Abstract
Receptor tyrosine kinases (RTK) and their ligands control critical biologic processes, such as cell proliferation, migration, and differentiation. Aberrant expression of these receptor kinases in tumor cells alters multiple downstream signaling cascades that ultimately drive the malignant phenotype by enhancing tumor cell proliferation, invasion, metastasis, and angiogenesis. As observed in human glioblastoma (hGBM) and other cancers, this dysregulation of RTK networks correlates with poor patient survival. Epidermal growth factor receptor (EGFR) and c-Met, two well-known receptor kinases, are coexpressed in multiple cancers including hGBM, corroborating that their downstream signaling pathways enhance a malignant phenotype. The integration of c-Met and EGFR signaling in cancer cells indicates that treatment regimens designed to target both receptor pathways simultaneously could prove effective, though resistance to tyrosine kinase inhibitors continues to be a substantial obstacle. In the present study, we analyzed the antitumor efficacy of EGFR inhibitors erlotinib and gefitinib and c-Met inhibitor PHA-665752, along with their respective small hairpin RNAs (shRNAs) alone or in combination with human umbilical cord blood stem cells (hUCBSCs), in glioma cell lines and in animal xenograft models. We also measured the effect of dual inhibition of EGFR/c-Met pathways on invasion and wound healing. Combination treatments of hUCBSC with tyrosine kinase inhibitors significantly inhibited invasion and wound healing in U251 and 5310 cell lines, thereby indicating the role of hUCBSC in inhibition of RTK-driven cell behavior. Further, the EGFR and c-Met localization in glioma cells and hGBM clinical specimens indicated that a possible cross talk exists between EGFR and c-Met signaling pathway.
Introduction
The aggressive nature and dismal prognosis of glioblastoma multiforme (GBM) highlights the need for novel therapeutic options. Recent advances in the molecular characterization of GBM have exposed new potential mechanisms for targeted therapeutic agents. Studies on patients with human GBM (hGBM) detailed molecular alterations in the epidermal growth factor receptor (EGFR), indicating its crucial role in the development and progression of glioblastoma [1,2]. Because EGFR and other receptor tyrosine kinases (RTKs) and their ligands control critically important biologic processes, such as cell proliferation, migration, and differentiation, aberrant expression of these receptor kinases by the tumor cells or by nonmalignant tumor-infiltrating cells alters multiple downstream signaling cascades [3]. These changes may ultimately drive the malignant phenotype by enhancing tumor cell proliferation, invasion, metastasis, and angiogenesis [4–6]. In addition to EGFR, c-Met expression is also correlated with tumor grade in different cancers, where it is known to have a similarly prominent role in cellular proliferation, motility, invasion, angiogenesis, and survival [7–10]. Overexpression of c-Met presages a poor prognosis and correlates with malignant grade in glial neoplasms [11]. EGFR and c-Met are often coexpressed in several malignancies, such as astrocytoma, lung, head and neck, breast, and colon cancers, and the convergent downstream signaling pathways of both kinases enhance a malignant phenotype [12–17]. Glioblastoma cells treated with hepatocyte growth factor (HGF) demonstrated increased tumorigenicity, whereas blockade of c-Met in vivo inhibited tumor formation, thereby implying its pivotal role in tumor formation [18].
Despite their intricacy, it is thought that cell surface receptors EGFR and c-Met elicit similar signal transduction pathways; therefore, their cross talk could affect the strength and duration of shared subsequent signaling pathways [19]. Furthermore, immunoprecipitation experiments conducted on SUM229 cells demonstrated physical and functional interactions between EGFR and c-Met [20]. The relatively high frequency of crossover between these tumor-promoting signaling pathways makes it worthwhile to study the clinical efficacy of their respective inhibitors. The tyrosine kinase inhibitors (TKIs) gefitinib, erlotinib, and PHA-665752 targeted to EGFR/c-Met have shown promising results in patients with glioblastoma [21–25]. Unfortunately, only a subpopulation of these patients responds clinically to the inhibitors, even though most patients with hGBM express EGFR/c-Met in their tumors [26]. Considering the apparent dominant role of EGFR in GBMs, targeted therapies that inhibit the functions of EGFR or c-Met may also have strong antitumor activity. However, since oncogenic tyrosine kinases orchestrate highly complex signaling pathways, the key drug-induced changes conferring sensitivity could be difficult to identify. Although agents against specific targets have shown modest activity in several clinical trials, there is a need to develop more effective strategies involving combined EGFR/c-Met-targeted therapy owing to the synergistic antitumor effects of combining EGFR and c-Met pathway inhibition.
Due to their tumor targeting properties, human umbilical cord blood stem cells (hUCBSCs) present a new therapeutic strategy. In our previous reports, we have shown that hUCBSCs have the capacity to induce apoptosis, regulate cell cycle progression, and inhibit in vivo tumor growth [27,33]. In the present study, we examined the antitumor efficacy of hUCBSC alone or in combination with well-known EGFR/c-Met inhibitors, such as shRNA to EGFR, erlotinib, gefitinib, and PHA-665752, in U251 and 5310 cells as well as in cells obtained from hGBM patient specimens. To further delineate the antitumor effects of combined targeting of EGFR and c-Met using hUCBSCs and inhibitors, we examined the effect of dual inhibition of both pathways on invasion and wound healing. We further determined the efficacy of combined treatment in decreasing the expression of important downstream signaling molecules intrinsic to both EGFR and HGF/c-Met pathways.
Materials and Methods
Antibodies and Reagents
We used the following antibodies: rabbit anti-EGFR, goat antipEGFR, mouse anti-EGFR, rabbit anti-c-Met, rabbit anti-p-c-Met, rabbit anti-Stat3, mouse anti-β-catenin, mouse anti-AKT, rabbit anti-p-AKT, goat anti-phosphoinositide 3-kinase (PI3K), goat anti-p-PI3K, mouse anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:200 dilution), and c-Met small-interfering RNA (siRNA) (sc-29397) were purchased from Santa Cruz Biotechnology Inc (Santa Cruz, CA). The c-Met inhibitor, PHA-665752, was obtained from Sigma-Aldrich (St Louis, MO). Erlotinib hydrochloride and gefitinib hydrochloride were obtained from Santa Cruz Biotechnology Inc. AG1478 was obtained from Calbiochem (San Diego, CA).
hUCBSC Culture
hUCBSCs were collected from healthy donors with informed consent according to the protocol approved by the University of Illinois College of Medicine at Peoria Institutional Review Board. hUCBSC were isolated using Ficoll-Paque (GE Healthcare, Piscataway, NJ) density gradient centrifugation. The isolated cells were plated in 100-mm plates in Dulbecco modified Eagle's medium (DMEM) knockout medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS, 10% knockout serum (Hyclone, Logan,UT), and 1% penicillin/streptomycin. When the adherent cells reached 20% to 30% confluence, they were supplemented with MesenCult medium (Stem Cell Technologies, Vancouver, Canada) containing human mesenchymal stem cell stimulatory supplements (Stem Cell Technologies) and 1% penicillin/streptomycin (Invitrogen). For coculture experiments, hUCBSC and glioma cells were cultured at a ratio of 1:1. Cocultures of hUCBSC and U251 were grown in DMEM; cocultures of hUCBSC and 5310 were grown in RPMI-1640 medium.
Glioma Cell Culture
U251 cells obtained from the National Cancer Institute (Frederick, MD) were grown in DMEM supplemented with 10% FBS (Hyclone) and 1% penicillin/streptomycin (Invitrogen). Xenograft cell line 5310 (a kind gift from Dr D. James of the University of California, San Francisco) was grown in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C. To visualize and identify two different populations of cells in the hUCBSC coculture studies with glioma cell lines U251 and 5310, we used standard flow cytometry technique as described previously [27].
Human Glioma Xenograft Cell Line
JKR-14 xenograft cells were isolated from a biopsy specimen from a 54-year-old male patient with glioblastoma. Xenografts were maintained in the flanks of athymic homozygous NU/J mice (Strain No. 2019 from Jackson Laboratory, Bar Harbor, ME), and early passages in RPMI-1640 medium were used to minimize genetic drift.
Construction of shRNA-Expressing Plasmid and Transfection of shEGFR
The plasmid vector, pSilencer 4.1-CMV (Ambion, Austin, TX), was used in the construction of the shRNA-expressing vector, and the human EGFR sequence AAGACTGCTAAGGCATAGGAA was the shRNA target sequence. Inverted repeat sequences were synthesized for EGFR. The inverted repeats were laterally symmetrical, making them self-complimentary with a 9-bp mismatch in the loop region that would aid in the loop formation of the shRNA. Oligonucleotides were heated in a boiling water bath in 6x SSC for 5 minutes and self-annealed by slow cooling to room temperature. The annealed oligonucleotides were ligated to pSilencer vector at BamHI and HindIII sites. Cells at 60% to 70% confluence in 100-mm tissue culture plates were transfected with 7 µg of shRNA-expressing plasmid constructs [shRNA specific for EGFR (shEGFR)] using FuGene HD following the manufacturer's instructions (Roche, Indianapolis, IN).
Single and Combination Treatments
U251 and 5310 cells were treated with either 5 µM erlotinib or gefitinib hydrochloride for 9 hours, 10 µM AG1478 for 1 hour, 0.5 µM PHA-665752 for 9 hours, or with 10 ng of recombinant EGF (rEGF; Millipore, Billerica, MA) for 1 hour. In all of the combination treatments, cells were pretreated with shEGFR, rEGF, or PHA-665752 for their respective time points and then were cocultured with hUCBSC (1:1 ratio) for 72 hours.
