response from the population. Interestingly, bimodal behavior required an intact copy of comS, which encodes the precursor of XIP. The comS-dependent capability for both unimodal and bimodal response suggests that a constituent – most likely peptides – of complex medium interacts with a positive feedback loop in the competence regulatory network.
Keywords: single-cell, bistability, quorum sensing, gene regulation, feedback, transformation
Introduction
Streptococcus mutans is a common member of the oral microbiome and the primary causative agent of dental caries. A number of factors contribute to the establishment, persistence and pathogenic potential of S. mutans, including its abilities to form biofilms on the tooth surface, to ferment a variety of carbohydrates, to tolerate low pH and to adapt rapidly to changes in available energy sources (Senadheera and Cvitkovitch, 2008). Many virulence attributes of S. mutans are connected to the capacity of the organism to develop natural genetic competence, i.e. the ability to take up exogenous DNA. S. mutans can enter a transient competent state during the early- or mid-exponential growth phase. A complex regulatory network controls competence and receives inputs from a number of environmental stimuli. The competence network in turn influences a number of other virulence behaviors, such as biofilm formation and bacteriocin production (Smith and Spatafora, 2012). The complex entanglement of genetic competence with other virulence factors in S. mutans raises the question of how the organism uses extracellular cues to regulate competence. The purpose of this work was to investigate and understand signal integration in S. mutans competence by analyzing – at the single cell level – how the network responds to precisely defined combinations of environmental inputs.
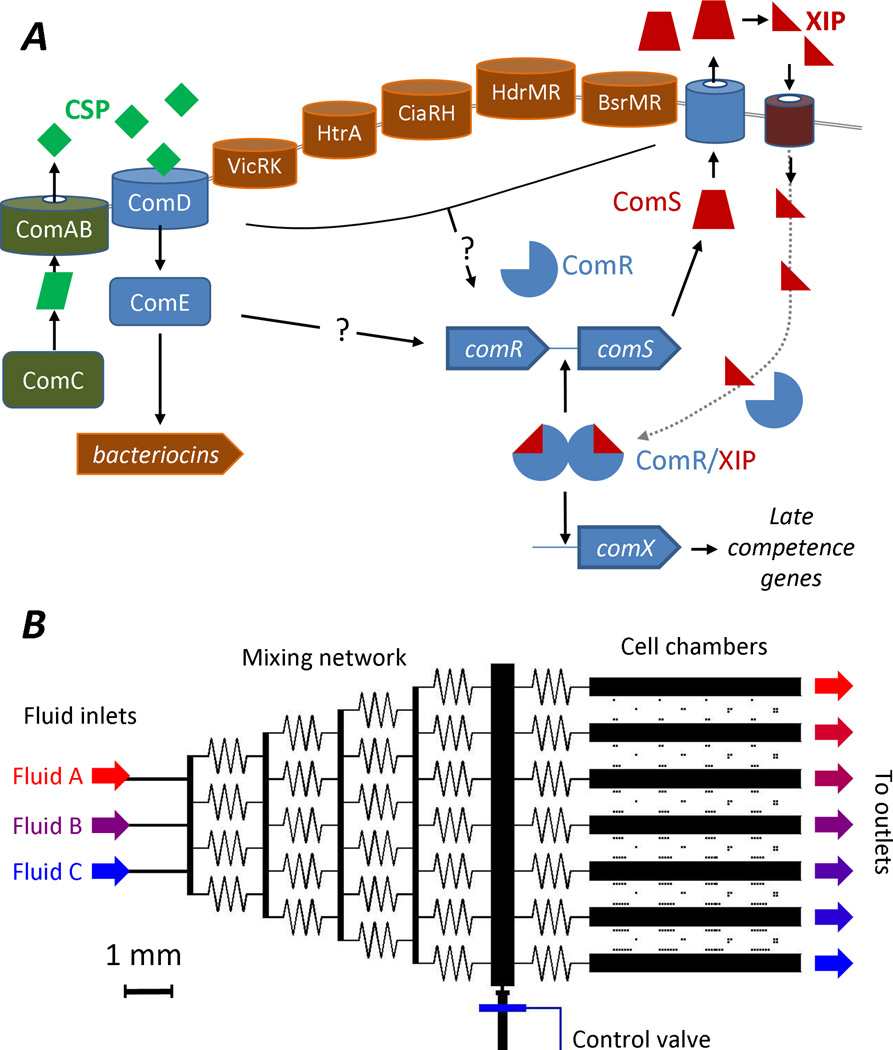
Genetic competence in S. mutans was first observed by Perry and Kuramitsu (Perry and Kuramitsu, 1981). The regulatory pathway has been shown to share some similarity with that of Streptococcus pneumoniae, which is also naturally transformable. Competence regulation in both S. pneumoniae and S. mutans employs a ComABCDE system, which produces the quorum signal peptide CSP (competence stimulating peptide) (Figure 1A). The 21 residue CSP of S. mutans (Cvitkovitch,2001; Li et al., 2001b; Suntharalingam and Cvitkovitch, 2005) is produced from its precursor ComC and exported by the ATP-binding cassette transporter ComAB. Environmental CSP is detected by a two-component system consisting of the membrane-bound histidine kinase sensor ComD and the intracellular response regulator ComE.
Figure 1.
(A) Schematic of competence regulation in S. mutans (Mashburn-Warren et al., 2010; Lemme et al., 2011; Senadheera et al., 2012; Smith and Spatafora, 2012). The 21-residue competence stimulating peptide (CSP) is detected by the ComDE two component signal transduction system which activates bacteriocin production and is believed to stimulate comR through an unknown mechanism. The comRS system in turn activates expression of comX, which encodes an alternative sigma factor for the genes required for DNA uptake and integration. The comRS system consists of a 17-residue peptide ComS that interacts with ComR to form the transcriptional activator for both comS and comX. The peptide XIP, which consists of residues 11–17 of ComS, can directly stimulate the comRS system and thereby activate comX, bypassing the CSP/ComDE mechanism. Competence also receives regulatory inputs from other gene products, including HtrA, CiaRH, and HdrMR. Hence, although ComR and ComS (or XIP) are required for transformability of S. mutans, ComDE is not required in all strains. (B) Design of microfluidic device for microscopy observation of S. mutans. Solutions of growth media and signal peptides (XIP or CSP) are injected at the inlets and mixed diffusively as they pass through the mixing network to the cell chambers at right (Jeon et al., 2000; Dertinger et al., 2001). In each chamber the adhered cells are continously perfused with a known admixture of media and/or signal peptides. An air-actuated control valve allows closure of a side channel, for control of cell loading.
Despite the fact that exogenous CSP dramatically increases transformation efficiency in S. mutans, ComABCDE in this organism are paralogs of the S. pneumoniae ComABCDE proteins, whereas they are orthologous to the bacteriocin regulation system BlpABCRH of S.pneumoniae (Martin et al., 2006). This leads to an important difference between the two organisms in the regulation of competence. In S. pneumoniae, phosphorylated ComE is a transcriptional activator for comX, which encodes an alternative sigma factor for the late competence genes required for DNA uptake and recombination. However, while S. mutans shares an orthologous comX, its ComE directly regulates production of several bacteriocins (Merritt and Qi, 2012). The factors that regulate expression and activity of comX in S. mutans are only beginning to be fully understood.
An essential component of comX activation in S. mutans was recently found to be the ComRS system (Mashburn-Warren et al., 2010). ComR is a transcriptional regulator of the Rgg family and regulates expression of ComS, is a 17-residue peptide that is produced by comS, located adjacent to comR. Although deletion of either comR or comS prevented transformation in S. mutans, transformability could be restored to the comS mutant by addition of exogenous, synthetic ComS. In particular, a seven-residue ComS fragment (residues 11–17), designated XIP, was especially effective at inducing competence. This suggested – and very recent work has confirmed (Desai et al., 2012; Khan et al., 2012) – that XIP is the mature form of ComS (Mashburn-Warren et al., 2010). XIP is most likely imported through the oligopeptide transporter Opp (Nepomuceno et al., 2007; Mashburn-Warren et al., 2010). As ComR/S was found to control transcription from a comX promoter, Mashburn-Warren et al. proposed that XIP entering the cell interacts directly with ComR to form a transcriptional activator for comX, and that this ComR/ComS complex is the immediate regulator of comX in S. mutans (Mashburn-Warren et al., 2010). ComDE presumably stimulates comRS expression, although the mechanism is unknown. Clearly, though, both XIP and CSP can activate genetic competence in S. mutans.
In addition to the existence of comRS, there are other intriguing differences between S. mutans and S. pneumoniae competence. One difference is in the population-wide distribution of the response to CSP. S. pneumoniae CSP induces comX uniformly across a population of cells, but only a sub-population of S. mutans responds to exogenous CSP (Aspiras et al., 2004; Perry et al., 2009; Lemme et al., 2011). When growing cultures of S. mutans were incubated with 0.2 or 2 µM CSP for 2.5 hours, the transformation efficiency was only 10% (Perry et al., 2009). In a growing biofilm of cells that contained a PcomX::gfp reporter, fewer than 1% of the cells expressed the gfp reporter, and the remainder showed no response (Aspiras et al., 2004). Surprisingly, when a PcomX::gfp reporting strain was incubated with CSP in a recent flow cytometry and transcriptome study, the comX-active (GFP+) and inactive (GFP−) cells expressed most (but not all – see Discussion) of the early com genes at fairly similar levels (Lemme et al., 2011).
The mechanism by which the CSP stimulus divides the population into responsive and unresponsive groups is not known. However, the heterogeneous and bimodal expression of comX suggests that noise and nonlinearity in gene expression may play a key role in the processing of this environmental signal. In this work we combine microfluidic methods with single-cell microscopy to examine the integration of environmental signals by the competence network. By placing S. mutans in microscopic flow chambers that maintain precise combinations of signal peptide concentrations and growth medium compositions, we are able to observe the effect of environmental conditions on heterogeneous and bimodal responses using a green fluorescent protein driven by the comX promoter in an otherwise wild-type genetic background.
