Abstract
The nutritional environment in which the mammalian fetus or infant develop is recognized as influencing the risk of chronic diseases, such as type 2 diabetes and hypertension, in a phenomenon that has become known as developmental programming. The late onset of such diseases in response to earlier transient experiences has led to the suggestion that developmental programming may have an epigenetic component, because epigenetic marks such as DNA methylation or histone tail modifications could provide a persistent memory of earlier nutritional states. One class of genes that has been considered a potential target or mediator of programming events is imprinted genes, because these genes critically depend upon epigenetic modifications for correct expression and because many imprinted genes have roles in controlling fetal growth as well as neonatal and adult metabolism. In this study, we have used an established model of developmental programming—isocaloric protein restriction to female mice during gestation or lactation—to examine whether there are effects on expression and DNA methylation of imprinted genes in the offspring. We find that although expression of some imprinted genes in liver of offspring is robustly and sustainably changed, methylation of the differentially methylated regions (DMRs) that control their monoallelic expression remains largely unaltered. We conclude that deregulation of imprinting through a general effect on DMR methylation is unlikely to be a common factor in developmental programming.
Keywords: DNA methylation, genomic imprinting, differentially methylated region, developmental programming, metabolism, nutrition
Introduction
Epidemiological and animal studies have shown that adverse environments or suboptimal nutritional conditions during pregnancy can alter the physiology of offspring and increase their predisposition to many diseases in adult life. This phenomena is referred to as developmental programming.1,2 One explanation advanced to account for long-term or later onset outcomes of early life experiences is epigenetic: that interventions, through adaptive or maladaptive responses, result in a permanent resetting of gene expression programs that is mediated by altered epigenetic marking (DNA methylation or posttranslational modifications of histones), which can persist long after the duration of the initiating factor or condition.3,4 This takes account of the fact that patterns of DNA methylation and histone modifications experience wholescale changes during embryogenesis, lineage specification and differentiation, and that there may be time-points particularly in pre- and early post-implantation development and lineage segregation during which epigenetic modifications could be especially vulnerable to abnormal nutritional states. Some epigenetic modifications, in particular DNA methylation, can be stably propagated through numerous cell divisions as a cellular memory of earlier events.
Nutritional and other environmental challenges could affect epigenetic marks in a variety of ways. They could change the availability of the common methyl donor, S-adenosylmethionine (SAM), used in DNA and histone methylation reactions, or alter the expression or activity of epigenetic modifiers. Accordingly, DNA methylation patterns have been reported to be sensitive to methyl deficient diets5-7 or to variations in other components of one-carbon metabolism, such as folate.8 Histone demethylases require flavin adenine dinucleotide or α-ketoglutarate,9 so that their activities are also potentially responsive to cellular energy status. In addition the Ten-11 translocation (Tet) proteins involved in the active removal of cytosine methylation by oxidation depend upon oxoglutarate. It is possible, therefore, that nutritional status could have global, non-specific effects on histone modifications and DNA methylation, or locus-specific effects that could be more adaptive in nature.
A set of genes that has received a great deal of interest as possible mediators of developmental programming effects are imprinted genes.10-12 This is because these genes are entirely dependent on long-term epigenetic marks for their normal expression. Imprinted genes are unusual in that the two alleles are expressed to different degrees depending on their parental origin; in many cases, there is complete silencing of one parental allele.13 Imprinting is determined by the establishment of different states of epigenetic modification in male and female gametes at imprinting control regions (ICRs), typified by differences in DNA methylation (giving rise to the term germline differentially methylated region or gDMR). Imprinting also depends on the faithful maintenance of these parental allele-specific marks throughout the lifetime of the individual. We have some understanding about how these gametic marks are created and then maintained in embryos,14,15 but the degree to which they can be eroded during the lifetime is less clear, although some studies have shown individual variation in DMR methylation levels in human populations.16-18 Most imprinted genes are clustered, with monoallelic expression of multiple imprinted genes in a domain being controlled in cis by a single ICR.19,20 The action of the ICR in the embryo results in elaboration of further epigenetic differences between the two alleles across the imprinted domain, including the establishment of additional DMRs (secondary or somatic DMRs) and allelic differences in histone modifications. A particular appeal of imprinted genes as possible targets of developmental programming is that they play important roles in controlling offspring growth, both pre- and post-natally, in early post-natal adaptations and in adult energy homeostasis.21,22 Owing to their clustered nature and shared dependence on single ICRs, changes in DNA methylation of an ICR would result in altered expression of multiple imprinted genes. Because of their monoallelic expression, imprinted genes are considered to be particularly dosage-sensitive,23,24 such that even modest deregulation of imprinting at single loci could have phenotypic consequences. Moreover, there could be additive effects from deregulation of several imprinted domains if their imprinting was similarly vulnerable to nutritional or other interventions. In addition, once perturbed, there is thought to be no mechanism to restore normal DNA methylation at ICRs during embryogenesis or later life, because methylation of these elements is specified primarily by mechanisms acting in gametogenesis.25 There is evidence for altered methylation of DMRs of imprinted genes in some developmental programming phenomena, such as the Dutch Hunger Winter,26,27 and methylation of some DMRs has been shown to be affected by pre- and peri-conceptional micronutrient supplementation.28 Nevertheless, the magnitude of reported effects is small and the analysis has necessarily been conducted in accessible tissues in which the physiological relevance of observed changes cannot easily be inferred.
In this study, we have specifically addressed whether expression at imprinted genes is altered in a paradigm of developmental programming and whether this is associated with altered methylation of their DMRs. Using an established model of developmental programming that employs protein restriction of maternal diets during gestation or lactation,29,30 we find that although changes in expression level of imprinted genes can be detected in liver of offspring from dietary-restricted female mice, DMR methylation appears to be robust.