Western Blot Analysis
Cells from various treatment groups were collected and lysed in RIPA buffer [50 mmol/ml Tris-HCl (pH 8.0), 150 mmol/ml NaCl, 1% IGEPAL (octylphenoxypolyethoxyethanol), 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS)] containing 1 mM sodium orthovanadate, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 10 µg/ml aprotinin, and 10 µg/ml leupeptin, and the lysates were then resolved using SDS-polyacrylamide gel electrophoresis (PAGE). After transfer onto nitrocellulose membranes, blots were blocked with 5% nonfat dry milk in phosphate-buffered saline (PBS) and 0.1% Tween-20. Blots were incubated with respective primary antibodies, followed by incubation with a horse radish peroxidase (HRP)-conjugated secondary antibody. Immunoreactive bands were visualized using chemiluminescence enhanced chemilumescent (ECL) Western blot detection reagents on Hyperfilm MP autoradiography film (Amersham, Piscataway, NJ). GAPDH antibody was used to verify equal loading of proteins in all lanes.
Wound Healing Assay
For wound healing migration assay, a straight scratch was made manually in subconfluent cell monolayers using a sterile 200-µl pipette tip in individual wells, and cells were allowed to migrate into the cell-free area. This point was considered the “0 hour,” and the width of the wound was immediately photographed under the light microscope. The cells were observed for ∼12 to 20 hours depending on the cell line [28].
Matrigel Invasion Assay
Matrigel invasion assay was used to assess cell invasive potential as described previously [29]. Cell culture medium supplemented with 10% FBS was added to the lower chamber to act as a chemoattractant. U251 and 5310 control cells, alone or in coculture with hUCBSC, were seeded at a density of 2 x 105 cells/well onto the upper inserts and then incubated at 37°C. After 24 hours, the noninvasive cells were removed from the upper surface of the separating membrane by gentle scrubbing with a cotton swab, and the invading cells were fixed in 100% methanol and stained with 2-hydroxyethyl methacrylate (Hema)-3. Similar experimental procedures were used when we investigated the inhibitory effects of EGFR shRNA-, PHA-665752-, and rEGF-mediated invasion.
Immunoprecipitation Analysis
Protein lysates were prepared from 3 to 5 mm3 pieces of frozen intracranial tumors. Approximately 200 to 400 µg of protein cell lysates were incubated at 4°C with 50 µl of Protein G/A beads (Miltenyi Biotec, Auburn, CA), followed by sequential additions of 10 µl (2 µg) of EGFR and c-Met antibodies with end-to-end rotation overnight. The immunoprecipitates were then loaded onto “µ” columns (Miltenyi Biotec) and rinsed with lysis buffer according to the manufacturer's instructions. The columns were washed twice with 200 µl of lysis buffer. Preheated (95°C) 1x SDS gel loading buffer was loaded onto the column matrix using a fresh pipette tip and incubated at room temperature for 5 minutes. After discharging the supernatant, 50 µlof 1x SDS gel loading buffer was added to the immunoprecipitates, and the supernatants were then collected and loaded into 8% to 12% SDS-PAGE followed by electrophoretic transfer to nitrocellulose membranes for further analysis.
Immunocytochemistry
Cells were grown in two-well chamber slides, washed with PBS, fixed, permeabilized with ice-cold methanol, and blocked with 10% goat serum for 1 hour. Cells were incubated with primary antibodies for either 2 hours at room temperature or overnight at 4°C, washed with PBS, and incubated with fluorescent-labeled, species-specific secondary antibodies (Alexa Fluor) for 1 hour at room temperature. Before mounting, the slides were washed with PBS, stained with 4′,6-diamidino-2-phenylindole (DAPI) for nuclear staining, and analyzed under a microscope (Olympus BX61 Fluoview, Minneapolis, MN).
Immunohistochemical Staining
For detection of EGFR and c-Met, 5-µmsections of formalin-fixed, paraffin-embedded tissues of U251 and 5310 controls or hUCBSC-treated or hGBM tissues were deparaffinized and subjected to antigen retrieval for 10 minutes at 90°C in 0.1 M sodium citrate buffer (pH 6.0). Next, sections were washed in PBS and blocked in 10% goat serum for 30 minutes. A 1:100 dilution of mouse anti-EGFR and rabbit anti-c-Met antibodies in 10% goat serum was added to sections, which were incubated overnight at 4°C. After washing in PBS, sections were incubated with HRP-conjugated goat anti-mouse and goat anti-rabbit immunoglobulin G (IgG), respectively. These sections were washed in PBS and developed with the DAB substrate (Sigma-Aldrich) to produce color. The specificity of EGFR and c-Met staining was confirmed by a lack of staining without primary antibody or an isotype-matched irrelevant antibody. After counterstaining with hematoxylin, sections were mounted, cleared, coverslipped, and examined using a confocal microscope. For immunofluorescence, sections were treated with primary antibodies overnight at 4°C and then treated with appropriate Alexa Fluor secondary antibodies at room temperature for 1 hour. Negative controls were maintained either without primary antibody or using respective IgG. All immunostained sections, which were stained with fluorescent antibodies, were counterstained with DAPI. The sections were blind reviewed by a neuropathologist.
Cell Cycle Analysis Using Flow Cytometry
Progression of glioblastoma through different cell cycle phases in varying concentrations of erlotinib and gefitinib was monitored by flow cytometric analysis. DNA content was analyzed by staining with propidium iodide and carried out with an FACS (FACSCalibur flow cytometer; Becton Dickinson, San Jose, CA). The percentage of cells within G1,S,G2, and M phases was determined using CellQuest software. Approximately 10,000 events were counted for each analysis, and three to four independent experiments performed in triplicate were conducted for each group (n = 3).
Intracranial Administration of Glioma Cells and hUCBSC in Nude Mice
The Institutional Animal Care and Use Committee of the University of Illinois College of Medicine at Peoria approved all surgical interventions and postoperative animal care. Glioma cells were injected intracerebrally into the right side of the brains of nude mice with 10-µl aliquots of U251 and 5310 (1 x 106) cells under isofluorane anesthesia with the aid of a stereotactic frame. hUCBSCs were injected left of the sagittal suture and bregma of mouse brain after a week of tumor implantation. The ratio of the hUCBSC to cancer cells was maintained at 1:4. Three weeks after tumor inoculation, six mice from each group were sacrificed by intracardiac perfusion, first with PBS and then with 4% formaldehyde in PBS. The removed brains were stored in 4% para-formaldehyde, processed, embedded in paraffin, and sectioned (5 µm thick) using a microtome. Paraffin-embedded sections were stained with hematoxylin and eosin to visualize tumor cells.
Statistical Analysis
Quantitative data from cell counts, FACS analysis, Western blot analysis, and other assays were evaluated for statistical significance using one-way analysis of variance (ANOVA). Data for each treatment group were represented as means ± SEM and compared with other groups for significance by one-way analysis of variance followed by Bonferroni post hoc test (multiple comparison tests) using GraphPad Prism version 3.02, a statistical software package. Results were considered statistically significant at P <.05.
Results
EGFR Knockdown Suppresses c-Met Signaling
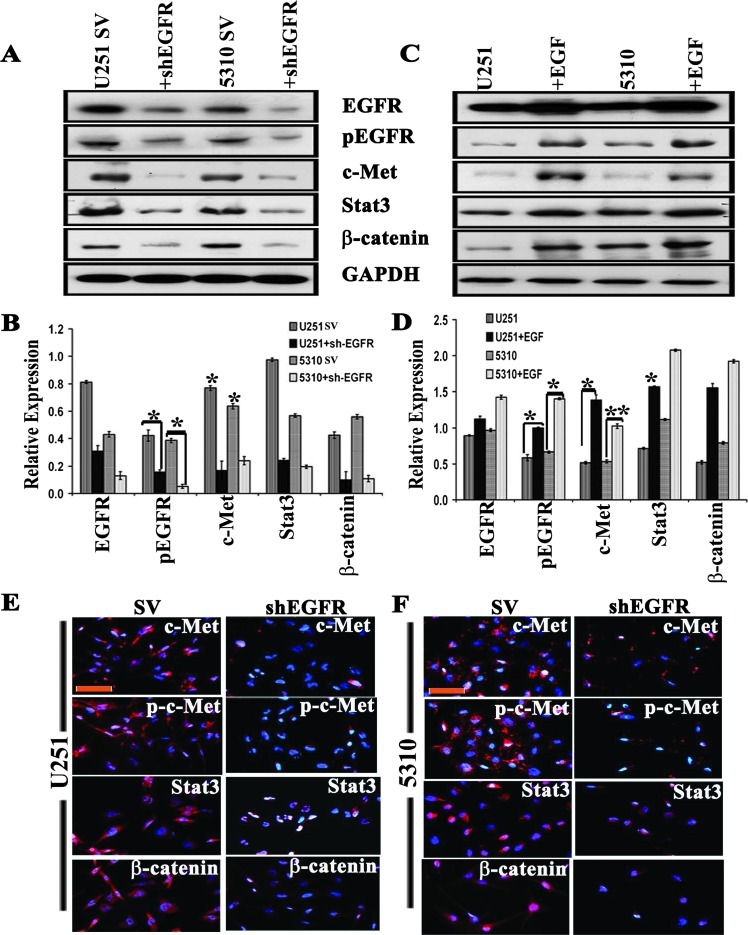
To determine the effect of EGFR expression on c-Met and c-Met-mediated cellular signaling networks, we transfected U251 and 5310 glioblastoma cells with shEGFR and performed Western blot analysis. The Western blot results revealed that the total EGFR was reduced to 28% in U251 and to 38% in 5310 control cells, whereas the phosphorylated EGFR (pEGFR) levels were reduced to 38% and 49% in U251 and 5310 cells, respectively. We also observed that c-Met expression levels correlated with EGFR levels, with 34% and 38% reductions in U251 and 5310 cells. Overexpression and activity of Stat3 and β-catenin have been linked to the invasion and metastasis of several cancers in humans [30,31] and their reliance on EGFR/c-Met signaling [32]. Further, the relative expression levels of EGFR/c-Met downstream signaling molecules such as Stat3 and β-catenin in shEGFR-transfected samples were attenuated to 23% and 16% in U251 cells and 21% and 19% in 5310 cells, respectively (Figure 1, A and B). Furthermore, the immunocytochemical analysis of shEGFR-transfected U251 and 5310 cells showed reduced expression levels of c-Met, p-c-Met, Stat3, and β-catenin (Figure 1, E and F). To confirm that the decreased expression of EGFR/c-Met signaling molecules is because of the EGFR knockdown, we serum starved the two cell lines for 3 hours and then substituted them with serum-starved medium containing rEGF (10 ng/ml) for 1 hour with subsequent immunoblot analysis. We observed that EGFR expression was enhanced by 25% in U251 cells and by 27% in 5310 cells, whereas the pEGFR levels were found to be increased by 69% and 71% in EGF-treated U251 and 5310 cells, respectively. Interestingly, the c-Met levels were increased to 59% and 68% in U251 and 5310 cells, respectively, and showed increase in the expression of downstream signaling molecules Stat3 and β-catenin (Figure 1, C and D). These results indicate that EGFR-mediated downstream signaling represents an integrated signaling cascade resulting from the coactivation of c-Met receptors.
