Abstract
Although radiotherapy represents one of the most effective treatment modalities for patients with cancer, inherent and/or acquired resistance of cancer cells to radiotherapy is often an impediment to effective treatment. Diverse strategies have been developed to improve the efficacy of radiotherapy. The ubiquitin-proteasome system (UPS) operates in numerous vital biologic processes by controlling the protein turnover in cells. Ubiquitination is central to the UPS pathway, and it relies on the E3 ubiquitin ligases to catalyze the covalent attachment of ubiquitin to its protein substrates. Cullin-based RING ligases (CRLs) are the largest family of E3 ligases that are responsible for the ubiquitination and destruction of numerous cancer-relevant proteins. Its deregulation has been linked to many human cancers, making it an attractive target for therapeutic intervention. This review discusses how targeting the ubiquitin-proteasome system, particularly CRLs, is an exciting new strategy for radiosensitization in cancer and, specifically, focuses on MLN4924, a recently discovered small-molecule inhibitor of the NEDD8-activating enzyme, which is being characterized as a novel radiosensitizing agent against cancer cells by inactivating CRL E3 ubiquitin ligases.
Introduction
Cancer is a large group of highly complex diseases with dramatically different biologic behaviors. Even within cancers of the same organ, the extent of therapeutic response varies considerably, making it unlikely that any single agent would cure all cancers or even cancers of a single organ. Radiation therapy represents one of the most clinically effective forms of treatment [1]. It is frequently applied as a single treatment modality with curative intent or, more often, combined with surgery and/or chemotherapy to maximize the therapeutic effect [2]. Treatment outcome of patients with cancer receiving radiotherapy has improved in recent decades, mainly because of optimized therapeutic plans and technological advancements in the precise delivery of radiation to the targeted tumor tissues [3]. However, in many patients, disease recurs locally after radiotherapy. Although some treatment failures can be explained by the traditionally accepted clinical factors, such as tumor stage and grade, many failures remain unexplained [1]. It is now increasingly recognized that multiple biologic factors of tumors may contribute to radioresistance and, thereby, have a potential role in determining treatment outcome of patients. Examples include the intrinsic radioresistance of tumor cells, the existence of radioresistant cancer stem cells, repopulation of surviving cells after radiotherapy, repair of radiation-induced damage, the vasculature, as well as the extent of hypoxia and inflammation within tumors [1]. These factors associated with radioresistance have been extensively studied in both the preclinical and clinical settings, leading to the development of diverse strategies, including targeted agents to overcome or modulate them with the goal of improving radiotherapy efficacy.
The ubiquitin-proteasome system (UPS) is responsible for the timely degradation of many regulatory proteins within the cell [4] and also mediates various nondegradative functions [5]. Abnormal regulation of UPS has been implicated in a growing number of human diseases, notably in cancer [6]. Ubiquitination plays a central role in the UPS pathway and relies on the E3 ligases to catalyze the covalent attachment of ubiquitin to its protein substrates, which usually confers a recognition signal for proteasome targeting [4,7]. Cullin-based RING ligases (CRLs) are the largest family of E3 ubiquitin ligases that control the ubiquitination and proteasomal degradation of numerous cancer-relevant proteins [8], thus representing potential therapeutic targets in cancer [9,10]. Here, we provide an overview of CRL E3 ligases and discuss how general targeting of the UPS as well as selective targeting of CRL E3 ligases are being used for radiosensitization of cancer cells.
Ubiquitin and CRL E3 Ligases
Posttranslational modification of proteins by ubiquitin or ubiquitin-like proteins (e.g., NEDD8, SUMO-1, SUMO-2, SUMO-3, FUBI, HUB1, ISG15, FAT10, URM1, UFM1, ATG12, and ATG8) represents one of the most prevalent mechanisms for regulating most aspects of cell physiology [4,7,11,12]. As a bona fide modifier, ubiquitin is a highly conserved protein of 76 amino acids that can be covalently attached to other proteins through a stepwise cascade of three enzymes, i.e., E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and finally E3 (ubiquitin ligase), thereby influencing protein fate and function [4]. Ubiquitination typically acts as a degradation signal for the 26S proteasome (poly-ubiquitylation) [13] and also serves nonproteolytic roles (Lys63-linked poly- or mono-ubiquitylation) in regulating the nuclear factor kappaB (NF-κB) signaling pathway [14,15], DNA replication and repair [16,17], as well as intracellular trafficking [13,18].
In humans, there are two E1 enzymes, at least 38 E2 enzymes [19], and hundreds of E3 enzymes [8]. The E3 ligases are responsible for substrate specificity [8] and are subdivided into two major classes characterized by the presence of either a HECT or a RING domain within them [4,8,20]. RING domain-containing E3 ligases have more than 600 members, comprising about 95% of human E3 ligases [8]. Among the RING-based E3 ligases, the CRLs are the largest family of multiunit ubiquitin ligases that control the turnover of approximately 20% of all ubiquitinated proteins through proteasome-mediated degradation [21]. Within the CRL complex, cullin serves as a molecular scaffold and interacts at its C terminus with the RING finger protein, creating the catalytic core of the ligase, whereas its N terminus interacts directly or indirectly (through an adapter protein) with the substrate-recognition subunit (SRS). It is this SRS that confers specificity toward its substrate proteins [22]. In human and mouse, there are eight cullins (cullins 1–3, 4A, 4B, 5, 7, and 9) [22] and two RING family members, RING box protein-1 (RBX1) and RBX2, also known as sensitive to apoptosis gene (SAG) [8,23,24]. Both RBX1 and RBX2 are capable of binding to six members of the human cullin family (cullins 1–3, 4A, 4B, and 5) under overexpressed conditions [25] and demonstrate in vitro E3 ubiquitin ligase activity when complexed with cullin 1 [26]. A potential difference between RBX1 and RBX2 lies in that RBX1 is constitutively expressed, whereas RBX2/SAG seems to be stress-induced [27]. Furthermore, RBX1 preferentially interacts with cullin 2, whereas RBX2 is selectively associated with cullin 5, under physiological conditions [28]. The best characterized CRL complex is the SKP1-cullin 1-F-box protein (SCF) E3 ligase [29–31]. SCF is also known as CRL1 [32], where cullin 1 tethers both the RING finger protein RBX1/RBX2 and the adaptor protein SKP1, which, in turn, binds to the F-box protein [e.g., FBXW7, beta-transducin repeat containing protein (β-TrCP) and SKP2] [33]. The human genome contains approximately 69 F-box proteins that can potentially form a complex with cullin 1 [34]. Conceivably, the availability of two RBX family members, along with eight cullins, hundreds of substrate receptors, and many adaptor proteins allows for the assembly of a multitude of CRLs in eukaryotic cells, imparting these enzymes with key regulatory functions in protein homeostasis.