Results
Expression of imprinted genes in offspring from maternal dietary restriction
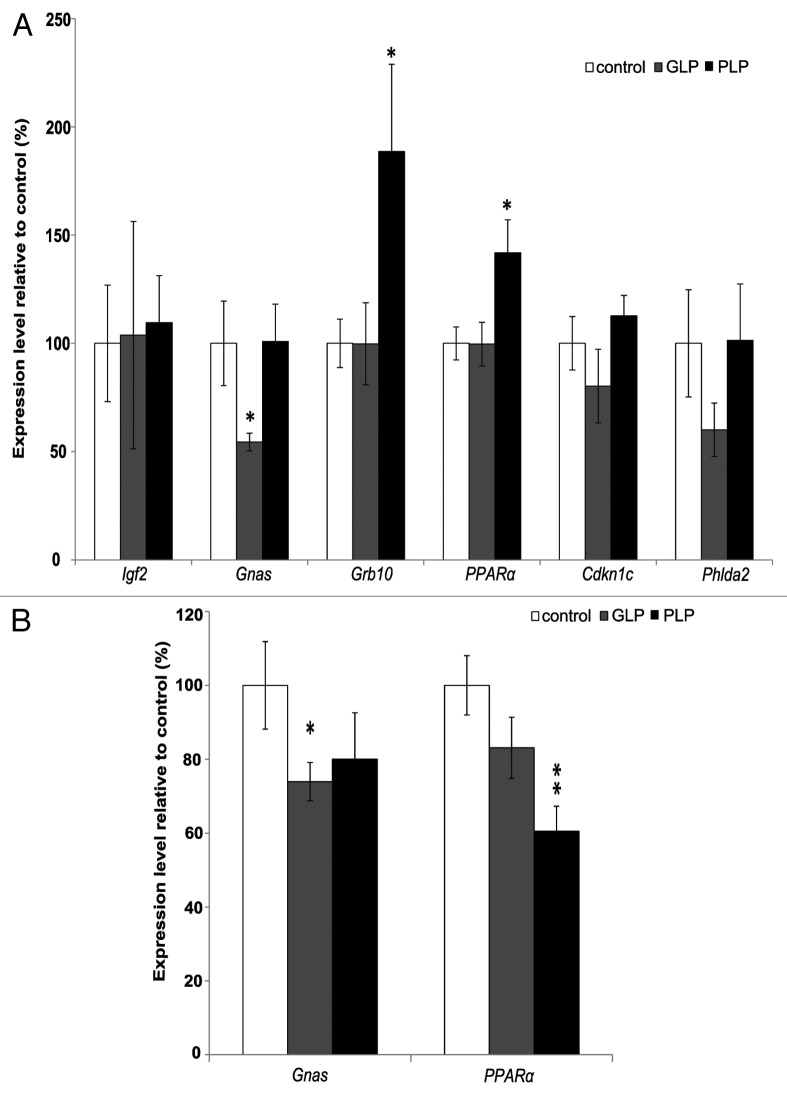
Quantitative reverse transcriptase PCR (RT-qPCR) assays were performed to ascertain if imprinted genes exhibited altered expression in liver of offspring of female mice fed an isocaloric low-protein diet during gestation (GLP) or lactation (PLP) and whether any observed gene expression changes were sustained beyond weaning. Five imprinted genes from four imprinted domains—Gnas, Grb10, Igf2, Cdkn1c and Phlda2—were chosen for analysis owing to their established or possible roles in fetal growth, liver development or physiology. Expression was determined first in liver at the end of postnatal week 3. Significant changes in transcript levels were detected for Gnas (reduced in GLP pups; p = 0.036) and Grb10 (elevated in PLP pups; p = 0.025) (Fig. 1A). Both Cdkn1c and Phlda2 showed a tendency toward reduced expression in the GLP group, but not significantly (Phlda2 expression levels were particularly low in general; Table S1); these genes were not assessed further. Igf2 expression was not changed in either of the groups compared with controls at this age. In addition, we examined expression of the peroxisome proliferator-activated receptor α gene (Pparα), which has been described to be modulated in other developmental programming paradigms, and found that it was upregulated in PLP pups (p = 0.014). To assess whether any of these expression changes was sustained into adult life, RT-qPCR was also performed on liver of mice sacrificed at the end of postnatal week 12 (Fig. 1B). The significant downregulation of Gnas in the GLP group was maintained at week 12 (p = 0.039), although the magnitude of the effect was reduced as compared with week 3. It was not possible to assess expression of Igf2 or Grb10, as these genes are known to be downregulated postnatally and expression was no longer detectable by RT-qPCR (Table S2). Surprisingly, the effect on hepatic Pparα expression was reversed at week 12, with transcript levels in the PLP group reduced in comparison with the control group (p = 0.002), suggesting dynamic regulation of this gene in pups nursed by dams with dietary protein restriction specifically during lactation.
Figure 1. Expression analysis of imprinted and other genes in offspring from dams experiencing dietary protein restriction during gestation (GLP) or lactation (PLP). (A) RT-qPCR analysis for the indicated genes at week 3 after birth. (B) RT-qPCR analysis for the indicated genes at week 12 after birth. mRNA levels are expressed as the percentage of the means of the control groups (set at 100%) following normalization for each gene against three housekeeping genes. Bars represent standard errors; *p < 0.05, **p < 0.01 compared with control.
DNA methylation analysis of imprinted genes in offspring from maternal dietary restriction
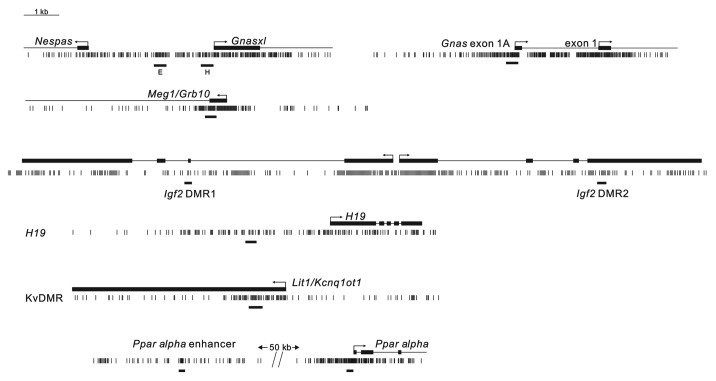
For the quantitative assessment of DMR methylation, we used the EpiTYPER system on the Sequenom platform. This method depends upon bisulphite conversion of DNA in which, as in conventional bisulphite sequencing, unmethylated cytosines (C) are converted to uracil (U) but methylated cytosines are resistant to conversion. Bisulphite treated DNA is subject to PCR amplification using primers that include T7 RNA promoter sequences. Amplification products are used as templates for in vitro transcription by T7 RNA polymerase, followed by digestion with RNase A, which cleaves at U and C corresponding to adenosine and guanine in the originating DNA strand. The methylation status of the input DNA will determine the presence of U or C in the RNase A cleaved fragments, affecting their mass, which is revealed with precision by mass spectrometry (MALDI-TOF). In addition, fragment peak height is a direct function of the absolute quantity in the sample, such that the relative heights of the corresponding “methylated” and “unmethylated” peaks provide a quantitative score of methylation of the corresponding “CpG unit.” The frequency of RNase A cleavage sites within the region analyzed is such that multiple CpG units can be scored; amplicons are typically designed to cover 300–500 bp. It should be noted, however, that this method does not necessarily provide information on all CpGs at single base resolution: some CpG units comprise multiple CpGs and some CpG units are outside of the effective size range for mass analysis. In addition, the assay provides a quantitative score for CpG unit methylation of the amplicon as a whole, not CpG methylation profiles of individual DNA strands, so it is not possible to discriminate methylation on maternal or paternal alleles by this method. Assays were designed for seven DMRs in four imprinted domains (Fig. 2). This included the maternal germline DMRs in the Gnas locus (the 1A DMR and two amplicons in the Nespas/Gnasxl DMR), in Grb10 and at Kcnq1ot1 (the KvDMR, which also regulates monoallelic expression of the Cdkn1c and Phlda2 genes), the paternal germline DMR at H19 (responsible for monoallelic expression of Igf2) and the paternally methylated, somatic DMRs in Igf2 (DMR1 and DMR2). Between 9 and 17 CpG units were scored for each amplicon, apart from the two, less CG-rich DMRs at Igf2, for which only three CpG units were assessed (Table S3). In addition, CpG islands at the Pparα promoter and an upstream enhancer31 were assayed, as well as B1 elements to provide a measure of global DNA methylation.