Figure 1.
EGFR knockdown by specific shRNA reduces c-Met expression. (A) Down-regulation of EGFR by an EGFR-specific shRNA results in loss of c-Met phosphorylation in both U251 and 5310 cells when transfected for 72 hours. Corresponding whole-cell lysates were immunoblotted with anti-Met, anti-Stat3, and β-catenin antibodies. GAPDH served as a loading control (SV, scrambled vector). (B) Quantitation of A. (C) U251 and 5310 cells were serum starved for 3 hours and then treated with 10 ng/ml rEGF for 1 hour. Total and phosphorylated EGFR, c-Met, Stat3, β-catenin, and β-actin levels were detected by Western blot analysis. The results shown are representative of three independent experiments. (D) Quantitation of C. The results presented in this study are the representative images of three independent experiments (n = 3) and are expressed as means ± SE; *P < .05; **P < .01. (E, F) Immunofluorescence staining using anti-Met, p-c-Met, anti-Stat3, and β-catenin antibodies followed by an Alexa Fluor 594-tagged secondary antibody was performed in shEGFR-transfected U251 and 5310 cells compared to the SV controls. Panels were photographed at 20x. Red indicates the expression levels of the respective proteins, whereas DAPI is used to stain the nucleus. Scale bars, 200 µm.
EGFR Inhibitors Annihilate c-Met Signaling Molecules
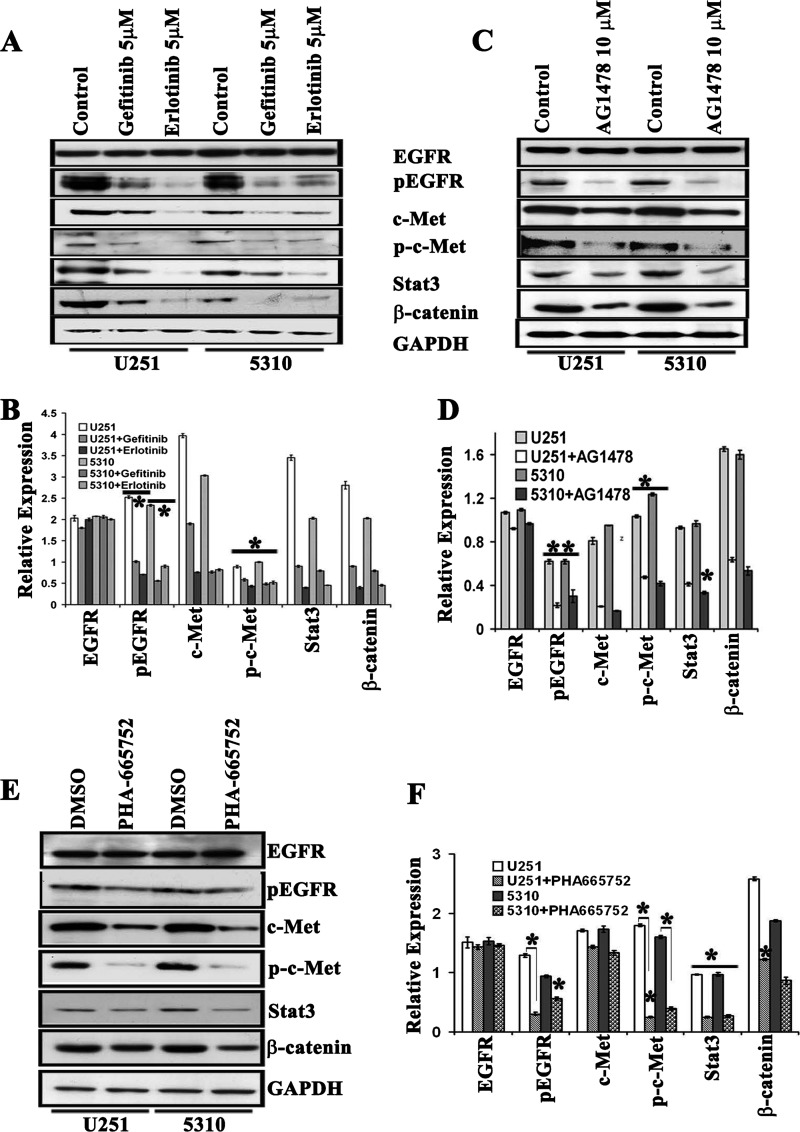
To analyze the sensitivity of U251 and 5310 cells to EGFR target-specific drugs, we studied the tolerance of two different EGFR kinase inhibitors, erlotinib and gefitinib. Initially, the GBM cells were treated with increasing concentrations (1, 5, and 10 µM) of erlotinib and gefitinib and were compared with untreated cells at different time points (3, 6, 9, and 24 hours). Both erlotinib and gefitinib showed time-dependent and dose-dependent inhibition of EGFR with maximal inhibition at 5 µM for 9 hours of treatment. Significant cell death was observed when the cells were incubated more than 24 hours. Following erlotinib treatment, pEGFR levels were observed to be reduced even at the lower concentrations, although no statistically significant effects on the cell proliferation were observed (Figure W1, A–C). Furthermore, at 5 µM concentration of erlotinib, the expression levels of c-Met were reduced along with its phosphorylated form, p-c-Met. Cosuppression of key ancillary molecules like Stat3 and β-catenin was also confirmed by Western blot analysis (Figure 2, A and B). Studies with 5 µM concentration of gefitinib demonstrated similar, but not identical, results. The expression levels of pEGFR, c-Met, p-c-Met, and β-catenin displayed a decrement compared to what was observed with erlotinib in U251 cells, whereas β-catenin expression was comparable in both erlotinib and gefitinib treatments (Figure 2, A and B). To examine if other EGFR inhibitors can also inhibit c-Met downstream signaling molecules, we treated the U251 and 5310 GBM cells with 10 µM AG1478. The levels of EGFR (total and phosphorylated forms) along with c-Met, Stat3, and β-catenin proteins were decreased in U251 and 5310 glioblastoma cell lines as determined by Western blot analysis (Figure 2, C and D). Similarly, to demonstrate that EGFR activation was a direct consequence of c-Met receptor, we treated U251 and 5310 cells with PHA-665752, a specific c-Met kinase inhibitor. Western blot analysis revealed a suppression of EGFR phosphorylation levels accompanied by a concomitant decrease in the phosphorylation status of c-Met and reduced Stat3 and β-catenin expression levels (Figure 2, E and F).
Figure 2.
RTK inhibitors, erlotinib, gefitinib, AG1478, and PHA-665752, reduce EGFR/c-Met cross activation in U251 and 5310 glioblastoma cells. (A) U251 and 5310 cells were grown in serum-starved medium containing 5 µM erlotinib and gefitinib for 9 hours. Cell lysates were evaluated for the expression levels of total and phosphorylated EGFR, c-Met, Stat3, and β-catenin. (B) Quantitative estimation of A. (C) To study the effect of EGFR phosphorylation of c-Met expression, we treated U251 and 5310 cells with 10 µM AG1478 for 1 hour in serum-starved medium. Expression levels of various molecules were subjected to Western blot analysis. (D) Quantitative estimation of C. (E) The phosphorylation of EGFR, c-Met, Stat3, and β-catenin is substantially reduced by 0.5 µM PHA-665752 in U251 and 5310 cells. Cells treated only with DMSO were treated as controls. Cells were lysed, and the indicated proteins were detected by immunoblot analysis with indicated antibodies. (F) Bar graph quantification values of E. Values are the means ± SE of three independent experiments; *P < .05; **P < .01. All the data represented in this panel are representative of two individual separate experiments. In all the experiments, the Western blots were normalized using GAPDH.
