Abstract
We have introduced a targeted mutation in SH2D1A/DSHP/SAP, the gene responsible for the human genetic disorder X-linked lymphoproliferative disease (XLP). SLAM-associated protein (SAP)-deficient mice had normal lymphocyte development, but on challenge with infectious agents, recapitulated features of XLP. Infection of SAP− mice with lymphocyte choriomeningitis virus (LCMV) or Toxoplasma gondii was associated with increased T cell activation and IFN-γ production, as well as a reduction of Ig-secreting cells. Anti-CD3-stimulated splenocytes from uninfected SAP− mice produced increased IFN-γ and decreased IL-4, findings supported by decreased serum IgE levels in vivo. The Th1 skewing of these animals suggests that cytokine misregulation may contribute to phenotypes associated with mutation of SH2D1A/SAP.
X-linked lymphoproliferative disease (XLP) is a genetic disorder characterized by immune dysregulation and lymphoproliferation on exposure to Epstein–Barr virus (EBV) (1, 2). EBV, which infects B cells, causes infectious mononucleosis and a marked proliferation of CD8+ cytotoxic T lymphocytes (CTL). In XLP, patients have an uncontrolled expansion of CD8+ cells that fails to effectively eliminate EBV-infected cells. The consequent infiltration and destruction of the liver and bone marrow leads to mortality early in life in over half of reported cases (2). Patients surviving infectious mononucleosis have variable clinical pictures postinfection and usually develop immune abnormalities, including lymphoproliferative states and dysgammaglobulinemia (3). Asymptomatic female carriers show random X-inactivation, suggesting that normal cells can rescue the phenotype and that cellular interactions may be an important component of the disease (4).
In 1998, three groups simultaneously identified the gene for XLP through both positional (5, 6) and functional cloning (7). The XLP gene-product, SH2D1A/DSHP/SAP (SLAM-associated protein, hereafter referred to as SAP), is a 128-residue protein that consists almost entirely of a single Src homology 2 (SH2) protein interaction domain. Over 25 mutations have been identified, and include missense, nonsense, frameshift, and splice-site mutations, as well as entire gene deletions, suggesting that the phenotype can be caused by a loss of function. At present, there is no correlation between clinical presentation and the type or position of mutation (3).
The SH2 domain of SAP binds a conserved tyrosine-containing motif found in the intracellular domains of CD2 family members CD150/SLAM, CD244/2B4, and CD84 (7–9). Although little is known about CD84, signaling lymphocytic activation molecule (SLAM) is a self-ligand expressed on both B and T cells that functions as a costimulatory molecule and is thought to be important for B–T cell interactions (10, 11). 2B4 is an activating receptor expressed on natural killer (NK) and cytotoxic T cells (12). Because SAP is expressed primarily in T and NK cells, both SLAM and 2B4 are attractive candidates for receptors involved in the pathogenesis of XLP. In vitro data have demonstrated that SAP competes for binding to phosphotyrosine motifs in SLAM and 2B4 with SHP1, SHP2, and SHIP, phosphatases that negatively regulate antigen receptor signaling (7, 8, 13). Evidence suggests that 2B4 signaling may be impaired in NK cells from XLP patients (14–17). In addition, a recent report indicates that SAP can also associate with the intracellular signaling intermediate, p62Dok (18), although the implications for Dok function are unclear. Thus, the consequences of loss of SAP on signaling from these receptors are not well understood.
Studies of SLAM also raise the possibility that cytokines may play a role in XLP pathogenesis. Stimulation of human T helper clones via SLAM increases activation and production of IFN-γ irrespective of their T helper phenotype (19). Similarly, ligation of SLAM on murine Th1 clones results in increased IFN-γ production (20). However, the contribution of cytokine regulation to the pathogenesis of XLP remains unclear.
Because XLP is rare, there are few preinfection studies and their findings are quite variable. Nonetheless, the existence of phenotypes before apparent EBV infection raises the possibility of a more basic immune dysfunction, independent of viral infection (21–23). To develop a model that may present insights into the pathogenesis of this disease, we mutated the XLP locus in mice. SAP-mutant mice were viable, fertile, and transmitted the gene in Mendelian fashion. However, challenge with the infectious agents Toxoplasma gondii and lymphocyte choriomeningitis virus (LCMV) revealed abnormal immune responses and evidence of T cell hyperactivation with increased IFN-γ production, decreased B cell function, and, in a model of chronic infection, increased morbidity and mortality. Furthermore, splenocytes cultured from uninfected mice also exhibited a bias toward production of Th1 cytokines. The initial characterization of these mice reveals a phenotype with characteristics of XLP and suggests that cytokine misregulation may contribute to the phenotype of this genetic disease.
Materials and Methods
Antibodies.
Anti-SAP C-terminal serum was raised against peptide GRGPQAPTGRRDSDI in rabbit. Anti-SAP N-terminal and anti-goat Ig HRP were obtained from Santa Cruz Biotechnology; anti-mouse CD3ɛ, CD4, CD8, TCR, CD19, IFN-γ, Fc Block, and DX5 were obtained from Becton Dickinson/PharMingen; anti-green fluorescent protein (GFP), anti-mouse Ig HRP, and anti-rabbit Ig horseradish peroxidase (HRP) were obtained from Roche Molecular Biochemicals; anti-mouse Ig isotype HRP was obtained from Zymed; anti-IL-4 was obtained from BioSource International (Camarillo, CA); anti-tubulin was obtained from ICN; anti-CD28 was a gift from J. Powell and R. Schwartz (National Institute of Allergy and Infectious Disease); and anti-SLAM was a gift from A. O'Garra (DNAX).