Figure 2. Representation of the location of the amplicons tested in relation to the DMRs within each imprinted domain. Each vertical line represents an individual CpG site. Thick bars above the CpG track represent exons of the indicated transcripts, with transcription start sites and direction of transcription indicated by arrows. The locations of the amplicons used for methylation analysis is given by the short bars under each DMR. In addition, the locations of the amplicons to assay methylation at the Pparα promoter and upstream enhancer are shown similarly.
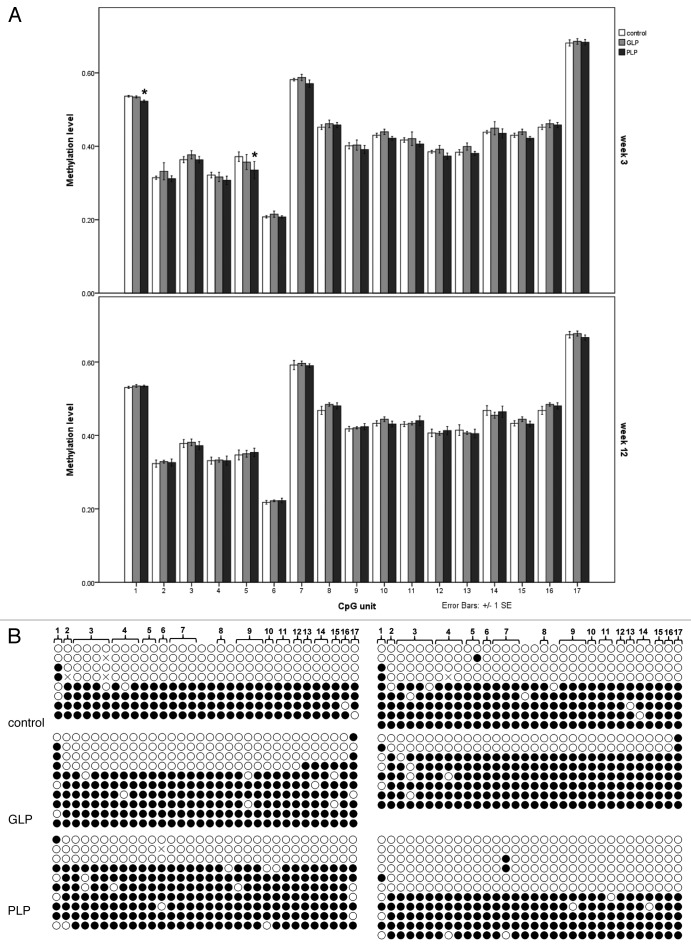
We found that the EpiTYPER method was quantitative and reproducible, but exhibited some idiosyncrasies that suggested that the methylation score does not necessarily correspond directly to the percentage methylation of given CpGs (or CpG units). This was apparent, for example, in analysis of the Gnas exon 1A DMR (Fig. 3). Although the mean methylation score across the amplicon as a whole was consistently between 42.1% and 44.2% (Table 1), close to the expected 50% for a faithfully maintained germline DMR, the individual methylation scores of the 17 CpG units analyzed ranged between 20% and 65% (Fig. 3A). However, the methylation profile was highly reproducible between samples (Fig. S1). Although it might be anticipated that the maternal allele is fully methylated and the paternal allele is fully unmethylated at this DMR, the pattern from EpiTYPER analysis could be interpreted to suggest that the CpG comprising CpG unit 6, for example, is substantially unmethylated also on the maternal allele and, conversely, the three CpGs corresponding to CpG unit 7 have a degree of methylation on the normally unmethylated paternal allele, because the methylation indices at these positions depart substantially and reproducibly from 50%. To test whether this was indeed the case, we cloned the corresponding PCR products for sequencing of individual clones. This revealed the expected distribution of near fully methylated and near fully unmethylated sequences that would be consistent with strict parental-allele-specific methylation levels (Fig. 3B), with no evidence for the skewed methylation level of the adjacent CpGs corresponding to CpG units 6 and 7 predicted by EpiTYPER. A similar analysis was conducted for the Grb10 DMR (data not shown) and here, again, a strong methylation peak predicted by EpiTYPER (CpG unit 2) was not found as a site of skewed methylation in cloned bisulphite products. Aside from the fact that the methylation profile reported by EpiTYPER does not appear to correspond to the absolute percentage methylation of individual CpGs, the strong sample-to-sample reproducibility (Fig. S1) suggests that it provides a reliable method to quantify methylation differences between samples.
Figure 3. Methylation analysis of the Gnas exon 1A DMR. (A) Methylation analysis by EpiTYPER of the DMR in control, GLP and PLP liver samples at week 3 (above) and week 12 (below) after birth. Methylation values on a scale 0.0 to 1.0 for each CpG unit that can be resolved by EpiTYPER are given. Values expressed are means and bars represent standard errors; n = 7–9. Where individual CpG units exhibited a significant difference in methylation level in treated group compared with control, * p < 0.05. (B) Bisulphite sequencing analysis of two representative samples from each of the control, GLP and PLP groups (week 3). Each row of circles represents a single, non-redundant bisulphite sequence clone. Individual CpGs are represented by circles, with open circles indicating unmethylated and filled circles methylated CpGs. The numbering at the top refers to the CpG units identified by EpITYPER analysis; note that some CpG units contain multiple CpGs (bracketed), while other CpGs are not scored by EpiTYPER analysis.
