Abstract
It has become appreciated over the last several years that protein phosphorylation within the cardiac mitochondrial matrix and respiratory complexes is extensive. Given the importance of oxidative phosphorylation and the balance of energy metabolism in the heart, the potential regulatory effect of these classical signaling events on mitochondrial function is of interest. However, the functional impact of protein phosphorylation and the kinase/phosphatase system responsible for it are relatively unknown. Exceptions include the well-characterized pyruvate dehydrogenase and branched chain α-ketoacid dehydrogenase regulatory system. The first task of this review is to update the current status of protein phosphorylation detection primarily in the matrix and evaluate evidence linking these events with enzymatic function or protein processing. To manage the scope of this effort, we have focused on the pathways involved in energy metabolism. The high sensitivity of modern methods of detecting protein phosphorylation and the low specificity of many kinases suggests that detection of protein phosphorylation sites without information on the mole fraction of phosphorylation is difficult to interpret, especially in metabolic enzymes, and is likely irrelevant to function. However, several systems including protein translocation, adenine nucleotide translocase, cytochrome c, and complex IV protein phosphorylation have been well correlated with enzymatic function along with the classical dehydrogenase systems. The second task is to review the current understanding of the kinase/phosphatase system within the matrix. Though it is clear that protein phosphorylation occurs within the matrix, based on 32P incorporation and quantitative mass spectrometry measures, the kinase/phosphatase system responsible for this process is ill-defined. An argument is presented that remnants of the much more labile bacterial protein phosphoryl transfer system may be present in the matrix and that the evaluation of this possibility will require the application of approaches developed for bacterial cell signaling to the mitochondria.
Keywords: kinase, phosphatase, citric acid cycle, oxidative phosphorylation, protein transport, mitochondria intermembrane space, phosphohistidine, adenine nucleotide translocase, pyruvate dehydrogenase, branched chain α-ketoacid dehydrogenase
this review article is part of a collection on Post-translational Protein Modification in Metabolic Stress. Other articles appearing in this collection, as well as a full archive of all Review collections, can be found online at http://ajpheart.physiology.org/.
Introduction
The heart is a tissue with one of the highest rates of energy conversion in the body, being critically dependent on mitochondrial oxidative phosphorylation as a major source of adenosine 5′-triphosphate (ATP). The remarkably fast turnover of ATP in the heart is in the order of 10 s at high workloads (12, 13), and a mismatch of just a few percent between the production and utilization rates can result in major disruptions in the potential energy available for function (12, 13). The dysregulation of the cardiac energy conversion process has been suggested to be one of the major elements in heart failure (144). Thus the precise orchestration of mitochondrial energy conversion is critical for normal heart function, though the mechanism for this regulatory process is still under investigation. Since enzyme protein phosphorylation [i.e., pyruvate dehydrogenase (PDH) and glycogen phosphorylase (GP)] was one of the first mechanism of modulating metabolic processes to match physiological needs, it was logical to further explore enzyme protein phosphorylation at other levels of energy metabolism of the heart. Therefore, the focus of the present article is to review the evidence, or lack thereof, supporting a role of protein phosphorylation in the regulation of mitochondrial oxidative phosphorylation. It should be pointed out that other post-translational modifications (PTMs) reviewed elsewhere, such as acetylation (125), S-nitrosylation (142), nitration (35), and glutathionylation (91), among others, have also been implicated in the regulation of mitochondrial metabolism.
Since the late 1960s, protein phosphorylation was implicated as one of the regulatory mechanisms balancing energy conversion with utilization in the heart. Protein phosphorylation of mitochondrial PDH, described almost simultaneously by Linn et al. (123) and Weiland and colleagues (219, 220), was one of the first examples of metabolic control by enzyme phosphorylation. This discovery and the previously described regulation of GP by protein phosphorylation (114) were two of the early examples of acute modulation of enzyme activity by protein phosphorylation events [see the early review (194)], and both of these reactions are important for cardiac energy conversion. PDH constitutes the major entry point of reducing equivalents from glycolysis to oxidative phosphorylation. Indeed, PDH molar activity via dephosphorylation increases with cardiac workload in the perfused heart (113, 132), consistent with its playing a role in the adaptation of metabolic conversion rates to cardiac workload. Similarly, glycogen breakdown, via GP, has been correlated with the early metabolic response to cardiac work transitions (72). Thus protein phosphorylation has already been demonstrated to play a key role in the regulation of cardiac energy metabolism by regulating the delivery of reducing equivalents to the citric acid cycle.