Our results demonstrate that the interaction between the two peptide signals and the medium composition is complex and unexpected. However the behavior can be understood in a model where bimodality originates in the positive feedback autoregulation of the ComR/S system, if the composition of the growth medium can modulate the strength of that feedback.
Results
Individual S. mutans were studied in microfluidic flow devices (Jeon et al., 2000; Dertinger et al., 2001). Each device (Figure 1B) consists essentially of seven parallel channels or rectangular grooves that are patterned into the surface of a slab of inert, transparent polymer and sealed by a glass coverslip window (Experimental Procedures). Cells loaded into the channels settle and adhere to the glass window, allowing their observation in an inverted microscope while a continuous flow of growth medium (containing XIP and/or CSP) is passed over them. The cells constitute a sparse layer on the window and do not form a biofilm on the experimental time scale. The mixing network on the device allows each channel to supply medium of a slightly different composition, so that one can quantitatively compare the response of comX in populations of cells that are subject to slightly different chemical environments. Because the device can supply a continuous flow of fresh medium over the cells at all times (refreshing the medium in each chamber every 2–3 seconds) it maintains a constant chemical environment for the cells. This prevents endogenous secretions from accumulating in the cellular environment and altering the response to the input media and its exogenous signal peptides.
The S. mutans used in the microfluidic studies contained a PcomX::gfp reporter plasmid in either a wild-type or a comS-deficient genetic background. Through this reporter the activity of PcomX could be monitored in response to different combinations of XIP and CSP peptides in a peptide-free, chemically defined medium (FMC) or in a complex medium (BHI). We also performed a separate series of bulk (population-averaged) experiments using a PcomX::lacZ reporter strain to confirm that the observed interplay between the growth medium and the sensitivity of comX to XIP and CSP peptides was not limited to the microfluidic environment.
Bimodal and comS-dependent response of comX in BHI medium
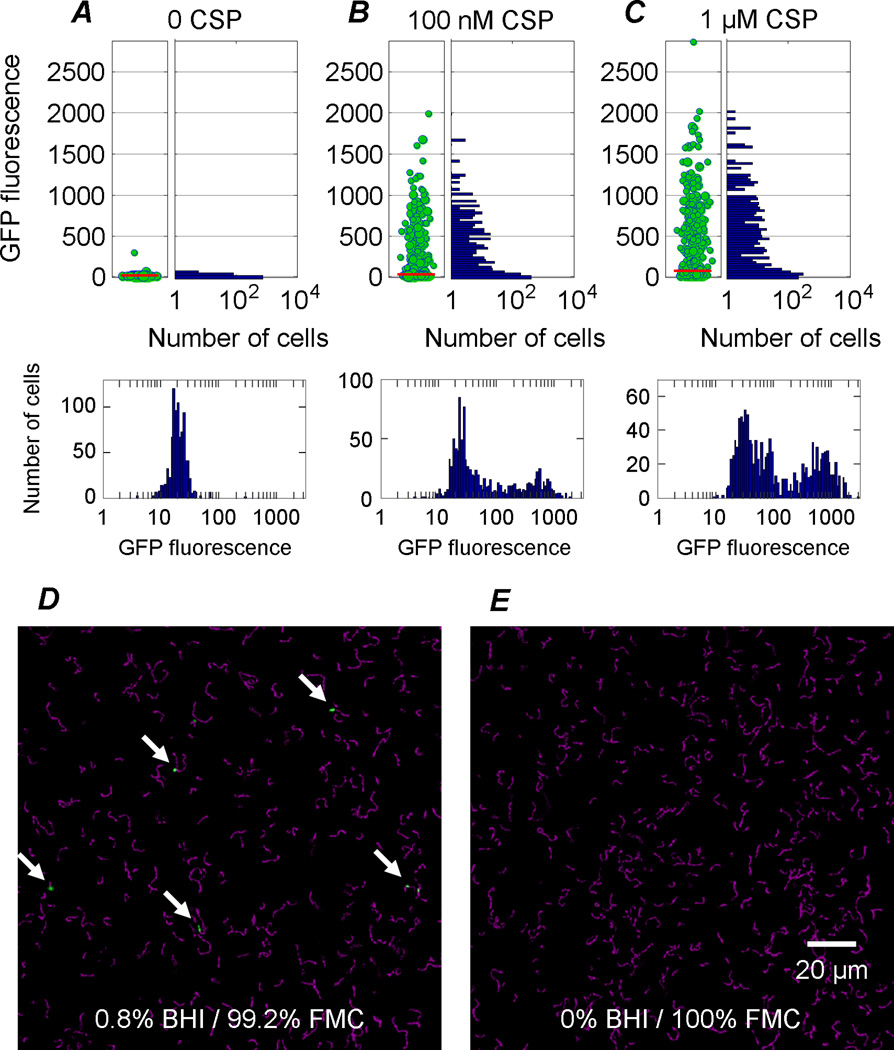
We first confirmed the expected bimodal response of the PcomX::gfp plasmid reporter strain to exogenous CSP (Aspiras et al., 2004; Perry et al., 2009; Lemme et al., 2011). Cells were observed in static (no flow) medium on a microscope slide. We incubated three samples of exponential-phase culture in BHI broth for 2.5 h in the presence of 0, 0.1 µM, and 1 µM CSP and then measured the GFP fluorescence per cell. Figure 2 shows that the response is strongly bimodal, as the median fluorescence is similar to baseline (the level of the untreated controls) and yet some cells fluoresce much more brightly than baseline. Approximately 30–35% of cells fluoresced in 100 nM CSP and approximately 50% of cells fluoresced in 1 µM CSP. The remainder of cells were dark. Therefore stimulation with CSP in BHI medium induced a bimodal comX response across a population of cells, as in prior work. (Microscopy images are shown in Figure S1.)
Figure 2.
(A–C) Activation of PcomX::gfp in S. mutans by exogenous CSP in static (no-flow) complex medium (BHI). Samples were imaged after 2.5 h incubation at CSP concentrations of (A) 0, (B) 100 nM, and (C) 1 µM. Each green circle indicates a per-cell GFP fluorescence level that was observed in the population, while the area of the circle represents the number of individual cells responding at that level. The red bar shows the median fluorescence of the population. The bars at right of each plot show a histogram on a logarithmic frequency scale. Beneath each plot is a histogram on a logarithmic fluorescence scale. The response to CSP is bimodal: While CSP activates PcomX in many cells, many cells do not respond and the median activation is low. (D–E) Overlay of GFP fluorescence (green) and phase contrast (magenta) microscopy images of S. mutans (with PcomX::gfp reporter) incubated with 300 nM CSP for 3 h. (D) Response in 0.8% BHI / 99.2% FMC (v:v) medium is bimodal, with only a small number of cells fluorescing brightly; (E) No response is observed in 100% FMC. Data for cells in undiluted BHI medium is not shown as pure BHI generates a very large autofluorescence background in the images.
We found unexpectedly that the response to CSP was highly sensitive to the composition of the medium. This is seen in Figures 2D and 2E, which show the stimulation of comX by CSP in a microfluidic device, where cells are subject to continuous flow of medium. Cells that were perfused with complex medium (BHI), or even BHI medium that was present as a dilute admixture with defined medium (FMC), exhibited a bimodal comX response to CSP, with a very small number of cells expressing bright GFP and most others remaining dark (Figure 2D). However cells perfused with FMC + CSP alone showed no induction of comX (Figure 2E), so the presence of some BHI was required for induction of comX by CSP.
In fact, we never observed CSP activation of comX in flowing FMC, even at CSP concentrations exceeding 1 µM. In static experiments (i.e. on a microscope slide) as well, we observed no comX response to CSP in FMC medium alone (Figure S2). The data imply that a comX response to CSP requires the presence of a constituent that is present in complex – but not defined – medium.
It was also of interest that a continuous flow of the complex/defined mixture (BHI/FMC) + CSP induced a bimodal response from comX, even in the absence of exogenous XIP. Mashburn-Warren et al. (Mashburn-Warren et al., 2010) reported that genetic competence in S. mutans requires the comRS system, so it might be anticipated that CSP activation of comX would require an extracellular accumulation of the ComS peptide (or its fragments such as XIP). However, this was not the case in the flow system, which raises the question of whether extracellular XIP is truly necessary for comX to respond to CSP.
We therefore tested whether bimodal response to CSP required an intact comS gene (Figure S3). We found that following incubation for ~2 h in static BHI containing 1 µM CSP, the PcomX::gfp reporter was activated only in the strain containing an intact comS gene, with no response in a comS mutant. Therefore the bimodal comX response to CSP requires an intact copy of comS.
Collectively, our data verify earlier findings that in a population of S. mutans, CSP induces a bimodal response from comX. However, the data also show that this behavior does not require an accumulation of extracellular ComS (or XIP), but it does require endogenous production of ComS, as well as a constituent of complex (BHI) medium that is absent in defined (FMC) medium.