Table 1. Effect of maternal low protein diet on DMR methylation in offspring.
| DMR | DMR status | Week | Group |
||||
|---|---|---|---|---|---|---|---|
| Control | GLP-control | p value | PLP-control | p value | |||
|
Gnas (Exon 1A) |
Maternal germline |
Week 3 |
42.2 ± 0.99 |
1.62 ± 1.39 |
0.399 |
-0.13 ± 1.34 |
0.993 |
| Week 12 |
44.2 ± 0.88 |
-0.71 ± 1.32 |
0.812 |
-0.15 ± 0.99 |
0.990 |
||
|
Grb10 |
Maternal germline |
Week 3 |
34.05 ± 1.11 |
3.92 ± 1.46 |
0.014 |
-0.54 ± 1.39 |
0.895 |
| Week 12 |
47.6 ± 1.15 |
-2.91 ± 1.49 |
0.094 |
-1.98 ± 1.43 |
0.285 |
||
|
H19 |
Paternal germline |
Week 3 |
31.6 ± 1.56 |
-0.17 ± 2.58 |
0.997 |
0.85 ± 2.43 |
0.914 |
| Week 12 |
35.9 ± 1.70 |
-1.29 ± 2.63 |
0.844 |
-0.29 ± 2.4 |
0.990 |
||
|
Igf2 (DMR1) |
Paternal somatic |
Week 3 |
65.7 ± 0.55 |
0.22 ± 1.06 |
0.971 |
-2.21 ± 1.14 |
0.106 |
| Week 12 |
57. Four ± 0.74 |
-0.41 ± 0.86 |
0.849 |
0.52 ± 0.84 |
0.763 |
||
|
Igf 2 (DMR2) |
Paternal somatic |
Week 3 |
43.3 ± 1.47 |
0.66 ± 2.08 |
0.933 |
1.09 ± 2.34 |
0.862 |
| Week 12 |
36.7 ± 1.33 |
1.48 ± 1.6 |
0.541 |
0.32 ± 1.54 |
0. 966 |
||
|
Kcnq1ot1 (KvDMR) |
Maternal germline |
Week 3 |
36.1 ± 0.98 |
-1.35 ± 1.5 |
0.574 |
1.71 ± 0.46 |
0.458 |
| Week 12 |
41.0 ± 0.74 |
1.66 ± 1.16 |
0.262 |
2.19 ± 1.08 |
0.081 |
||
|
Gnas Nespas/Gnasxl (Amplicon E) |
Maternal germline |
Week 3 |
68.7 ± 1.6 |
4.28 ± 1.86 |
0.041 |
0.69 ± 1.75 |
0.890 |
| Week 12 |
64.4 ± 1.5 |
2.77 ± 2.3 |
0.388 |
3.63 ± 2.26 |
0.197 |
||
|
Gnas Nespas/Gnasxl (Amplicon H) |
Maternal germline |
Week 3 |
61.7 ± 0.86 |
-1.86 ± 1.39 |
0.310 |
1.51 ± 1.44 |
0.478 |
| Week 12 |
42.3 ± 0.56 |
1.20 ± 0.76 |
0.200 |
-1.56 ± 0.72 |
0.055 |
||
|
Pparα promoter |
Not applicable |
Week 3 |
6.08 ± 1.07 |
0.22 ± 1.86 |
0.990 |
0.55 ± 1.9 |
0.938 |
| Week 12 |
6.38 ± 1.27 |
-0.17 ± 1.71 |
0.993 |
-0.57 ± 1.7 |
0.916 |
||
|
Pparα enhancer |
Not applicable |
Week 3 |
60.3 ± 0.86 |
0.4 ± 1.2 |
0.633 |
3.74 ± 1.6 |
0.048 |
| Week 12 |
45.3 ± 1.95 |
-3.05 ± 2.1 |
0.243 |
1.18 ± 2.3 |
0.799 |
||
| B1 repetitive elements |
Not applicable | Week 3 |
40.8 ± 6.27 |
-1.02 ± 9.18 |
0.991 |
4.19 ± 9.34 |
0.867 |
| Week 12 | 40.4 ± 7.95 | -1.52 ± 12.4 | 0.989 | -5.11 ± 11.5 | 0.868 | ||
Overall DMR methylation levels in mice at week 3 and 12 of age born to dams fed a low protein diet during gestation (GLP) or lactation (PLP). The columns GLP-control and PLP-control give the difference in absolute methylation percentage between the treated groups and control group. n = 4–10 per group; data are expressed as means ± standard error. p-values were obtained from Dunnett’s post-hoc tests following 1-way ANOVAs.
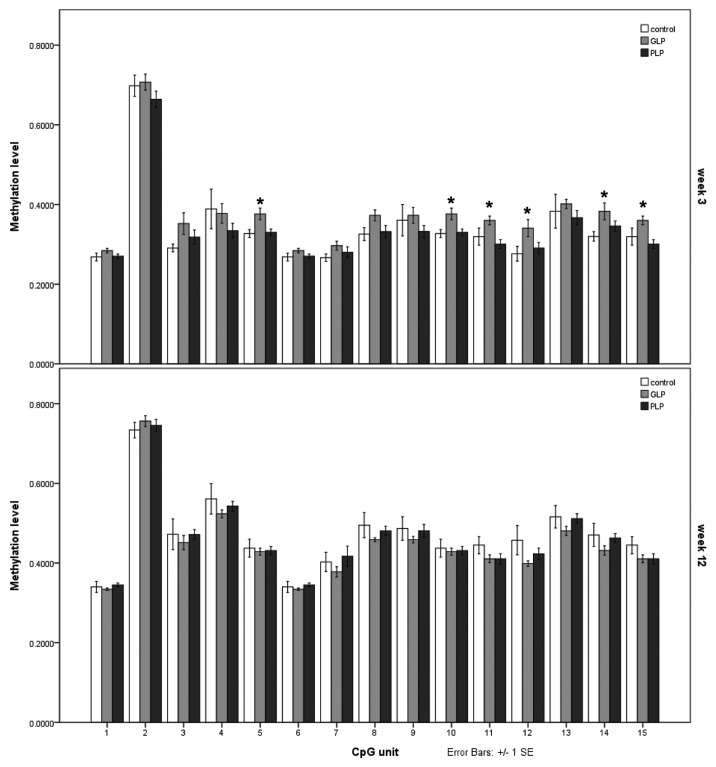
To quantify methylation across the DMRs and at the other loci tested using EpiTYPER, in the first place methylation levels for each CpG unit were averaged per group and the total methylation level of the DMR obtained as the mean of the means. The data are summarized in Table 1. Considering methylation status at weaning (week 3), most of the DMRs were not significantly altered in either GLP or PLP groups compared with controls. This applies to maternal germline DMRs (Gnas exon 1A and the KvDMR), a paternal germline DMR (H19 DMD) as well as to somatically acquired DMRs (Igf2 DMR1 and DMR2). In the GLP group (offspring from dams protein restricted during gestation) we observed an increase in methylation in the Grb10 maternal germline DMR (34.05 ± 1.11 in control compared with 37.97 ± 1.46 in the PLP group; p = 0.014; Table 1). However, at the CpG-unit level, the difference was significant, or only reached borderline significance, at six of the 15 CpG units scored (Fig. 4). Similarly, there was an increase in methylation in the maternal germline DMR at Nespas/Gnasxl amplicon E (control = 68.7 ± 1.60, PLP = 72.98 ± 1.86; p = 0.041; Table 1). However, only three of 16 CpG units in this amplicon had a significant difference (data not shown), and the overall methylation level of a second amplicon tested in the Nespas/Gnasxl DMR, amplicon H, showed no significant difference between groups. When tested at week 12, no significant differences between the groups were detected at any of the DMRs (Table 1). There was no evidence for greater variation in DMR methylation levels in the GLP or PLP groups; nor was there a general increase in variation of DMR methylation between week 3 and week 12 (data not shown). We also assessed methylation of the promoter and upstream enhancer of Pparα, in view of the significantly altered expression detected in the PLP group both at week 3 and 12. Methylation of the promoter was low and not significantly altered in either experimental group at either time point (Table 1). On the other hand, there was an overall increase in methylation of the upstream enhancer region of Pparα at week 3 in the PLP group (from 60.3 ± 0.86 to 64.0 ± 1.6; p = 0.048). The overall genome methylation level was not found to be affected in offspring by maternal diet as assessed by methylation of B1 repetitive elements (Table 1).