Notably, most CRLs, if not all, are activated by neddylation through attachment of ubiquitin-like protein (NEDD8) to cullin, therefore preventing the inhibitory binding by the cullin-associated neddylation-dissociated protein (CAND1) [35–38]. The conformation-based mechanisms that explain these activating roles of neddylation have been discussed previously [36,38,39]. Neddylation, through a process analogous to ubiquitination, involves an enzymatic cascade through the sequential activity of E1, E2, and E3, resulting in the covalent attachment of NEDD8 to its substrates. The NEDD8 cascade is currently known to contain a single E1, NEDD8-activating enzyme (NAE), two E2s, UBE2M (also known as UBC12), UBE2F [40–43], and a few candidate E3s, such as RBX1 [44,45], SCCRO (DCN1) [46], and IAP [47]. Owing to the critical role of neddylation in the activation of CRLs, it provides an alternative approach to modulate CRL activity by controlling the NEDD8 cascade, as discussed below.
Targeting the UPS and Radiosensitization
Aberrant UPS function has been strongly associated with cancer, and its pharmacologic inhibition has proved efficacious in the treatment of cancers [48]. Bortezomib (Velcade, PS-341) is the first commercially available proteasome inhibitor approved for clinical use in treating selected human cancers [49,50], whereas next-generation compounds, such as carfizomib, MLN9708, and CEP18770, are in clinical development [51]. By blocking the active sites in the 20S proteasome, bortezomib disrupts the entire UPS. Although bortezomib has demonstrated clinical efficacy as a single agent and in combination with chemotherapy in multiple myeloma and mantle cell lymphoma, its overall success has been limited because of a lack of response in other malignancies and drug-associated toxicity [52–54].
In the setting of radiation therapy, general proteasome inhibition has been shown to impart tumor cell radiosensitization in many preclinical models and is thought to involve modulation of proteins involved in apoptosis, cell cycle, and DNA double-strand break repair [55–57] (see Table 1). In particular, bortezomib has demonstrated radiosensitizing properties in a number of tumor cell models and in association with stabilization of p38 mitogen-activated protein kinase (MAPK) [58] and inhibition of NF-κB [59] or cancerous inhibitor of protein phosphatase 2A [60]. Other proteasome inhibitors in clinical (marizomib) and preclinical studies (MG-132) also showed radiosensitizing activity in glioma, prostate, and lung cancer cells [61–63] (Table 1). Whereas bortezomib combined with chemoradiation is an active area of clinical investigation, initial studies suggest that toxicity is a significant concern [64,65]. Taken together, these studies with general proteasome inhibitors have provided proof-of-concept that proteasome inhibition is a worthwhile strategy for sensitizing tumor cells to radiation and chemotherapy but underscore the importance of developing agents with better selectivity.
Table 1.
Radiosensitization of Human Cancer Cells by UPS Inhibitors.
| Developmental Stages | Radiosensitization Activity | Cancer Types | References | |
| UPS inhibitors | ||||
| Bortezomib | Clinical use | Yes | Prostate, gastric, cervical, rectal, esophageal, lung, and liver cancers; lymphoma; and central nervous system malignancies | [58–60,64,116–118,129] |
| Marizomib | Phase 1 | Yes | Glioma | [63] |
| MG-132 | Preclinical | Yes | Prostate and lung cancers; Hodgkin's lymphoma; and melanoma | [55,61,62,130,131] |
| CRL inhibitor | ||||
| MLN4924 | Phase 1 | Yes | Pancreatic, lung, and breast cancers | [91,119] |
One approach to circumvent the toxicity associated with general proteasome inhibitors is to directly target the E3 ligases, because each E3 ligase is responsible for a subset of cellular protein substrates. In contrast to general proteasome inhibition that has a broad impact on total cellular proteolysis, specific E3 ligase inhibition is expected to selectively stabilize a subset of cellular proteins, thus avoiding unwanted stabilization of other cellular proteins that may have deleterious effects on normal cells. Therefore, it is expected that a greater therapeutic window could be achieved with agents that target specific components of the UPS rather than the entire UPS.
Targeting CRL Components and Radiosensitization
CRLs represent the largest known class of E3 ubiquitin ligases and are fundamental in controlling protein homeostasis, thus regulating various biologic processes including cell cycle progression, gene transcription, signal transduction, and DNA replication among others [8,32,66]. Not surprisingly, deregulation of CRL has been associated with uncontrolled proliferation, genomic instability, and cancer [66]. Among the components of CRL, some have been defined as oncogenes (e.g., SKP2) that are frequently amplified and/or overexpressed in cancers, whereas others act as tumor suppressors (e.g., FBXW7) that are often found to be mutated in cancer [48,67,68]. The oncogenic properties of some CRLs make them potential targets for therapeutic intervention. The CRL components with attractive potential as radiosensitizing targets in cancer cells are discussed below.
RBX1/RBX2
Our previous and recent studies showed that RBX1 and RBX2, two family members of the RING component of CRL found in human and mouse, are frequently overexpressed in many types of human cancer [69–71]. In multiple human cancer cell lines, knockdown of either RBX1 or RBX2 suppresses cancer cell growth and survival [70,71]. This impaired growth and survival in response to RBX1 or RBX2 knockdown appears to involve the induction of apoptosis and senescence or only apoptosis in the case of RBX2 [70,71]. Similarly, ectopic expression of RBX2 protects cells from apoptosis induced by a variety of stresses including metal ions and redox compounds [23,72], nitric oxide [73], neurotoxin and 1-methyl-4-phenylpyridinium [74], heat shock [75], UV irradiation [76], and ischemia/hypoxia both in vitro [77] and in vivo [78,79]. Taken together, these results support the notion that cancer cells are more reliant on RBX1/RBX2 overexpression for their survival, thus more sensitive to RBX1/RBX2 targeting. Because every individual member of CRL ligase family requires either RBX1 or RBX2 for activity, targeted inhibition of RBX1/RBX2 would lead to general inactivation of the entire family of CRL ligases, thus having broader anticancer effects.