Unimodal and comS-independent response of comX in a chemically-defined medium
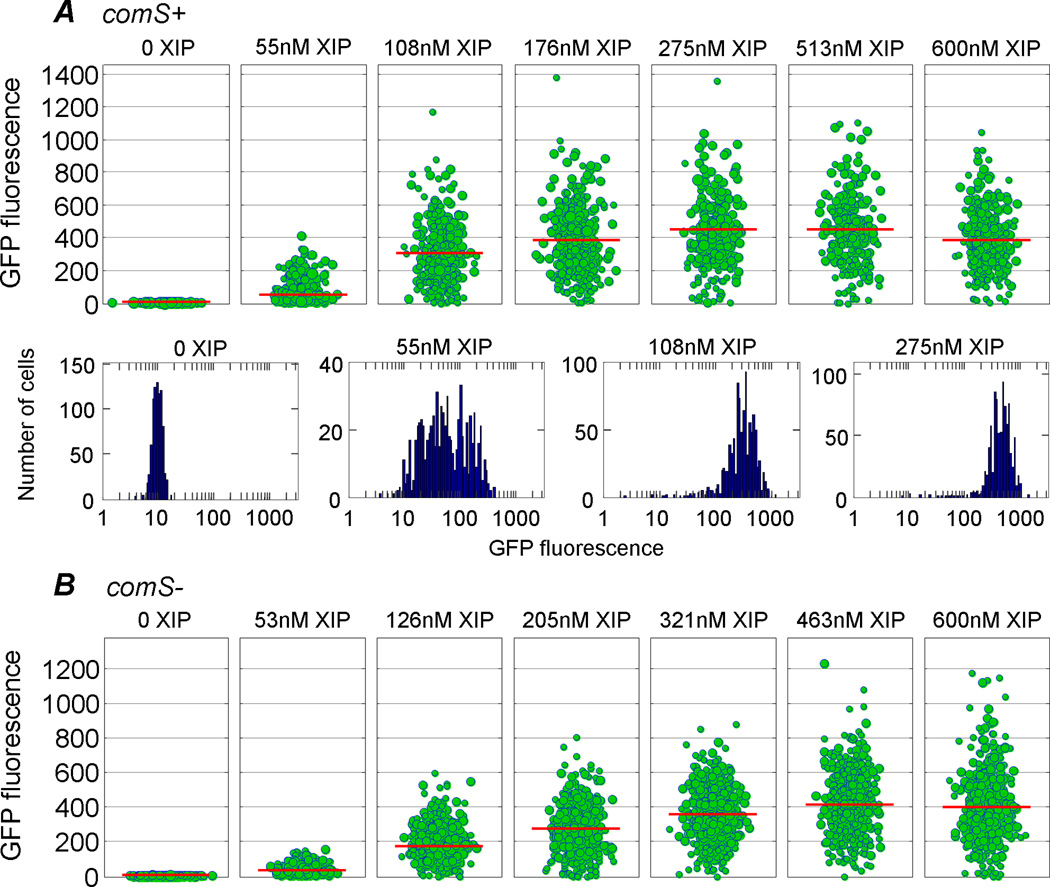
The pattern of comX expression in response to exogenous XIP was very different than was seen for CSP. Figure 3A shows the response of the PcomX::gfp reporter when cells were supplied with flowing FMC that contained XIP concentrations ranging from 0 to 600 nM. We observed a sensitive response, with GFP levels per cell rising above background for XIP concentrations of about 30 nM, and saturation at about 300 nM. The kinetics of this response to XIP are shown in Figure S4. Notably, the response of the comS mutant (Figure 3B) was virtually identical to that of the wild-type genetic background (Figure 3A). These data are consistent with the findings of Mashburn-Warren et al., who found that the transformability of the WT and comS mutant strains was enhanced at XIP concentrations exceeding 100 nMin a defined medium (Mashburn-Warren et al., 2010). However unlike the bimodal response to CSP, the response to XIP is best described as graded or unimodal: the level of comX activity in Figure 3A and 3B is broadly heterogeneous across the population, but virtually all cells are activated to some extent. The median single-cell fluorescence rises continuously above the baseline as the XIP concentration increases.
Figure 3.
(A) PcomX reponse of S. mutans with a wild-type genetic background subjected to a continuous flow of chemically defined (FMC) medium containing variable concentrations of exogenous XIP peptide. No CSP was provided. As in Figure 2, green circles indicate expression levels observed in the population. The red bar indicates the median expression level. Histograms (on a logarithmic fluorescence scale) are also shown for select XIP concentrations. Unlike CSP, the XIP peptide induces a unimodal response: roughly 98% of cells activate PcomX when the XIP concentration exceeds 100 nM. (B) As above, except with a comS mutant. The unimodal response of comX to XIP in FMC medium does not require comS.
Curiously however, we find that XIP activates comX expression only in FMC medium. In BHI or in BHI/FMC mixtures, exogenous XIP did not activate comX to any detectable level, as no fluorescent cells were observed. These results are detailed in Figure S5. (See also Figure 4 and Figure 7 below.) In this regard, the response of comX to XIP is virtually the opposite of its response to CSP: activation of comX by XIP is unimodal, occurs only in the absence of BHI, and does not require a copy of comS.
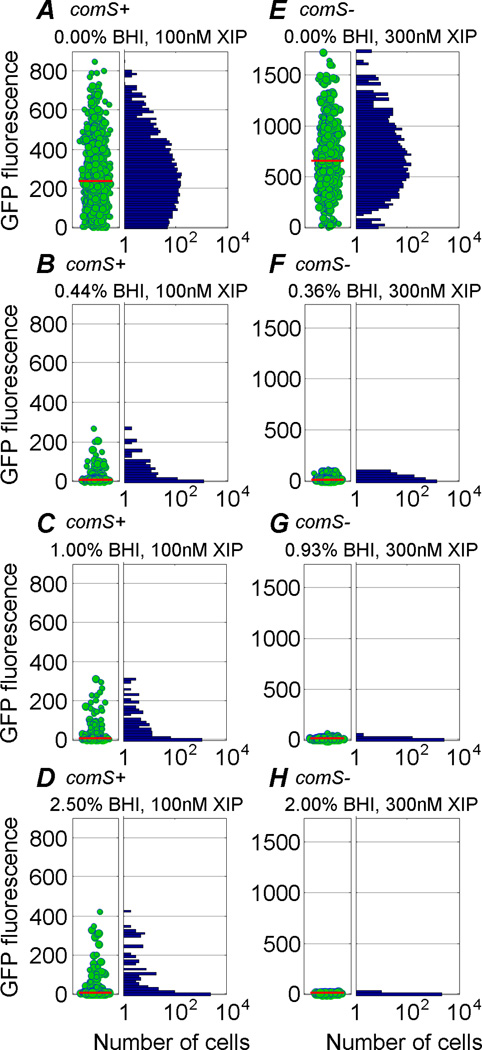
Figure 4.
(A–D) Response of wild-type cells to defined mixtures of complex (BHI) and defined (FMC) medium. All cells were subjected to a continuous flow containing 100 nM XIP and an admixture of BHI and FMC. No CSP was provided. (A) In the absence of BHI a broad unimodal response to XIP is observed. (B–D) Introduction of small amounts of BHI suppresses the response of most, but not all, cells. A bimodal distribution of responses is then observed, in which the median excitation is very low but a few cells continue to express strongly. In pure FMC roughly 98% of cells activate comX, while only 9–10% activate comX in the presence of 0.44% or 1% BHI. Additional data are shown in Figure S7.
(E–H) Response of the comS mutant strain in the same experiment. Cells were provided admixtures of complex and defined medium, all of which contained 300 nM XIP and no CSP. Addition of small amounts of BHI suppresses the response of all cells: No bimodal distribution is observed.
Figure 7.
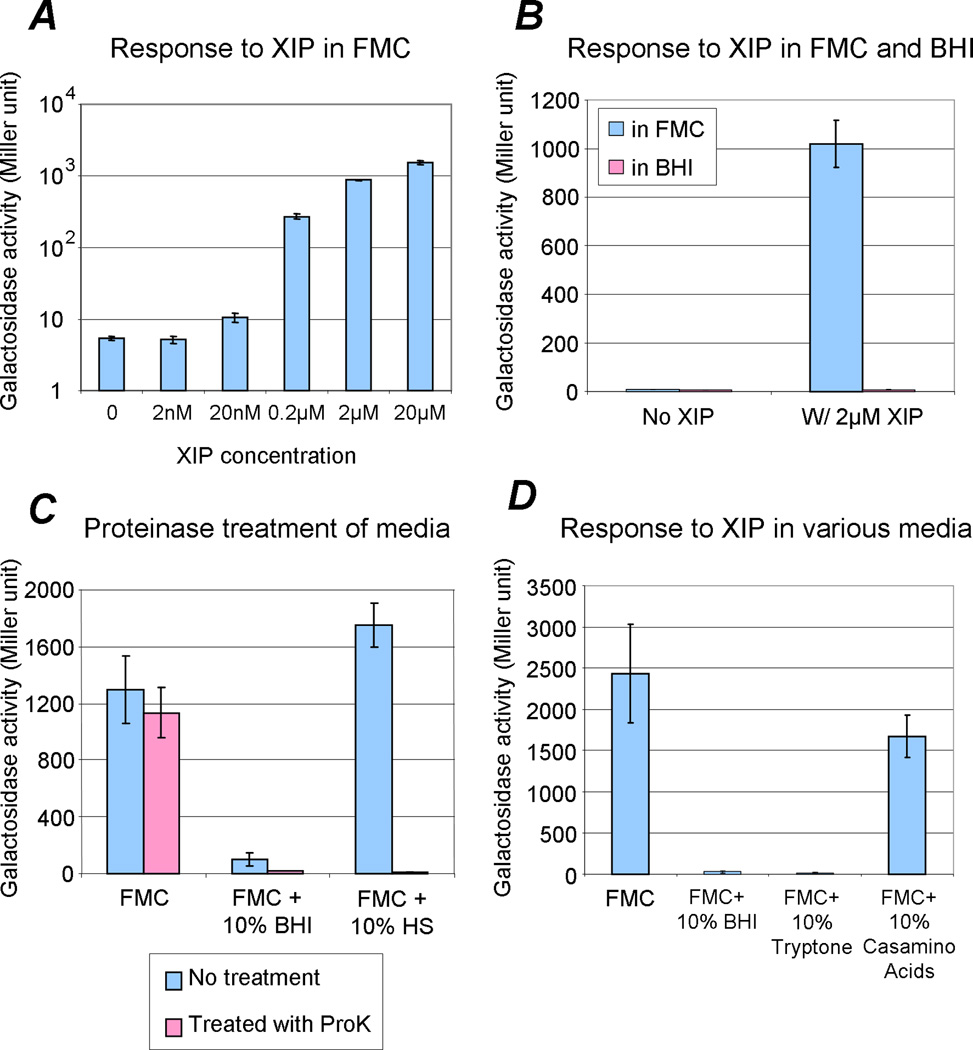
Bulk studies of PcomX response of S. mutans. The PcomX:lacZ reporter strain was grown to an optical density of 0.15-0.2 in FMC [except for (B)] and the components indicated in (A)–(D) were then added. The culture was then incubated for 1 hr prior to the LacZ assay. (A) PcomX induction at different concentrations of XIP (0, 2nM, 20nM, 0.2µM, 2µM); (B) Inhibition of XIP response by BHI. The strain was grown in FMC or BHI and then LacZ activity was measured in the absence or presence of XIP; (C) Effects of treatment of the medium with proteinase K. The strain was grown in FMC supplemented with 10% BHI, 10% horse serum (HS), or its protease K-digested products. Digests were produced by incubating 2 ml of HS or BHI with 10 µl of proteinase-K stock (20 mg ml−1) overnight at 37°C, and then adding phenylmethanesulfonylfluoride (PMSF) to inhibit the proteinase. (D) Inhibition of XIP response by protein and peptide-rich media. The strain was grown in FMC supplemented with 10% BHI, 10% tryptone (a trypsin digest of caseins), or 10% casamino acids (acid hydrolyzed-product of caseins). The bars represent the means and standard deviations (error bars) for three independent biological replicates. Adding 10% BHI or tryptone significantly depressed comX expression relative to a pure FMC medium (both p ≈0.002). In FMC + 10% casamino acids there was a moderate but less significant depression of comX response (p ≈ 0.11).