Figure 4. Methylation analysis of the Grb10 DMR. EpiTYPER analysis of the DMR in control, GLP and PLP liver samples at week 3 (above) and week 12 (below) after birth. Methylation values for each CpG unit that can be resolved by EpiTYPER are given. Values expressed are means and bars represent standard errors. n = 7–9; *p ≤ 0.055 compared with control.
Discussion
In the current study, we investigated germline and somatic DMR methylation and imprinted gene transcript levels in offspring of female mice that had experienced a low protein diet during gestation (GLP) or the early postnatal period. Early nutritional deficits can potentially affect DNA methylation of the developing organism through one-carbon mechanism, which provides the methyl groups for all biological methylation reactions and depends on dietary methyl donors. Epigenetic marks could be particularly vulnerable during early stages of embryonic development, which is a critical period for their establishment and maintenance.32 Despite a number of reports concerning the effects of parental or early life dietary status on DNA methylation,26,27,33-39 it is still unclear the extent to which manipulating maternal diet can lead to substantial and sustained methylation changes in offspring and what the underlying mechanisms might be.
We investigated DMR methylation at five imprinted loci and found that dietary protein deficiency either during gestation or the early postnatal period did not have a substantial impact on the methylation levels of imprinted gene DMRs in the liver of offspring. It is possible that there are tissue-specific effects on the methylation of one or more of the DMRs, but our results indicate that there are no constitutive effects in response to major maternal nutritional deprivation. Therefore, DMRs do not appear to be especially sensitive to early dietary manipulations, and it might be that mechanisms that faithfully maintain methylation at DMRs have evolved to help ensure that imprinted genes are normally regulated within quite narrow boundaries and that imprinted genes are not especially plastic to nutritional or environmental states. We did observe slight gain of methylation in the Grb10 and Nespas/Gnasxl DMRs in 3-week old offspring from females protein-restricted during gestation, but the changes affected a limited number of CpGs, were not present throughout the DMR in the case of Nespas/Gnasxl, and were not sustained into adulthood. Such transient changes in methylation could be explained in a variety of ways. If gain of methylation on the normally unmethylated allele was mosaic, it might be of insufficient density to ensure maintenance by DNA methyltransferase 1 (Dnmt1) at DNA replication, or insufficient to recruit repressive complexes that in turn recruit DNA methyltransferase activities. Alternatively, as the liver experiences continual replacement of cell populations, the cell populations with altered methylation might have been replaced by week 12 from progenitor cell pools in which DMR methylation was normal. The magnitude of the effects we observe are similar to the findings of Tobi et al.27 in blood samples from individuals from the Dutch Hunger Winter who were exposed to malnutrition in utero, as well as those from Cooper et al.28 who studied an African cohort receiving periconceptional micronutrient supplementation, or Kovacheva et al.,38 who report changes in Igf2 DMR2 methylation in response to altered maternal dietary choline. In each case, as with the current study, only isolated DMRs demonstrated altered methylation and the magnitude of change is small.
In contrast to the relatively small changes in DMR methylation, we detected more robust changes in expression of some imprinted genes as a consequence of maternal dietary protein restriction. Substantial upregulation of Igf2 mRNA in liver of pups on the day of birth without corresponding changes in DMR methylation have also been reported in a rat maternal low protein model model.39 We found a significant increase in Grb10 expression in liver of 3-week-old offspring from females experiencing protein restriction during lactation, but not in the GLP group in which offspring were assessed three weeks after the end of the maternal dietary restriction. Radford et al.40 also reported upregulation of Grb10 transcripts in liver of e16.5-old fetuses of dams experiencing caloric restriction late in gestation, suggesting that upregulation of Grb10 expression is a consistent, but transient, response to nutritional insufficiency. Grb10 is an adaptor protein that interacts with a number of receptor tyrosine kinases that regulate cell metabolism, development and growth.41 Analysis of mice deficient in Grb10 has demonstrated a role of Grb10 in attenuating insulin receptor signaling and in glucose metabolism in the liver around birth,42,43 so the effect we observe could be physiologically relevant, at least at that time, although Grb10 expression in liver is declining in the immediate postnatal period and does not persist to adulthood.43 If increased expression reflected a degree of loss of imprinting of Grb10 caused by altered methylation of its DMR, one would anticipate it to be accompanied by gain in methylation, as methylation is normally present on the expressed maternal allele and the unmethylated copy of the DMR is proposed to operate as a silencer.44,45 However, we did not detect significantly altered Grb10 DMR methylation in the PLP group, rather in the GLP group, showing that adaptive changes in imprinted gene expression can occur independently of changes in DMR methylation. For Gnas, we detected downregulation of expression in liver of 3-week as well as 12-week-old offspring from females experiencing gestational protein restriction. The stimulatory G-protein subunit Gsα, encoded by Gnas, mediates responses to glucagon and catecholamines in the liver, via the cAMP-potein kinase A pathway, including transcriptional activation of genes encoding gluconeogenetic enzymes. Liver-specific ablation of Gsα results in increased insulin sensitivity, increased hepatic glycogen storage and blunted induction of gluconeogenic enzymes but, paradoxically, little impairment of gluconeogenesis on fasting, probably owing to a compensatory increase in extra-hepatic sites of gluconeogenesis.46 Considering its role in glucose mobilization in fasting, the reduction in Gnas expression in livers of the GLP group might reflect a rebound effect in offspring that have come out of gestational deprivation and have experienced normal maternal diet during lactation. Gnas expression is normally biallelic in liver, so changes in expression would presumably be immune to altered methylation of the 1A DMR, which has major importance in regulating allelic expression of Gnas in tissues with imprinted expression.47,48 In this case, therefore, there are adaptive and sustained changes in imprinted gene expression independent of changes in DNA methylation. The notion that transcription factor dependent changes in imprinted gene expression rather than epigenetic deregulation occur in developmental programming paradigms was also noted by Radford et al.,40 in their study of offspring from calorie-restricted dams, in which they found altered expression of the imprinted gene Peg3 in liver of offspring with no detectable effect on the methylation level of the Peg3 DMR. Similarly, the upregulation of Igf2 mRNA detected in liver at birth39 without corresponding effects on DMR methylation noted above could reflect a transcriptional response to altered maternal physiology at a time that gene expression patterns in the liver are being substantially reprogrammed as the pup adapts to the transition from placental supply of nutrients to gastric supply of milk, rather than a specific deregulation of Igf2 imprinting.