It is well established that radiation induces DNA damage and that G2 arrest is a crucial response to DNA damage in most cancer cells [80]. On the basis of the finding that RBX1 silencing triggers DNA double-strand breaks, leading to G2 arrest [70], it is conceivable that knockdown of RBX1 may sensitize otherwise resistant cancer cells to radiotherapy by redistributing them to G2, a more radiosensitive phase of cell cycle. This hypothesis is supported by our finding that RBX1 silencing indeed sensitizes human cancer cells to radiation [81]. The underlying mechanism for RBX1 silencing-mediated radiosensitization is likely attributable to the accumulation of DNA replication licensing proteins CDT1 and ORC1, two known CRL substrates [32], which leads to DNA double-strand breaks, DNA damage response, and G2 arrest, rendering cancer cells more sensitive to radiation [81].
RBX2 is a dual-function protein with CRL-independent antioxidant activity, when acting alone, or CRL-dependent E3 ligase activity, when forming a complex with other CRL components [24,27,82]. Analogous to RBX1, RBX2 silencing also sensitizes otherwise resistant cancer cells to radiation [71]. However, distinct from RBX1, RBX2 silencing-mediated radiosensitization in human cancer cells appears to be mechanistically linked with accumulation of the proapoptotic protein, NOXA [71]. However, as shown in an Rbx2 knockout model, complete elimination of Rbx2 expression sensitized mouse embryonic stem cells to radiation-induced cell killing through mechanisms involving an increase in steady-state levels of intracellular reactive oxygen species because of the abrogation of antioxidant activity of Rbx2, as well as the decreased NF-κB activation associated with accumulation of inhibitor κB (IκB) [83]. These findings further support the notion that RBX2 plays a protective role in response to DNA damage and, when absent, sensitizes cells to radiation-induced cell death, suggesting its potential as a novel target for radiosensitization.
Cullins
The cullins are a family of eight members, which do not have intrinsic catalytic activity, when acting alone, but instead are molecular scaffolds that facilitate the assembly of modular CRL complexes and mediate the transfer of ubiquitin from the E2-conjugating enzyme to the substrate proteins [22]. Cullin 1 is overexpressed in 40% of lung cancers [84], whereas cullin 4A expression is elevated in multiple cancer types, such as breast [85–87], hepatocellular [88], and mesothelioma [89]. Overexpression of cullin 4A in MCF10A cells abrogated the G2/M cell cycle checkpoint in response to radiation-induced DNA damage [90]. Because the biologic effects of the cullin proteins are reliant upon their SRSs [F-box, Bric-a-Brac, Tramtrack Broad-complex (BTB), von Hippel-Lindau (VHL) and suppressor of cytokine signaling (SOCS) proteins] and corresponding substrates, cullins themselves are not conventional oncoproteins or tumor suppressors. However, cullin overexpression could increase CRL activity in cancer cells, promoting uncontrolled proliferation. Thus, cullin 1 and cullin 4A are potential anticancer targets that when inhibited may shift cells to a more controlled growth state. Given that CDT1 and WEE1 are the substrates of cullin 1- and cullin 4A-based CRL and their accumulation is responsible for radiosensitization in pancreatic cancer cells [91], targeting either cullin 1 or cullin 4A might be a potential sensitization strategy for radiotherapy.
Substrate-recognition Subunit
SRSs recognize and recruit target substrates to the CRL complexes. Different cullins are known to use distinct classes of SRS, such as F-box proteins for SCF/CRL1, VHL-box for cullin 2, BTB proteins for cullin 3, DCAF proteins for cullin 4A/B, and SOCS-box proteins for cullin 5 [22]. The human genome contains about 69 F-box proteins that provide specificity for the particular substrate to be degraded [34]. Among them, only three are well characterized, namely, oncogenic SKP2, tumor suppressive FBXW7, and β-TrCP, which is considered either oncogenic or tumor suppressive in a substrate-dependent manner [67,68]. Given that SKP2 and β-TrCP have documented oncogenic activities, we focus on these two F-box proteins as potential therapeutic targets in cancer.
SKP2. SKP2 is the SRS of the SCFSKP2 ubiquitin ligase complex and mediates the degradation of several negative cell cycle regulators including p27, p21, p130, and p57, thus, positively regulating the G1/S transition [67]. Extensive studies have defined the oncogenic role of SKP2 in many human cancer types, including gastric [92], colon [93], and breast [94] cancers. Overexpression of SKP2 is associated with decreased p27 levels, which is an indicator of poor prognosis [66]. Elevated expression of SKP2 was shown to promote the radioresistance of esophageal squamous cell carcinoma, which negatively correlated with the survival of patients undergoing radiotherapy [95]. Likewise, depletion of SKP2 through genetic approaches inhibits the growth of many cancer cell lines [9] and also sensitizes esophageal squamous cell carcinoma to radiation-induced cell death [95]. These findings suggest that pharmacological inhibition of the SKP2 pathway may have therapeutic efficacy in cancer. Consistently, using high-throughput screening, Chen et al. recently identified an agent, compound A, which inhibits SCFSkp2 by preventing incorporation of SKP2 into the SCFSkp2 ligase [96]. Compound A treatment caused accumulation of SCFSKP2 substrates (e.g., p27) and consequently induced G1 cell cycle arrest as well as SCFSkp2- and p27-dependent cell killing [96]. It is conceivable that compound A in combination with radiation may have a therapeutic benefit, although it remains to be determined experimentally.
β-TrCP. β-TrCP, with two family members of β-TrCP1 and β-TrCP2 (also known as HOS), acts as the SRS of the SCFβ-TrCP complex and promotes ubiquitination and degradation of various cellular proteins [66,67]. However, whether β-TrCP is an oncoprotein or a tumor suppressor seems to be substrate-dependent. In some tissues, β-TrCP acts as an oncoprotein for proteasomal degradation of tumor suppressors (e.g., IκB, PDCD4, and BimEL1) [9]. Thus, it is anticipated that, in transgenic mice, overexpression of β-TrCP1 in mammary gland, intestine, liver, and kidney would stimulate tumor formation [97,98]. Consistent with its role in promoting tumorigenesis, β-TrCP1 overexpression was found in human breast cancers and β-TrCP1 inhibition sensitizes breast cancer cells to chemotherapy [99]. Similarly, up-regulation of β-TrCP1 increased NF-κB activity and chemoresistance, whereas β-TrCP1 knockdown decreased NF-κB activity and chemoresistance in pancreatic cancer cells [100] and sensitized cervical cancer cells to apoptosis [101]. Given the important role of NF-κB in mediating tumor radioresistance [102], targeting β-TrCP might represent an effective strategy for radiosensitization. Indeed, inhibition of β-TrCP2 was found to sensitize human melanoma cells to apoptosis induced by various anticancer agents, including ionizing radiation [103]. However, the development of inhibitors that selectively disrupt the binding between β-TrCP and tumor suppressive substrates, but not oncogenic substrates, is likely to be a challenge.