hUCBSC Treatment Suppresses EGFR/c-Met-Mediated Signaling
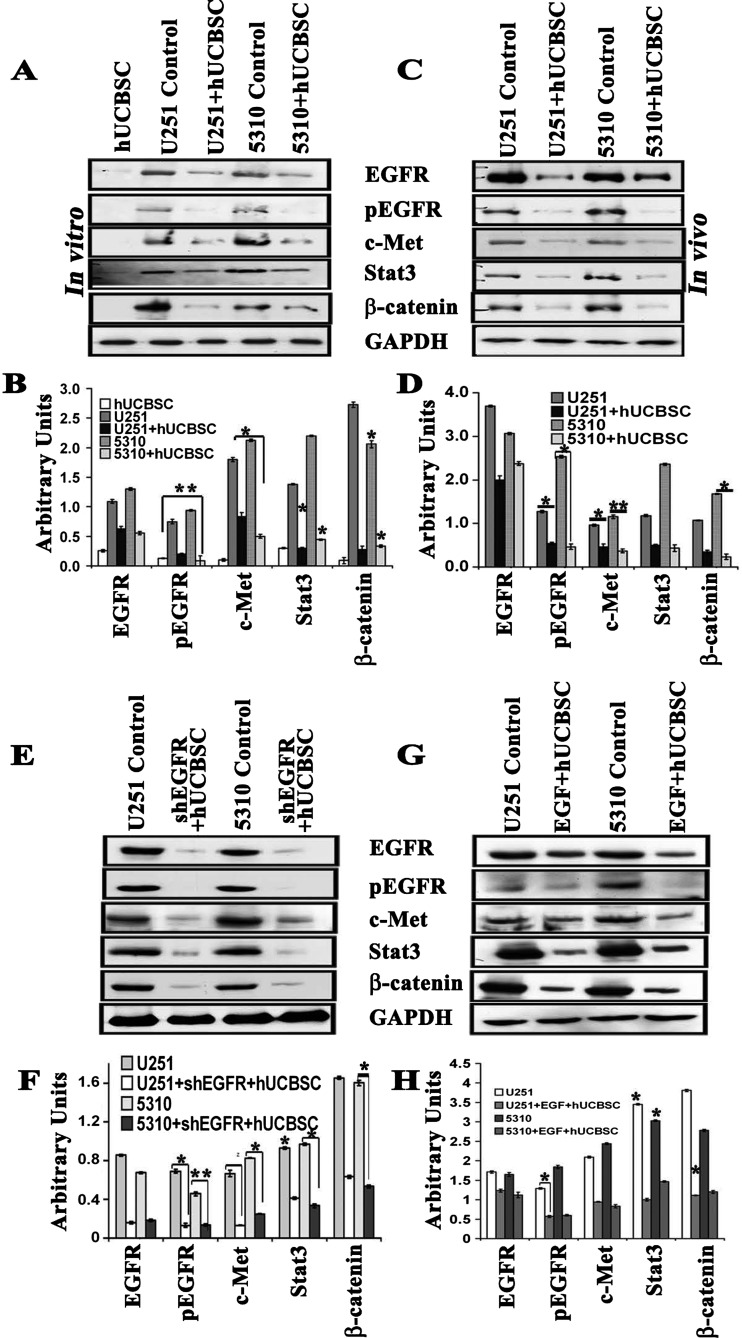
Oncogenic tyrosine receptor kinases sit at the top of multiple signaling pathways that are constitutively activated upon increased phosphorylation of these RTKs. Coculture treatment of U251 and 5310 cells with hUCBSC for 72 hours has demonstrated reduced expression of total and phosphorylated forms of EGFR. Apart from EGFR, c-Met, Stat3, and β-catenin levels were also reduced by more than 50% in both GBM cell lines (Figure 3, A and B). Though the protein expression levels varied in both the cell lines tested, the end effect was similar and comparable to the immunoblot analysis of mouse xenograft samples (Figure 3, C and D). To further validate the specificity of the treatments, we cocultured shEGFR-treated cells with hUCBSC. The expression levels of EGFR/c-Met signaling molecules were significantly reduced when compared to the small molecule inhibitor treatments alone or hUCBSC-alone treatments (Figure 3, E and F). In separate experiments, glioma cells were subjected to serum starvation for 3 hours and then treated with rEGF (100 ng/ml) for 1 hour. The cells that were given EGF treatment were cocultured with hUCBSC in 1:1 ratio for 72 hours. Western blot analysis performed on these lysates demonstrated that addition of rEGF increased the EGFR/c-Met signaling molecules, and further, this increase was suppressed noticeably with treatments (Figure 3, G and H), thereby indicating the hUCBSC potential role in down-regulation of EGF-induced EGFR/c-Met signaling pathway in glioma cells.
Figure 3.
hUCBSC controls the expression levels of EGFR and c-Met. We performed immunoblot analysis to study the expression of EGFR and its downstream signaling molecules in the (A) cell lysates (in vitro) and (C) tissue lysates of brains extracted from nude mice (in vivo). Orthotopic intracranial tumors were established in nude mice by injecting glioma cells (U251 and 5310) and then treating with hUCBSC. Equal amounts of proteins (40 µg) from untreated (control) and hUCBSC-treated cell lysates and tissue lysates were loaded onto 8% to 14% gels and transferred onto nitrocellulose membranes, which were then probed with respective antibodies. GAPDH was used a positive loading control. (B and D) The relative band intensities were measured by densitometry and normalized against the respective GAPDH signals. (E) U251 and 5310 cells were transfected with shEGFR and then allowed to coculture with hUCBSC for 72 hours at a 1:1 ratio. (G) U251 and 5310 cells were treated with rEGF and then subjected to coculture. In both experiments, cell lysates were analyzed for the expression levels of EGFR, pEGFR, c-Met, Stat3, and β-catenin in various treatment groups. (F and H) Quantification of E and G. Values are the means ± SE of three independent experiments, each with three samples per experimental treatment; *P < .05; **P < .01.
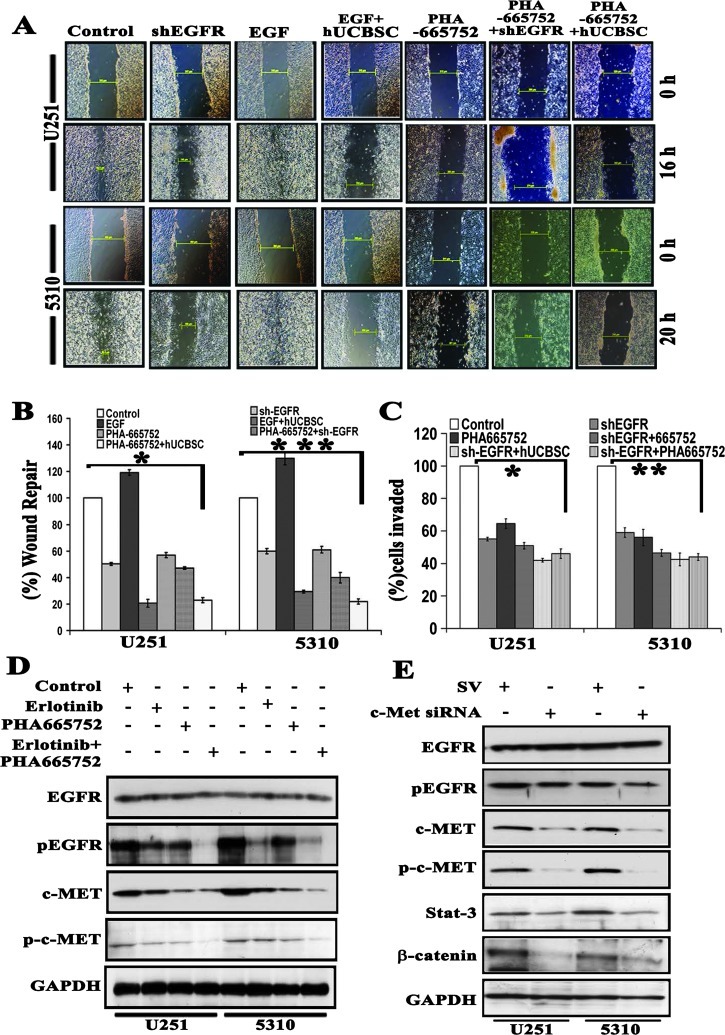
Combination of shEGFR and PHA-665752 with hUCBSC Coculture Treatments Showed Enhanced Inhibition of GBM Cell Invasion and Wound Healing
The overexpression of EGFR with other cell surface receptors like c-Met has been reported independently to affect the efficacy and offers resistance to targeted therapies [17]. On the basis of these findings, we hypothesized that combined targeting of both EGFR and c-Met pathways with hUCBSC treatment might increase antitumor efficacy in U251 and 5310 cells. Further, we used the clinically relevant c-Met kinase inhibitor PHA-665752 in combination with shEGFR and hUCBSC coculture treatments to study the wound healing ability of U251 and 5310 cells. We have previously shown that hUCBSC inhibits the wound closure in glioma cells [33]. In the present study, we observed that wound healing was inhibited by 50% and 57% with shEGFR, 80% and 71% with rEGF (10 ng/ml) + hUCBSC, 57% and 61% with PHA-665752, 67% and 74% with PHA-665752 + shEGFR, and 82% and 81% with PHA-665752 + hUCBSC treatments in U251 and 5310 cells, respectively. Wound closure in untreated U251 and 5310 cells was measured between 16 and 24 hours, whereas in EGF-treated cells wound closure was observed between 12 and 16 hours (Figure 4A). Statistically significant differences in all the treatments compared to the control cells are displayed as a bar graph (Figure 4B). We next investigated the role of EGFR and c-Met inhibitors on GBM cell invasion. We performed the Matrigel invasion assay using U251 and 5310 glioma cells treated with shEGFR, PHA-665752, and EGF alone or in combination with hUCBSC. In both of the cell lines tested, the shEGFR and PHA-665752 treatments significantly reduced the number of invaded cells versus controls, and this reduction was further decreased in combination treatments (Figures 4C and W2A). Further to substantiate our findings that combination treatments were more effective on glioma cells, we treated U251 and 5310 cells with erlotinib, PHA-665752, and PHA-665752 in combination with erlotinib. We found that erlotinib treatment reduces pEGFR expression effectively when compared to the total and phosphorylated forms of c-Met. PHA-665752, the c-Met inhibitor, effectively decreases the expression of c-Met and phosphorylated forms, compared to the EGFR expression. Interestingly, U251 and 5310 cells treated with EGFR and c-Met inhibitors demonstrated additive reduction in the EGFR and c-Met expression (Figure 4D). In another experiment, we attempted to examine the c-Met silencing using siRNA specific to c-Met. Western blot analysis revealed that c-Met siRNA showed a small reduction in the expression of pEGFR levels, whereas Stat3 and β-catenin levels were reduced to maximum, pointing to the fact that c-Met operate through the downstream signaling molecules, Stat3 and β-catenin (Figure 4E). Together, these data suggest that both EGFR and c-Met pathways play an important role in the glioma cell migration and invasion; regulating these pathways by means of hUCBSC treatment may lead to a greater reduction in invasive and migratory capabilities, which is of utmost importance to prevent the infiltration of healthy nontumor brain tissue by glioblastoma cells.