Expression Plasmids.
pSX-Lck and pCEFL were gifts from J. S. Gutkind (National Institute of Dental Research, Bethesda); pEBG-SLAM and pEBG-CD84 are described in ref. 9. Full-length SH2D1A/SAP was generated by PCR from a mouse embryo day-15.5 Gene-Trapper library (using a 3′ primer that mutated the stop codon to encode a Thr), sequenced, and subcloned into pEGFP-N1 (CLONTECH). Truncated SAP-GFP was generated by PCR (incorporating the T68I mutation in the 3′ primer), sequenced, and ligated into pCEFL followed by a GFP variant (24).
Generation of Mutant Mice.
The targeting construct was derived from a 129Sv/Ev Lambda phage library (Stratagene) screened with EST Image Clone 719479 (Research Genetics, Huntsville, AL). The 5′ arm of the construct was generated by PCR using a 3′ primer to incorporate the T68I mutation (atgtactCtagatgctatctggaa). Genomic DNA arms and a variant of GFP (24) were inserted sequentially into pPNT (25), and introduced into TC1 ES cells. Homologous recombinants were screened by Southern blots of genomic DNA digested with XbaI and probed sequentially with a 3′ flanking probe and GFP. Correct targeting was verified by KpnI digestion and a 5′ internal probe. Chimeric mice were bred to either 129Sv/Ev/Tac or C57BL/6 mice. Northern blots of total thymic RNA (25 μg) were probed with full-length SH2D1A/SAP and GFP.
Glutathione S-Transferase (GST) Pulldown Assays.
293T cells were transfected by Ca3(PO4)2 with 5 μg of each plasmid, harvested, and lysed in GST Lysis buffer (25 mM Hepes, pH 7.0, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10% glycerol plus protease inhibitors, 1 mM NaF, and 1 mM Na orthovanadate). A portion of lysates were analyzed for protein expression and the rest for precipitation of GST chimeras and associated proteins with 10 μl glutathione Sepharose beads. Beads were washed four times in lysis buffer before Western analysis (9).
Western Analysis.
Cells or glutathione agarose pellets were boiled in SDS/PAGE reducing sample buffer, resolved by SDS/PAGE, transferred to nitrocellulose and probed with appropriate primary and secondary antibodies (anti-SAP-C-terminal 1:500 with anti-rabbit Ig HRP 1:10,000; anti-SAP-N-terminal, 0.4 μg/ml with anti-Goat Ig HRP 1:2,000; anti-GFP 0.4 μg/ml with anti-mouse Ig HRP 1:10,000; anti-tubulin 1:5,000 with anti-rabbit Ig HRP), followed by enhanced chemiluminescence (ECL, Amersham Pharmacia).
Cell Culture, Proliferation, and Flow Cytometry.
Thymi, lymph nodes, and spleens from age matched mice were teased in RPMI medium 1640 plus 10% FBS (HyClone) and splenic red blood cells lysed in NH3Cl. T or CD4+ cells were purified by negative selection columns (R & D Systems). Splenocytes (5 × 105 per 96 well) were stimulated with antibodies or surface toxo antigen (STAg) and pulsed for 8 h with [3H]thymidine (1 μCi per well; 1 Ci = 37 GBq) 24–72 h after stimulation. Alternatively, 5 × 104 purified T cells per well were stimulated by using plate-bound antibodies. Proliferations were performed in triplicate. For flow cytometry, 106 cells were stained as described (26), data collected on a Becton Dickinson FacScan or FacsCalibur and analyzed with FLOJO (TreeStar Software, San Carlos, CA).
Cytokine and Ig Assays.
Cell culture supernatants were harvested at 24 h (IL-2), 48 h (IL-4), or 72 h (IFN-γ) and cytokines determined by ELISA (R & D Systems or Cytokine Core Laboratory, University of Maryland, Baltimore). Alternatively, splenocytes from uninfected mice were stimulated 24 to 48 h with anti-CD3 (1 μg/ml) plus anti-CD28 (1:5,000), incubated with Brefeldin A for 3 h, and stained for intracellular cytokines (27). Serum Ig levels and anti-STAg Ig isotypes were determined by ELISA (Anilytics, Gaithersburg, MD; ref. 28).
In Vitro Polarization.
Purified CD4+ or sorted naive CD4+62L+ cells from wild-type (WT) and SAP− mice were stimulated with anti-CD3 and anti-CD28 plus irradiated T-depleted WT or IL-4 deficient splenocytes under Th1 (IL-12 plus anti-IL-4), Th2 (IL-4 plus anti-IL-12), or null (anti-IL-4, anti-IL-12, plus anti-IFN-γ) conditions (29). Cells were restimulated with anti-CD3 and CD28 and cell culture supernatants were analyzed at 48 h for IL-4 and 72 h for IFN-γ.
T. gondii Infections.
Mice were injected i.p. with 20 T. gondii cysts (ME49 strain) and serum, spleens, and brains were harvested at 31 days postinfection (pi) for analysis (28).
LCMV Infections.
Mice (4–5 months old) were infected with LCMV Armstrong strain [2 × 105 plaque-forming units (pfu) i.p.] or Clone 13 (2–5 × 106 pfu i.v.) and killed for analyses at 8, 30, or 37 days pi (26). Cytotoxic activity of splenocytes was tested in a standard 6 h [51Cr] release assay by using LCMV-infected MC57 cells as targets. Splenocytes were stained with LCMV-specific tetramers as described (26). For intracellular cytokines, splenocytes (106 cells per 96 flat-bottom well) from LCMV infected animals were stimulated in the presence of Brefeldin A for 5 h in the presence or absence of LCMV-specific peptides and stained as above. Total and LCMV-specific antibody-secreting cells (ASC) were quantitated by the ELISPOT technique using nitrocellulose-bottomed 96-well plates (Millipore) coated with virus (30).