The qualitatively different (i.e. bimodal vs unimodal) effects of stimulating comX with CSP versus XIP raised the question of how comX might respond to stimulation by both peptides. We monitored comX expression in flowing FMC medium that contained 50 nM XIP and CSP concentrations ranging from 5–600 nM. Qualitatively the activation of comX in this experiment was not much affected by the CSP: all cells expressed GFP and we observed no bimodal response at any CSP concentration. Quantitatively however, several hours of exposure to CSP appeared to cause a moderate (<30%) reduction in the median comX expression level, at the highest CSP concentrations (> 400 nM). The data are shown in Figure S6. Nevertheless, we conclude that in pure FMC medium the addition of XIP always induces a unimodal comX response, regardless of whether CSP is present, and regardless of whether an intact copy of comS is present in the chromosome.
In summary, our microfluidic studies verify that XIP and CSP can both stimulate comX in a continuous flow environment, that CSP induces a (comS dependent) bimodal response, that XIP induces a (comS independent) unimodal response, and that the composition of the medium (±BHI) determines whether the CSP/bimodal or XIP/unimodal behavior will be observed.
Transition to bimodal comX response through addition of BHI
To further explore the role of medium constituents, we studied the effect of admixing small proportions of BHI into FMC that contained the XIP peptide. In this case, all channels of the microfluidic device were supplied with 100 nM XIP, but the growth medium in different channels consisted of different proportions of BHI and FMC, ranging from 5% BHI in FMC (v:v) to 0% BHI in FMC. Remarkably, even small proportions of BHI suppressed the population-wide comX response to XIP (Figure 4 and Figure S7). The presence of 0.26% BHI in the FMC medium reduced the median comX response by nearly tenfold, relative to 0% BHI. Even more surprisingly, the shape of the expression distribution changed qualitatively as the average expression level declined. As BHI was added, the distribution took on a bimodal character, with most cells remaining dark but a small minority continuing to express bright GFP fluorescence. In fact, in 1–2.5% BHI the median fluorescence of the population was indistinguishable from the baseline, yet the few cells that continued to express GFP were fully as bright (200–400 fluorescence units/cell) as in pure FMC (median 200–250 units/cell). The introduction of small proportions of BHI suppressed the unimodal, XIP-like response of comX and elicited a bimodal, CSP-like response, even in the absence of CSP.
Figure 4 and Figure S7 also show the results of the same experiment using a comS mutant. Here the introduction of BHI sharply suppressed the unimodal response to XIP, as in the wild-type genetic background. However, comX response was suppressed across the population and bimodality was not observed. Therefore, whether by addition of CSP or BHI, we were unable to induce a bimodal comX response in a strain lacking comS.
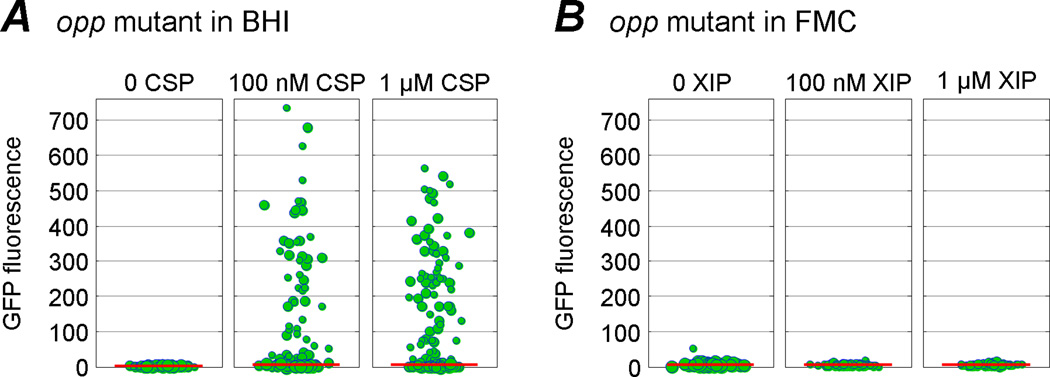
We also tested the effect of an opp mutation on the bimodal and unimodal response of comX. The Opp permease is required for internalization of environmental ComS or XIP (Mashburn-Warren et al., 2010). Figure 5 shows that the PcomX-gfp reporter exhibits a bimodal response to stimulation by exogenous CSP in an Opp-deficient strain, as it does in the wild type background. Hence the bimodal behavior of comX does not require import of extracellular ComS or XIP peptide through the Opp permease. Presumably therefore it is driven by an intracellular accumulation of ComS or XIP. However Figure 5 also shows that unimodal response to exogenous XIP is completely absent in the opp mutant, confirming that unimodal response to XIP requires entry of extracellular XIP into the cell via the permease.
Figure 5.
Effect of an opp deletion on PcomX response to CSP and XIP. (A) When CSP is provided in BHI medium, PcomX in the Opp-deficient mutant exhibits bimodal activation, as in wild type. (B) In contrast, PcomX in the opp mutant is not activated above its baseline by exogenous XIP in FMC medium. The unimodal response of comX to XIP requires the peptide permease, but the bimodal response to CSP does not.
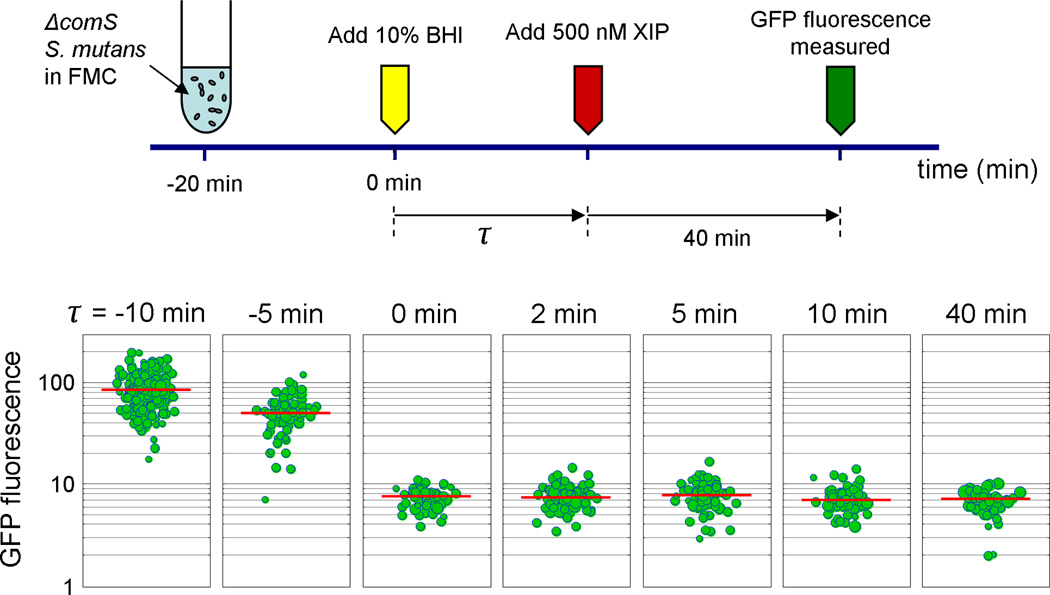
Figure 6 shows an additional experiment that tested the kinetics of BHI suppression of the response to XIP. We divided a culture of S. mutans (comS-deficient) growing in FMC into seven samples. At time t = 0, we added 10% BHI to each sample. At time τ, we added 500 nM XIP to each sample, where τ ranged from negative (XIP before BHI) to positive (XIP after BHI). Each sample was then incubated for 40 minutes and the GFP fluorescence of the cells was measured. Figure 6 shows that when XIP was added prior to BHI (τ <0), a strong (unimodal) comX response was observed. However when XIP was added concurrently with or following BHI (τ ≥ 0), comX activity remained at baseline levels. These data suggest that BHI acts through a very rapid (< 2 minutes) mechanism to block activation of comX by exogenous XIP, but that this mechanism is largely bypassed if XIP is provided just 5–10 minutes prior to BHI.