Figure 4.
Combined treatment with shEGFR and PHA-665752 maximally inhibits wound healing and invasion comparable to hUCBSC coculture treatments. (A) Wounds were generated in confluent U251 and 5310 cells in six-well plates using a sterile 200-µl pipette tip. Cells were then exposed to different treatments: shEGFR, 10 ng/ml EGF, 10 ng/ml EGF-treated cells cocultured with hUCBSC, 0.5 µM PHA-665752, 0.5 µM PHA-665752-treated cells cocultured with hUCBSC, and 0.5 µM PHA-665752 cells transfected with hUCBSC. Wound closure was assessed under a microscope at 10x magnification. (B) Cell migration distance was measured for each wound and compared with baseline measurements and was plotted as a bar graph. Values are the means ± SE of three independent experiments; *P < .05; **P < .01; ***P < .001. (C) Serum-starved cells using the above conditions were plated in Biocoat Matrigel Transwell chambers. Cells were allowed to invade for 24 hours, fixed, stained, and counted at 10x magnification. Values are the means ± SE of three independent experiments; *P < .05; **P < .01. (D) U251 and 5310 cells were grown in serum-starved medium containing 5 µM erlotinib, 0.5 µM PHA-665752, and 5 µM erlotinib + 0.5 µM PHA-665752 for 9 hours. Cell lysates were evaluated for the expression levels of total and phosphorylated forms of both EGFR and c-Met. (E) Both U251 and 5310 cells were transfected for c-Met-specific shRNA for 72 hours. Corresponding whole-cell lysates were immunoblotted for total and phosphorylated forms of EGFR and c-Met, Stat3, and β-catenin antibodies with respective antibodies. GAPDH served as a loading control.
hUCBSC Inhibits Physical Association of EGFR and c-Met in Glioblastoma Cells
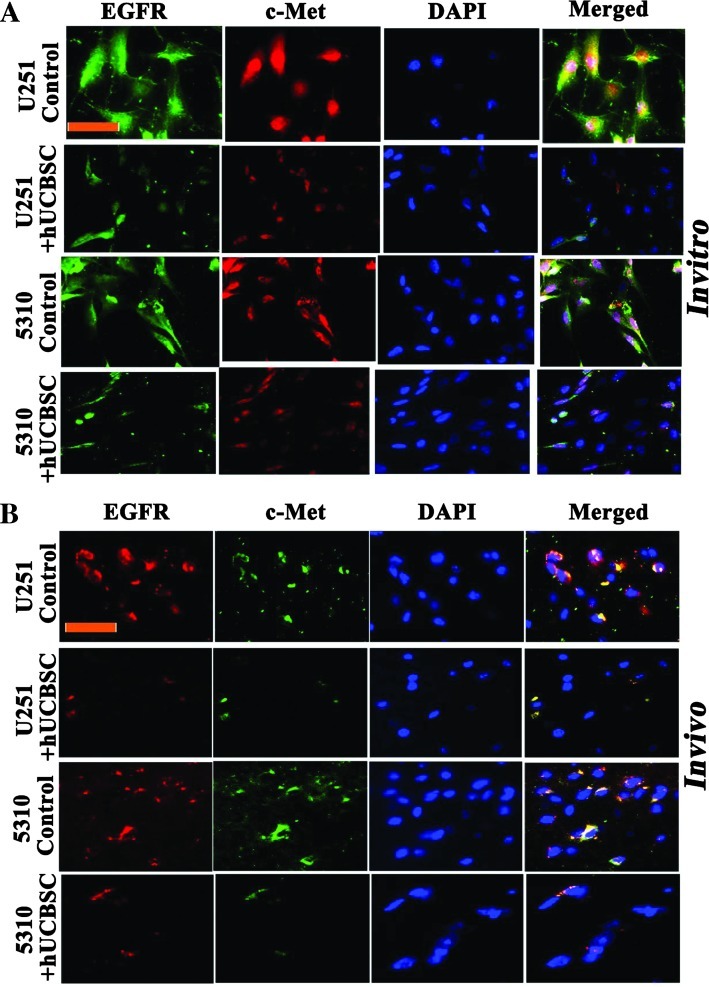
The expression of EGFR and c-Met in U251 and 5310 cells was examined by immunocytochemical and immunohistochemical analyses. We observed that the EGFR and c-Met overexpressed in U251 and 5310 cells and were entirely colocalized; EGFR immunoreactivity was superimposed with that of c-Met in U251 and 5310 glioma cells. In contrast, in hUCBSC cocultured cells, EGFR and c-Met expressions were reduced to a minimum and revealed minimal colocalization (Figure 5A). To corroborate in vitro immunocytochemical results, we processed U251 and 5310 tissue sections using the same experimental protocols. Figure 5B shows the fluorescence micrographs of immunostained U251 and 5310 control tissue sections, with EGFR strongly overlapping with c-Met, demonstrating their strong association in glioma cells.
Figure 5.
hUCBSC coculture reduces EGFR/c-Met localization. (A) We carried out immunocytochemistry of U251 and 5310 cells either alone or in coculture with hUCBSC to study the colocalization of EGFR/c-Met. EGFR is conjugated with Alexa Fluor 594 (red), and c-Met is conjugated with Alexa Fluor 488 (green). Scale bar, 100 µm. (B) Similarly, dual immunohistochemical staining for colocalization in U251 and 5310 controls and hUCBSC-treated mouse xenografts was conducted with anti-EGFR and anti-c-Met antibodies followed by the secondary antibodies conjugated with fluorophores for red and green fluorescence, respectively. Representative merged images show the cells expressing EGFR/c-Met. Scale bar, 100 µm. Each experiment was performed in triplicate with each sample (n = 3).
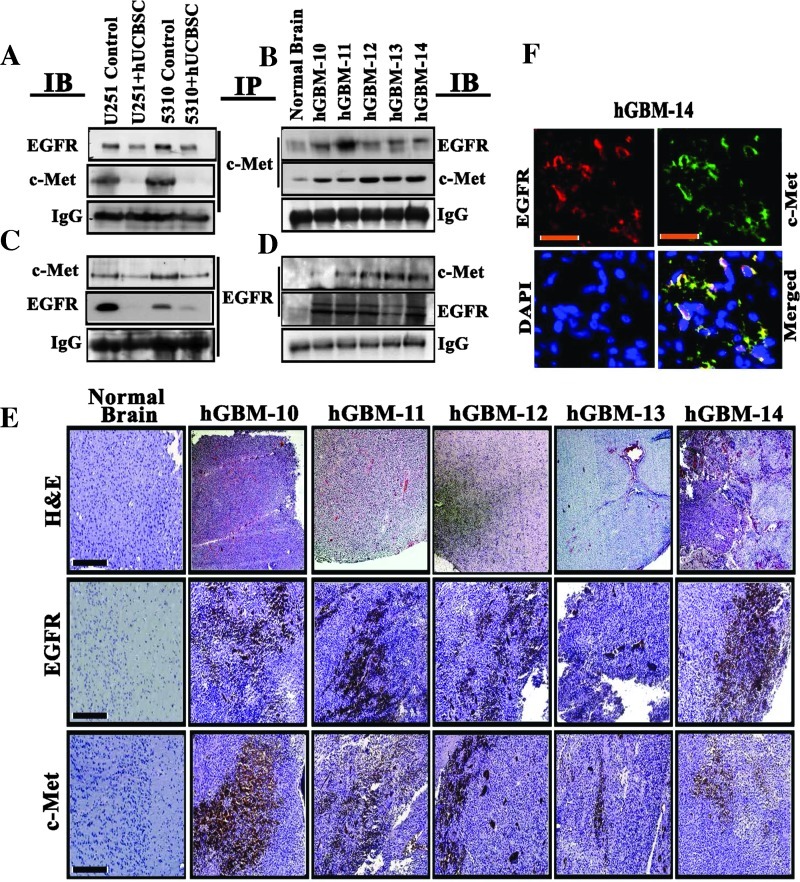
EGFR Coprecipitates with c-Met in Mouse Xenografts and hGBM Patient Tissue Specimens
EGFR may be phosphorylated by itself or by a different receptor family member through heterodimerization. Alternatively, constitutive phosphorylation of c-Met by addition of exogenous EGF in A431 cells suggested the heterodimeric association between the two EGFR and c-Met receptors [8]. To elucidate this cross talk, we immunoprecipitated tissue lysates from U251 and 5310 controls along with the hUCBSC treatments or tissue lysates obtained from hGBM patient specimens (hGBM10-hGBM14) using anti-EGFR antibodies and immunoblotted against anti-EGFR and c-Met antibodies. We observed coimmunoprecipitation of these proteins in control U251 and 5310 and hGBM lysates, whereas hUCBSC-treated U251 and 5310 cells showed reduced interaction of EGFR and c-Met proteins (Figure 6, A and B). The reverse pull-down assay using a c-Met antibody also confirmed their association (Figure 6, C and D). EGFR and c-Met did not coprecipitate in normal human brain samples. The glioma control tissue lysates and hGBM tissue lysates demonstrated high levels of EGFR and c-Met expression. Further, in a separate experiment, Western blot analysis of these patient-derived samples demonstrated increased expression levels of total and phosphorylated forms of EGFR and c-Met, along with their downstream signaling molecules, Stat3 and β-catenin (Figure W2B). Earlier, Lee et al. [34] demonstrated that tumors derived from patients with hGBM mirror genotypic and phenotypic characteristics of standard glioma cell lines. In the present study, we observed that EGFR and c-Met were coexpressed in five hGBM patient-derived specimens. Furthermore, the high EGFR and c-Met expression levels were also confirmed in the patient specimens when compared to normal brain specimens using immunohistochemistry (Figure 6E). Following this confirmation, we tested their association in hGBM14 specimen. Figure 6F demonstrates that EGFR is associated with c-Met, thus implying the existence of EGFR/c-Met mechanism in the hGBM subjects.