Results
Generation and Characterization of Mice Deficient in SAP.
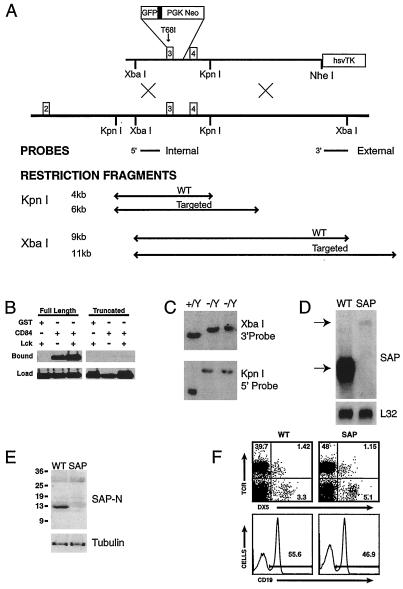
To introduce a mutation in the mouse XLP locus, we engineered a construct based on a splice-site mutation that truncated SH2D1A/SAP at the beginning of the third exon, fusing GFP in frame to follow expression of the mutant protein. However, because GFP could theoretically stabilize the protein, we incorporated an additional XLP patient mutation, T68I, that disrupts SAP binding to SLAM (Fig. 1A; refs. 1 and 5). To confirm that this mutation impairs binding properties of SAP, expression constructs for the truncated SAP-GFP and a full-length SAP-GFP chimera were generated and coexpressed in 293T cells with GST fusions to the intracellular domains of either SLAM or CD84. The full-length SAP fusion bound both GST chimeras, whereas the truncated SAP fusion showed barely detectable binding to CD84, even after Lck phosphorylation, which increases SAP binding affinity (ref. 9; Fig. 1B). Binding to SLAM was also severely compromised (data not shown), suggesting that the predicted mutant protein was functionally impaired. The mutation was introduced into mouse embryonic stem cells (Fig. 1C), and chimeric mice were obtained that transmitted the mutation to their progeny. However, Northern analysis of thymus RNA from mutant mice demonstrated only a trace (2% of WT) of a slowly migrating mRNA species reactive with both SH2D1A/SAP and GFP probes (Fig. 1D and data not shown), suggesting that this mutation destabilized the mRNA. A lack of SAP protein was confirmed by Western analyses with antibodies reactive against either the N or C terminus of SAP, or against GFP (Fig. 1E and data not shown). Thus, although the GFP tag could not be used to follow cell fates, this mutant appeared to eliminate expression of functional protein.
Figure 1.
Generation of mice deficient in SAP. (A) Targeting construct and corresponding gene structure are shown with XbaI and KpnI restriction fragments and probes used to distinguish WT and targeted alleles. (B) The mutant form of SAP shows decreased binding to the intracellular domain of CD84. Full-length and truncated forms of SAP were fused to GFP and coexpressed in 293T cells with GST, or GST fused to the intracellular domain CD84 without or with coexpression of the tyrosine kinase, Lck. Glutathione agarose beads were used to extract GST fusions and associated proteins from cell lysates. Cell lysates (Lower) and adsorbed proteins were immunoblotted with an anti-GFP antibody to determine the relative abundance of SAP proteins. (C) Southern analysis of ES cell clone DNA by using a 3′ probe following XbaI digestion, and a 5′ probe following KpnI digestion to confirm targeting. Two clones were positive for recombination. (D) Northern analysis of total thymic RNA from WT and SAP− mice, hybridized to a full-length SH2D1A/SAP cDNA probe. Equivalent loadings were confirmed by rehybridization to L32. (E) Analyses of SAP in total thymocyte protein. 1.5 × 106 cell equivalents per lane were blotted sequentially with anti-SAP N terminus and anti-tubulin. (F) Splenic lymphocyte populations show grossly normal profiles of T, B, and NK cells. (Upper) T cell (TCR+ DX5−) and NK cell (TCR− DX5+) proportions. (Lower) Percentages of B cells (CD19+) cells.
Mice deficient in SAP were born in normal numbers, were viable and fertile, and showed no overt phenotypes. Lymphocyte populations were also grossly normal, although occasional mutant animals showed a higher percentage of T and NK cells and a lower percentage of B cells in the spleen (2 of 21 animals, ranging from 8 to 18 weeks age; Fig. 1F). Additionally, thymocyte populations also appeared grossly normal (data not shown). Thus, as expected from the phenotype of XLP, SAP is not required for lymphocyte development.
SAP-Deficient Mice Show Altered Responses to LCMV.
Patients with XLP often manifest symptoms only after infection with EBV. Because mice are not susceptible to this virus, we examined responses to LCMV, a virus that elicits certain responses similar to those seen with EBV, including a large CD8+ CTL response (26). Wild-type and mutant mice were infected with two strains of LCMV, the Armstrong strain, which causes acute infection, and Clone 13, a more virulent strain that can establish a chronic infection. On infection with either strain, SAP-deficient mice demonstrated clear differences in their immune responses.