Targeting the CRL and Radiosensitization
Underscoring the importance of CRL E3 ligases as potential therapeutic targets, abnormal activation of CRL E3 ligases has been demonstrated in many types of cancer, resulting in the aberrant turnover of numerous cancer-related proteins [10,66]. Efforts to identify specific small-molecule inhibitors of CRL E3 ligases are well underway and three such inhibitors have recently been reported [96,104,105], although the anticancer properties of these newly identified inhibitors remain to be determined [104,105]. Importantly, the discovery of MLN4924 as a small-molecule inhibitor of NAE, capable of inactivating CRL through blocking cullin neddylation [21], has opened up an alternative strategy for targeting CRL activity. Mechanistically, MLN4924 binds to NAE to form a tight-binding NEDD8-MLN4924 adduct, which resembles the first intermediate of the reaction catalyzed by the NAE, but cannot be further used in subsequent intraenzyme reactions, thus inhibiting the activity of the NEDD8 E1 enzyme [106]. In contrast to bortezomib, MLN4924 appears to be more specific because it does not inhibit bulk proteasomal degradation [21]. In preclinical studies, MLN4924, by inactivating CRL E3 ubiquitin ligase, causes the accumulation of several SCF E3 substrates to induce apoptosis [21,107–110] and senescence [111–113], thus inhibiting growth of a variety of human cancer cells both in vitro and in vivo. Importantly, MLN4924 was found to inhibit cancer cell growth but was well tolerated under various dose levels and treatment regimens in several mouse xenograft models [21,91], suggesting cancer cell selectivity. MLN4924 is currently being evaluated in a number of phase I clinical trials against some solid tumors and hematologic malignancies [114,115]. Most recently released trial results on cancer patients with metastatic melanoma and other solid tumors showed that MLN4924 indeed targets CRL ligases and leads to disease stabilization with mostly grade 1 or 2 adverse effects, including fatigue, diarrhea, nausea, vomiting, and anemia (http://www.takeda.com/press/article_41890.html).
Given that general proteasome inhibition using bortezomib has been demonstrated to sensitize tumor cells to radiation [58,59,116–118], whether MLN4924 can radiosensitize in a tumor cell-selective manner is an important question. Our recent studies showed that knockdown of RBX1/RBX2, which mimics inhibition of CRL activity, induced tumor cell radiosensitization [71,81], thus suggesting that MLN4924 may act as a radiosensitizing agent. We, therefore, tested this hypothesis and found that indeed MLN4924 possesses potent radiosensitizing activity in pancreatic, lung, and breast cancer cells but, importantly, not in normal lung fibroblasts [91,119], demonstrating the tumor cell selectivity of MLN4924-mediated radiosensitization. The radiosensitizing mechanisms of MLN4924 are causally related to the accumulation of a subset of CRL substrates within cells [91,119]. In pancreatic cancer cells, MLN4924 treatment caused the accumulation of several CRL substrates, including CDT1, WEE1, and NOXA, in parallel with an enhancement of radiation-induced DNA damage, aneuploidy, G2/M phase cell cycle arrest, and apoptosis [91]. Knockdown of CDT1 and WEE1 partially rescued MLN4924-induced aneuploidy, G2/M arrest, and radiosensitization, indicating a causal role of CDT1 and WEE1 accumulation in MLN4924-mediated radiosensitization. Similarly, MLN4924 displayed potent radiosensitizing activity in a human pancreatic tumor xenograft model with minimal toxicity [91]. However, the radiosensitization effect of MLN4924 on breast cancer cells appears to mainly depend on the accumulation of p21, a well-known CRL substrate associated with cell growth arrest, apoptosis, and DNA damage response. This is supported by the finding that p21 accumulates in response to combined MLN4924 and radiation treatment and that transient silencing of p21 partially rescues MLN4924-induced G2/M arrest and radiosensitization [119]. Taken together, these findings suggest that the mechanisms of MLN4924-mediated radiosensitization may be dependent on specific tumor cell types.
Conclusions and Perspectives
The data summarized in this review clearly show that blockage of global protein degradation by general proteasome inhibitors (such as bortezomib) or inactivation of CRL E3s by siRNA silencing of CRL components or small-molecule inhibitors (e.g., MLN4924) can achieve radiosensitization in various human cancer cells (Figure 1). Although MLN4924 should be less toxic than bortezomib because of its selective inactivation of one type of E3 ligase rather than general inhibition of proteolysis, for the future development of MLN4924 as an anticancer or radiosensitizing agent, some intrinsic specificity issues are worth considering. First, MLN4924 is an NAE inhibitor and would likely inhibit, in addition to cullins, other cellular neddylation reactions [37,38], although cullins are the only known physiological substrates [36,38]. Second, in addition to causing accumulation of some tumor suppressors (e.g., p21, p27, IκBα, DEPTOR, NOXA, or PDCD4), our unpublished data also showed that MLN4924 could cause accumulation of some oncogenic proteins (e.g., c-Jun, cyclin D1, c-Myc, or Notch1), all of which are known CRL substrates [66,120] in a cell line-dependent manner. Thus, the net outcome of MLN4924 action will depend on the interaction of these substrates in a cell context, temporal, and spatial dependent manner. Third, two recent studies showed that cancer cells could develop resistance to MLN4924 by selecting rare clones with heterozygous mutations in the targeting enzyme NAEβ [121,122]. Nevertheless, given the fact that human cancer harbors multiple mutations with alterations in multiple signaling pathways [123,124], it is unlikely that drugs that target a single mutated gene product/single pathway would be effective. Because MLN4924 targets multiple signaling pathways by inactivating CRL E3s, it would be likely more efficacious as a single anticancer or radiosensitizing agent. Finally, quantitative proteomic analysis at the unbiased global level in a variety of MLN4924-treated cancer cells [125–128], when performed in combination with radiation, would likely identify potential targets as well as biomarkers for radiosensitization. Thus, future mechanistic characterization of MLN4924 or other CRL E3 inhibitors and development of these inhibitors as a novel class of radiosensitizers would eventually benefit cancer patients by enhancing the efficacy of radiotherapy.
