Abstract
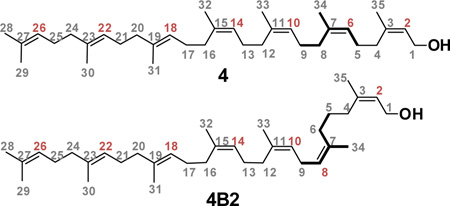
We describe a practical, multi-gram synthesis of (2Z,6Z,10Z,14Z,18E,22E)-3,7,11,15,19,23,27-heptamethyl-2,6,10,14,18,22,26-octacosaheptaen-1-ol [(Z4,E2,ω)-heptaprenol, 4] using the nerol-derived sulfone 8 as the key intermediate. Sulfone 8 is prepared by the literature route, and is converted in five additional steps (18% yield from 8) to (Z4,E2,ω)-heptaprenol 4. The use of Eu(hfc)3 as an NMR shift reagent not only enabled confirmation of the structure and stereochemistry of 4, but further enabled the structural assignment to a major side-product from a failed synthetic connection. The availability by this synthesis of (Z4,E2,ω)-heptaprenol 4 in gram quantities will enable preparative access to key reagents for the study of the biosynthesis of the bacterial cell envelope.
INTRODUCTION
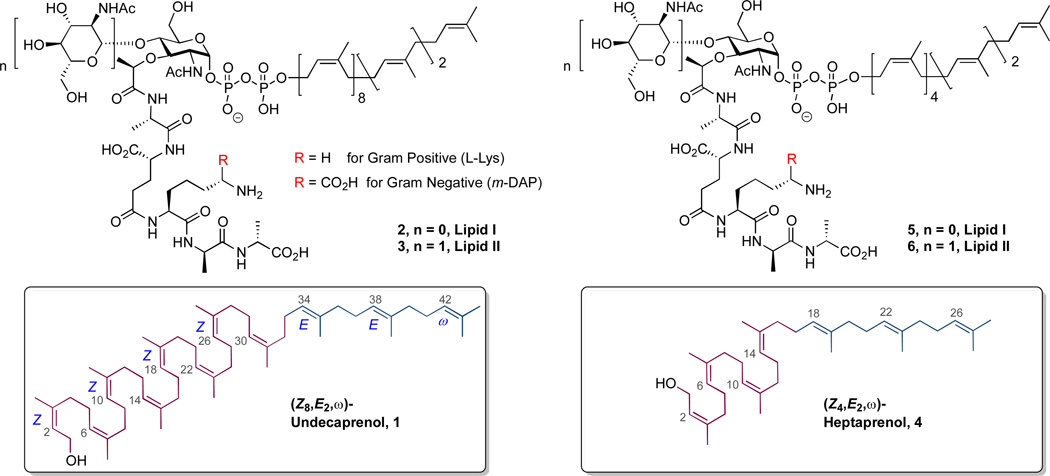
The five-carbon isoprene segment is foundational to the biosynthetic pathways of both primary and secondary metabolism.1 One class of isoprenoids, the polyprenols, is used catalytically in primary metabolism as a membrane-integral carrier of hydrophilic structure: of the wall teichoic acids in Gram-positive bacteria; of the saccharides destined for the lipopoly-saccharide of the outer membrane of Gram-negative bacteria; of the peptidoglycan monomers used by bacteria to assemble their cell wall; and of the saccharides destined for posttranslational modification of prokaryotic and eukaryotic proteins.2, 3 The discovery in 1964 that a lipid intermediate was required for bacterial peptidoglycan synthesis4, 5 was followed in 1967 by its identification as a C55 undecaprenol containing eleven C=C double bond functional groups.6–8 As a result of the extensive overlap of resonances in the polyprenols,9 the stereochemical assignment to the bacterial undecaprenol as the (Z8,E2,ω)-undecaenol [(2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E,38E,42)-3,7,11,15,19,23,27,31,35,39,43-undecamethyl-2,6,10,14,18,22,26,30,34,38,42-tetratetracontaundecaen-1-ol: compound 1 in Chart 1] rests upon exquisite mechanistic studies pertaining to its biosynthesis. These studies show that a single enzyme, undecaprenol diphophate synthase, synthesizes 1 from farnesyl diphosphate by eight-fold cis-prenyl homologation using isopentenyl diphosphate as the prenyl donor.10–13
Chart 1.
The structures of the undecaprenol-containing Lipid I and Lipid II used in bacterial peptidoglycan biosynthesis and their heptaprenol analogs.
Nonetheless, the low pool levels of the bacterial lipid intermediates,14 their limited accumulation in cell-free systems, and the tedious effort involved in their isolation and purification have restricted their availability for the study of the peptidoglycan pathway.15 Several efforts toward the synthesis of the Lipid I, II, and IV structures and their analogues, using both chemical and chemoenzymatic methods, have been reported.15–22 These syntheses are demanding, and indeed several pivotal Lipid I/II structures (2 and 3 in Chart 1), such as those having the meso-diaminopimelic sub-structure characteristic of Gram-negative Lipid II, have yet to be completed. Moreover, syntheses of prenol-containing structures often use the plant-derived (Z7,E3,ω)-undecaprenol as the lipid component. While the evidence thus far indicates that the (Z7,E3,ω)-undecaprenol and (Z8,E2,ω)-undecaprenol are functionally equivalent as the lipid component of Lipid II, these plant polyprenols are themselves rare. Given the important role of the lipid component in many aspects of biological recognition17 and the scarcity of the natural polyprenols, the development of practical synthetic routes to these structures is important. The chemical synthesis of oligo- and polyprenols for use in prenol-containing structures is recent.23, 24 Previous syntheses of polyprenols include dolichol-20,25 (Z8,E2,ω)-undecaprenol,26 (Z7,E3,ω)-undecaprenol,27 and (Z4,E2,ω)-heptaprenol.24, 26, 28
Walker, Kahne, and colleagues19, 20 showed that the Lipid II analog 6 having the shorter C35 (Z4,E2,ω)-heptaprenol (Chart 1) is accepted in vitro as a component of the Lipid II substrate of the penicillin-binding protein (PBP) enzymes. Accordingly, a general synthesis of heptaprenols—and no less importantly, a versatile spectroscopic means of confirming their structure—would be especially useful. As a demonstration of both of these objectives, we report the synthesis of (2Z,6Z,10Z,14Z,18E,22E)-3,7,11,15,19,23,27-heptamethyl-2,6,10,14,18,22,26-octacosaheptaen-1-ol (Z4,E2, ω)-heptaprenol, 4, and its complete 1H and 13C NMR assignment using chiral shift reagent-facilitated analysis of its homo- and heteronuclear 2D spectra.