Figure 6.
EGFR and c-Met interact and colocalize in hGBM patient specimens. Eight hundred micrograms of total soluble protein from (A) U251 and (B) 5310 control and hUCBSC-treated tissue lysates or from (C) hGBM patient specimens was immunoprecipitated with EGFR, and the samples were immunoblotted against c-Met. The same blots were stripped and reprobed with anti-EGFR antibody. In a reverse experiment, above tissue lysates were immunoprecipitated with c-Met and were immunoblotted against EGFR. The same blots were stripped and reprobed with anti-c-Met antibody. (E) Representative immunohistochemical staining of EGFR and c-Met in human GBM patient specimens and normal brain. Scale bars, 200 µm. Strong positivity of EGFR and c-Met was detected in all the five specimens (hGBM10, hGBM11, hGBM12, hGBM13, and hGBM14) tested. Negative staining was seen in normal human brain samples. (F) Representative micrographs of colocalization studies of EGFR with c-Met in the hGBM14 sample. Red indicates EGFR, whereas green indicates c-Met. Scale bars, 200 µm.
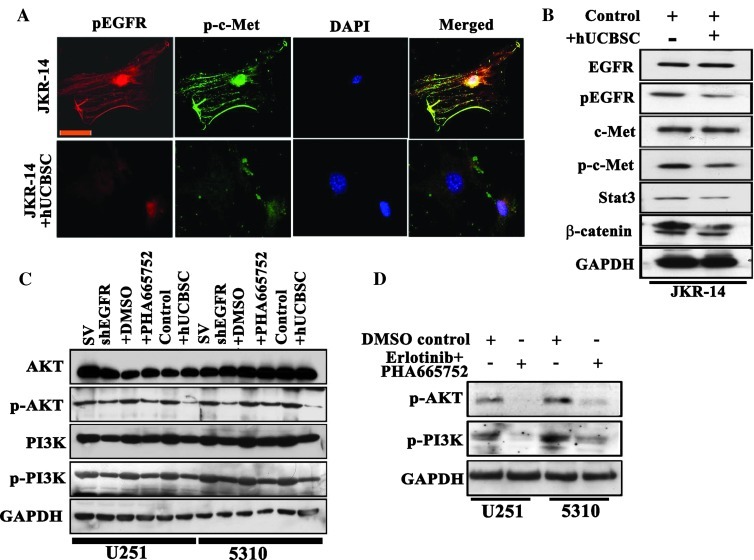
hUCBSC Treatment Regulates EGFR/c-Met Expression in Primary Glioma JKR-14 Cells; EGFR and c-Met Signal through Common PI3K/Akt Axis
To prove that our laboratory coculture studies on established GBM lines U251 and 5310 conform to preclinical standards, we isolated cells (JKR-14) from a 54-year-old male patient with GBM. Using immunocytochemical analysis, untreated JKR-14 cells demonstrated increased expression of both EGFR and c-MET, whereas the hUCBSC coculture treatment reduced the expression of both EGFR and c-Met and further suppressed their association (Figure 7A). Moreover, the decreased expression of total and phosphorylated forms of EGFR and c-Met signaling molecules in JKR-14 as examined by Western blot analysis substantiates our immunocytochemical results (Figure 7B). Bowers et al. [35] demonstrated that HGF/c-Met activates the PI3K/Akt pathway in U-373 human malignant glioma cells. PI3K activates a number of cell signaling pathways that include Akt activation through phosphorylation at Thr308 and Ser473 [36]. Activated PI3K/Akt pathway provides potent survival signals in GBM [37]. Activated PI3K/Akt elicits a downstream cascade that activates the transcription of several promoting factors essentially to hGBM survival, growth, and motility [38]. Although our results point to a relationship between EGFR and c-Met signaling, the relevance of these findings would be ideal if we could identify EGFR/c-Met synergism with a pertinent PI3K/Akt pathway. Total and phosphorylated forms of both Akt and PI3K levels were observed to be decreased in shEGFR, PHA-665752, and hUCBSC treatments (Figure 7C). Combination treatments of EGFR and c-Met inhibitors, erlotinib and PHA-665752, reduced the expression of the phosphorylated forms of both Akt and PI3K when compared to their individual treatments (Figure 7D). Our studies demonstrate the additive potential therapeutic role of hUCBSC in downregulating EGFR/c-Met pathway through the suppression of PI3K/Akt axis in glioblastoma cell lines.
Figure 7.
hUCBSC treatment reduces pEGFR/p-c-Met interactions in JKR-14 cells. (A) Colocalization analysis of pEGFR and p-c-Met. After hUCBSC coculture, both control and hUCBSC cells were fixed, permeabilized, and incubated with pEGFR and p-c-Met antibodies. Cells were further incubated with Alexa Fluor 594 (red) and Alexa Fluor 488 (green) at room temperature for 1 hour. Digital images were obtained by confocal microscopy. (B) Effect of total and phosphorylated forms of EGFR, c-Met, Stat3, and β-catenin in control and hUCBSC-treated JKR-14 cells. Cell lysates were prepared as described in Materials and Methods. (C) EGFR and MET function through the PI3K/Akt axis. The expression pattern of total and phosphorylated forms of Akt and PI3K was examined using standard immunoblot analysis of the whole-cell lysates of U251 and 5310 cells obtained under the following conditions: shEGFR transfections, c-Met inhibitor PHA-665752 treatments, and hUCBSC cocultures. (D) Combination treatments of erlotinib and PHA-665752 on p-AKT and p-PI3K expression. GAPDH served as a loading control.
Discussion
Dysregulation of tyrosine-kinase receptors is one plausible mechanism by which malignant cancer cells confer resistance to novel anticancer therapies. In this study, we evaluated the possible interactions of two known receptor kinases, EGFR and c-Met, in glioma cells. The most common hallmark of GBM biology is aberrant EGF/EGFR signaling, a potent driver of glioma proliferation [39,40]. Aberrant EGFR signaling in glioblastoma increases secreted HGF/scatter factor binding to Met tyrosine kinase receptors, displaying their increased association in malignancy and invasion [26]. Apart from EGFR dysregulation in human lung, gastric carcinomas, and rhabdomyosarcomas, the c-Met pathway is also an absolute requirement for their progression [15,41]. Earlier Engelman et al. [18] in lung cancer demonstrated the possible involvement of c-Met-related signal transduction in offering resistance to EGFR-targeting agents. Even with combinatorial anti-EGFR-targeting and anti-c-Met-targeting agents, acquired resistance was observed during the cancer treatment illustrating the need to identify other molecular therapeutic agents in EGFR-related and c-Met-related cellular events [24]. In light of these facts, we focused our present work on developing strategies to block the c-Met pathway in combination with known anti-EGFR-targeting agents like erlotinib, gefitinib, shEGFR, or c-Met inhibitor PHA-665752 and consider future perspectives of this rationale in cancer therapeutics. In this study, we have evaluated the relevance of EGFR-mediated and c-Met-mediated signaling in glioma cell proliferation and survival. In addition, we also analyzed whether EGFR and c-Met are functionally dependent on each other for controlling such responses. Our results show that both EGFR and c-Met demonstrate positive cross talk in glioma cells to promote their proliferation, invasion, and migration. Our results also indicate that EGFR-mediated downstream signaling represents an integrated signaling cascade resulting from the coactivation of c-Met receptors in glioblastoma.