Many models of the regulation of cardiac mitochondrial oxidative phosphorylation suggest a distributed control of enzymatic activity (11, 237). Therefore, it is a reasonable extrapolation, based on PDH and GP regulation, to look first at mitochondrial protein phosphorylation events that might contribute to metabolic regulation. Today, due to the explosion of proteomic screening methods with remarkable sensitivity, the number of potentially important mitochondrial protein phosphorylation sites is staggering. It is also important to note that the original results on PDH and GP were derived by initially observing acute alterations in enzymatic activity in vitro and then determining the PTMs responsible. Because of the current explosion in phosphoprotein analysis, this proven and time-honored strategy has been generally reversed. That is, many PTMs are being constantly discovered in the mitochondrial proteome that are yet to be established as functionally significant, rather than mining for PTMs as sources of demonstrated alterations in extracted enzyme molar activities. This reversed strategy may be an inefficient approach for many reasons. First, many protein amino acids, and likely associated PTMs, are not conserved and apparently not critical for enzyme function. Thus PTMs on these amino acids, and even on some that are conserved, will likely not influence function if they are not in a critical region of the enzyme. In addition, the actual energetic cost of most cellular protein PTMs, assuming a slow turnover, is very low compared with the demands of differentiated function, such as muscle contraction, especially in active tissues like the heart. This energetic analysis suggests that the effect of neutral PTM events on the natural selection of PTM-generating systems (i.e., kinases) might be very low. Indeed, it is likely more “expensive” to “tune” a kinase for a specific set of active sites than to allow some neutral phosphorylation sites to be populated at negligible cost in energy or function. These neutral sites may also contribute to signaling variation when one of these sites becomes “active” during mutation alterations in protein sequence.
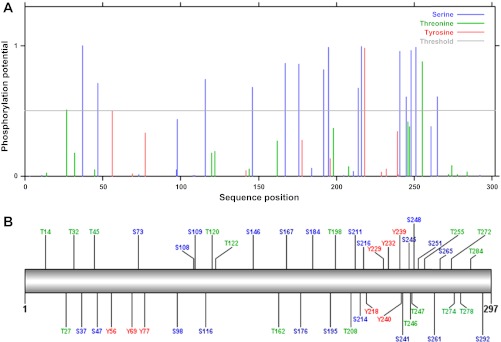
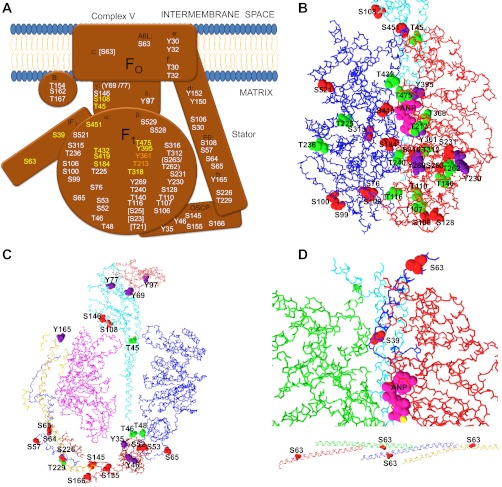
What is the absolute selectivity of known protein kinases? Several protein 32P phosphorylation screening methods [see KESTREL (111), for example] show that a given kinase catalyzes the phosphorylation of many proteins in cell extracts, consistent with a low absolute specificity. Indeed, as stated by the authors of KESTREL, a significant challenge in this type of study is deciphering the kinase phosphorylations that impact function versus neutral sites. Taking this concept further, several web-based tools (NETPHOS, NETPHOSK, and GPS) are available to predict the vulnerability of a given protein amino acid to kinase activity. These probabilities are based on a variety of criteria (22, 224, 226). Using the human mitochondrial complex V γ-subunit as an example (sp_P36542–2_ATPG_HUMAN), the GPS and NETPHOS predicted >46 phosphorylation sites (see Fig. 1) from numerous kinases based on the consensus recognition of these enzymes even using the highest threshold of detection. Similar results were obtained with NETPHOSK. These results support the notion that protein kinases are rather “nonspecific,” depending on what threshold is applied. In general, some threshold of “phosphorylation potential” is selected to determine the likelihood of phosphorylation by a given kinase. This is illustrated by the gray line in Fig. 1A. The issue with many of today's methods of detecting phosphorylated peptides is that no threshold, in terms of mole fraction or “occupancy,” is given for the protein phosphorylations reported. Indeed, in most cases the mole fraction of a phosphorylated peptide is simply not known. Thus one of the reasons for the explosion in detection of protein phosphorylation sites is that we are simply dropping the threshold for detection with our sensitive methods revealing potentially less specific and possibly neutral phosphorylation sites. This is analogous to bringing the gray line in Fig. 1A closer to zero as the methods get more sensitive detecting more of the “low specificity” sites of protein kinase activity.