Figure 6.
Kinetics of stimulation of comX by XIP and suppression by BHI. The comS mutant was grown to an optical density of 0.15 in FMC and then divided into seven samples. At time t = 0, we added 10% BHI to each sample. At time τ we added 500 nM XIP to each sample, where τ ranged from −10 minutes (XIP before BHI) to +40 minutes (XIP after BHI). Each sample was then incubated until t = τ + 40 minutes and then the GFP fluorescence of the cells was measured on a microscope slide. A strong, unimodal comX response as observed when XIP was added prior to BHI (τ <0). However when XIP was added concurrently with or following BHI (τ ≥ 0), comX activity remained at baseline levels.
Bulk studies – the active constituent of BHI medium
We performed additional experiments to verify that the effect of growth medium on the response to XIP and CSP was not an artifact of the microfluidic environment. We studied XIP and CSP response of S. mutans containing a PcomX::lacZ plasmid in a wild-type background, in bulk (planktonic) cultures growing in defined (FMC) versus complex (BHI) media. Figure 7A shows the results of a LacZ assay following incubation of the PcomX::lacZ S. mutans with XIP for 1 h in FMC medium. As in the microfluidic experiments, PcomX is strongly activated at XIP concentrations exceeding roughly 100 nM. Furthermore, this response is absent in BHI medium; incubation with 2 µM XIP caused no increase in comX activity in BHI medium (Figure 7B). We also tested the effect of mixing BHI and FMC and found that the addition of 10% BHI into FMC suppressed the response of comX to 2 µM exogenous XIP (Figure 7C). This finding is consistent with the microfluidic results in (e.g.) Figure 4. Experiments with the PcomX::lacZ strain also found no response to exogenous CSP in FMC medium, consistent with our microfluidic data (data not shown).
We also investigated the nature of the constituent in BHI that suppresses the response of PcomX to XIP. The fact that PcomX response declines even at 0.2% BHI (by volume) in FMC, whereas BHI broth itself is prepared at 3.7% or 37 g L−1, suggests either that the mechanism of suppression is very sensitive to a single constituent of BHI, or that the response to XIP is affected by a class of constituents (such as peptides and proteins) that is abundant in BHI. If the peptides in BHI are typically 7 residues in length (840 Da) then the peptide concentration of pure BHI is roughly 30–40 mM. At 0.2% BHI in FMC the concentration of such peptides would be close to 60–80 µM. Even if only 10% of these peptides were capable of competing with XIP for importation by the transporter Opp, then nanomolar XIP would encounter significant competition in 0.2% BHI.
Therefore we studied whether other protein or peptide-rich media suppressed XIP response in a fashion similar to BHI. We also tested whether digestion of these media by proteinase-K affected response to XIP. Figure 7C shows that adding horse serum (HS) into FMC had only a weak positive effect on the PcomX response to XIP. However, the products of overnight proteinase-K digestion of HS sharply suppressed XIP response when added to FMC. Proteinase-K digestion of BHI also greatly enhanced its suppression of the response to XIP. Tryptone, a tryptic digest of caseins that contains an assortment of peptides, had an effect similar to that of BHI. However casamino acids, an acid hydrolysate of caseins that contains primarily amino acids and some small peptides, did not effectively suppress the XIP response. (Figure 7D). Collectively these data suggest that small peptides (but not free amino acids) that are present in BHI and in enzymatic digests of BHI or HS may be responsible for quenching the response of comX to XIP.
To test whether this quenching might be due to competition for (e.g.) the Opp permease, we tested whether high concentrations of exogenous XIP could overcome the presence of 0.15% BHI in FMC and restore the comX response. The result (Figure S8) show that comX in this medium does become activated once the environmental XIP concentration approaches 1 µM, consistent with a competition mechanism for inhibition of XIP activity in complex media.
Discussion
We found that although the S. mutans competence system responds to two different peptide signals XIP and CSP, the composition of the growth medium determines which peptide is actually effective in stimulating a comX response. The medium also determines whether the comX response is shared throughout the population (unimodal), or is limited to a subpopulation of cells (bimodal). In the presence of an intact comS gene, modulating the proportion of complex medium alone was sufficient to tune the system between unimodal and bimodal behavior.
S. mutans regulates genetic competence through a complex network that receives inputs from multiple environmental signals and provides outputs to multiple other genes. In addition to the XIP and CSP peptide signals investigated here, expression and activity of the com system is influenced by a variety of gene products, including HdrM, HtrA, and the two-component systems CiaRH and VicRK. More broadly the competence regulon is linked to acid tolerance, biofilm formation, and oxidative stress and is intimately connected to the production of bacteriocins, some of which modulate autolytic behavior (Li et al., 2001a; Li et al., 2002; Ahn et al., 2005; Ahn et al., 2006; Merritt et al., 2007; Senadheera et al., 2012).
S. mutans competence induction by CSP has been shown to exhibit bimodal behavior. Lemme et al. found that not only did only a fraction of S. mutans cells activate comX in the presence of CSP, but an even smaller subpopulation actually demonstrated competence as assessed by the uptake of a Cy3-labeled DNA fragment (Lemme et al., 2011). These data indicate that activation of comX may not necessarily result in development of the capacity to take up exogenous DNA (transformation) and that additional regulatory decisions occur downstream of comX. Evidence for such regulatory mechanisms was provided recently by Seaton et al., who showed that mutants with altered expression levels of the rcrRPQ genes, encoding a MarR-like transcriptional regulator and two ABC efflux pumps, could express high levels of comX but that activation of comYA differed depending on the presence or absence and expression levels of the RcrRPQ gene products (Seaton et al., 2011). Therefore, similar to S. pneumoniae and Bacillus subtilis, there may be factors that govern the activity or stability of ComX of S. mutans that ultimately control whether this organism translates the signals for competence development into activation of the machinery for DNA internalization (Berg et al., 2012).
Here we have set aside most of these details in order to study how a limited set of inputs – the peptides XIP and CSP and the composition of the growth medium – affects heterogeneity in the expression of comX. Our single-cell studies reveal a curious interaction between complex medium and comX expression, where the presence or absence of complex medium can determine whether comX exhibits a bimodal or unimodal response to peptide stimuli. Interestingly, only the bimodal behavior requires an intact copy of comS.
The link between comS and bimodality is intriguing because bimodal gene expression is a signature behavior of bistable, positive feedback regulated systems (Smits et al., 2006; Raj and van Oudenaarden, 2008), and comS as described by Mashburn-Warren et al. regulates itself through positive feedback. ComS or its processed form (such as XIP) is believed to interact with ComR to form the transcriptional activator of comS as well as comX (Mashburn-Warren et al., 2010; Desai et al., 2012; Khan et al., 2012). Bimodality can arise in the presence of feedback because a gene that produces its own transcriptional activator can potentially maintain itself in either a switched ON or a switched OFF state. The actual level of expression in any particular cell depends on highly variable factors specific to that cell, including its past history and stochasticity (noise) in basal expression or in the regulatory inputs (Smits et al., 2006; Raj and van Oudenaarden, 2008). The result may be bistability, where the gene can remain indefinitely in the active or inactive state, or excitability, where intermittent switching between the two states is observed. In either case a population of cells will show a bimodal distribution of expression activity for that gene.
Among the best studied examples of bimodal expression arising from positive feedback is, interestingly, the regulation of genetic competence in B. subtilis. The com genes in B. subtilis comprise an excitable system that exhibits transient, bimodal switching. As a culture approaches stationary phase, roughly 10–15% of B. subtilis enter the competent state (Dubnau,1991; Miller and Bassler, 2001; Maamar et al., 2007). The B. subtilis system receives input from a quorum signal peptide ComX via a two component system (ComP / ComA). Phosphorylated ComA activates the expression of B. subtilis ComS, which inhibits degradation of ComK, the master regulator of the competence genes (Miller and Bassler, 2001). The bimodal behavior of this system arises in the positive feedback regulation of ComK; because ComK acts as its own transcriptional activator, noise in its expression is amplified. This allows transient excitations into the competent state (Maamar and Dubnau, 2005; Smits et al., 2005; Suel et al., 2006; Maamar et al., 2007). Therefore positive feedback regulation provides a mechanism by which a population of cells can spontaneously subdivide into two phenotypically distinct groups (James E,2002).
The example of B. subtilis raises the question of whether autofeedback regulation in comRS could account for the medium-induced switching between bimodal and unimodal comX expression in S. mutans. Strong positive feedback could allow autoactivation of comRS in at least some cells, leading to accumulation of ComS (or its processed form XIP) sufficient to activate comX in a subpopulation. Our data would require that the complex medium favor this autoactivation behavior in two ways. First, the constituents (probably peptides) of complex medium would have to promote accumulation of ComS (or XIP) within the cell. This could occur if peptides present in the medium enter the cell and compete with the enzymes that turn over intracellular ComS, or if they simply block export of ComS or XIP, or else stimulate comR expression via some other input such as IrvA, Hdr, or CiaRH. This is necessary because the bimodal behavior is observed only in the presence of the complex medium. Second, the complex medium would have to block the sensitivity of comX to exogenous XIP. This is necessary because we find that XIP elicits no response in the presence of complex medium. This inhibition could arise through competition between medium peptides and XIP at the permease Opp, or through (for example) a two-component system responsive to other environmental inputs. Overall, though, our data are consistent with a model where complex medium favors bistability by enhancing the tendency of comRS to autoactivate and by isolating it from environmental XIP.
By the same argument, the unimodal expression of comX in FMC is consistent with weaker autoactivation of comRS, accompanied by greater sensitivity to environmental XIP. If defined medium led to diminished feedback (e.g. by more efficient degradation or export of endogenous ComS or XIP, and/or downregulation of comR), while permitting exogenous XIP to enter the cell and interact with ComR, then endogenous production of ComS would be less important than environmental XIP levels in determining the activity of comX inside each cell. Exogenous XIP would then elicit a comX response across the entire population. Therefore, if defined medium reduces autoactivation of comRS and also permits the intracellular XIP concentration to approach environmental concentrations, then we expect unimodal activation of comX as observed in our experiments.