For testing EGFR-targeted anticancer therapy, the selective EGFR-TKIs gefitinib and erlotinib were administered to a cohort of patients with lung cancer with a proven survival rate. These TKI drugs exert their mode of action by competing with ATP for binding to the receptor kinase pocket of the receptor, thereby blocking the respective receptor activation. In patients with hGBM, clinical trials conducted using these small-molecule EGFR kinase inhibitors demonstrated significant recurrent tumor regression by about 15% to 20% [42]. Although the EGFR gene is the most commonly amplified in patients with glioblastoma, only a small subgroup seems to benefit from the EGFR kinase inhibitors, not correlating with tolerance to EGFR kinase inhibitors [43]. This again illustrates a complex interplay of cell surface signaling and downstream signaling pathways. In our studies using EGFR kinase inhibitors, we observed that c-Met in particular was found to be inhibited along with its phosphorylated form. Other key signaling molecules like Stat3 and β-catenin were also observed to be reduced in their expression levels. Similarly, using c-Met inhibitor, we observed reduced activation of EGFR. These results suggest possible cross talk between EGFR and c-Met and its essential role in various biologic activities of glioma cells.
Despite rapid advances in EGFR oncological therapeutics over the past decade, substantial room for progress still remains. Most patients with cancer do not respond to EGFR inhibitor therapy, which implies intrinsic resistance. It is observed that even in patients who demonstrate clear tumor response to EGFR inhibitors will eventually show disease progression, implying acquired resistance [44]. It is well known that oncogenic tyrosine kinases initiate multiple signaling pathways that can constitutively activate a large downstream network upon their phosphorylation. Recently, it is observed that the c-Met is activated in the setting of EGFR resistance or blockade. Various researchers have suggested that although c-Met and EGFR may not interact directly, c-Met activation provides greater resistance to EGFR inhibitors. Moreover, silencing of EGFR is compensated by c-Met signaling. We focused our work to address the hypothesis that activation of c-Met contributes to glioblastomal tumorigenesis and to resistance to EGFR inhibition where c-Met targeting strategies might have therapeutic benefit alone or in combination with EGFR inhibitors. In uveal melanoma, it is reported that EGFR and c-Met knockdown reduces proliferative effect, whereas elevated EGFR and c-Met enhances to their greatest migration potential. It is also reported that the pathogenesis of uveal melanoma is categorized by levels of c-Met and EGFR expression, which are associated with migratory/invasiveness responses to soluble factors present at high levels in the liver providing biologic relevance for its clinical behavior with potential therapeutic implications [45]. In another report, Martinez-Palacian et al. [46] investigated the significance of the EGFR signaling and potential functional cross talk between the c-Met and the EGFR pathways in oval cell proliferation and survival and found that EGFR induced proliferation and survival equally in Met (flx/flx) and Met (-/-), suggesting that EGFR signaling does not rely on c-Met activity and EGFR functions independent of c-Met. In contrast, Liu et al. demonstrated that activated c-Met positively regulates the EGFR activity and this activation is reversed upon the inhibition of c-Met in SNU-5, SNU-1, U-87MG, 786-O, A549, H441, H596, H1437, H1993, BT474, A549, and HT-29 cell lines, suggesting the pleotrophic effects of c-Met on multiple signaling pathways. Recently, in non-small cell lung cancer, it is observed that c-Met activation is associated with EGFR resistance to inhibition, suggesting a possible interaction [47]. In the squamous cell carcinoma, it is reported that combining inhibitors for EGFR and c-Met lead to the greatest decrease in cell proliferation and invasion, compared to the single inhibitor treatments [48]. In another study, delayed c-Met activation in non-small lung cell lung carcinoma was initiated by wild-type EGFR. EGFR phosphorylates c-Met and this occurs without the supplementation of growth factors, suggesting the nonligand activation and interdependence of EGFR and c-Met pathways [49].
As an alternative to direct TKI inhibition, we tested if the hUCBSC coculture treatment decreases the constitutive activation of EGFR/c-Met pathway and the resultant effect on the downstream signaling pathway. Coculture treatments of hUCBSC with glioma cells demonstrated reduced expression levels of total and phosphorylated forms of EGFR and pEGFR. In addition, the attenuation in EGFR, c-Met, Stat3, and β-catenin levels was also observed to be reduced by more than 50%. These results were confirmed using shEGFR-treated cells, which were cocultured with hUCBSC. The expression levels of the aforementioned proteins were reduced and are comparable to the TKI treatments. We also proved that hUCBSC were efficient against exogenously supplied EGF treatments of glioma cells [50].
RTK coactivation is an alternative mechanism by which GBM mediates chemoresistance to EGFR inhibitor monotherapy [22]. Although in vitro studies suggest that inhibition of individual RTKs is insufficient to eliminate downstream oncogenic pathways, chemical inhibition or genetic depletion of multiple RTKs (such as c-Met and PDGF receptor) in combination with EGFRvIII inhibition led to enhanced GBM cell death [22,23,51]. These studies have provided insight into the contribution of RTK coactivation to GBM chemoresistance, but the mechanisms underlying this process and the clinical relevance of integrated signaling networks that result from the activation of multiple RTKs remain poorly understood [22]. In our studies with hUCBSC alone or in combination with other inhibitors, we have shown that hUCBSCs are capable of inhibiting the coactivation of RTKs in glioma cells, thereby inhibiting their migratory and invasive capacities. Targeting both EGFR and c-Met resulted in significantly more inhibition of invasion, wound healing, and downstream signaling. Finally, targeting these two signaling pathways in glioma cells isolated from an hGBM patient specimen and treating them with hUCBSC demonstrated antitumor effects, providing pre-clinical support that targeting EGFR/c-Met axis using hUCBSC might be a promising therapeutic strategy for treating glioblastoma.
On the basis of our previous reports, we have continuously observed that hUCBSC reduced the expression of several molecules implicated in glioblastoma and that exposure of hUCBSC reduces glioma cell migration and invasion. However, the actual mechanism underlying this reduction in glioblastomal malignancy when treated with hUCBSC needs to be studied. With the development of hUCBSC-based targeted therapies, it will be possible to study the oncogenic mechanisms of glioblastoma. Studies on cells isolated from hGBM patient biopsies and their treatment with hUCBSC will allow us to create patient-tailored combination therapies. It will be critical to screen these treatments for toxicity and adverse effects, given their potential to be therapeutic agents. A favorable strategy ostensibly might be a combinatorial therapy. In the future, as we accumulate more human biopsies and treat them with hUCBSC at laboratory levels, the ability to completely shut off deregulated pathways in cancer cells will hopefully lead to less drug resistance and increased patient survival. Our present study concludes the high interdependency between the expression of activation of both EGFR and c-Met receptors contributing to glioblastoma malignancy. Knockdown or kinase inhibition of EGFR suppresses c-Met expression and signaling, whereas combination of EGFR and c-Met silencing leads to greater antitumor effects. Finally, we conclude that coculture of GBM cells and xenografts with hUCBSC inhibits EGFR and c-Met expression, signaling, and association, and their combination treatment with EGFR and c-Met inhibitors showed enhanced additive antitumor effect.
Supplementary Material
Acknowledgments
We thank Noorjehan Ali for her technical assistance, Alicia Woodworth for manuscript preparation, and Diana Meister and Sushma Jasti for manuscript review.
Footnotes
This research is supported by a grant from the Illinois Neurological Institute (A.J.T.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no conflict of interest exists with this manuscript.
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.transonc.com.