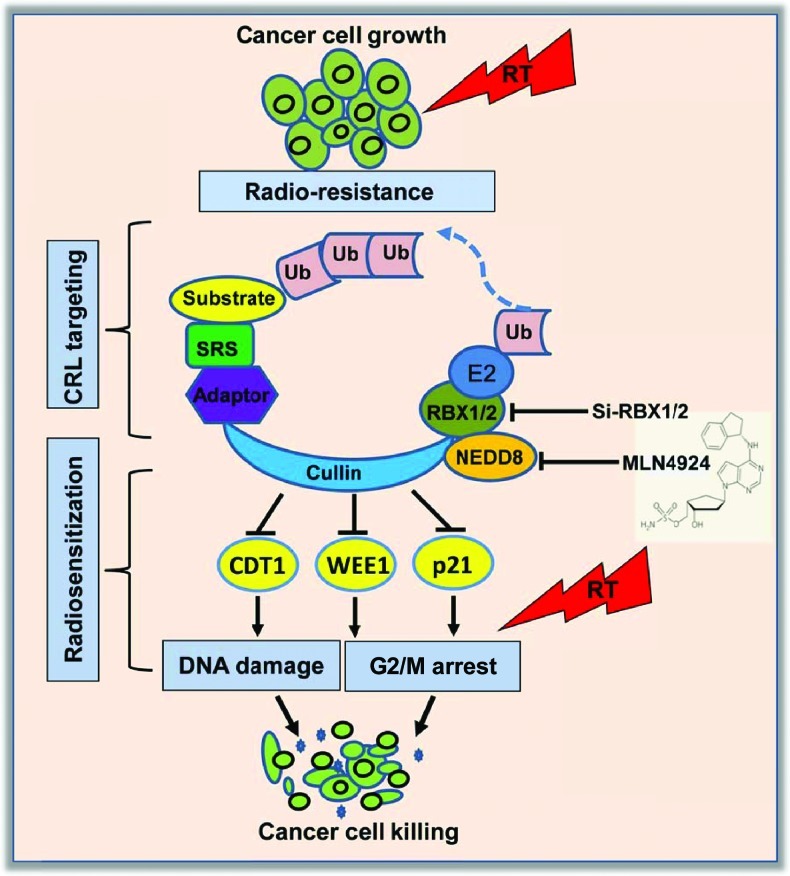
Figure 1.
Cancer cell radiosensitization by inactivation of CRL E3: Inactivation of CRL ligase activity by siRNA silencing of its components (e.g., RBX1/RBX2) or by the small-molecule MLN4924 which inhibits cullin neddylation, causes accumulation of CRL substrates. Accumulation of some of these substrates, such as CDT1, WEE1, and p21, leading to altered DNA damage response and G2/M arrest, was found to be causally related to MLN4924-mediated radiosensitization in a cancer cell type-dependent manner.
References
- 1.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11:239–253. doi: 10.1038/nrc3007. [DOI] [PubMed] [Google Scholar]
- 2.Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer. 2005;104:1129–1137. doi: 10.1002/cncr.21324. [DOI] [PubMed] [Google Scholar]
- 3.Bhide SA, Nutting CM. Recent advances in radiotherapy. BMC Med. 2010;8:25. doi: 10.1186/1741-7015-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Keating SE, Bowie AG. Role of non-degradative ubiquitination in interleukin-1 and toll-like receptor signaling. J Biol Chem. 2009;284:8211–8215. doi: 10.1074/jbc.R800038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edelmann MJ, Nicholson B, Kessler BM. Pharmacological targets in the ubiquitin system offer new ways of treating cancer, neurodegenerative disorders and infectious diseases. Expert Rev Mol Med. 2011;13:e35. doi: 10.1017/S1462399411002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strieter ER, Korasick DA. Unraveling the complexity of ubiquitin signaling. ACS Chem Biol. 2012;7:52–63. doi: 10.1021/cb2004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 9.Jia L, Sun Y. SCF E3 ubiquitin ligases as anticancer targets. Curr Cancer Drug Targets. 2011;11:347–356. doi: 10.2174/156800911794519734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang C, Yi GS. Identification of ubiquitin/ubiquitin-like protein modification from tandem mass spectra with various PTMs. BMC Bioinformatics. 2011;12(suppl 14):S8. doi: 10.1186/1471-2105-12-S14-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 13.Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 14.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 16.Bennett EJ, Harper JW. DNA damage: ubiquitin marks the spot. Nat Struct Mol Biol. 2008;15:20–22. doi: 10.1038/nsmb0108-20. [DOI] [PubMed] [Google Scholar]
- 17.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 18.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 22.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan H, Wang Y, Aviram M, Swaroop M, Loo JA, Bian J, Tian Y, Mueller T, Bisgaier CL, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3:635–650. doi: 10.1089/15230860152542989. [DOI] [PubMed] [Google Scholar]
- 25.Ohta T, Michel JJ, Xiong Y. Association with cullin partners protects ROC proteins from proteasome-dependent degradation. Oncogene. 1999;18:6758–6766. doi: 10.1038/sj.onc.1203115. [DOI] [PubMed] [Google Scholar]
- 26.Swaroop M, Wang Y, Miller P, Duan H, Jatkoe T, Madore S, Sun Y. Yeast homolog of human SAG/ROC2/Rbx2/Hrt2 is essential for cell growth, but not for germination: chip profiling implicates its role in cell cycle regulation. Oncogene. 2000;19:2855–2866. doi: 10.1038/sj.onc.1203635. [DOI] [PubMed] [Google Scholar]
- 27.Wei D, Sun Y. Small RING finger proteins RBX1 and RBX2 of SCF E3 ubiquitin ligases: the role in cancer and as cancer targets. Genes Cancer. 2010;1:700–707. doi: 10.1177/1947601910382776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deshaies RJ. SCF and cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 30.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 31.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 32.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 33.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 34.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 36.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 38.Deshaies RJ, Emberley ED, Saha A. Control of cullin-RING ubiquitin ligase activity by Nedd8. In: Groettrup M, editor. Conjugation and Deconjugation of Ubiquitin Family Modifiers: Subcellular Biochemistry. Vol. 54. NY: Springer; 2010. pp. 41–56. [DOI] [PubMed] [Google Scholar]
- 39.Merlet J, Burger J, Gomes JE, Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci. 2009;66:1924–1938. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 41.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301:392–398. doi: 10.1016/s0006-291x(02)03051-6. [DOI] [PubMed] [Google Scholar]
- 46.Yang X, Zhou J, Sun L, Wei Z, Gao J, Gong W, Xu RM, Rao Z, Liu Y. Structural basis for the function of DCN-1 in protein neddylation. J Biol Chem. 2007;282:24490–24494. doi: 10.1074/jbc.C700038200. [DOI] [PubMed] [Google Scholar]
- 47.Broemer M, Tenev T, Rigbolt KT, Hempel S, Blagoev B, Silke J, Ditzel M, Meier P. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell. 2010;40:810–822. doi: 10.1016/j.molcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 48.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 49.Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11:239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8:508–513. doi: 10.1634/theoncologist.8-6-508. [DOI] [PubMed] [Google Scholar]