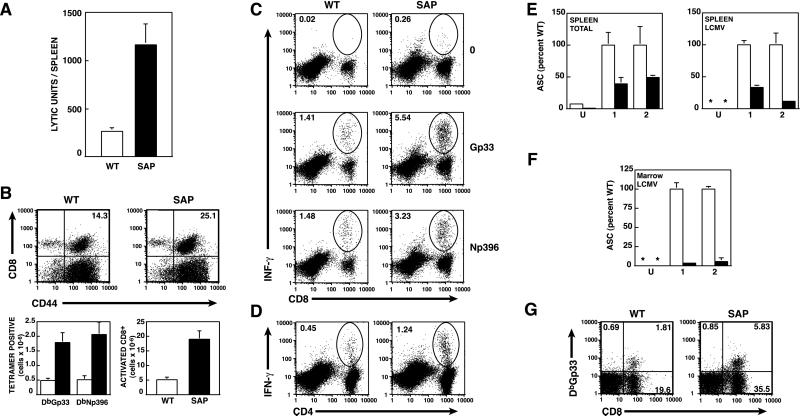
First, both WT and SAP− mice cleared the LCMV Armstrong infection within 1 week [<50 plaque-forming units (pfu) per ml in serum]. However, SAP− mice showed evidence of increased T cell responses (Fig. 2 and Tables 1–3, which are published as supplemental data on the PNAS web site, www.pnas.org). At 8 days pi, CTL activity directed against LCMV-pulsed target cells was significantly elevated in spleens of SAP− mice (Fig. 2A). This result was seen in two independent experiments comparing a total of six SAP− mice to four WT mice and correlated with an increase in the numbers of activated (CD44+) CD8+ cells that reacted with LCMV-specific tetramers in the spleens of infected SAP− mice (Fig. 2B). Furthermore, SAP− mice exhibited elevated percentages of CD8+ splenic T cells expressing IFN-γ or TNF-α when stimulated with LCMV-specific peptides as determined by intracellular cytokine staining (Fig. 2C and data not shown), confirming that these LCMV-specific CD8+ cells were functional. Percentages of CD4+ positive cells that expressed IFN-γ or IL-2 on stimulation with an LCMV-specific peptide were also elevated 2–3-fold compared with WT, and the amount of IFN-γ produced per cell was also greater [mean fluorescent intensity 728 (±45) for SAP− and 493 (±12.8) for WT; Fig. 2D). Furthermore, elevated T cell responses were still seen 30 days pi (Tables 2 and 3).
Figure 2.
Altered responses to LCMV in SAP− mice. (A–F) Mice were infected with LCMV Armstrong strain. WT, open bars; SAP, filled bars. (A) Elevated CTL activity against LCMV infected targets in SAP− mice 8 days pi. A lytic unit (LU) is defined as the number of lymphocytes required for a specific lysis of 35%. Means and standard error (SEM) for three SAP− and two WT mice are shown. (B) Elevated antigen-specific and activated CD8+ T splenocytes. (Upper) Splenocytes from animals on day 8 pi stained for CD8 and anti-CD44. (Lower) Numbers of antigen-specific CD8+ cells as determined by MHC Class I tetramers (DbGp33 or DbNp396) and total activated (CD44+) CD8+ cells per spleen (means of three SAP− and two WT mice ±SEM). Similar results were obtained in a second independent experiment (see supplemental data). (C) Splenocytes were unstimulated (0) or stimulated with LCMV-specific peptides (Gp33–41 or Np396–404) and the percentage of CD8+ IFN-γ producing cells was determined by intracellular cytokine staining. (D) Increased IFN-γ production by SAP− CD4 T cells. Splenocytes from WT and SAP− animals on day 8 pi with LCMV-Armstrong were stimulated with the class II A-b restricted peptide Gp61 and IFN-γ production was measured by intracellular staining. An average increase in Gp61-specific CD4 cells was 2–3-fold in two independent experiments using three SAP− mice each. The average mean fluorescent intensity for IFN-γ was 728 for SAP− (±45 SEM) and 493 (±12.8) for WT. (E) Spleens were harvested from uninfected mice (U) or mice infected with Armstrong strain LCMV at day 8 pi and the number of total IgG (Left) or LCMV-specific IgG (Right) ASCs were determined by ELISPOT. The graph compares the ASCs per million splenocytes from two independent experiments (1 and 2) examining a total of four WT and six SAP− mice (WT mean converted to 100%). WT, open bars; SAP, filled bars; *, below detection. (F) LCMV-specific ASCs in bone marrow cells from uninfected and Armstrong strain-infected mice 30–37 days pi. (G) Increased CD8+ T cells reactive with LCMV-specific tetramer DbGp33 after chronic infection with LCMV Clone 13 strain.
In contrast, ASCs were reduced in SAP− mice after infection with LCMV. The numbers of both total and LCMV-specific ASCs, determined by ELISPOT, were decreased in the spleens of mutant relative to WT mice, both on days 8 and 30 pi (Fig. 2E and data not shown). Notably, the number of total and LCMV-specific ASCs observed in the bone marrow on day 30 pi, a reflection of the number of plasma cells generated, was markedly suppressed (less than 5% WT levels for bone marrow LCMV-specific ASCs; Fig. 2F). In contrast, neither splenic B cell numbers nor sIgG levels were markedly reduced. Together, these data suggest marked abnormalities in immune responses to LCMV in SAP− animals.
To further explore responses to LCMV, we challenged mice with the more virulent Clone 13 strain, which causes a more disseminated infection and can establish chronic infections in adult mice (31). In addition, Clone 13 infection is often characterized by considerable T cell-mediated immunopathology during the first few weeks. In two separate experiments (n = 3 in Experiment 1 and n = 6 in Experiment 2), SAP− mice infected with Clone 13 exhibited increased morbidity (ruffled fur, hunched posture, and lethargy) compared with WT mice. In the second experiment, using a higher dose of Clone 13 [5 × 106 plaque-forming units (pfu)], one SAP− mouse died on day 8 and the remaining five mice were very ill. Three of these were killed on day 8 to analyze antiviral T cell responses and the two remaining SAP− mice died the next day. In contrast, only 1/6 of the WT mice died. Analysis of T cell responses showed that Clone 13-infected SAP− mice also had higher numbers of LCMV reactive CD8+ T cells compared with WT mice (Fig. 2G). Thus, under conditions of a chronic viral infection, SAP− mice may be more susceptible to LCMV-induced morbidity, perhaps because of their increased T cell responses.