Offspring of female rats fed a protein-deficient diet have been reported as having decreased methylation at the Pparα promoter. Pparα is a transcription factor involved in the regulation of numerous metabolic processes and abundantly expressed in the liver. The absolute difference in methylation reported was relatively small, but was associated with a large change in Pparα expression,33 and the observed reduction in methylation was transmitted from F1 to F2 generations.49 In our study, we did not detect any significant changes in Pparα promoter methylation at week 3 or 12 of postnatal development in either GLP or PLP groups compared with controls; however, Pparα gene expression was affected. Postnatal maternal protein restriction was associated with upregulation of Pparα transcripts at week 3 in offspring. This could be explained by the physiological state of mice rather than changes in promoter methylation, as it is known that Pparα expression is activated by fasting.50 Conversely, Pparα expression was found to be downregulated at postnatal week 12 in the same group but, again, this was not associated with any changes in promoter methylation. In addition to effects at the promoter, altered methylation of an enhancer element for Pparα in liver of offspring of male mice subjected to a protein-restricted diet has been reported.31 However, although we detected increased methylation of this element at weaning in the PLP group, the gain in methylation was small, correlated unexpectedly with increased transcript levels and was not sustained to the later time point when Pparα expression is downregulated, again indicating a disconnect between methylation and expression.
Altered gene expression, including imprinted genes, in the offspring of diet manipulated females could be brought about and perpetuated by epigenetic mechanisms other than DNA methylation. For example, maternal protein restriction results in reduced expression of the Hnf4a gene in pancreatic islets of adult rat offspring. This change is accompanied by altered levels of histone modifications, e.g., a reduction in histone H3 lysine-4 monomethylation (H3K4me1) and elevation of the repressive modification H3K9me2 at an enhancer element, and by a reduced interaction of this enhancer with the pancreatic-specific promoter.36 In another model of developmental programming, intrauterine growth restriction induced by uterine artery ligation in pregnant rats is associated with reduced expression of the Pdx1 gene in islets, also accompanied by altered histone modification state.37 In the growth restricted fetuses, there is increased binding of histone deacetylases and reductions in H3 and H4 acetylation at the Pdx1 promoter, effects on H3K4me3 and H3K9me2 are detected after birth, and only in adults does the promoter become DNA methylated. Despite these important observations, the mechanisms by which early life experiences program longer-term changes in gene expression, and whether the observed epigenetic modifications by themselves are able to sustain altered expression of the associated genes in the absence of ongoing, perhaps unidentified, physiological signals, are still unclear. Our study, together with other recent results,40 does not single out imprinted genes and relaxation of imprinting as particular purveyors of developmental programming. The challenges for future work are to identify the biochemical signals linking nutritional deficiency and epigenetic changes and the degree to which epigenetic outcomes are selected or global.
Materials and Methods
Animals and tissues
The animal model used in this study has been described previously.51 Mice (C57BL/6J) were bred at a designated animal unit (University or Cambridge) following approval from the Local Ethics Committee and in accordance with the Home Office Animals (Scientific Procedures) Act 1986. Adult females were individually housed and maintained at 22°C on a 12h:12h light:dark cycle. At six weeks of age mice were mated and pregnancy established by visual appearance of a vaginal plug. Pregnant females were divided into two groups with the first fed ad libitum (20% protein) and the second maintained on isocaloric low protein diet (8%) during gestation. Both diets had identical contents of methyl donor folic acid and choline. To establish the final experimental groups, pups were cross-fostered at three days of age, as follows. The first experimental group comprised pups born and nursed by normal fed dams (control group); the second group were pups born from ad libitum mothers and nursed by females fed low protein diet during lactation (postnatal low protein group, PLP); the third group were pups born from protein restricted dams and nursed by control dams (recuperated or gestational low protein group, GLP). Effects in offspring of prenatal or postnatal maternal dietary protein restriction were studied at 21 d after birth (week 3, weaning) and 12 weeks after birth. Prior to tissue collection, food was withdrawn overnight from mice. Liver samples were snap frozen in liquid nitrogen and stored at -80°C before processing. Experimental groups for all analysis reported here were 8–9 animals from separate litters.
Real-time quantitative RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen). Total RNA, 2 µg, was reverse-transcribed using SuperScript II (Invitrogen). Real-time quantitative PCR (qPCR) was performed using a Biorad CFX-96 system, according to standard protocols, using SYBR Green Master Mix (Applied Biosystems). PCR reactions (12 µl) contained 2 µl of 1:20 diluted cDNA and each sample was assessed in triplicate. Expression levels were calculated by the standard curve method52 using three housekeeping genes (β-actin, β-microglobulin and Gapdh) for normalization. Primer sequences for Gnas, Igf2, Grb10, Cdkn1c, Phlda2 and Pparα transcripts, as well as the housekeeping genes, are listed in Table S4.
DNA methylation analysis
Genomic DNA was isolated from liver using All Prep DNA/RNA kits (Invitrogen). DNA, 1 µg, was bisulphite converted using the EZ-DNA Methylation Gold kit (Zymo Research). DNA methylation at the indicated loci was measured using a mass-spectrometry based method (EpiTYPER, Sequenom). Pre-analysis sample treatment was performed according to the manufacturer’s specifications. After conversion, DNA was eluted in 50 µl and PCR was performed on 20 ng of bisulphite treated DNA. PCR primers specific for bisulphite converted DNA for the DMRs in the imprinted loci Gnas (Nespas/Gnasxl and Exon 1A DMRs), Igf2 (DMR1 and 2), Grb10, H19 and Kcnq1ot1 (KvDMR), as well as regions of the Pparα promoter and enhancer, and B1 repetitive elements are listed in Table S3. All samples were analyzed in duplicate, and all experimental and control samples for a given time point were analyzed on a single plate. Data quality filtering included removal of CpG units from analysis for which the measurement success rate was below 90%, which was typically caused by low or high masses of fragments. For methylation analysis the difference in methylation score for each CpG unit between technical replicates was expressed in percentages, the mean technical variability (difference) and confidence intervals were calculated for each amplicon, and replicates between which the difference was above the upper boundary were removed from the analysis. Selected amplicons were also analyzed by conventional bisulphite sequencing and cloning, as described in.53 Amplicons were cloned into the vector pGEM-T Easy (Promega) and 12 clones per sample were sequenced. Analysis of sequences was done using BiQ Analyzer.54 Sequences with < 90% conversion based on conversion rate of non-CpGs and duplicate sequences based on identical patterns of non-CpG non-conversion events were excluded.