RESULTS AND DISCUSSION
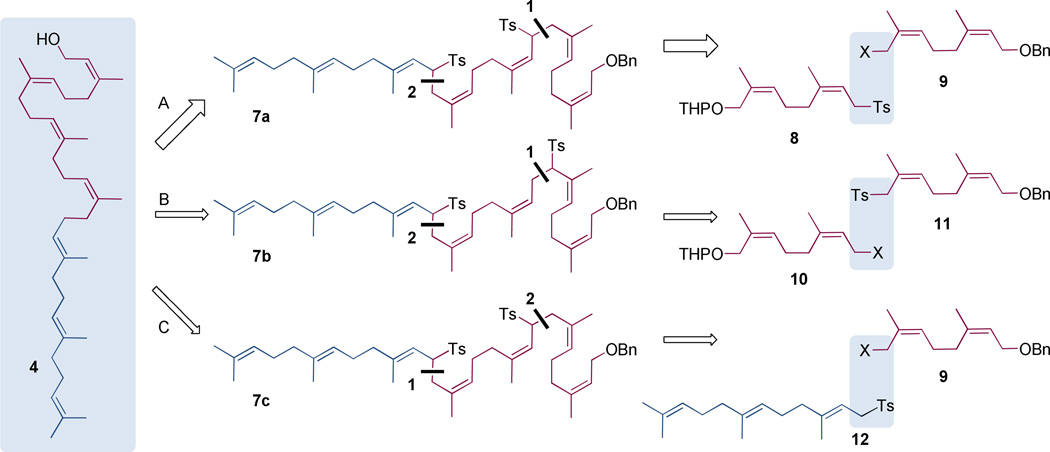
Three different retrosynthetic disconnections toward 4 were considered, corresponding to structures 7a, 7b, and 7c as key intermediates (Scheme 1). These three intermediates coincide with the reaction of different sulfone-stabilized carbanions (8, 11, and 12) with a diprenyl halide (9 or 10, X = Cl, Br). Although the use of sulfone-stabilized carbanions is a standard method for polyprenol synthesis, a systematic evaluation of the most suitable reaction pairing in terms of yield and stereochemical fidelity was absent from the literature. The most frequently used route is A of Scheme 1. Our interest in the alternative routes B and C emerged from our recognition, as we examined these reactions, that the reagent pair 10 and 11 was easier to prepare on scale than the reagent pair 8 and 9. While routes A and B synthesize the Z4-tetraprenol for coupling with the E,E-farnesol-derived sulfone 12, route C reacts 12 with the nerol-derived halide 9, followed by a second addition of 9 to complete the synthesis. Routes A and C have the advantage of using the most readily prepared neryl reagent (9, compared to the three other neryl derivatives 8, 10, and 11 shown in Scheme 2) as a starting material, but route C is dis-advantaged relative to routes A and B by its requirement for two separate desulfonylation reactions (Scheme 3: 22 to 23 and 7c to 4c).
Scheme 1.
The retrosynthetic disconnections evaluated as a practical synthesis of (Z4,E2,ω)-heptaprenol.
Scheme 2.
Syntheses of the four neryl units 8–11.
Scheme 3.
Synthetic routes evaluated as a practical synthesis of the (Z4,E2,ω)-heptaprenol
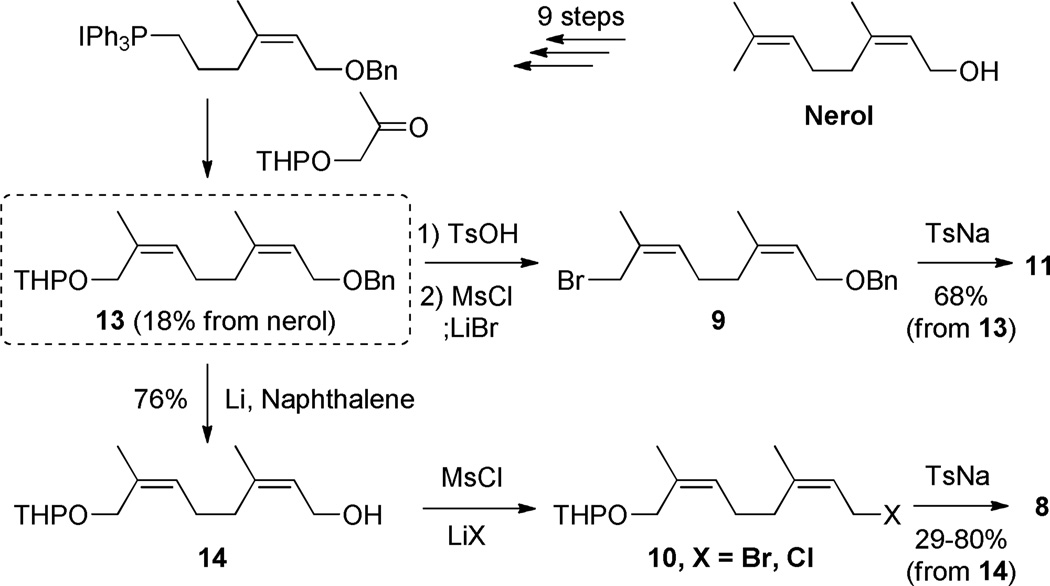
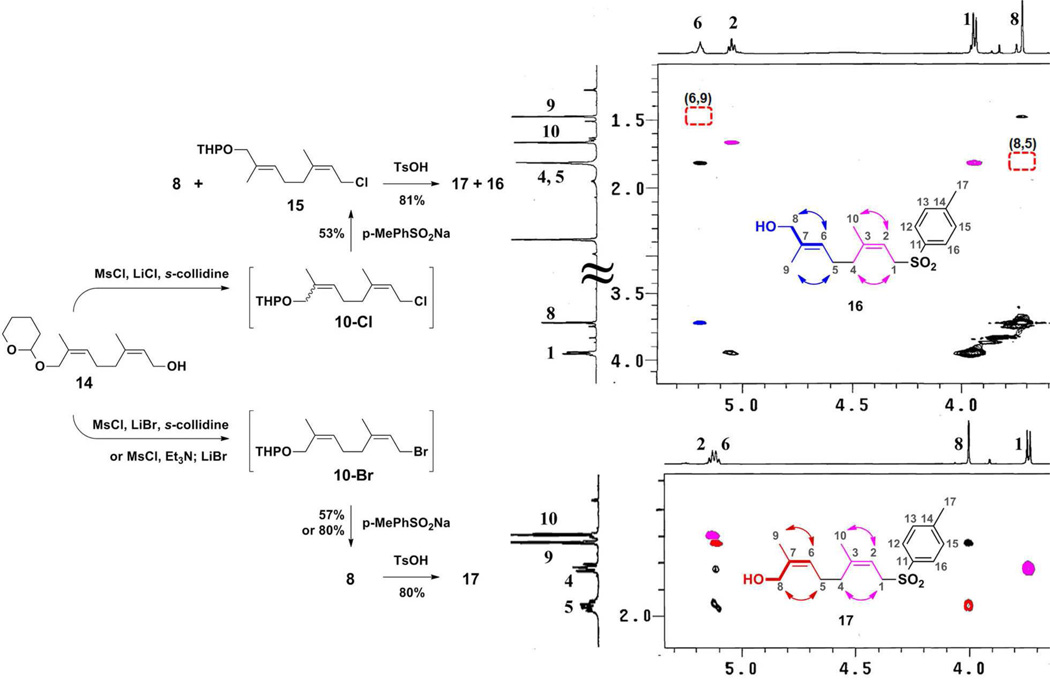
We converted (commercially available) nerol to the pivotal intermediate 13 in ten steps (including epoxide formation from the bromohydrin, epoxide hydrolysis, oxidative cleavage, reduction, iodination, triphenylphosphonium iodide formation, and Wittig reaction with the hydroxyketone) using the literature route.29 Although this route appears laborious, it is quite amenable to scale (we have prepared 13 on a 50-g scale). Compound 13 was separately transformed into the four neryl derivatives (8–11) of Scheme 2. The syntheses of two (9 and 11) were straightforward. The syntheses of 8 and 10 were not. The synthesis of sulfone 8 ultimately involved comparison of two different halides (10-Br and 10-Cl) for the sulfone alkylation (Figure 1). Using 10-Br as a reagent, we isolated 8 as the major product. In contrast, the analogous reaction using 10-Cl as a reagent gave a 5:4 diastereomeric mixture of 8 and 15 as assessed by 1H NMR analysis. These structures were assigned by extensive NMR analyses, done after removal of the O-THP protecting group in order to both simplify the NMR spectra and facilitate chromatographic purification.