Altered Responses to T. gondii.
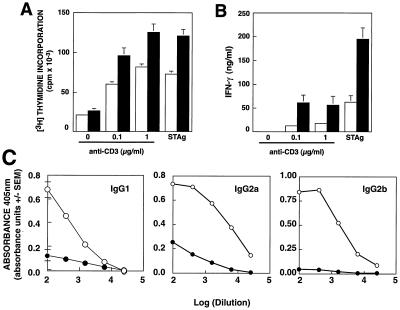
To determine whether changes in immune responses are limited to viral infection, we infected SAP− mice with T. gondii. Control of this intracellular parasite depends on production of IFN-γ, both by NK cells initially and then, as the adaptive immune response develops, by T lymphocytes (28). SAP-deficient mice survived the initial stages of the infection and, like WT mice, successfully established chronic infection. Significantly, however, splenocytes harvested from SAP− mice on day 31 pi showed elevated T cell proliferative responses (Fig. 3A) with increased secretion of IFN-γ after stimulation with either anti-CD3ɛ or soluble Toxoplasma antigen (STAg), despite normal T cell numbers (Fig. 3B and data not shown). Furthermore, SAP− mice exhibited lower titers of multiple Ig isotypes directed against STAg (Fig. 3C). Together, these data provide evidence for more generalized immune dysfunction in these mice.
Figure 3.
SAP-deficient mice show elevated T cell proliferation and IFN-γ production following infection with T. gondii. WT (open bars and circles) and SAP− mice (filled bars and circles) were infected with T. gondii cysts and at day 31 splenocytes were stimulated with anti-CD3ɛ or 10 μg/ml STAg. Proliferation (A) and production of IFN-γ (B) of splenocytes were assessed at 48 h of culture. The mean ± SEM of cultures from five individual mice for each genotype are shown. (C) Serum Ig directed against STAg 30 days pi.
Altered T Cell Responses in Uninfected SAP− Mice.
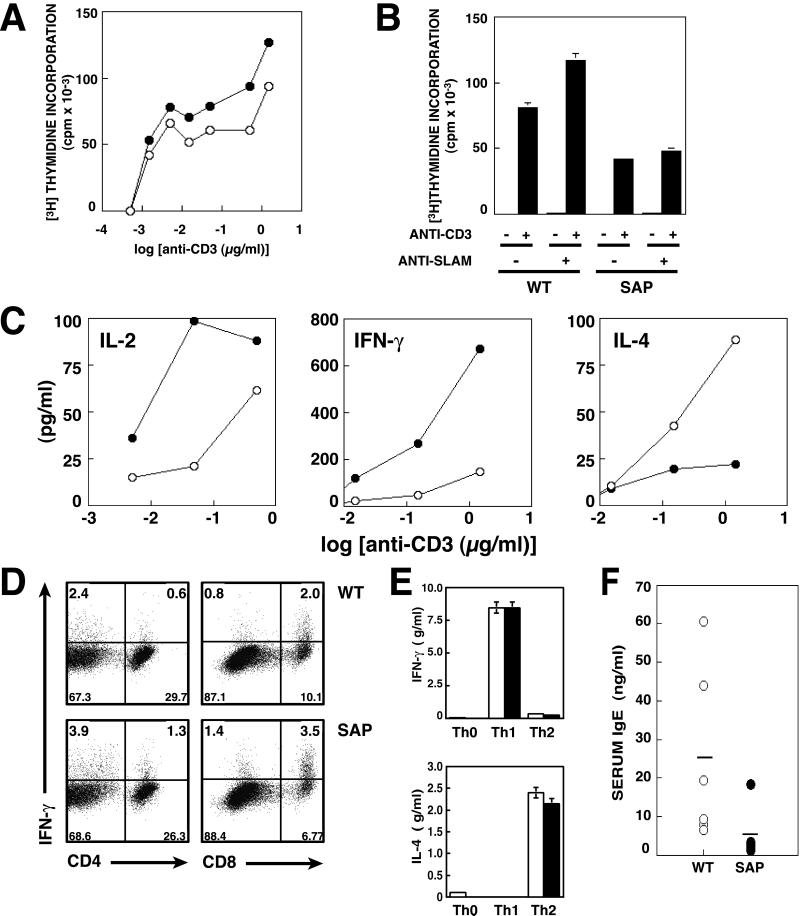
To determine whether altered T cell responses occur before challenge with pathogens, we examined T cell function from uninfected mice. Consistent with our findings postinfection, anti-CD3ɛ stimulation of splenocytes from SAP− mice resulted in mild elevations in T cell proliferation, although this increase was less robust and more variable than seen postinfection with T. gondii (Fig. 4A and data not shown).
Figure 4.