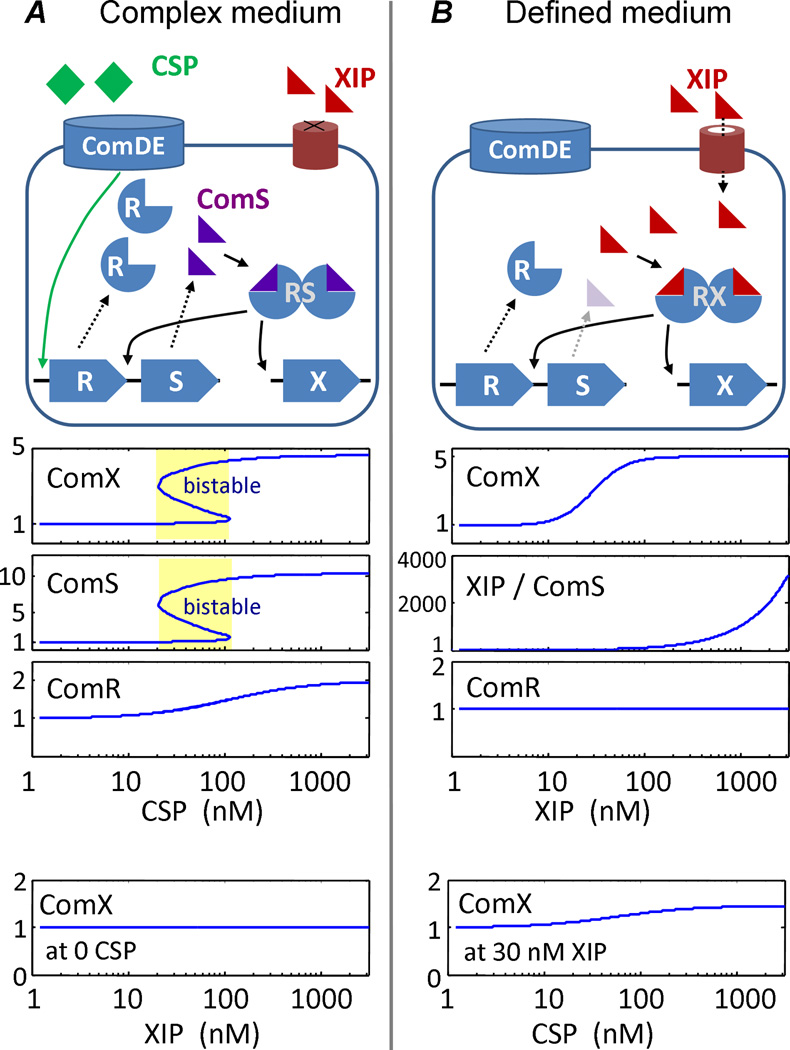
We cannot outline a precise mechanism by which complex medium could both stimulate autofeedback and reduce sensitivity to environmental XIP, as the competence regulon responds to numerous environmental inputs in ways not completely understood. Even the pathway through which CSP and ComDE stimulate comRS, and consequently comX, remains unknown. We can however demonstrate that a comRS autofeedback mechanism is a plausible interpretation of our data by analyzing the reductive model that is shown in Figure 8. The model assumes that (1) in the presence of complex medium only, ComS (or XIP) tends to accumulate within the cell rather than be degraded or exported; (2) in defined medium only, environmental XIP can interact with comRS and comX; and (3) exogenous CSP stimulates (indirectly) expression of comR. (While it is possible that CSP stimulates comR in complex medium only, the model does not require this.)
Figure 8.
Computer simulation of a model for bimodal comX activation via positive feedback regulation in comRS. The figure presents a simple model with properties (1), (2), and (3) described in the main text. The model assumes that complex medium favors positive feedback in comRS by (1) allowing ComS (or XIP) to accumulate intracellularly (rather than being exported or degraded) and (2) impeding import of extracellular XIP. In defined medium, endogenous ComS (or XIP) is not retained in the cell, although extracellular XIP can enter. The model also assumes (3) that exogenous CSP slightly (~2-fold) enhances comR expression. Therefore, ComR will interact with endogenous ComS or exogenous XIP in complex medium or defined medium, respectively. This interaction forms the multimeric transcriptional activator for comX and comS. The mathematical implementation of the model is described in Experimental Procedures. The lower panels show the simulated fold-increases in expression of the com genes in response to exogenous XIP or CSP, under conditions of complex medium (A) vs. defined medium (B). In both cases the model parameters are those of Table 1. (A) In complex medium, endogenous ComS is retained in the cell but exogenous XIP is excluded. Environmental CSP stimulates expression of ComR (indicated by R), which interacts with basal levels of ComS to form a transcriptional activator complex (indicated by RS) (Mashburn-Warren et al., 2010), allowing comS to amplify its own expression. The simulation finds that autofeedback can readily lead to 10-fold upregulation of comS, even if comR is only upregulated 2× by the CSP signal. However the dynamics of autofeedback are bistable, as the expression of comS and comX can be either high or low when the exogenous CSP concentration lies within a transitional range (indicated in yellow). Hence the simulation predicts bimodality in the population-wide comX response to CSP. In the absence of CSP, comX is unresponsive to exogenous XIP (bottom panel). (B) In defined medium endogenously produced ComS cannot accumulate in the cell but environmental XIP can enter. Environmental XIP interacts with ComR to form the transcriptional activator complex (indicated by RX) that induces expression of comX (and comS). The response of comX is not bimodal however, as endogenous ComS does not accumulate to stimulate its own expression. Therefore, comX is regulated not by comS autofeedback but by environmental XIP, and so the model predicts that comX responds in a graded (unimodal) fashion to the exogenous XIP concentration. This comX response is insensitive to stimulation of comR by exogenous CSP (bottom panel). The bimodal response to CSP shown in (A) requires comS, but the unimodal response to XIP shown in (B) is independent of comS.
Figure 8 shows the results of a Matlab simulation of the model (see Experimental Procedures). Details are given in the figure legend. Consistent with the experimental data, the model exhibits a graded or unimodal comX response to exogenous XIP in a defined medium and a bimodal response to exogenous CSP in complex medium. Whether the simulated comX behaves unimodally or bimodally depends on the medium, not the signal peptide (XIP or CSP). Further, the simulation gives no comX response to XIP in complex medium and no response to CSP in defined medium, as observed. Finally, bimodality in the model requires the presence of comS, although unimodality does not. Therefore, with relatively few assumptions an autofeedback model centered on comRS is able to capture the most remarkable qualitative observations of our study.
Recently Lemme et al. combined flow cytometry with transcriptome analysis to investigate the origin of bimodality in S. mutans competence. They incubated a PcomX::gfp reporter strain with CSP and then separated the cells into two groups according to their level of comX expression (i.e. GFP+ vs. GFP−) (Lemme et al., 2011). Comparing the two subpopulations in a microarray analysis, they found that the GFP+ and GFP− cells differed only modestly in expression of most of the com genes: comR was only 1.4× higher and comDE only ~2× higher in the GFP+ vs GFP− subpopulations. The exception was the comS region, which was ~12× more strongly expressed in the high-comX cells than in the low-comX cells. The authors interpreted their findings within a model in which comE is the origin of bimodality in S. mutans competence (Perry et al., 2009; Lemme et al., 2011). In this model ComE participates in a positive feedback loop of autoregulation, via additional elements that are unknown, and stimulation by CSP combines with noise in the expression of comE to induce the bimodal response of comX.
However in a model where bimodality originates from positive feedback in ComE, it may be difficult to interpret our finding that the composition of the growth medium shapes the distribution of expression levels and the sensitivity to exogenous XIP or CSP. Our data would require that complex medium allows CSP to activate the putative positive feedback loop in ComE while simultaneously blocking exogenous XIP from activating comRS which is downstream of ComE. It may also be difficult to rationalize the effect of introducing complex medium (Figure 4), which would have to stimulate the bimodal ComE autofeedback loop in the absence of CSP, but only if XIP (which acts downstream) is present. Finally it would be difficult to explain the absence of bimodality in the comS mutant.
Furthermore, we note that S. mutans and S. pneumoniae both possess similar ComABCDE systems and yet only S. mutans shows bimodality in competence. However only S. mutans possesses comRS. Therefore our findings suggest that comRS – rather than comE –may give rise to bimodality in S. mutans competence. The results of our simulations based on this idea (Figure 8) are quantitatively consistent with the fold changes in gene expression that were observed by Lemme et al.: The gene that shows the strongest bimodality, or variation between the GFP+ vs GFP− subpopulations (following CSP stimulus), is comS, as expected for the gene that is at the center of bistability. By contrast, other genes such as comR and comDE, which show much weaker bimodality (~2×) in the transcriptome data, provide inputs to the bistable circuit but are not expected to be bistable themselves. The positive feedback model allows a relatively modest change in comR expression to control much larger changes in comS expression.
Finally we note that Mashburn-Warren et al. have proposed that other com system inputs such as HdrM, HtrA and CiaRH all act upstream of comRS (Mashburn-Warren et al., 2010). In this case we would not expect any such inputs to induce a unimodal response from comX. Rather they would all trigger, at best, a bimodal response. It will be interesting to explore such questions in future work.