Figure 1.
The key correlations in the 1H ROESY NMR spectra used to assign the alcohol diastereomers 16 (top spectrum) and 17 (bottom spectrum). Empty boxes in top spectrum indicate the absence of correlations from 6Z (H-6,H-9) and (H-5,H-8). Instead, the ROESY spectrum for 16 shows correlation between H-6 and H-8 (colored in blue), demonstrating a 6E configuration. The relevant synthetic transformations are summarized on the left.
The 1H and 13C spectra of alcohols 17 (from 8) and 16 (from 15) are too similar to allow definitive structural assignment by themselves. Following resonance assignments from 2D homo- and heteronuclear correlation spectra, analysis of the corresponding ROESY spectra (Figure 1) identified the difference. Compound 17 (arising from reaction with 10-Br) had the desired (2Z,6Z) stereochemistry whereas 16 (from reaction with 10-Cl) was the (2Z,6E)-stereoisomer. While E,Z scrambling is well known to occur during the synthesis of many allyl halides from allyl alcohols, we are at a loss to suggest the critical experimental difference accounting for the contrast between 10-Br compared to 10-Cl, and as well as to account for the unanticipated regiochemistry of the isomerization. Nonetheless, the discovery of this unexpectedly facile (and reproducible) isomerization under one set of standard halogenation conditions, but not under another, set the requirement for full verification of structure and stereochemistry, following each key step of our synthesis.
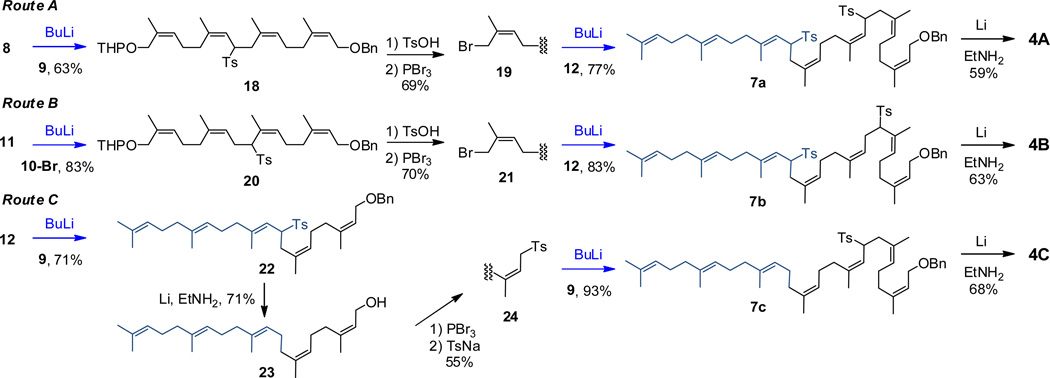
From 8, three routes toward heptaprenol 4 were compared (Scheme 3). Routes A and B used different neryl derivatives to give the nerylneryl derivative 18 (Route A) and nerylneryl derivative 20 (Route B), respectively. Each was separately coupled with farnesylsulfone 12. Route C used repetitive coupling of neryl derivative 9. In contrast to the final prenol structures, the penultimate synthetic intermediates give well-dispersed and easily interpretable NMR spectra, as exemplified by the complete assignment of 18-OH, 20-OH, and 7b (18-OH was obtained after removal of O-THP in 18, Supporting Information). Last, intermediates 7a, 7b, and 7c were deprotected by sequential reductive desulfonylation and reductive debenzylation to give 4 (identified in Scheme 3 by their routes of synthesis: 4A, 4B, and 4C).