Fig. 1.
Predicted phosphorylation sites in the human mitochondrial complex V γ-subunit. The P-Ser (green), P-Thr (blue), and P-Tyr (red) sites predicted by the NETPHOS (A) and GPS (B) program (224) on the human γ-subunit sequence (sp_P36542–2_ATPG_HUMAN). A: graphical presentation of phosphorylation potential as a function of position. Gray vertical line indicates a 0.5 potential. B: graphical presentation of GPS model with the “high” threshold. Agreement between methods was good; similar results were obtained with NETPHOSK.
The lack of absolute specificity of kinases together with the increased sensitivity of current phosphopeptide detection methods might explain the finding of a number of phosphorylation sites in the presequence of mitochondrial proteins, including NADH dehydrogenase ubiqinone flavoprotein (NDUF)S4 (208) and NDUFS8 (48, 94) of complex I, electron transfer flavoprotein (ETF) α-subunit (ETFA) (227), succinate dehydrogenase subunit A (flavoprotein) (SDHA) in complex II (151), ubiquinol-cytochrome c reductase binding protein (UQCRB) in complex III (24), and subunits-β (151) and c (217) of complex V. Some of these sites were found in isolated mitochondria, where the amount of preassembled precursors is very low compared with the mature and assembled proteins. These sites are likely irrelevant in terms of the function of the enzymatic complexes. However, it could be argued that these phosphorylation sites are important for protein import and, consequently, affect steady-state levels of mitochondrial complexes. Except for a few isolated cases, such as NDUFS4 at a site within the mature protein (54, 55) and several subunits of the translocase of the outer membrane (TOM) complex (170, 191), phosphorylation has not been correlated with protein import to mitochondria. An interesting example is the β-subunit of complex V, where a significant amount of phosphorylated precursor was detected when cAMP levels were increased by treatment of mouse lymphoma cells with isoproterenol (202). However, import was not altered by this PTM, and the phosphorylated precursor was either rapidly degraded or dephosphorylated before assembly into the mature complex. The conclusion of this study was that phosphorylation of the nascent subunit-β polypeptide by PKA in the cytosol was merely accidental and physiologically irrelevant, although tolerated. Results like this highlight that unspecific phosphorylation events do occur in experimental settings and not just in algorithm predictions, although not all sites found in silico will necessarily be found experimentally. To complicate matters further, very low levels of nonenzymatic phosphorylation of Tyr, Ser, and even Thr residues can be achieved when proteins are warmed in the presence of divalent cations (189), so the greater instrument sensitivity achieved requires additional precautions in sample preparation to avoid detection of deeply substoichiometric events that are completely unrelated to biology. Because of the nonquantitative nature of most phosphoprotein detection schemes, we rarely know the mole fraction of a phosphopeptide, so the extent of this problem is still unknown and a major concern of these authors.
In this review, we will attempt to initially provide an updated list of protein phosphorylation sites within the mitochondrial matrix enzymes involved in energy metabolism. These proteins are divided into membrane transporters, intermediary metabolism enzymes, and the complexes of oxidative phosphorylation. We also review the current understanding of the matrix kinase/phosphatase system. In some cases, because of the lack of information, data outside of the heart will be included. A critical evaluation will be provided of which of these modifications have clearly been linked to alterations in activity using the criteria outlined below. In addition, we will attempt to characterize the sites with regard to the probability that they may influence function based on our understanding of the molecular mechanisms within the oxidative phosphorylation complexes. Since this exciting field is moving very quickly, we apologize in advance for any literature omissions in this survey that we are sure will occur at our error.