- 51.Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- 53.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu Rev Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 54.Shah MA, Power DG, Kindler HL, Holen KD, Kemeny MM, Ilson DH, Tang L, Capanu M, Wright JJ, Kelsen DP. A multicenter, phase II study of bortezomib (PS-341) in patients with unresectable or metastatic gastric and gastroesophageal junction adenocarcinoma. Invest New Drugs. 2011;29:1475–1481. doi: 10.1007/s10637-010-9474-7. [DOI] [PubMed] [Google Scholar]
- 55.Pajonk F, Pajonk K, McBride WH. Apoptosis and radiosensitization of Hodgkin cells by proteasome inhibition. Int J Radiat Oncol Biol Phys. 2000;47:1025–1032. doi: 10.1016/s0360-3016(00)00516-2. [DOI] [PubMed] [Google Scholar]
- 56.Russo SM, Tepper JE, Baldwin AS, Jr, Liu R, Adams J, Elliott P, Cusack JC., Jr Enhancement of radiosensitivity by proteasome inhibition: implications for a role of NF-κB. Int J Radiat Oncol Biol Phys. 2001;50:183–193. doi: 10.1016/s0360-3016(01)01446-8. [DOI] [PubMed] [Google Scholar]
- 57.Motegi A, Murakawa Y, Takeda S. The vital link between the ubiquitin-proteasome pathway and DNA repair: impact on cancer therapy. Cancer Lett. 2009;283:1–9. doi: 10.1016/j.canlet.2008.12.030. [DOI] [PubMed] [Google Scholar]
- 58.Lioni M, Noma K, Snyder A, Klein-Szanto A, Diehl JA, Rustgi AK, Herlyn M, Smalley KS. Bortezomib induces apoptosis in esophageal squamous cell carcinoma cells through activation of the p38 mitogen-activated protein kinase pathway. Mol Cancer Ther. 2008;7:2866–2875. doi: 10.1158/1535-7163.MCT-08-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamer S, Ren Q, Dicker AP. Differential radiation sensitization of human cervical cancer cell lines by the proteasome inhibitor velcade (bortezomib, PS-341) Arch Gynecol Obstet. 2009;279:41–46. doi: 10.1007/s00404-008-0667-7. [DOI] [PubMed] [Google Scholar]
- 60.Huang CY, Wei CC, Chen KC, Chen HJ, Cheng AL, Chen KF. Bortezomib enhances radiation-induced apoptosis in solid tumors by inhibiting CIP2A. Cancer Lett. 2012;317:9–15. doi: 10.1016/j.canlet.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 61.Grimes KR, Daosukho C, Zhao Y, Meigooni A, St Clair W. Proteasome inhibition improves fractionated radiation treatment against non-small cell lung cancer: an antioxidant connection. Int J Oncol. 2005;27:1047–1052. [PubMed] [Google Scholar]
- 62.Pajonk F, van Ophoven A, Weissenberger C, McBride WH. The proteasome inhibitor MG-132 sensitizes PC-3 prostate cancer cells to ionizing radiation by a DNA-PK-independent mechanism. BMC Cancer. 2005;5:76. doi: 10.1186/1471-2407-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vlashi E, Mattes M, Lagadec C, Donna LD, Phillips TM, Nikolay P, McBride WH, Pajonk F. Differential effects of the proteasome inhibitor NPI-0052 against glioma cells. Transl Oncol. 2010;3:50–55. doi: 10.1593/tlo.09244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Neil BH, Raftery L, Calvo BF, Chakravarthy AB, Ivanova A, Myers MO, Kim HJ, Chan E, Wise PE, Caskey LS, et al. A phase I study of bortezomib in combination with standard 5-fluorouracil and external-beam radiation therapy for the treatment of locally advanced or metastatic rectal cancer. Clin Colorectal Cancer. 2010;9:119–125. doi: 10.3816/CCC.2010.n.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edelman MJ, Burrows W, Krasna MJ, Bedor M, Smith R, Suntharalingam M. Phase I trial of carboplatin/paclitaxel/bortezomib and concurrent radiotherapy followed by surgical resection in stage III non-small cell lung cancer. Lung Cancer. 2010;68:84–88. doi: 10.1016/j.lungcan.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 66.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 67.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 69.Huang Y, Duan H, Sun Y. Elevated expression of SAG/ROC2/Rbx2/Hrt2 in human colon carcinomas: SAG does not induce neoplastic transformation, but antisense transfection inhibits tumor cell growth. Mol Carcinog. 2001;30:62–70. [PubMed] [Google Scholar]
- 70.Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69:4974–4982. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun Y. Alterations of SAG mRNA in human cancer cell lines: requirement for the RING finger domain for apoptosis protection. Carcinogenesis. 1999;20:1899–1903. doi: 10.1093/carcin/20.10.1899. [DOI] [PubMed] [Google Scholar]
- 73.Yang ES, Park JW. Regulation of nitric oxide-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Res. 2006;40:279–284. doi: 10.1080/10715760500511500. [DOI] [PubMed] [Google Scholar]
- 74.Kim SY, Kim MY, Mo JS, Park JW, Park HS. SAG protects human neuroblastoma SH-SY5Y cells against 1-methyl-4-phenylpyridinium ion (MPP+)-induced cytotoxicity via the downregulation of ROS generation and JNK signaling. Neurosci Lett. 2007);413:132–136. doi: 10.1016/j.neulet.2006.11.074. [DOI] [PubMed] [Google Scholar]
- 75.Lee SJ, Yang ES, Kim SY, Shin SW, Park JW. Regulation of heat shock-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Biol Med. 2008;45:167–176. doi: 10.1016/j.freeradbiomed.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 76.He H, Gu Q, Zheng M, Normolle D, Sun Y. SAG/ROC2/RBX2 E3 ligase promotes UVB-induced skin hyperplasia, but not skin tumors, by simultaneously targeting c-Jun/AP-1 and p27. Carcinogenesis. 2008;29:858–865. doi: 10.1093/carcin/bgn021. [DOI] [PubMed] [Google Scholar]
- 77.Chanalaris A, Sun Y, Latchman DS, Stephanou A. SAG attenuates apoptotic cell death caused by simulated ischaemia/reoxygenation in rat cardiomyocytes. J Mol Cell Cardiol. 2003;35:257–264. doi: 10.1016/s0022-2828(03)00003-8. [DOI] [PubMed] [Google Scholar]