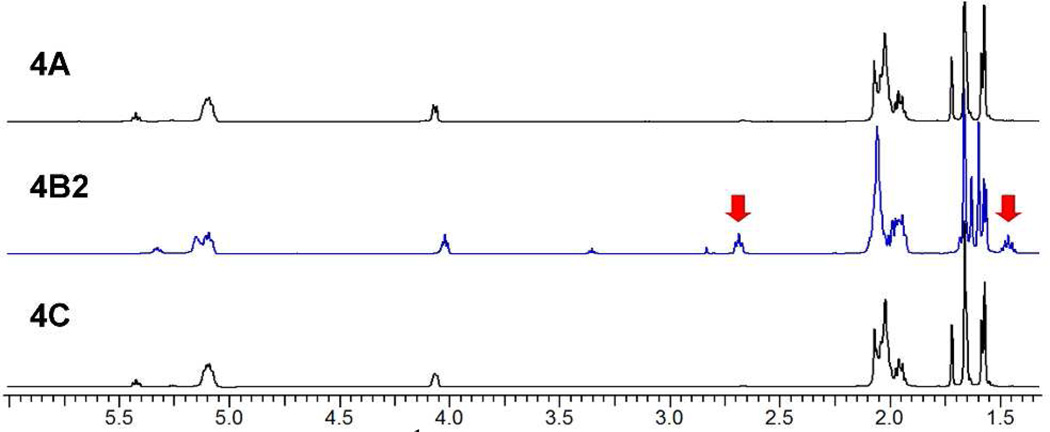
The final products 4A and 4C gave the identical 1H NMR spectrum, characterized by eight methyl resonances in a 4:2:1 Z/E/ω pattern (Figure 2). Further NMR analysis (see below) verified the product stereochemistry as the desired (Z4,E2, ω)-heptaprenol 4. The purity of the heptaprenol 4 isolated by silica flash chromatography (following the reductive desulfonylation step) exceeded 90% by 1H NMR analysis (Supporting Information). The analytical data for 4 matched the published data given by Chang et al.28 In contrast, the heptaprenol from route B was a 1:3 diasteromeric mixture of 4 and an isomer designated 4B2. The mixture was separated by column chromatography. Although the 1H NMR spectra of 4 and 4B2 are very similar (and show extensive overlap of signals), the 1H spectrum of 4B2 has two distinguishing resonances (Figure 2, red arrows). These new resonances are diagnostic of an undesirable isomerization.
Figure 2.
Comparison of 1H NMR spectra (500 MHz) of the heptaprenol isomers 4A, 4B2, and 4C in CDCl3.
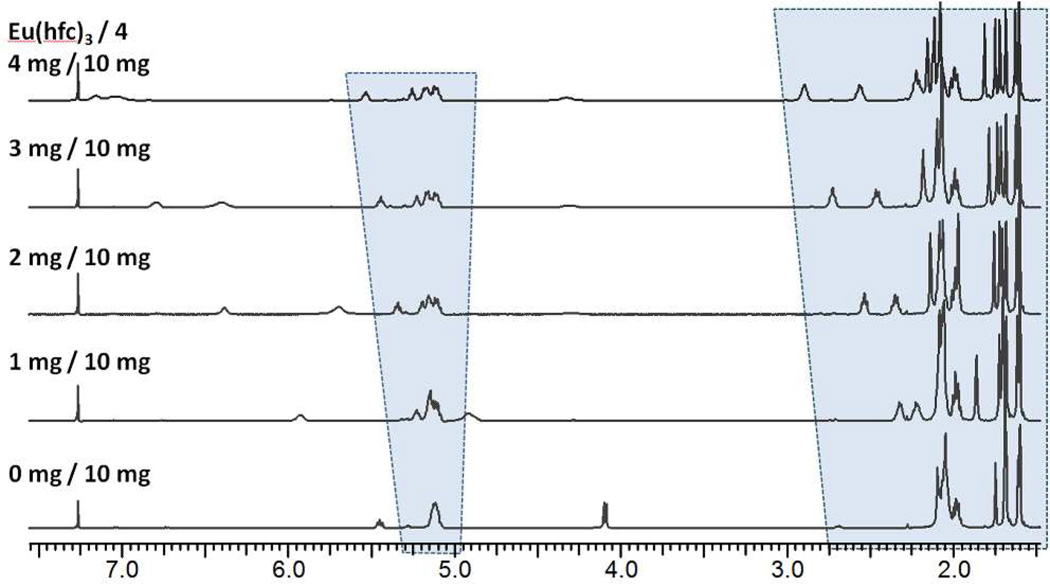
The extensive resonance overlaps for 4 and 4B2, and the requirement for a definitive verification of the structure of 4, prompted a search for a general NMR method to facilitate the structural assignment of prenol derivatives. Proof of structure by NMR of a polyprenol (and of the synthetic intermediates leading to the polyprenol) is rare. Indeed, there is a 45-year gap between the discovery of polyprenol involvement in bacterial primary metabolism and a full NMR-based structural assignment of a polyprenol, that of the plant-derived (Z6,E3, ω)-decaprenol [(2Z,6Z,10Z,14Z,18Z,22Z,26E,30E,34E,38)-3,7,11,15,19,23,27,31,35,39-decamethyl-2,6,10,14,18,22,26,30,34,38-tetracontadecaen-1-ol] accomplished in 2009 using 3D-NMR spectra analyses.30 The absence of this characterization is understandable, due to the extensive overlap of resonances even at high magnetic field (~18.79 T). Nonetheless (and to our very pleasant surprise) an exploration of several common chiral shift reagents (including Eu(tcf)3, Eu(hcf)3, Pr(hcf)3, Yt(hcf)3, and Resolve-Al AgFOD) demonstrated promise. Of these reagents, Eu(hcf)3 was best. The use of chiral shifts to disperse the resonances of terpenoids for the purpose of structure assignment31, 32 or for enantiomeric ratio determination33–36 is known, but is infrequent. Optimization of the molar ratio of Eu(hcf)3 to 4 for the purpose of structure assignment is shown in Figure 3. The structure of heptaprenol 4 was verified by analysis of homonuclear (DQF-COSY, TOCSY, ROESY) and heteronuclear (1H-13C HSQC, HSQC-TOCSY, HMBC) spectra obtained using 0.13 equivalents of Eu(hcf)3 to disperse the 1H resonances. 1H connectivities were established by analysis of the DQF-COSY and TOCSY spectra.
Figure 3.
1H NMR spectra (500 MHz) of 4 in the presence of different molar ratios of Eu(hcf)3 (from bottom to top, molar ratios of 0, 0.04, 0.09, 0.13, and 0.17 equivalents of shift agent to 4). The optimal balance between resonance dispersion and line broadening occurs at 0.13 equivalents of Eu(hcf)3 (3 mg Eu(hfc)3 to 10 mg of 4 in CDCl3).