Uninfected mice show alterations in cytokine production and serum IgE. Splenocytes from WT (open circles or bars) and SAP− mice (filled circles or bars) were cultured with increasing concentrations of anti-CD3ɛ. (A) Proliferation of anti-CD3ɛ-stimulated splenocytes assessed by incorporation of [3H]thymidine after 24 h of culture. (B) Purified T cells were unstimulated or stimulated with platebound 1 μg/ml anti-CD3ɛ plus 10 μg/ml nonimmune IgG or anti-mouse SLAM. Proliferation was assessed at 72 h. Means and SEM of triplicate samples are shown. (C) Supernatants from splenocyte cultures stimulated with anti-CD3ɛ were assayed for IL-2 at 24 h, IFN-γ at 48 h, and IL-4 at 72 h by ELISA. (D) Splenocytes stimulated with 3 μg/ml anti-CD3ɛ plus 1:5,000 anti-CD28 for 24 h were stained for surface CD4 and CD8 and intracellular IFN-γ. (E) Purified naive CD4+ splenocytes from WT and SAP− mice were stimulated with anti-CD3 and anti-CD28 plus irradiated T-depleted splenocytes under Th1 (IL-12 plus anti-IL-4), Th2 (IL-4 plus anti-IL-12) or nonpolarizing conditions. Cells were restimulated with anti-CD3 and CD28 and supernatants assayed at 48 h for IL-4 and 72 h for IFN-γ. (F) Serum IgE levels from uninfected mice. WT, open circles; SAP−, filled circles. Horizontal bars indicate means for each genotype.
Interestingly, when purified T cells were stimulated with platebound antibodies (Fig. 4B), T cells from mutant mice proliferated less well, unlike the splenocyte cultures (Fig. 4A). This observation suggests that the increased response to anti-CD3ɛ stimulation was not intrinsic to the mutant T cells, but may be secondary to cellular interactions in the total splenocyte cultures. Furthermore, proliferation of purified SAP− T cells was not increased by SLAM ligation, suggesting that these cells fail to respond appropriately to this receptor (Fig. 4B).
Nonetheless, cytokines assayed from culture supernatants of CD3-stimulated splenocytes indicated that cells from SAP− mice were biased toward increased production of the Th1 cytokines IL-2 and IFN-γ (Fig. 4C). Intracellular cytokine staining of anti-CD3-stimulated splenocytes from uninfected mice demonstrated increased percentages of both CD4+ and CD8+ T cells producing IFN-γ (Fig. 4D). Furthermore, a decrease in the Th2 cytokine, IL-4, correlated inversely with the increase in IFN-γ produced by SAP− cells (Fig. 4C). To rule out the possibility that mutant T cells were biased toward a Th1 phenotype because they were unable to respond to Th2 cytokines, purified naive CD4+ cells were stimulated under polarizing conditions in which IL-12 and anti-IL-4 were included to induce Th1 differentiation or IL-4 and anti-IL-12 to induce Th2 differentiation. The mutant cells responded equally well to both Th1 and Th2 polarizing conditions, producing IFN-γ and IL-4, respectively, on restimulation (Fig. 4E), suggesting that SAP− cells do not exhibit defective cytokine responsiveness.
Finally, to examine whether SAP− mice show evidence of in vivo cytokine alteration, we examined patterns of serum immunoglobulins (Igs). In contrast to their pi phenotype, uninfected SAP− mice had normal levels of serum IgM, IgG, and IgA. Significantly, however, we observed lower levels of the IL-4-dependent Ig class, IgE, in the SAP− mice, providing an in vivo corroboration of the cytokine findings from cultured T cells (Fig. 4F). Thus, SAP-deficient mice show evidence of a Th1 bias and immune dysregulation even in the absence of infection.
Discussion
XLP is a complex disorder characterized by alterations in multiple compartments of the immune system. Although usually triggered by EBV, immune dysfunction, including recurrent infections, hypogammaglobulinemia, and lymphoproliferative disorders may also be seen in the absence of evidence of EBV infection, suggesting a more basic level of immune dysregulation (2).
The phenotype of mice with a mutant XLP locus supports a model for impaired function and altered homeostasis of multiple components of the immune system. Like XLP patients, before infection SAP− mice have grossly normal lymphocyte profiles, indicating that SAP is not critical for lymphocyte development. However, after infection with LCMV, SAP− mice recapitulate several aspects of XLP, including higher numbers of activated T cells, decreased ASC, and in a chronic infection, increased morbidity and mortality. The reduction in ASC, particularly those directed against LCMV in the bone marrow 30 days pi, is perhaps one of the most striking findings and strongly reminiscent of the hypogammaglobulinemias associated with XLP. Additionally, the increase in CTL activity and IFN-γ secreting cells suggests deregulation of multiple arms of the immune system, including both T and B cells. This abnormal response is not limited to viral infection, because exposure to T. gondii results in elevated T cell proliferation, increased IFN-γ production, and reduced serum Ig titers against STAg. Indeed, lymphocytes from uninfected mice also show increased Th1 and decreased Th2 cytokine production. Thus, as in XLP, immune dysfunction can occur independent of viral infection.
The altered cytokine production observed in these mice suggests that misregulation of Th1 and Th2 responses may contribute to the phenotype of XLP. The role of cytokines in XLP is unclear, although changes in Ig isotype profiles in some XLP patients before EBV infection suggests that cytokine-mediated regulation of class switching might be altered (22). Our findings suggest that mutation of SAP leads to a Th1 skewing of cytokine production both in vitro and, importantly, in vivo, as evidenced by the decreased serum IgE in uninfected mice. Furthermore, the observation that SAP− cells respond to both Th1 and Th2 polarizing conditions suggests that the defects in SAP− cells are due to altered patterns of primary cytokine secretion, not differences in cytokine responsiveness. It is interesting to speculate that such altered cytokine patterns may lead to a more cellular immune response at the expense of humoral mediated immunity. Indeed, reductions in Ig secretion have been observed from XLP B cell lymphoblastoid lines cocultured with autologous T cells (32), suggesting roles for secreted factors and/or cellular interactions in this abnormal response. The notion that cellular interactions are important for XLP may also be consistent with the differences we observe in proliferation between anti-CD3-stimulated splenocytes and purified T cells.