Our analysis is motivated largely by the observation of switching between unimodal and bimodal behavior – a phenomenon that is observable only at the single cell level. Furthermore the ability to finely manipulate and control the composition of the environment is essential to our experiments. Our results show how microfluidic studies of S. mutans can provide valuable insight into the mechanisms and dynamics of competence regulation. One question arising from this study is why the presence of peptides, such as those in BHI, might alter the behavior of S. mutans via ComRS signaling. Along these lines, we have observed that adding saliva to FMC does not block the comX response to XIP, nor does it induce a bistable comX response in cells growing in FMC with CSP, even at relatively high concentrations of saliva (data not shown). Thus, the different behaviors of S. mutans vis-à-vis ComRS activation of comX may reflect adaptations to different environmental conditions in the oral biofilm. For example, in relatively immature dental plaque the system may behave in a unimodal manner, perhaps allowing for optimization of colonization and competition with commensals. Conversely, in mature dental plaque, protelolytic activity may release higher concentrations of peptides that can accumulate in the biofilm matrix and trigger a bimodal character in the ComRS/CSP pathways. Such behavior may enhance the survival of, or influence the cariogenic potential and physiology of, S. mutans in the more densely-packed biofilms that are more conducive to caries development. Future studies will focus on determining how specific environmental factors that influence the homeostasis and pathogenic potential of human oral biofilms impact the behavior of, and virulence expression by, S. mutans at the individual cell level.
Experimental Procedures
Bacterial strains, growth conditions and reagents
Streptococcus mutans UA159 and its derivative strains were maintained in BHI (Difco Laboratories) at 37°C in 5% CO2 and 95% air with antibiotics used at the following concentrations: kanamycin (Km, 1 mg ml−1), erythromycin (10 µg ml−1) or spectinomycin (1 mg ml−1), when necessary. The lacZ assays were performed by a modified Miller protocol (Zubay et al., 1972), as previously described (Liu et al., 2009).
Construction of reporter strains
To construct the PcomX::gfp reporter strains for microfluidics studies, a 200-bp region comprising PcomX and sequences 5’ proximal was amplified with primers that incorporated HindIII and SpeI sites, and a gene, encoding a new GFP reporter with improved brightness, was amplified with primers that contained SpeI and EcoRI sites from the plasmid pCM11 (Lauderdale et al., 2010), kindly provided by Dr. Horswill AR (University of Iowa). Both PCR products were digested with SpeI and ligated to each other with T4 DNA ligase (Invitrogen). The PcomX::gfp ligation product was re-amplified and cloned into the HindIII/EcoRI site of pDL278 (Spr), a shuttle vector of oral streptococcal origin (LeBlanc et al., 1992). The resulting construct was transformed into a wild-type (WT) UA159 to generate strain SJ380 (PcomX-gfp/WT). To create the PcomX::gfp reporter strain in the comS-defective background (SJ406), the comS gene in the SJ380 was replaced by a non-polar erythromycin (NPEm) resistance cassette using PCR ligation mutagenesis (Lau et al., 2002). Construction of the opp mutant has been described previously (Nepomuceno et al., 2007). To construct the PcomX::lacZ reporter strain, the promoter region of the comX gene was amplified with primers containing SacI and BamHI sites, and cloned into a pMC340B-based lacZ reporter vector (Liu et al., 2009) that carries a kanamycin resistance gene (Kmr). The vector has sequence homology to the mtlA and phnA genes flanking the cloning sites, which allowed for double-crossover homologous recombination and integration of the PcomX::lacZ cassette DNA in a single copy in the S. mutans chromosome. The lacZ-promoter fusion was transformed into the wild-type UA159 strain. All engineered strains were verified by PCR and DNA sequencing.
Microfluidic experiments used both chemically defined and complex media. The defined medium was FMC (Terleckyj et al., 1975) prepared with 16 mM glucose and, if needed, with 1 mg ml−1 spectinomycin, and adjusted to pH 7.0. The complex medium was brain-heart infusion (BHI) obtained commercially from BD (catalog #211059). Horse serum was obtained from MP (catalog #2921249). Tryptone (catalog #211705) and casamino acids (catalog #223050) were obtained from BD.
CSP was synthesized by the ICBR facility at University of Florida and its purity was confirmed by HPLC. The XIP peptide GLDWWSL, corresponding to residues 11–17 of ComS, was synthesized and purified to 96% by NeoBioSci, Inc.
For microfluidic experiments, overnight cultures of S. mutans harboring the PcomX::gfp vector were grown in fresh BHI broth with antibiotic selection: 1 mg ml−1 spectinomycin for strains with an intact copy of comS, and spectinomycin and erythromycin (10 µg ml−1) for comS mutant strains. Cultures were then diluted 30-fold in medium containing only spectinomycin and were incubated at 37 °C for 3–4 hrs to bring them to an exponential growth phase (OD600 = 0.3). Cells were mildly sonicated at 25% amplitude for 30 seconds using an ultrasonic dismembrator (Model FB120, Fisher Scientific) to dechain the cells before loading into the device. All single-cell experiments were conducted using spectinomycin as the only antibiotic present, so that antibiotic pressure on all strains was consistent between organisms and experiments.
Microfluidic design and fabrication
Microfluidic mixing devices were fabricated from an inert transparent silicon elastomer (PDMS, polydimethylsiloxane) at the University of Florida through the method of soft lithography (Sia and Whitesides, 2003). Each microfluidic mixing device consists of seven parallel channels that are typically 15 µm deep and 400 µm wide, and are sealed by an uncoated glass coverslip window. Two or three solutions containing different media or different concentrations of signal peptides are pumped into a mixing network that is also patterned into the elastomer block. The flow of medium exiting the mixing network generates seven streams containing different admixtures of the input solutions (Figure 1B), and these streams flow into the seven channels that contain S. mutans. Each chamber receives a spatially and temporally constant medium, but the different chambers span a gradient in medium composition.
The devices employ a two-layer (control channel + fluid channel) design (Unger et al., 2000) that allows rapid and precise control of fluid flows during both the setup and observation stages of an experiment. The top layer contains all the fluid flow channels and is adjacent to the glass window. Beneath that layer is an (isolated) layer of channels for N2 flow: pressurizing or depressurizing channels in this layer closes or opens the fluid channels in the top layer, allowing switching of fluid flow.
We used conventional lithographic methods to fabricate the mixing devices. Channel patterns were designed in AutoCAD (Autodesk), printed onto a 20,000 dpi transparency mask, and then transferred to a 4” silicon wafer coated with Shipley S1813 photoresist. Developed wafers were then processed with deep reactive ion etching to give fluid flow channels of ~15 µm height. The etched wafer acts as a master or mold for the casting of the elastomer devices themselves. Flow and control layers for the mixing devices were then cast from degassed PDMS using a covered mold (Pfeil et al., 2009). After assembly of the two layers each device was cured in a 70°C oven for 3–4 hrs to bond the layers, and the device was then sealed with a O2-plasma-treated 24×50 mm coverslip and baked in a 40–50°C oven overnight to ensure a permanent seal.
The microfluidic device is clamped to a custom PTFE holder that provides all fluid connections for the experiment. Fluids are supplied to the holder through HPLC fittings and enter the elastomer block through a set of O-ring seals (Pfeil et al., 2009). The holder and flow chamber together are mounted to the stage of an inverted microscope (TE2000U, Nikon) that is enclosed in a custom-built Lexan chamber. This enclosure uses electronic temperature control and continuous gas purging to maintain an anaerobic environment at 33°C during all measurements.
Microfluidic experiments
An exponential phase culture of S. mutans is sonicated briefly and then injected into the exit side of the microfluidic device, which is isolated from signals and media entering the mixing network by a gas-actuated valve in the control layer of the device. This valve ensures that the cells are not prematurely exposed to the stimulus. After sufficient cells have settled to the glass window, the gas- actuated valve is opened and the signals or media flow into the cell chambers. This marks the beginning (time t =0) of a microfluidic experiment. The stability of the flow, and the relative concentrations of signal peptides or other constituents passing through the device channels, can be confirmed and quantified by measuring the fluorescence of a red dye tracer (sulforhodamine 101, Acros Organics, at 50–100 ng ml−1) in one of the input media.
Cells in the device were imaged at 20× (CFI Plan Fluor DLL, NA 0.5, Nikon) onto a cooled CCD camera (CoolSNAP HQ2, Photometrics). Images were collected in phase contrast, GFP fluorescence (using a Nikon C-FL GFP HC HISN zero shift filter cube), or red fluorescence (C-FL Y-2E/C Texas Red filter cube, Nikon). A set of three images of each channel are collected at intervals of one hour, for periods up to four hours. Fluorescence images are collected in 3–5 s exposures at the lowest intensity setting of the excitation source (Intensilight C-HGFI, Nikon), which was sufficient to prevent significant photobleaching. By imaging the cells over intervals as long as 4 hours, we were able to monitor the development of the comX response in hundreds to thousands of individual cells that were subjected to different but well-defined, stable chemical and physical environments.
Image analysis
The expression of PcomX::gfp in individual cells was quantified by analyzing phase contrast and GFP fluorescence images of individual cells and small (2–3 cells) clusters (Kwak et al., 2012). Briefly, the method compares phase contrast and fluorescence images of the same scene and applies a phase-shift model to determine a parameter for each cell, characterizing the mean concentration of GFP for that cell. This parameter was used as the measure of PcomX activity.
Modeling
In order to test whether modulation of autofeedback in comRS regulation and sensitivity to environmental XIP could account for our data, we constructed a mathematical model based on the comX activation scheme shown in Figure 8. The model simulates the expression of comR, comS, and comX to produce R, S, and X respectively. The transcriptional activator of both comS and comX is a multimeric complex of R with either S or XIP. For simplicity the model treats intracellular S and exogenous XIP as equally effective in forming the transcriptional activator complex (which is denoted by M below). For comR, the transcriptional activator is modeled simply as CSP, i.e. setting aside the complexity of the actual ComDE two-component system pathway and the fact that its actual mechanism for stimulating comR/comS is not known.