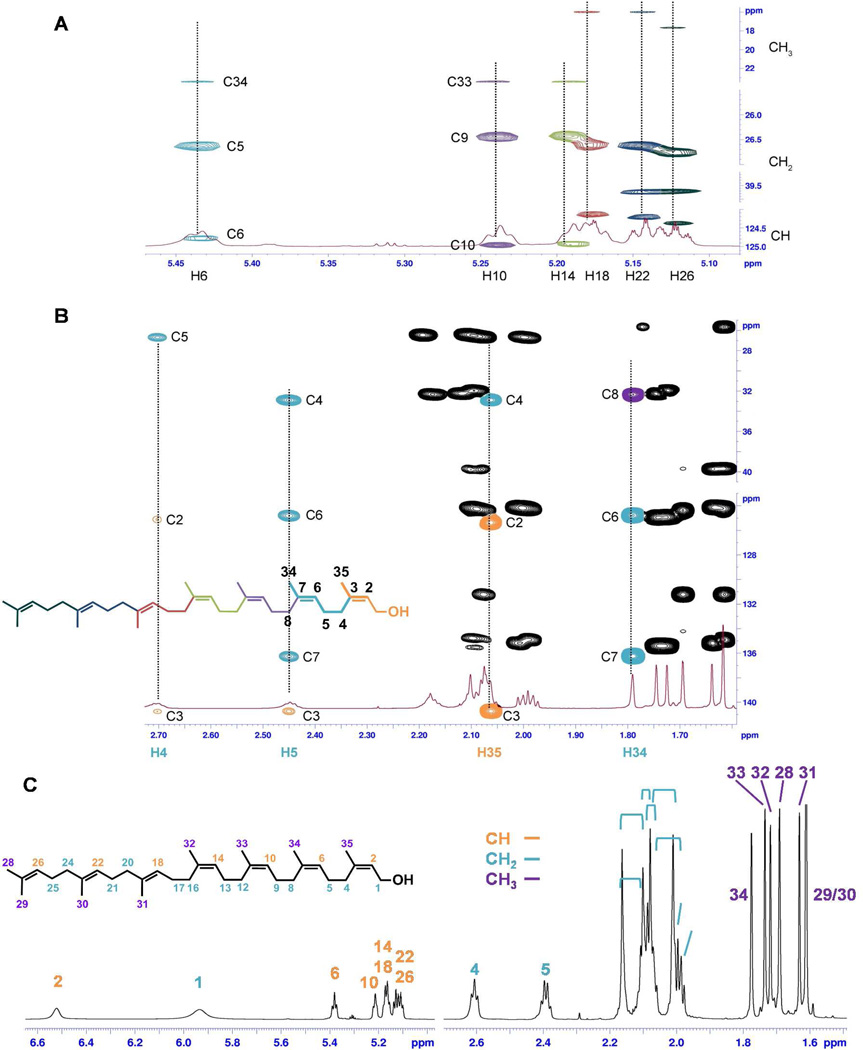
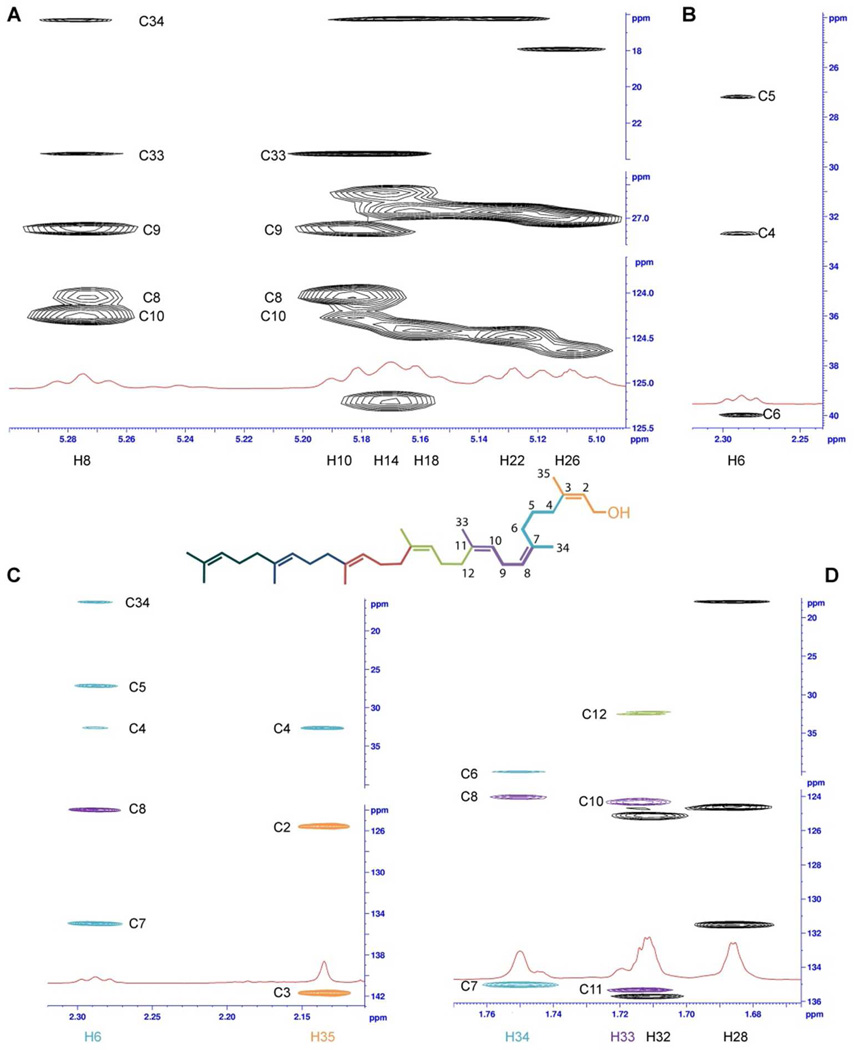
Resonances of all carbons with directly attached protons were assigned using the HSQC and HSQC-TOCSY spectra. For example, Figure 4A shows the correlation of the vinyl hydrogen and carbon resonances in the HSQC-TOCSY spectrum. The HMBC spectrum was then employed to assign the quaternary carbon resonances and to verify the correctness of the connectivities established by the analysis of the other spectra. Figure 4B shows the connectivities of the first two prenyl units (colored as orange and turquoise, respectively) deduced from the HMBC spectrum. The H-5 resonance is identified in the HMBC spectrum by its correlation with the three carbon resonances (C-4, C-6, and C-7) of its prenyl unit (colored in turquoise), and by its additional correlation with the C-3 signal of the neighboring prenyl unit (colored in orange). The H-4 resonance also correlates with the C-2 and C-3 resonances from the neighboring prenyl unit (colored in orange). The signal of H-34 correlates with the signal of C-8 (colored in purple). In this way, connectivity among the prenyl units was established. Finally, the assignment to each C=C double bond of an E or Z configuration was determined by analyzing the ROESY spectrum. Figure 4C summarizes the 1H resonance identities for 4. The complete 1H and 13C NMR resonance assignments for 4 are given in Table S2 in the Supporting Information.