References
- 1.Cancer Genome Atlas Research Network Collaborators, author. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wikstrand CJ, Reist CJ, Archer GE, Zalutsky MR, Bigner DD. The class III variant of the epidermal growth factor receptor (EGFRvIII): characterization and utilization as an immunotherapeutic target. J Neurovirol. 1998;4:148–158. doi: 10.3109/13550289809114515. [DOI] [PubMed] [Google Scholar]
- 3.Abounader R, Laterra J. Scatter factor/hepatocyte growth factor in brain tumor growth and angiogenesis. Neuro Oncol. 2005;7:436–451. doi: 10.1215/S1152851705000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benvenuti S, Comoglio PM. The MET receptor tyrosine kinase in invasion and metastasis. J Cell Physiol. 2007;213:316–325. doi: 10.1002/jcp.21183. [DOI] [PubMed] [Google Scholar]
- 5.Jin X, Yin J, Kim SH, Sohn YW, Beck S, Lim YC, Nam DH, Choi YJ, Kim H. EGFR-AKT-Smad signaling promotes formation of glioma stem-like cells and tumor angiogenesis by ID3-driven cytokine induction. Cancer Res. 2011;71:7125–7134. doi: 10.1158/0008-5472.CAN-11-1330. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Zhou M, Shi B, Zhang Q, Jiang H, Sun Y, Liu J, Zhou K, Yao M, Gu J, et al. Identification of an exon 4-deletion variant of epidermal growth factor receptor with increased metastasis-promoting capacity. Neoplasia. 2011;13:461–471. doi: 10.1593/neo.101744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 8.Habib AA, Hognason T, Ren J, Stefansson K, Ratan RR. The epidermal growth factor receptor associates with and recruits phosphatidylinositol 3-kinase to the platelet-derived growth factor β receptor. J Biol Chem. 1998;273:6885–6891. doi: 10.1074/jbc.273.12.6885. [DOI] [PubMed] [Google Scholar]
- 9.Trusolino L, Comoglio PM. Scatter-factor and semaphorin receptors: cell signalling for invasive growth. Nat Rev Cancer. 2002;2:289–300. doi: 10.1038/nrc779. [DOI] [PubMed] [Google Scholar]
- 10.Yasmeen A, Bismar TA, Al Moustafa AE. ErbB receptors and epithelial-cadherin-catenin complex in human carcinomas. Future Oncol. 2006;2:765–781. doi: 10.2217/14796694.2.6.765. [DOI] [PubMed] [Google Scholar]
- 11.Nabeshima K, Shimao Y, Sato S, Kataoka H, Moriyama T, Kawano H, Wakisaka S, Koono M. Expression of c-Met correlates with grade of malignancy in human astrocytic tumours: an immunohistochemical study. Histopathology. 1997;31:436–443. doi: 10.1046/j.1365-2559.1997.3010889.x. [DOI] [PubMed] [Google Scholar]
- 12.Han CB, Ma JT, Li F, Zhao JZ, Jing W, Zhou Y, Zou HW. EGFR and KRAS mutations and altered c-Met gene copy numbers in primary non-small cell lung cancer and associated stage N2 lymph node-metastasis. Cancer Lett. 2012;314:63–72. doi: 10.1016/j.canlet.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Hochgrafe F, Zhang L, O'Toole SA, Browne BC, Pinese M, Porta CA, Lehrbach GM, Croucher DR, Rickwood D, Boulghourjian A, et al. Tyrosine phosphorylation profiling reveals the signaling network characteristics of basal breast cancer cells. Cancer Res. 2010;70:9391–9401. doi: 10.1158/0008-5472.CAN-10-0911. [DOI] [PubMed] [Google Scholar]
- 14.Lo HW. EGFR-targeted therapy in malignant glioma: novel aspects and mechanisms of drug resistance. Curr Mol Pharmacol. 2010;3:37–52. doi: 10.2174/1874467211003010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, Gibbs JB, Pan BS. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67:2081–2088. doi: 10.1158/0008-5472.CAN-06-3495. [DOI] [PubMed] [Google Scholar]
- 16.Pathi SS, Jutooru I, Chadalapaka G, Sreevalsan S, Anand S, Thatcher GR, Safe S. GT-094, a NO-NSAID, inhibits colon cancer cell growth by activation of a reactive oxygen species-microRNA-27a: ZBTB10-specificity protein pathway. Mol Cancer Res. 2011;9:195–202. doi: 10.1158/1541-7786.MCR-10-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, Stabile LP, Gubish CT, Gooding WE, Grandis JR, Siegfried JM. Dual blockade of EGFR and c-Met abrogates redundant signaling and proliferation in head and neck carcinoma cells. Clin Cancer Res. 2011;17:4425–4438. doi: 10.1158/1078-0432.CCR-10-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 19.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 20.Mueller KL, Yang ZQ, Haddad R, Ethier SP, Boerner JL. EGFR/Met association regulates EGFR TKI resistance in breast cancer. J Mol Signal. 2010;5:8. doi: 10.1186/1750-2187-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci USA. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 23.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. Targeting MET as a strategy to overcome crosstalk-related resistance to EGFR inhibitors. Lancet Oncol. 2009;10:709–717. doi: 10.1016/S1470-2045(09)70137-8. [DOI] [PubMed] [Google Scholar]
- 25.Kris MG, Natale RB, Herbst RS, Lynch TJ, Jr, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 26.Koochekpour S, Jeffers M, Rulong S, Taylor G, Klineberg E, Hudson EA, Resau JH, Vande Woude GF. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57:5391–5398. [PubMed] [Google Scholar]
- 27.Velpula KK, Dasari VR, Tsung AJ, Gondi CS, Klopfenstein JD, Mohanam S, Rao JS. Regulation of glioblastoma progression by cord blood stem cells is mediated by downregulation of cyclin D1. PLoS One. 2011;6:e18017. doi: 10.1371/journal.pone.0018017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asuthkar S, Nalla AK, Gondi CS, Dinh DH, Gujrati M, Mohanam S, Rao JS. Gadd45a sensitizes medulloblastoma cells to irradiation and suppresses MMP-9-mediated EMT. Neuro Oncol. 2011;13:1059–1073. doi: 10.1093/neuonc/nor109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velpula KK, Dasari VR, Tsung AJ, Dinh DH, Rao JS. Transcriptional repression of MAD-MAX complex by human umbilical cord blood stem cells downregulates extracellular signal-regulated kinase in glioblastoma. Stem Cells Dev. 2011;21:1779–1793. doi: 10.1089/scd.2011.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kermorgant S, Parker PJ. Receptor trafficking controls weak signal delivery: a strategy used by c-Met for STAT3 nuclear accumulation. J Cell Biol. 2008;182:855–863. doi: 10.1083/jcb.200806076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Chen L, Bao Z, Li S, You G, Yan W, Shi Z, Liu Y, Yang P, Zhang W, et al. Inhibition of STAT3 reverses alkylator resistance through modulation of the AKT and β-catenin signaling pathways. Oncol Rep. 2011;26:1173–1180. doi: 10.3892/or.2011.1396. [DOI] [PubMed] [Google Scholar]
- 32.Mimeault M, Batra SK. Complex oncogenic signaling networks regulate brain tumor-initiating cells and their progenies: pivotal roles of wild-type EGFR, EGFRvIII mutant and hedgehog cascades and novel multitargeted therapies. Brain Pathol. 2011;21:479–500. doi: 10.1111/j.1750-3639.2011.00505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dasari VR, Kaur K, Velpula KK, Gujrati M, Fassett D, Klopfenstein JD, Dinh DH, Rao JS. Upregulation of PTEN in glioma cells by cord blood mesenchymal stem cells inhibits migration via downregulation of the PI3K/Akt pathway. PLoS One. 2010;5:e10350. doi: 10.1371/journal.pone.0010350. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Lee TK, Poon RT, Yuen AP, Ling MT, Kwok WK, Wang XH, Wong YC, Guan XY, Man K, Chau KL, et al. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin Cancer Res. 2006;12:5369–5376. doi: 10.1158/1078-0432.CCR-05-2722. [DOI] [PubMed] [Google Scholar]
- 35.Bowers DC, Fan S, Walter KA, Abounader R, Williams JA, Rosen EM, Laterra J. Scatter factor/hepatocyte growth factor protects against cytotoxic death in human glioblastoma via phosphatidylinositol 3-kinase- and AKT-dependent pathways. Cancer Res. 2000;60:4277–4283. [PubMed] [Google Scholar]
- 36.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter GF, Holmes AB, Gaffney PR, Reese CB, McCormick F, Tempst P, et al. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh MK, Sharma P, Harbor PC, Rahaman SO, Haque SJ. PI3KAKT pathway negatively controls EGFR-dependent DNA-binding activity of Stat3 in glioblastoma multiforme cells. Oncogene. 2005;24:7290–7300. doi: 10.1038/sj.onc.1208894. [DOI] [PubMed] [Google Scholar]
- 38.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 39.Demuth T, Berens ME. Molecular mechanisms of glioma cell migration and invasion. J Neurooncol. 2004;70:217–228. doi: 10.1007/s11060-004-2751-6. [DOI] [PubMed] [Google Scholar]
- 40.Hoelzinger DB, Demuth T, Berens ME. Autocrine factors that sustain glioma invasion and paracrine biology in the brain microenvironment. J Natl Cancer Inst. 2007;99:1583–1593. doi: 10.1093/jnci/djm187. [DOI] [PubMed] [Google Scholar]
- 41.Taulli R, Scuoppo C, Bersani F, Accornero P, Forni PE, Miretti S, Grinza A, Allegra P, Schmitt-Ney M, Crepaldi T, et al. Validation of met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res. 2006;66:4742–4749. doi: 10.1158/0008-5472.CAN-05-4292. [DOI] [PubMed] [Google Scholar]
- 42.Rich JN, Rasheed BK, Yan H. EGFR mutations and sensitivity to gefitinib. N Engl J Med. 2004;351:1260–1261. [PubMed] [Google Scholar]
- 43.Toth J, Egervari K, Klekner A, Bognar L, Szanto J, Nemes Z, Szollosi Z. Analysis of EGFR gene amplification, protein over-expression and tyrosine kinase domain mutation in recurrent glioblastoma. Pathol Oncol Res. 2009;15:225–229. doi: 10.1007/s12253-008-9082-4. [DOI] [PubMed] [Google Scholar]
- 44.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors—impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X, Zhou J, Rogers AM, Janne PA, Benedettini E, Loda M, Hodi FS. c-Met, epidermal growth factor receptor, and insulin-like growth factor-1 receptor are important for growth in uveal melanoma and independently contribute to migration and metastatic potential. Melanoma Res. 2012;22:123–132. doi: 10.1097/CMR.0b013e3283507ffd. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Palacian A, del Castillo CG, Herrera B, Fernandez M, Roncero C, Fabregat I, Sanchez A. EGFR is dispensable for c-Met-mediated proliferation and survival activities in mouse adult liver oval cells. Cell Signal. 2012;24:505–513. doi: 10.1016/j.cellsig.2011.09.031. [DOI] [PubMed] [Google Scholar]
- 47.Sequist LV, von Pawel PJ, Garmey EG, Akerley WL, Brugger W, Ferrari D, Chen Y, Costa DB, Gerber DE, Orlov S, et al. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol. 2011;29:3307–3315. doi: 10.1200/JCO.2010.34.0570. [DOI] [PubMed] [Google Scholar]
- 48.Xu H, Stabile LP, Gubish CT, Gooding WE, Grandis JR, Siegfried JM. Dual blockade of EGFR and c-Met abrogates redundant signaling and proliferation in head and neck carcinoma cells. Clin Cancer Res. 2011;17:4425–4438. doi: 10.1158/1078-0432.CCR-10-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dulak AM, Gubish CT, Stabile LP, Henry C, Siegfried JM. HGF-independent potentiation of EGFR action by c-Met. Oncogene. 2011;30:3625–3635. doi: 10.1038/onc.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dasari VR, Velpula KK, Alapati K, Gujrati M, Tsung AJ. Cord blood stem cells inhibit epidermal growth factor receptor translocation to mitochondria in glioblastoma. PLoS One. 2012;7:e31884. doi: 10.1371/journal.pone.0031884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.