As described in the Discussion, our data suggest that comRS operates as an autofeedback system where complex medium has two effects, reducing the sensitivity to environmental XIP and preventing intracellular accumulation of endogenous ComS. Formally we can model these effects by introducing a hypothetical channel through which exogenous XIP can enter the cell and endogenous ComS can exit. In defined medium the channel is presumed open, so that intracellular and extracellular levels of XIP and/or ComS become equalized. In complex medium the channel is presumed closed, so that exogenous XIP is excluded, while endogenous ComS can accumulate and stimulate the comRS system. Whether this accurately describes the role of the complex medium is not important to the mathematical modeling.
In the equations of the model, R,S,X,M denote the copy numbers of those molecules within the cell, while CSP and Z represent the environmental concentrations (nM) of CSP and XIP, respectively. The model is deterministic and takes no account of stochasticity in expression. This is sufficient for establishing whether single vs. multiple values for X are possible (i.e. unimodal vs. bimodal behavior) when the system operates in defined vs. complex medium respectively. The model is described by:
Here a and b represent peak synthesis rates, KM and KCSP describe binding equilibria of the transcriptional activators, KRS is the association constant for the multimer M, n is the degree of association, β is the rate of protein degradation or dilution, and v is the cellular volume. D characterizes the rate of equilibration between intracellular and extracellular ComS or XIP through the hypothetical channel. We take D = 0 to represent a complex medium (closed channel) and we take an arbitrarily large D = 500 to represent defined medium (open channel).
We select constitutive expression parameters aR = 1, aS = 0.091, aX = 0.25 for comR, comS, comX respectively so that the fold-changes in expression upon activation are approximately 2×, 12×, and 5×, consistent with prior data (Lemme et al., 2011). We select n = 2.5 to capture the proposed dimeric – or higher multimer – character of the RS complex (Mashburn-Warren et al., 2010). We also note that the steady states of the system are sensitive to the ratios a/β and b/β, rather than to a, b, β individually. The free parameters in the model are then KM, KCSP, KRS, a/β b/β, v.
We programmed the model equations into MATLAB (Mathworks, Inc.) and adjusted its free parameters until comX exhibited bimodal activation at CSP concentrations near 100 nM (in complex medium) and unimodal activation at XIP concentrations near 30 nM (in defined medium), as shown in Figure 8. We find that the capacity for bimodal/unimodal switching over this range of stimulus concentrations is a fairly generic property of the model as it occurs over a range of parameter values. Table 1 indicates approximate parameter ranges. The model could be extended to include a more complete or accurate description of the CSP-ComDE stimulation mechanism, differences in the affinity of ComS (unprocessed) vs. XIP for ComR, more mechanistic detail on the role of complex medium, links to other inputs and outputs, etc. However we do not expect these changes would change the model’s qualitative property of unimodal/bimodal switching in response to changes in growth medium.
Table 1.
Parameter values for the simulation model (Experimental procedures – Modeling) of autofeedback in comRS, consistent with bimodal switching at 100 nM CSP in complex medium and unimodal switching at 30 nM exogenous XIP in defined medium. A range of parameter values are consistent with this behavior, with ~80% of those values falling within the range indicated in the table. Results from simulation using the typical parameters are shown in Figure 8.
| Parameter | Typical | 10–90% range | Units |
|---|---|---|---|
| KCSP | 112 | 112–113 | nM |
| kM1/n | 1.1 × 106 | (0.81 – 1.4) × 106 | nM |
| a/β | 180 | 130–270 | dimensionless |
| b/β | 330 | 270–420 | dimensionless |
| KRS | 27 | 19 – 34 | (nM)−1 |
| ν | 7.4 | 6.1 – 9.5 | (nM)−1 |
Supplementary Material
Acknowledgments
The authors thank Don Morrison for sharing unpublished information on comRS. The authors also thank Inhae Kwak for assistance with cell image analysis. This work is supported by the NIH under NIDCR awards 5R21DE18826 and RO1DE13239.
References
- Ahn S, Wen ZT, Burne RA. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun. 2006;74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S, Lemos JAC, Burne RA. Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol. 2005;187:3028–3038. doi: 10.1128/JB.187.9.3028-3038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspiras MB, Ellen RP, Cvitkovitch DG. ComX activity of Streptococcus mutans growing in biofilms. FEMS Microbiol Lett. 2004;238:167–174. doi: 10.1016/j.femsle.2004.07.032. [DOI] [PubMed] [Google Scholar]
- Berg KH, Biørnstad TJ, Johnsborg O, Håvarstein LS. Properties and biological role of streptococcal fratricins. Appl Environ Microbiol. 2012;78:3515–3522. doi: 10.1128/AEM.00098-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitkovitch D. Genetic competence and transformation in oral streptococci. Crit Rev Oral Biol M. 2001;12:217–243. doi: 10.1177/10454411010120030201. [DOI] [PubMed] [Google Scholar]
- Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Generation of gradients having complex shapes using microfluidic networks. Anal Chem. 2001;73:1240–1246. [Google Scholar]
- Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. Development of competence for genetic transformation by Streptococcus mutans in a chemically defined medium. J Bacteriol. 2012 doi: 10.1128/JB.00337-12. Online early. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Genetic competence in Bacillus subtilis. Microbiol Rev. 1991;55:395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James EFJ. Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Generation of solution and surface gradients using microfluidic systems. Langmuir. 2000;16:8311–8316. [Google Scholar]
- Khan R, Rukke HV, Ricomini Filho AP, Fimland G, Arntzen MØ, Thiede B, Petersen FC. Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J Bacter. 2012 doi: 10.1128/JB.00624-12. Online early. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak IH, Son M, Hagen SJ. Analysis of gene expression levels in individual bacterial cells without image segmentation. Biochem Biophys Res Commun. 2012;421:425–430. doi: 10.1016/j.bbrc.2012.03.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PCY, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: Application and efficiency. J Microbiol Methods. 2002;49:193–205. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res. 2010;28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid. 1992;28:130–145. doi: 10.1016/0147-619x(92)90044-b. [DOI] [PubMed] [Google Scholar]
- Lemme A, Grobe L, Reck M, Tomasch J, Wagner-Dobler I. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol. 2011;193:1863–1877. doi: 10.1128/JB.01363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hanna M, Svensater G, Ellen R, Cvitkovitch D. Cell density modulates acid adaptation in Streptococcus mutans: Implications for survival in biofilms. J Bacteriol. 2001a;183:6875–6884. doi: 10.1128/JB.183.23.6875-6884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lau P, Lee J, Ellen R, Cvitkovitch D. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001b;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tang N, Aspiras M, Lau P, Lee J, Ellen R, Cvitkovitch D. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol. 2002;184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zeng L, Burne RA. AguR is required for induction of the Streptococcus mutans agmatine deiminase system by low pH and agmatine. Appl Environ Microb. 2009;75:2629–2637. doi: 10.1128/AEM.02145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B, Quentin Y, Fichant G, Claverys J. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 2006;14:339–345. doi: 10.1016/j.tim.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Qi F. The mutacins of Streptococcus mutans: Regulation and ecology. Molec Oral Microbiol. 2012;27:57–69. doi: 10.1111/j.2041-1014.2011.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Zheng L, Shi W, Qi F. Genetic characterization of the hdrRM operon: A novel high-cell-density-responsive regulator in Streptococcus mutans. Microbiology. 2007;153:2765–2773. doi: 10.1099/mic.0.2007/007468-0. [DOI] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Ann Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Nepomuceno RSL, Tavares MB, Lemos JA, Griswold AR, Ribeiro JL, Balan A, et al. The oligopeptide (opp) gene cluster of Streptococcus mutans: Identification, prevalence, and characterization. Oral Microbiol Immunol. 2007;22:277–284. doi: 10.1111/j.1399-302X.2007.00368.x. [DOI] [PubMed] [Google Scholar]
- Perry D, Kuramitsu HK. Genetic-transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Lévesque CM. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol. 2009;72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeil SH, Wickersham CE, Hoffmann A, Lipman EA. A microfluidic mixing system for single-molecule measurements. Rev Sci Instrum. 2009;80 doi: 10.1063/1.3125643. 055105. [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton K, Ahn S, Sagstetter AM, Burne RA. A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J Bacteriol. 2011;193:862–874. doi: 10.1128/JB.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senadheera DB, Cordova M, Ayala EA, Chávez de Paz LE, Singh K, Downey JS, et al. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol. 2012 doi: 10.1128/JB.06071-11. Online early. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senadheera D, Cvitkovitch DG. Quorum sensing and biofilm formation by Streptococcus mutans. In: Utsumi R, editor. Bacterial Signal Transduction: Networks and Drug Targets. New York: Springer; 2008. pp. 178–188. [DOI] [PubMed] [Google Scholar]
- Sia SK, Whitesides GM. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis. 2003;24:3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- Smith EG, Spatafora GA. Gene regulation in S. mutans. J Dent Res. 2012;91:133–141. doi: 10.1177/0022034511415415. [DOI] [PubMed] [Google Scholar]
- Smits WK, Kuipers OP, Veening J. Phenotypic variation in bacteria: The role of feedback regulation. Nat Rev Micro. 2006;4:259–271. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- Smits WK, Eschevins CC, Susanna KA, Bron S, Kuipers OP, Hamoen LW. Stripping bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol. 2005;56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- Suntharalingam P, Cvitkovitch D. Quorum sensing in streptococcal biofilm formation. Trends Microbiol. 2005;13:3–6. doi: 10.1016/j.tim.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Terleckyj B, Willett NP, Shockman GD. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun. 1975;11:649–655. doi: 10.1128/iai.11.4.649-655.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- Zubay G, Morse DE, Schrenk WJ, Miller JHM. Detection and isolation of the repressor protein for the tryptophan operon of Escherichia coli. P Natl Acad Sci USA. 1972;69:1100–1103. doi: 10.1073/pnas.69.5.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.