Figure 4.
(A) Use of the HSQC-TOCSY spectrum for 13C (CH3, CH2, and CH) resonance correlation. Each prenyl unit is shown in a different color. (B) The interpretive use of the HBMC is exemplified by the correlations for the two terminal prenyl units (colored in orange and turquoise). (C) A pictorial summary of the 1H resonance assignments for 4 (800.13 MHz spectrum, obtained using a solution of 10 mg of 4 and 3 mg of Eu(hcf)3 in 600 µL of CDCl3).
Eu(hcf)3 likewise facilitated the NMR assignment of structure to 4B2 (derived from 7b). While the dispersion effect of Eu(hcf)3 was less than that seen for 4, it was nonetheless sufficient for the NMR assignment of structure to 4B2.
The structural difference occurs from C-5 to C-9 in 4B2, giving two distinct and atypical units: one having three continuous CH2 carbons (C-4, C-5, and C-6) and the second having a single CH2 between two vinyl units (C-9 between C-8 and C-10). The two distinguishing resonances in the 1H spectrum of 4B2 (Figure 2) are those of H-5 and H-9. As a prenyl segment has a single vinyl hydrogen, there is always only one crosspeak in the vinyl 13C region of HSQC-TOCSY between vinyl proton and carbon resonances, which corresponds to a given vinyl CH group (see for example the crosspeak for the CH-6 vinyl group shown in Figure 5A). In contrast, the vinyl H-8 resonance of 4B2 shows in the same type of spectrum two crosspeaks; one with the vinyl C-8 resonance and the other with vinyl the C-10 resonance (Figure 5A). Similarly, the vinyl H-10 resonance also shows two crosspeaks; one with the vinyl C-10 resonance and the other with vinyl C-8 resonance Both H-8 and H-10 resonances then exhibit a crosspeak with the C-9 resonance at δ = 27.19 ppm, defining the first atypical unit. The second unit contains three consecutive CH2 (correlations are shown in Figure 5B). The HMBC spectrum confirmed the proposed connectivities of the two atypical units: both connected to each other and between the first and fourth prenyl units (Figures 5C and D). Hence the terminal prenyl (colored in orange) is connected to -(CH2)3- segment (colored in turquoise) as evidenced by the HMBC correlation of the H-35 resonance with the C-4 resonance. Correlations of the H-6 and H-34 resonances with the C-8 resonance confirmed connection of the the -(CH2)3- segment to the prenyl segment colored in purple. Finally, the crosspeak between the H-33 (purple unit) and C-10 resonances confirmed connectivity to the prenyl unit colored in green. The 1H and 13C resonance assignments for 4B2 are given in Table S2 in the Supporting Information.
Figure 5.
The HSQC-TOCSY (A and B) and HBMC spectra (C and D) of two atypical prenyl units of 4B2 (800.13 MHz, obtained using a solution of 10 mg of 4B2 and 3 mg of Eu(hcf)3 in 600 µL of CDCl3).
An overall 30% yield of the 3:1 mixture of 4B2 and 4 (from prenyl sulfone 11) characterized the failed Route B. The two successful routes A and C to 4 gave comparable overall yields of 20% for route A and 18% for route C relative to their respective prenyl sulfones (8 and 12) as starting materials. In terms of heptaprenol yield the two routes are equivalent. A clear choice between the two routes emerges for the synthesis of longer polyprenols. After the first coupling reaction, route A requires transformation of the coupling product 18 to halide 19, via O-THP deprotection, followed by bromination. Route C requires transformation of the coupling product 22 to sulfone 24 via debenzylation/detosylation, followed by bromination and subsequent tosylation. Overall, these three steps of route C are lower yielding (39%), as compared to the overall yield (69%) for the comparable steps of route A. For longer prenols (polyprenols) route C is uncompetitive compared to route A, as route C will require iterative repetition of the lower yielding steps.
CONCLUSION
Polyprenols are the membrane-bound component of such a diversity of biosynthetic intermediates that they have been termed “superlipids”.37 This diversity is exemplified by the recent syntheses, using both chemical and chemoenzymatic means, of the GlcNAc-UDP intermediate of bacterial cell wall biosynthesis;38 of the glycosyl intermediates of bacterial N-linked glycosylation;27, 39 of bacterial teichoic acid biosynthesis;40 of the Gram-positive bacterial S-layer glycan;41 of the bacterial polysialic capsule;42 and of the mycobacterial arabinan glyconjugates.43, 44 In all of these syntheses the prenol is a mass-limiting reagent.
Herein we report a practical, multi-gram synthesis of the (Z4,E2,ω)-heptaprenol 4 as a key enabling material for the synthesis of these biosynthetic intermediates, and also including the Lipid II intermediate for peptidoglycan biosynthesis. The purity of 4 we obtain (>90%) is directly comparable to the purity of 4 that may be obtained by solid-phase synthesis.28 The key advantage of our solution synthesis is its potential for scale. The pivotal intermediate 8 is prepared easily on a 30-g scale, and may be transformed to secure 4 on a > 2 g scale.