Statistical analysis
All data are expressed as means ± standard error. The effects of maternal diet manipulation on gene expression and DNA methylation were assessed by one-way ANOVA or two-way ANOVA followed by Dunnet’s post-hoc tests, performed with PASW Statistics v.18.
Supplementary Material
Acknowledgments
Research in Gavin Kelsey’s group is supported by the Biotechnology and Biological Sciences Research Council and the Medical Research Council of the United Kingdom and by the European Union. This work in Susan Ozanne’s group was supported by the British Heart Foundation (Senior Fellowship to SEO) and the Biotechnology and Biological Sciences Research Council (reference B/E002161/1). We should like to thank Christel Krueger for advice with the Sequenom MassARRAY system, and Shinichi Tomizawa for advice on DMRs. We also acknowledge the expert technical assistance of Adrian Wayman and Delia Hawkes.
Glossary
Abbreviations:
- ANOVA
analysis of variance
- cAMP
cyclic adenosine monophosphate
- Cdkn1c
cyclin-dependent kinase inhibitor 1c gene
- CpG
dinucleotide CG
- DMR
differentially methylated region
- Dnmt1
DNA methyltransferase 1
- Gapdh
glyceraldehyde-3-phosphate dehydrogenase gene
- GLP
maternal gestational low protein diet
- Gnas
guanine nucleotide binding protein, alpha stimulating, complex locus
- Grb10
growth factor receptor-bound protein 10 gene
- H19
H19 gene
- H3K4me1
histone H3 monomethylated at lysine 4
- H3K4me3
histone H3 trimethylated at lysine 4
- H3K9me2
histone H3 dimethylated at lysine 9
- Hnf4a
hepatic nuclear factor 4 alpha gene
- ICR
imprinting control region
- Igf2
insulin-like growth factor II gene
- Kcnq1ot1
Kcnq1 opposite strand/antisense transcript 1 (non-protein coding) gene
- KvDMR
DMR in the Kcnq1ot1 locus
- MALDI-TOF
matrix-assisted laser desorption/ionization-time of flight
- Nespas/Gnasxl
neuroendocrine secretory protein antisense transcript/Gnas extra large DMR
- PCR
polymerase chain reaction
- Pdx1
pancreatic and duodenal homeobox 1 gene
- Peg3
paternally expressed gene-3
- Phlda2
pleckstrin homology-like domain, family A, member 2 gene
- PLP
maternal postnatal low protein diet
- Pparα
peroxisome proliferator-activated receptor α
- RT-qPCR
reverse transcriptase-quantitative PCR
- Tet
ten-eleven translocation protein
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/22141
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/22141
References
- 1.Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br Med Bull. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Ozanne SE. Metabolic programming in animals. Br Med Bull. 2001;60:143–52. doi: 10.1093/bmb/60.1.143. [DOI] [PubMed] [Google Scholar]
- 3.Hochberg Z, Feil R, Constancia M, Fraga M, Junien C, Carel JC, et al. Child health, developmental plasticity, and epigenetic programming. Endocr Rev. 2011;32:159–224. doi: 10.1210/er.2009-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabory A, Attig L, Junien C. Developmental programming and epigenetics. Am J Clin Nutr. 2011;94(Suppl):1943S–52S. doi: 10.3945/ajcn.110.000927. [DOI] [PubMed] [Google Scholar]
- 5.Ghoshal K, Li X, Datta J, Bai S, Pogribny I, Pogribny M, et al. A folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. J Nutr. 2006;136:1522–7. doi: 10.1093/jn/136.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pogribny IP, Karpf AR, James SR, Melnyk S, Han T, Tryndyak VP. Epigenetic alterations in the brains of Fisher 344 rats induced by long-term administration of folate/methyl-deficient diet. Brain Res. 2008;1237:25–34. doi: 10.1016/j.brainres.2008.07.077. [DOI] [PubMed] [Google Scholar]
- 7.Nohara K, Baba T, Murai H, Kobayashi Y, Suzuki T, Tateishi Y, et al. Global DNA methylation in the mouse liver is affected by methyl deficiency and arsenic in a sex-dependent manner. Arch Toxicol. 2011;85:653–61. doi: 10.1007/s00204-010-0611-z. [DOI] [PubMed] [Google Scholar]
- 8.Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr. 2012;3:21–38. doi: 10.3945/an.111.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12:321–7. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Junien C, Nathanielsz P. Report on the IASO Stock Conference 2006: early and lifelong environmental epigenomic programming of metabolic syndrome, obesity and type II diabetes. Obes Rev. 2007;8:487–502. doi: 10.1111/j.1467-789X.2007.00371.x. [DOI] [PubMed] [Google Scholar]
- 11.Curley JP, Mashoodh R. Parent-of-origin and trans-generational germline influences on behavioral development: the interacting roles of mothers, fathers, and grandparents. Dev Psychobiol. 2010;52:312–30. doi: 10.1002/dev.20430. [DOI] [PubMed] [Google Scholar]
- 12.Keverne EB. Epigenetically regulated imprinted genes and foetal programming. Neurotox Res. 2010;18:386–92. doi: 10.1007/s12640-010-9169-z. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–75. doi: 10.1038/nrg3032. [Erratum in: Nat Rev Genet 2011;12:663] [DOI] [PubMed] [Google Scholar]
- 14.Arnaud P. Genomic imprinting in germ cells: imprints are under control. Reproduction. 2010;140:411–23. doi: 10.1530/REP-10-0173. [DOI] [PubMed] [Google Scholar]
- 15.Kelsey G, Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Phil Trans Royal Society B. Biol Sciences. doi: 10.1098/rstb.2011.0336. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandovici I, Leppert M, Hawk PR, Suarez A, Linares Y, Sapienza C. Familial aggregation of abnormal methylation of parental alleles at the IGF2/H19 and IGF2R differentially methylated regions. Hum Mol Genet. 2003;12:1569–78. doi: 10.1093/hmg/ddg167. [DOI] [PubMed] [Google Scholar]
- 17.Woodfine K, Huddleston JE, Murrell A. Quantitative analysis of DNA methylation at all human imprinted regions reveals preservation of epigenetic stability in adult somatic tissue. Epigenetics Chromatin. 2011;4:1. doi: 10.1186/1756-8935-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy SK, Huang Z, Hoyo C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS One. 2012;7:e40924. doi: 10.1371/journal.pone.0040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barlow DP. Genomic imprinting: a mammalian epigenetic discovery model. Annu Rev Genet. 2011;45:379–403. doi: 10.1146/annurev-genet-110410-132459. [DOI] [PubMed] [Google Scholar]
- 20.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3:1–18. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frontera M, Dickins B, Plagge A, Kelsey G. Imprinted genes, postnatal adaptations and enduring effects on energy homeostasis. Adv Exp Med Biol. 2008;626:41–61. doi: 10.1007/978-0-387-77576-0_4. [DOI] [PubMed] [Google Scholar]