The gene responsible for the human disease XLP encodes a small SH2-containing protein, SAP, with obscure function(s). In vitro data describe SAP binding to the intracellular domain of a subset of the CD2 family, including SLAM and 2B4 (7, 8, 33). Although studies of cells from XLP patients suggest that 2B4-mediated NK cell cytolytic activity is impaired (15–17, 34), preliminary evidence suggests this may not be true in the mouse (D. W. McVicar, M.J.C., and P.L.S., unpublished observations). Indeed, SAP− mice survived early stages of infection with T. gondii, suggesting that innate immune responses are intact in these mice. In contrast, we do observe defective T cell responses to SLAM stimulation. Thus, in the mouse, SLAM may contribute more to the phenotypes we observe.
SLAM, a costimulatory molecule expressed on T lymphocytes and activated B cells, may provide a potential link between XLP and alterations in cytokine production. In addition to increasing T cell proliferation, stimulation of T cell clones via SLAM results in increased production of IFN-γ (19, 20). How loss of SAP affects signaling from SLAM and other receptors is unclear. Whereas SAP-deficient cells fail to increase proliferation in response to SLAM, splenocytes have increased production of IFN-γ, a response that is normally associated with SLAM engagement. Biochemical studies argue that SAP binds competitively to the same site on SLAM as phosphatases SHP-1, SHP-2, and SHIP-1 (7, 13). If SAP is required to disassociate phosphatases from SLAM for signaling, loss of SAP may prevent such signals, consistent with our observation that SAP− cells do not respond to SLAM engagement. However, if SLAM functions as a costimulatory molecule by sequestering inhibitory phosphatases away from antigen receptors, loss of SAP may result in a constitutive or prolonged signal from SLAM and potentially amplify TCR signals. Such a model would be consistent with the increased IFN-γ secretion and increased T cell responses we observe from CD3-stimulated splenocytes and might suggest that SAP− cells function independent of SLAM stimulation. Nonetheless, because SAP is expressed in multiple cell types, including some B cells (6), and binds to multiple receptors as well as other signaling molecules, interpretations of these data are complex and will require additional cellular and biochemical studies.
The development of mice mutant at the XLP locus may provide an important tool not only for understanding SAP function, but also for understanding normal responses to infection. In particular, our studies raise the possibility that cytokines may contribute to manifestations of XLP. Whether SAP− mice will recapitulate other features of this disease, including development of lymphomas, will be of interest for future studies.
Supplementary Material
Acknowledgments
We thank S. Anderson for flow cytometry assistance, M. Chetana and the Transgenic Core for mouse husbandry, J. Puck for critical reading of the manuscript, D. Jankovic, A. Veillette, and the Schwartzberg lab for invaluable advice and reagents, and A. O'Garra for the anti-SLAM antibody. E.N.K. was supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation. P.L.S. is supported in part by funds from the Searle Scholar's Program/Chicago Community Trust.
Abbreviations
- XLP
X-linked lymphoproliferative disease
- SLAM
signaling lymphocytic activation molecule
- SAP
SLAM-associated protein
- WT
wild type
- CMV
lymphocyte choriomeningitis virus
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HRP
horseradish peroxidase
- CTL
cytotoxic T lymphocyte
- EBV
Epstein–Barr virus
- SH2
Src homology 2
- pi
postinfection
- ASC
antibody-secreting cells
- NK
natural killer
Note Added in Proof.
After acceptance of this paper, another group also reported the generation of a SAP-deficient mouse strain (35).
References
- 1.Morra M, Howie D, Grande M S, Sayos J, Wang N, Wu C, Engel P, Terhorst C. Annu Rev Immunol. 2001;19:657–682. doi: 10.1146/annurev.immunol.19.1.657. [DOI] [PubMed] [Google Scholar]
- 2.Schuster V, Kreth H W. In: Primary Immunodeficiencies. Ochs H, Smith C A, Puck J M, editors. New York: Oxford Univ. Press; 1999. pp. 222–232. [Google Scholar]
- 3.Sumegi J, Huang D, Lanyi A, Davis J D, Seemayer T A, Maeda A, Klein G, Seri M, Wakiguchi H, Purtilo D T, et al. Blood. 2000;96:3118–3125. [PubMed] [Google Scholar]
- 4.Conley M E, Sullivan J L, Neidich J A, Puck J M. Clin Immunol Immunopathol. 1990;55:486–491. doi: 10.1016/0090-1229(90)90133-b. [DOI] [PubMed] [Google Scholar]
- 5.Coffey A J, Brooksbank R A, Brandau O, Oohashi T, Howell G R, Bye J M, Cahn A P, Durham J, Heath P, Wray P, et al. Nat Genet. 1998;20:129–135. doi: 10.1038/2424. [DOI] [PubMed] [Google Scholar]