Our optimized synthesis uses the classical retrosynthetic route (Route A) for Z-prenyl homologation. Notwithstanding the precedence for Route A, an unexpectedly facile and unusual rearrangement was encountered during allylic chlorination of a key intermediate, but not seen during allylic bromination of this same intermediate under otherwise identical experimental conditions. Our optimism that a better synthesis of 4 might be attained using a more accessible reagent pair in an alternate route (Route B) was dashed by a facile isomerization that occurred during the reductive desulfonylation of a late-stage intermediate. These observations emphasize the necessity for rigorous proof of structure in prenol synthesis, even following seemingly straightforward synthetic transformations. Here, proof of the latter isomerization and verification of the final (Z4,E2,ω)-heptaprenol structure of 4 used extensive two-dimensional NMR correlations in the presence of the chiral shift reagent Eu(hfc)3 for resonance dispersion. The advantageous use of chiral shift reagents and high magnetic field strength as seen here for these prenols suggests a general value to this approach for NMR structural assignment.
EXPERIMENTAL SECTION
NMR Spectroscopy
The structures of prenol 4 and 4B2 [10 mg of each was dissolved in 600 µL CDCl3 containing 3 mg Eu(hfc)3] were determined by interpretation of the homonuclear DQF-COSY, TOCSY, ROESY and heteronuclear 1H-13C HSQC, HSQC-TOCSY, HMBC NMR spectra. Proton connectivities were derived from the DQF-COSY and TOCSY spectra. 13C resonances corresponding to carbons with directly attached protons were assigned using HSQC and HSQC-TOCSY spectra. HMBC spectra were used to assign resonances of the quaternary carbons and to validate the connectivities established by the other spectra. ROESY spectra were utilized to establish the E or Z configurations for each C=C double bond. Experiments were performed at 25 °C using a Bruker AVANCE II spectrometer equipped with a TCI cryoprobe and operating at a 1H resonance frequency of 800.13 MHz. Standard pulse sequences were used.45–50 Time domain data (t2 and t1) for 2D experiments were recorded as 2048 × 1024 complex matrices with 16 and 32 scans per t1 increment for homonuclear and heteronuclear spectra, respectively. Relaxation delay between individual scans and spin-lock time for TOCSY experiments was 1.4 s and 60 ms, respectively. For ROESY experiments, a relaxation delay of 4 s and a spin-lock time of 400 ms gave 2048 × 512 complex time domain points. Linear prediction to 1024 complex points was applied in the t1 domain. The data were zero filled to obtain final 2048 × 2048 complex matrices. In all other homonuclear 2D experiments, zero filling was used only in the t1 domain to obtain final 2048 × 2048 complex time domain data. In heteronuclear 2D experiments, linear prediction to 2048 complex data points was employed in the t1 domain, which was zero filled to 4096 to get final 2048 × 4096 complex time domain data. Shifted sine bell weighting functions were applied in both domains prior to double Fourier transformation. In the 3D 1H-13C-HSQC-TOCSY experiments, 1024 × 48 × 128 complex time domain points in t3, t2, and t1 domains were collected with 16 scans per time increment, relaxation delay of 1.3 s, and 60 ms spin-lock. Linear predictions to 128 and 256 complex points were utilized in the t2 and t1 domains. Zero filling gave 2048 × 256 × 512 complex matrices. Shifted sine bell weighting functions were applied in all three domains prior to triple Fourier transformation. Spectra were processed using Bruker TopSpin 2.1 software. 1H spectra, the 1H dimension in 2D heteronuclear spectra, and the 1D 13C{1H} spectra were referenced to solvent (DMSO, δH 2.5 and δC 39.5 ppm; CDCl3, δH 7.27 and δC 77.23). The 13C dimension in the 2D heteronuclear spectra was referenced indirectly.51
Supplementary Material
ACKNOWLEDGMENT
We thank Prof. Po-Huang Liang (Academia Sinica) for the clarification of the stereochemistry of the endogenous bacterial undecaprenol. This work was supported by a grant from the National Institutes of Health. The Mass Spectrometry & Proteomics Facility of the University of Notre Dame is supported by grant CHE-0741793 from the National Science Foundation.
Footnotes
ASSOCIATED CONTENT
Supporting Information available. Experimental procedures and NMR data for all new compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Oldfield E, Lin F-Y. Angew. Chem. Int. Ed. 2012;51:1124–1137. doi: 10.1002/anie.201103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartley M, Imperiali B. Arch. Biochem. Biophys. 2012;517:83–97. doi: 10.1016/j.abb.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hug I, Feldman M. Glycobiology. 2011;21:138–151. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee AN, Park JT. Proc. Natl. Acad. Sci. U.S.A. 1964;51:9–16. doi: 10.1073/pnas.51.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meadow PM, Anderson JS, Strominger JL. Biochem. Biophys. Res. Commun. 1964;14:382–387. doi: 10.1016/s0006-291x(64)80014-0. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JS, Matsuhashi M, Haskin MA, Strominger JL. J. Biol. Chem. 1967;242:3180–3190. [PubMed] [Google Scholar]
- 7.Higashi Y, Strominger JL, Sweeley CC. Proc. Natl. Acad. Sci. U.S.A. 1967;57:1878–1884. doi: 10.1073/pnas.57.6.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feeney J, Hemming FW. Anal. Biochem. 1967;20:1–15. doi: 10.1016/0003-2697(67)90258-8. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y, Hirasawa H. Chem. Phys. Lipids. 1989;51:183–189. [Google Scholar]