- 22.Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes. 2007;14:3–12. doi: 10.1097/MED.0b013e328013daa2. [DOI] [PubMed] [Google Scholar]
- 23.Charalambous M, Ferron SR, da Rocha ST, Murray AJ, Rowland T, Ito M, et al. Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 2012;15:209–21. doi: 10.1016/j.cmet.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Rebollo E, Maeda A, Reyes M, Turan S, Fröhlich LF, Plagge A, et al. Loss of XLαs (extra-large αs) imprinting results in early postnatal hypoglycemia and lethality in a mouse model of pseudohypoparathyroidism Ib. Proc Natl Acad Sci U S A. 2012;109:6638–43. doi: 10.1073/pnas.1117608109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tucker KL, Beard C, Dausmann J, Jackson-Grusby L, Laird PW, Lei H, et al. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 1996;10:1008–20. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- 26.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–53. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper WN, Khulan B, Owens S, Elks CE, Seidel V, Prentice AM, et al. DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: results of a pilot randomized controlled trial. FASEB J. 2012;26:1782–90. doi: 10.1096/fj.11-192708. [DOI] [PubMed] [Google Scholar]
- 29.Snoeck A, Remacle C, Reusens B, Hoet JJ. Effect of a low protein diet during pregnancy on the fetal rat endocrine pancreas. Biol Neonate. 1990;57:107–18. doi: 10.1159/000243170. [DOI] [PubMed] [Google Scholar]
- 30.Metges CC. Early nutrition and later obesity: animal models provide insights into mechanisms. Adv Exp Med Biol. 2009;646:105–12. doi: 10.1007/978-1-4020-9173-5_11. [DOI] [PubMed] [Google Scholar]
- 31.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–96. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 33.Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br J Nutr. 2008;100:278–82. doi: 10.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–6. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 35.van Straten EM, Bloks VW, Huijkman NC, Baller JF, van Meer H, Lütjohann D, et al. The liver X-receptor gene promoter is hypermethylated in a mouse model of prenatal protein restriction. Am J Physiol Regul Integr Comp Physiol. 2010;298:R275–82. doi: 10.1152/ajpregu.00413.2009. [DOI] [PubMed] [Google Scholar]
- 36.Sandovici I, Smith NH, Nitert MD, Ackers-Johnson M, Uribe-Lewis S, Ito Y, et al. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci U S A. 2011;108:5449–54. doi: 10.1073/pnas.1019007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–24. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovacheva VP, Mellott TJ, Davison JM, Wagner N, Lopez-Coviella I, Schnitzler AC, et al. Gestational choline deficiency causes global and Igf2 gene DNA hypermethylation by up-regulation of Dnmt1 expression. J Biol Chem. 2007;282:31777–88. doi: 10.1074/jbc.M705539200. [DOI] [PubMed] [Google Scholar]
- 39.Gong L, Pan YX, Chen H. Gestational low protein diet in the rat mediates Igf2 gene expression in male offspring via altered hepatic DNA methylation. Epigenetics. 2010;5:619–26. doi: 10.4161/epi.5.7.12882. [DOI] [PubMed] [Google Scholar]
- 40.Radford EJ, Isganaitis E, Jimenez-Chillaron J, Schroeder J, Molla M, Andrews S, et al. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS Genet. 2012;8:e1002605. doi: 10.1371/journal.pgen.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen H, Svensson U, Zhu J, Laviola L, Giorgino F, Wolf G, et al. Interaction between the Grb10 SH2 domain and the insulin receptor carboxyl terminus. J Biol Chem. 1996;271:8882–6. doi: 10.1074/jbc.271.15.8882. [DOI] [PubMed] [Google Scholar]
- 42.Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc Natl Acad Sci U S A. 2003;100:8292–7. doi: 10.1073/pnas.1532175100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith FM, Holt LJ, Garfield AS, Charalambous M, Koumanov F, Perry M, et al. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol Cell Biol. 2007;27:5871–86. doi: 10.1128/MCB.02087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnaud P, Monk D, Hitchins M, Gordon E, Dean W, Beechey CV, et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum Mol Genet. 2003;12:1005–19. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
- 45.Shiura H, Nakamura K, Hikichi T, Hino T, Oda K, Suzuki-Migishima R, et al. Paternal deletion of Meg1/Grb10 DMR causes maternalization of the Meg1/Grb10 cluster in mouse proximal Chromosome 11 leading to severe pre- and postnatal growth retardation. Hum Mol Genet. 2009;18:1424–38. doi: 10.1093/hmg/ddp049. [DOI] [PubMed] [Google Scholar]
- 46.Chen M, Gavrilova O, Zhao WQ, Nguyen A, Lorenzo J, Shen L, et al. Increased glucose tolerance and reduced adiposity in the absence of fasting hypoglycemia in mice with liver-specific Gs alpha deficiency. J Clin Invest. 2005;115:3217–27. doi: 10.1172/JCI24196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Chen M, Deng C, Bourc’his D, Nealon JG, Erlichman B, et al. Identification of the control region for tissue-specific imprinting of the stimulatory G protein alpha-subunit. Proc Natl Acad Sci U S A. 2005;102:5513–8. doi: 10.1073/pnas.0408262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williamson CM, Ball ST, Nottingham WT, Skinner JA, Plagge A, Turner MD, et al. A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat Genet. 2004;36:894–9. doi: 10.1038/ng1398. [DOI] [PubMed] [Google Scholar]
- 49.Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr. 2007;97:435–9. doi: 10.1017/S0007114507352392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–98. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen JH, Tarry-Adkins JL, Heppolette CA, Palmer DB, Ozanne SE. Early-life nutrition influences thymic growth in male mice that may be related to the regulation of longevity. Clin Sci (Lond) 2010;118:429–38. doi: 10.1042/CS20090429. [DOI] [PubMed] [Google Scholar]
- 52.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruf N, Bähring S, Galetzka D, Pliushch G, Luft FC, Nürnberg P, et al. Sequence-based bioinformatic prediction and QUASEP identify genomic imprinting of the KCNK9 potassium channel gene in mouse and human. Hum Mol Genet. 2007;16:2591–9. doi: 10.1093/hmg/ddm216. [DOI] [PubMed] [Google Scholar]
- 54.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–8. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.