- 6.Nichols K E, Harkin D P, Levitz S, Krainer M, Kolquist K A, Genovese C, Bernard A, Ferguson M, Zuo L, Snyder E, et al. Proc Natl Acad Sci USA. 1998;95:13765–13770. doi: 10.1073/pnas.95.23.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo M G, et al. Nature (London) 1998;395:462–469. [Google Scholar]
- 8.Tangye S G, Lazetic S, Woollatt E, Sutherland G R, Lanier L L, Phillips J H. J Immunol. 1999;162:6981–6985. [PubMed] [Google Scholar]
- 9.Lewis, J., Eiben, L. J., Nelson, D. L., Cohen, J. I., Nichols, K. E., Ochs, H., Notrangelo, L. D. & Duckett, C. S. (2001) Clin. Immunol., in press. [DOI] [PubMed]
- 10.Aversa G, Carballido J, Punnonen J, Chang C C, Hauser T, Cocks B G, de Vries J E. Immunol Cell Biol. 1997;75:202–205. doi: 10.1038/icb.1997.30. [DOI] [PubMed] [Google Scholar]
- 11.Cocks B G, Chang C C, Carballido J M, Yssel H, de Vries J E, Aversa G. Nature (London) 1995;376:260–263. doi: 10.1038/376260a0. [DOI] [PubMed] [Google Scholar]
- 12.Mathew P A, Garni-Wagner B A, Land K, Takashima A, Stoneman E, Bennett M, Kumar V. J Immunol. 1993;151:5328–5337. [PubMed] [Google Scholar]
- 13.Mikhalap S V, Shlapatska L M, Berdova A G, Law C L, Clark E A, Sidorenko S P. J Immunol. 1999;162:5719–5727. [PubMed] [Google Scholar]
- 14.Benoit L, Wang X, Pabst H F, Dutz J, Tan R. J Immunol. 2000;165:3549–3553. doi: 10.4049/jimmunol.165.7.3549. [DOI] [PubMed] [Google Scholar]
- 15.Parolini S, Bottino C, Falco M, Augugliaro R, Giliani S, Franceschini R, Ochs H D, Wolf H, Bonnefoy J Y, Biassoni R, et al. J Exp Med. 2000;192:337–346. doi: 10.1084/jem.192.3.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakajima H, Cella M, Bouchon A, Grierson H L, Lewis J, Duckett C S, Cohen J I, Colonna M. Eur J Immunol. 2000;30:3309–3318. doi: 10.1002/1521-4141(200011)30:11<3309::AID-IMMU3309>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Tangye S G, Phillips J H, Lanier L L, Nichols K E. J Immunol. 2000;165:2932–2936. doi: 10.4049/jimmunol.165.6.2932. [DOI] [PubMed] [Google Scholar]
- 18.Sylla B S, Murphy K, Cahir-McFarland E, Lane W S, Mosialos G, Kieff E. Proc Natl Acad Sci USA. 2000;97:7470–7475. doi: 10.1073/pnas.130193097. . (First Published June 13, 2000; 10.1073/pnas.130193097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aversa G, Chang C C, Carballido J M, Cocks B G, de Vries J E. J Immunol. 1997;158:4036–4044. [PubMed] [Google Scholar]
- 20.Castro A G, Hauser T M, Cocks B G, Abrams J, Zurawski S, Churakova T, Zonin F, Robinson D, Tangye S G, Aversa G, et al. J Immunol. 1999;163:5860–5870. [PubMed] [Google Scholar]
- 21.Purtilo D T. Arch Pathol Lab Med. 1981;105:119–121. [PubMed] [Google Scholar]
- 22.Grierson H L, Skare J, Hawk J, Pauza M, Purtilo D T. Am J Med Genet. 1991;40:294–297. doi: 10.1002/ajmg.1320400309. [DOI] [PubMed] [Google Scholar]
- 23.Brandau O, Schuster V, Weiss M, Hellebrand H, Fink F M, Kreczy A, Friedrich W, Strahm B, Niemeyer C, Belohradsky B H, et al. Hum Mol Genet. 1999;8:2407–2413. doi: 10.1093/hmg/8.13.2407. [DOI] [PubMed] [Google Scholar]
- 24.Stauber R H, Horie K, Carney P, Hudson E A, Tarasova N I, Gaitanaris G A, Pavlakis G N. BioTechniques. 1998;24:462–471. doi: 10.2144/98243rr01. [DOI] [PubMed] [Google Scholar]
- 25.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 26.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J, Zajac A J, Miller J D, Slansky J, Ahmed R. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 27.Jankovic D, Kullberg M C, Noben-Trauth N, Caspar P, Paul W E, Sher A. J Immunol. 2000;164:3047–3055. doi: 10.4049/jimmunol.164.6.3047. [DOI] [PubMed] [Google Scholar]
- 28.Yap G S, Sher A. Immunobiology. 1999;201:240–247. doi: 10.1016/S0171-2985(99)80064-3. [DOI] [PubMed] [Google Scholar]
- 29.Hu-Li J, Huang H, Ryan J, Paul W E. Proc Natl Acad Sci USA. 1997;94:3189–3194. doi: 10.1073/pnas.94.7.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slifka M K, Matloubian M, Ahmed R. J Virol. 1995;69:1895–1902. doi: 10.1128/jvi.69.3.1895-1902.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed R, Salmi A, Butler L D, Chiller J M, Oldstone M B, Slifka M K, Matloubian M. J Exp Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasuda N, Lai P K, Rogers J, Purtlo D T. Clin Exp Immunol. 1991;83:10–16. doi: 10.1111/j.1365-2249.1991.tb05580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sayos J, Nguyen K B, Wu C, Stepp S E, Howie D, Schatzle J D, Kumar V, Biron C A, Terhorst C. Int Immunol. 2000;12:1749–1757. doi: 10.1093/intimm/12.12.1749. [DOI] [PubMed] [Google Scholar]
- 34.Benoit L, Wang X X, Pabst H F, Dutz J, Tan R. J Immunol. 2000;165:3549–3553. doi: 10.4049/jimmunol.165.7.3549. [DOI] [PubMed] [Google Scholar]
- 35.Wu C, Nguyen K B, Pien G C, Wang N, Gullo C, Howie D, Sosa M R, Edwards M J, Borrow P, Satoskar A R, et al. Nat Immunol. 2001;2:410–414. doi: 10.1038/87713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.