- 10.Chen Y, Chen A, Chen C, Wang A, Liang P. J. Biol. Chem. 2002;277:7369–7376. doi: 10.1074/jbc.M110014200. [DOI] [PubMed] [Google Scholar]
- 11.Lu Y, Liu H, Liang P. Biochem. Biophys. Res. Commun. 2009;379:351–355. doi: 10.1016/j.bbrc.2008.12.061. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y, Liu H, Teng K, Liang P. Biochem. Biophys. Res. Commun. 2010;400:758–762. doi: 10.1016/j.bbrc.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Teng K, Liang P. Bioorg. Chem. 2012;43:51–57. doi: 10.1016/j.bioorg.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Barreteau H, Magnet S, El Ghachi M, Touze T, Arthur M, Mengin-Lecreulx D, Blanot D. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009;877:213–220. doi: 10.1016/j.jchromb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 15.van Heijenoort J. Microbiol. Mol. Biol. Rev. 2007;71:620–635. doi: 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auger G, Crouvoisier M, Caroff M, van Heijenoort J, Blanot D. Lett. Pept. Sci. 1997;4:371–376. [Google Scholar]
- 17.Dumbre S, Derouaux A, Lescrinier E, Piette A, Joris B, Terrak M, Herdewijn P. J. Am. Chem. Soc. 2012;134:9343–9351. doi: 10.1021/ja302099u. [DOI] [PubMed] [Google Scholar]
- 18.VanNieuwenhze MS, Mauldin SC, Zia-Ebrahimi M, Winger BE, Hornback WJ, Saha SL, Aikins JA, Blaszczak LC. J. Am. Chem. Soc. 2002;124:3656–3660. doi: 10.1021/ja017386d. [DOI] [PubMed] [Google Scholar]
- 19.Ye X-Y, Lo M-C, Brunner L, Walker D, Kahne D, Walker S. J. Am. Chem. Soc. 2001;123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Fechter EJ, Wang T-SA, Barrett D, Walker S, Kahne DE. J. Am. Chem. Soc. 2007;129:3080–3081. doi: 10.1021/ja069060g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz B, Markwalder JA, Wang Y. J. Am. Chem. Soc. 2001;123:11638–11643. doi: 10.1021/ja0166848. [DOI] [PubMed] [Google Scholar]
- 22.VanNieuwenhze MS, Mauldin SC, Zia-Ebrahimi M, Aikins JA, Blaszczak LC. J. Am. Chem. Soc. 2001;123:6983–6988. doi: 10.1021/ja016082o. [DOI] [PubMed] [Google Scholar]
- 23.Liu C-Y, Guo C-W, Chang Y-F, Wang J-T, Shih H-W, Hsu Y-F, Chen C-W, Chen S-K, Wang Y-C, Cheng T-JR, Ma C, Wong C-H, Fang J-M, Cheng W-C. Org. Lett. 2011;12:1608–1611. doi: 10.1021/ol100338v. [DOI] [PubMed] [Google Scholar]
- 24.Gampe CM, Tsukamoto H, Wang T-SA, Walker S, Kahne D. Tetrahedron. 2011;67:9771–9778. doi: 10.1016/j.tet.2011.09.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue S, Kaneko T, Takahashi Y, Miyamoto O, Sato K. J. Chem. Soc. Chem. Commun. 1987:1036–1037. [Google Scholar]
- 26.Sato K, Miyamoto O, Inoue S, Matsuhashi Y, Koyama S, Kaneko T. J. Chem. Soc. Chem. Commun. 1986:1761–1762. [Google Scholar]
- 27.Lee J, Ishiwata A, Ito Y. Tetrahedron. 2009;65:6310–6319. [Google Scholar]
- 28.Chang Y-F, Liu C-Y, Guo C-W, Wang Y-C, Fang J-M, Cheng W-C. J. Org. Chem. 2008;73:7197–7203. doi: 10.1021/jo8010182. [DOI] [PubMed] [Google Scholar]
- 29.Sato K, Miyamoto O, Inoue S, Furusawa F, Matsuhashi Y. Chem. Lett. 1983:725–728. [Google Scholar]
- 30.Misiak M, Koźmiński W, Kwasiborska M, Wójcik J, Ciepichal E, Swiezewska E. Magn. Reson. Chem. 2009;47:825–829. doi: 10.1002/mrc.2473. [DOI] [PubMed] [Google Scholar]
- 31.Heldman D, Gilde HG. J. Chem. Educ. 1980;57:390–391. [Google Scholar]
- 32.Wenzel TJ, Chisholm CD. Prog. Nucl. Magn. Reson. Spectrosc. 2011;59:1–63. doi: 10.1016/j.pnmrs.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Baldovini N, Tomi F, Casanova J. Magn. Reson. Chem. 2001;39:621–624. [Google Scholar]
- 34.Blanc M-C, Bradesi P, Casanova J. Magn. Reson. Chem. 2005;43:176–179. doi: 10.1002/mrc.1523. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh I, Kishi Y, Tomoda H, Omura S. Org. Lett. 2004;6:4719–4722. doi: 10.1021/ol048060n. [DOI] [PubMed] [Google Scholar]
- 36.Lanfranchi D, Blanc M-C, Vellutini M, Bradesi P, Casanova J, Tomi F. Magn. Reson. Chem. 2008;46:1188–1194. doi: 10.1002/mrc.2330. [DOI] [PubMed] [Google Scholar]
- 37.Surmacz L, Swiezewska E. Biochem. Biophys. Res. Commun. 2011;407:627–632. doi: 10.1016/j.bbrc.2011.03.059. [DOI] [PubMed] [Google Scholar]
- 38.Al-Dabbagh B, Blanot D, Mengin-Lecreulx D, Bouhss A. Anal. Biochem. 2009;391:163–165. doi: 10.1016/j.ab.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 39.Hartley M, Larkin A, Imperiali B. Bioorg. Med. Chem. 2008;16:5149–5156. doi: 10.1016/j.bmc.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown S, Zhang Y, Walker S. Chem. Biol. 2008;15:12–21. doi: 10.1016/j.chembiol.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holkenbrink A, Koester D, Kadchel J, Werz D. Eur. J. Org. Chem. 2011;2011:6233–6239. [Google Scholar]
- 42.Shpirt A, Kononov L, Maltsev S, Shibaev V. Carbohydr. Res. 2011;346:2849–2854. doi: 10.1016/j.carres.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Liav A, Ciepichal E, Swiezewska E, Bobovská A, Dianišková P, Blaško J, Mikušová K, Brennan P. Tetrahedron Lett. 2009;50:2242–2244. [Google Scholar]
- 44.Zhang J, Angala S, Pramanik P, Li K, Crick D, Liav A, Jozwiak A, Swiezewska E, Jackson M, Chatterjee D. ACS Chem. Biol. 2011;6:819–828. doi: 10.1021/cb200091m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bax A, Morris GA. J. Magn. Reson. 1981;42:501–505. [Google Scholar]
- 46.Bax A, Summers MF. J. Am. Chem. Soc. 1986;108:2093–2094. [Google Scholar]
- 47.Bodenhausen G, Freeman R, Niedermeyer R, Turner DL. J. Magn. Reson. 1977;26:133–164. [Google Scholar]
- 48.Griesinger C, Otting G, Wüthrich K, Ernst RR. J. Am. Chem. Soc. 1988;110:7870–7872. [Google Scholar]
- 49.Sattler M, Schwalbe H, Griesinger C. J. Am. Chem. Soc. 1992;114:1126–1127. [Google Scholar]
- 50.Wijmenga SS, Hallenga K, Hilbers CW. J. Magn. Reson. 1989;84:634–642. [Google Scholar]
- 51.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.