- 78.Kim DW, Lee SH, Jeong MS, Sohn EJ, Kim MJ, Jeong HJ, An JJ, Jang SH, Won MH, Hwang IK, et al. Transduced Tat-SAG fusion protein protects against oxidative stress and brain ischemic insult. Free Radic Biol Med. 2010;48:969–977. doi: 10.1016/j.freeradbiomed.2010.01.023. [DOI] [PubMed] [Google Scholar]
- 79.Yang GY, Pang L, Ge HL, Tan M, Ye W, Liu XH, Huang FP, Wu DC, Che XM, Song Y, et al. Attenuation of ischemia-induced mouse brain injury by SAG, a redox-inducible antioxidant protein. J Cereb Blood Flow Metab. 2001;21:722–733. doi: 10.1097/00004647-200106000-00010. [DOI] [PubMed] [Google Scholar]
- 80.Wilson GD. Radiation and the cell cycle, revisited. Cancer Metastasis Rev. 2004;23:209–225. doi: 10.1023/B:CANC.0000031762.91306.b4. [DOI] [PubMed] [Google Scholar]
- 81.Jia L, Bickel JS, Wu J, Morgan MA, Li H, Yang J, Yu X, Chan RC, Sun Y. RBX1 (RING box protein 1) E3 ubiquitin ligase is required for genomic integrity by modulating DNA replication licensing proteins. J Biol Chem. 2011;286:3379–3386. doi: 10.1074/jbc.M110.188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Swaroop M, Bian J, Aviram M, Duan H, Bisgaier CL, Loo JA, Sun Y. Expression, purification, and biochemical characterization of SAG, a RING finger redox-sensitive protein. Free Radic Biol Med. 1999;27:193–202. doi: 10.1016/s0891-5849(99)00078-7. [DOI] [PubMed] [Google Scholar]
- 83.Tan M, Zhu Y, Kovacev J, Zhao Y, Pan ZQ, Spitz DR, Sun Y. Disruption of Sag/Rbx2/Roc2 induces radiosensitization by increasing ROS levels and blocking NF-κB activation in mouse embryonic stem cells. Free Radic Biol Med. 2010;49:976–983. doi: 10.1016/j.freeradbiomed.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salon C, Brambilla E, Brambilla C, Lantuejoul S, Gazzeri S, Eymin B. Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: relationships with CAND1 and cyclin E protein levels. J Pathol. 2007;213:303–310. doi: 10.1002/path.2223. [DOI] [PubMed] [Google Scholar]
- 85.Chen LC, Manjeshwar S, Lu Y, Moore D, Ljung BM, Kuo WL, Dairkee SH, Wernick M, Collins C, Smith HS. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–3683. [PubMed] [Google Scholar]
- 86.Melchor L, Saucedo-Cuevas LP, Munoz-Repeto I, Rodriguez-Pinilla SM, Honrado E, Campoverde A, Palacios J, Nathanson KL, Garcia MJ, Benitez J. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 2009;11:R86. doi: 10.1186/bcr2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schindl M, Gnant M, Schoppmann SF, Horvat R, Birner P. Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res. 2007;27:949–952. [PubMed] [Google Scholar]
- 88.Yasui K, Arii S, Zhao C, Imoto I, Ueda M, Nagai H, Emi M, Inazawa J. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology. 2002;35:1476–1484. doi: 10.1053/jhep.2002.33683. [DOI] [PubMed] [Google Scholar]
- 89.Hung MS, Mao JH, Xu Z, Yang CT, Yu JS, Harvard C, Lin YC, Bravo DT, Jablons DM, You L. Cul4A is an oncogene in malignant pleural mesothelioma. J Cell Mol Med. 2011;15:350–358. doi: 10.1111/j.1582-4934.2009.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gupta A, Yang LX, Chen L. Study of the G2/M cell cycle checkpoint in irradiated mammary epithelial cells overexpressing Cul-4A gene. Int J Radiat Oncol Biol Phys. 2002;52:822–830. doi: 10.1016/s0360-3016(01)02739-0. [DOI] [PubMed] [Google Scholar]
- 91.Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, Sun Y. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 2012;72:282–293. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Masuda TA, Inoue H, Sonoda H, Mine S, Yoshikawa Y, Nakayama K, Mori M. Clinical and biological significance of S-phase kinase-associated protein 2 (Skp2) gene expression in gastric carcinoma: modulation of malignant phenotype by Skp2 overexpression, possibly via p27 proteolysis. Cancer Res. 2002;62:3819–3825. [PubMed] [Google Scholar]
- 93.Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer. 2005;103:1336–1346. doi: 10.1002/cncr.20917. [DOI] [PubMed] [Google Scholar]
- 94.Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, Monti F, Loda M, Pagano M. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J Clin Invest. 2002;110:633–641. doi: 10.1172/JCI15795. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Wang XC, Tian LL, Tian J, Jiang XY. Overexpression of SKP2 promotes the radiation resistance of esophageal squamous cell carcinoma. Radiat Res. 2012;177:52–58. doi: 10.1667/rr2679.1. [DOI] [PubMed] [Google Scholar]
- 96.Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kudo Y, Guardavaccaro D, Santamaria PG, Koyama-Nasu R, Latres E, Bronson R, Yamasaki L, Pagano M. Role of F-box protein betaTrcp1 in mammary gland development and tumorigenesis. Mol Cell Biol. 2004;24:8184–8194. doi: 10.1128/MCB.24.18.8184-8194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Belaidouni N, Peuchmaur M, Perret C, Florentin A, Benarous R, Besnard-Guerin C. Overexpression of human β TrCP1 deleted of its F box induces tumorigenesis in transgenic mice. Oncogene. 2005;24:2271–2276. doi: 10.1038/sj.onc.1208418. [DOI] [PubMed] [Google Scholar]
- 99.Tang W, Li Y, Yu D, Thomas-Tikhonenko A, Spiegelman VS, Fuchs SY. Targeting β-transducin repeat-containing protein E3 ubiquitin ligase augments the effects of antitumor drugs on breast cancer cells. Cancer Res. 2005;65:1904–1908. doi: 10.1158/0008-5472.CAN-04-2597. [DOI] [PubMed] [Google Scholar]
- 100.Muerkoster S, Arlt A, Sipos B, Witt M, Grossmann M, Kloppel G, Kalthoff H, Folsch UR, Schafer H. Increased expression of the E3-ubiquitin ligase receptor subunit βTRCP1 relates to constitutive nuclear factor-κB activation and chemoresistance in pancreatic carcinoma cells. Cancer Res. 2005;65:1316–1324. doi: 10.1158/0008-5472.CAN-04-1626. [DOI] [PubMed] [Google Scholar]
- 101.Tan M, Gallegos JR, Gu Q, Huang Y, Li J, Jin Y, Lu H, Sun Y. SAG/ROC-SCFβ-TrCP E3 ubiquitin ligase promotes pro-caspase-3 degradation as a mechanism of apoptosis protection. Neoplasia. 2006;8:1042–1054. doi: 10.1593/neo.06568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deorukhkar A, Krishnan S. Targeting inflammatory pathways for tumor radiosensitization. Biochem Pharmacol. 2010;80:1904–1914. doi: 10.1016/j.bcp.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Soldatenkov VA, Dritschilo A, Ronai Z, Fuchs SY. Inhibition of homologue of Slimb (HOS) function sensitizes human melanoma cells for apoptosis. Cancer Res. 1999;59:5085–5088. [PubMed] [Google Scholar]
- 104.Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, et al. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 107.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-κB-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 108.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M, Nawrocki ST, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 109.Tan M, Li Y, Yang R, Xi N, Sun Y. Inactivation of SAG E3 ubiquitin ligase blocks embryonic stem cell differentiation and sensitizes leukemia cells to retinoid acid. PLoS One. 2011;6:e27726. doi: 10.1371/journal.pone.0027726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 2011;71:3042–3051. doi: 10.1158/0008-5472.CAN-10-2122. [DOI] [PubMed] [Google Scholar]
- 111.Jia L, Li H, Sun Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia. 2011;13:561–569. doi: 10.1593/neo.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70:10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 conjugation pathway and its relevance in cancer biology and therapy. Genes Cancer. 2010;1:708–716. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15:3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 116.Goktas S, Baran Y, Ural AU, Yazici S, Aydur E, Basal S, Avcu F, Pekel A, Dirican B, Beyzadeoglu M. Proteasome inhibitor bortezomib increases radiation sensitivity in androgen independent human prostate cancer cells. Urology. 2010;75:793–798. doi: 10.1016/j.urology.2009.07.1215. [DOI] [PubMed] [Google Scholar]
- 117.Fu DX, Tanhehco Y, Chen J, Foss CA, Fox JJ, Chong JM, Hobbs RF, Fukayama M, Sgouros G, Kowalski J, et al. Bortezomib-induced enzyme-targeted radiation therapy in herpesvirus-associated tumors. Nat Med. 2008;14:1118–1122. doi: 10.1038/nm.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kubicek GJ, Werner-Wasik M, Machtay M, Mallon G, Myers T, Ramirez M, Andrews D, Curran WJ, Jr, Dicker AP. Phase I trial using proteasome inhibitor bortezomib and concurrent temozolomide and radiotherapy for central nervous system malignancies. Int J Radiat Oncol Biol Phys. 2009;74:433–439. doi: 10.1016/j.ijrobp.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang D, Tan M, Wang G, Sun Y. The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PLoS One. 2012;7:e34079. doi: 10.1371/journal.pone.0034079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Skaar JR, D'Angiolella V, Pagan JK, Pagano M. SnapShot: F box proteins II. Cell. 2009;137:1358.e1–1358.e2. doi: 10.1016/j.cell.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 121.Milhollen MA, Thomas MP, Narayanan U, Traore T, Riceberg J, Amidon BS, Bence NF, Bolen JB, Brownell J, Dick LR, et al. Treatment-emergent mutations in NAEβ confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer Cell. 2012;21:388–401. doi: 10.1016/j.ccr.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 122.Toth JI, Yang L, Dahl R, Petroski MD. A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell Rep. 2012;1:309–316. doi: 10.1016/j.celrep.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis M, Olson JJ, Mikkelsen T, Lehman N, Aldape K, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liao H, Liu XJ, Blank JL, Bouck DC, Bernard H, Garcia K, Lightcap ES. Quantitative proteomic analysis of cellular protein modulation upon inhibition of the NEDD8-activating enzyme by MLN4924. Mol Cell Proteomics. 2011;10:M111.009183. doi: 10.1074/mcp.M111.009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee JE, Sweredoski MJ, Graham RL, Kolawa NJ, Smith GT, Hess S, Deshaies RJ. The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol Cell Proteomics. 2011;10:M110.006460. doi: 10.1074/mcp.M110.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jung J, Kim EJ, Chung HK, Park HJ, Jeong SY, Choi EK. c-Myc down-regulation is involved in proteasome inhibitor-mediated enhancement of radiotherapeutic efficacy in non-small cell lung cancer. Int J Oncol. 2012;40:385–390. doi: 10.3892/ijo.2011.1205. [DOI] [PubMed] [Google Scholar]
- 130.Warren G, Grimes K, Xu Y, Kudrimoti M, St Clair W. Selectively enhanced radiation sensitivity in prostate cancer cells associated with proteasome inhibition. Oncol Rep. 2006;15:1287–1291. [PubMed] [Google Scholar]
- 131.Munshi A, Kurland JF, Nishikawa T, Chiao PJ, Andreeff M, Meyn RE. Inhibition of constitutively activated nuclear factor-κB radiosensitizes human melanoma cells. Mol Cancer Ther. 2004;3:985–992. [PubMed] [Google Scholar]