The criteria for establishing the functional impact of a protein phosphorylation event are highly variable depending on the genetic control of the system, as well as the cellular tools available to modify the protein and the cellular physiology. In general, the simple correlation of protein phosphorylation with enzymatic activity in vivo and/or in vitro is the first line of evidence and is an absolute requirement as an initial step. Ideally, these initial correlations should be accomplished with physiological manipulations that reflect the operation of the heart in vivo. Another important element is to demonstrate that a significant mole fraction of the enzyme is affected by protein phosphorylation. This is particularly important when dealing with cardiac energy conversion enzymes since at “resting” metabolic rates most of the enzymes are present in excess and only approach Vmax conditions at maximum workloads (140, 163). Thus, to significantly impact energy metabolism along the energy conversion pathway, a significant fraction of the enzyme must be modified to have an impact on the metabolic rate. That is, if only 2% of a given complex subunit is phosphorylated, this will have little or no impact on the net flux through the system where flux alterations approach 500% from rest to maximum workloads. Naturally, any site with a 2% PTM mole fraction could grow to 100% under different experimental conditions. Thus low mole fraction PTMs do not rule out a site as being important under all conditions, just under the condition evaluated. Indeed, detection of the phosphorylation site confirms the presence of a potent regulating kinase. The importance of PTM mole fraction is not as critical for signaling molecules, such as kinases, acetylases, deacetylases, phosphatases, or ion channels, where the modification of a few copies can influence numerous downstream events or the membrane potential. This requirement is often overlooked when using high-sensitivity methods for detecting phosphorylated proteins, including phosphopeptide enhancement mass spectrometry (MS) (24, 58, 121, 165, 236), radiolabeling (6, 29, 128), and high-sensitivity MS (156, 175), all of which permit the detection of a few copies of phosphorylated peptides in the enzyme protein pool. Another line of evidence for the regulatory role of a protein phosphorylation site may include changes in enzyme activity upon specific amino acid substitution that simulate or eliminate the PTM (1, 101, 159). Though this might be a powerful approach in some circumstances, the substitution frequently leads to false-positive results because of changes in protein structure or assembly induced by the amino acid substitution that are seductive but could alter activity in a manner that does not actually occur upon phosphorylation of the wild-type residue. Finally, direct structural simulation studies demonstrating alteration of enzyme active sites or other critical structural parameters by phosphorylation provide the most reliable and definite confirmation of the functional relevance. Regrettably, as this review will illustrate, the vast majority of protein phosphorylation sites detected in the mitochondrial matrix proteome, which increase in number by the month, have not even been correlated with enzymatic activity, limiting the evaluation of their relevance to the examination of available protein structures to obtain a glimpse of which sites could in theory affect function.
A particular phosphorylation site can be safely assumed to have a potential impact in enzymatic function if it is located in the active site and if phosphorylation would sterically hinder the binding of substrates or interact with residues that participate in catalysis. The same can be said of sites that are in contact with a redox group such as a heme, an iron sulfur center, a copper atom, or a flavin molecule and that should significantly decrease the midpoint potential of the cofactor by the presence of a negative charge upon phosphorylation. However, it is more difficult to predict the impact of phosphorylation at sites that are farther away from a redox group, given that conformational changes can have a large role in determining the electrochemical properties of the cofactor (97). In this regard, experiments in which charges have been introduced by mutagenesis at different distances from a redox group suggest that electrostatic interactions become greatly attenuated at distances larger than ∼10 Å because of the high dielectric constant in the interior of most proteins (124). The potential functional role of phosphorylation sites found on the interface of subunits within a protein complex cannot be generalized, for it would depend on the particular enzyme, but it can be assumed that when movement is required at subunit interfaces during the catalytic cycle, one or more phosphorylation events could potentially block activity. Phosphorylation sites close to the entry or exit points for protons in respiratory enzymes are also potentially relevant if they can block access to protons channels or affect the pKa of residues that are sequentially protonated and deprotonated to facilitate proton movement (135). We will attempt to point out phosphorylation sites that have been found in heart or at least in muscle, taking into consideration what is known about the particular mechanism of each enzymatic complex and its structure even when there is an absence of functional data. We omit from the discussion sites that have been reported in other tissues, unless they are located in a structurally relevant region of the protein or have been associated with kinases or modifications of enzymatic activity. A significant number of sites have been reported in cancer cell lines or tumors (33, 37, 39, 61, 73, 95, 107, 131, 141, 150, 151, 176, 223, 231) but are not discussed further in this review under the assumption that metabolic regulation in these cells is in disarray and is therefore largely unrelated to the physiology of differentiated tissues like the heart.
Phosphorylation of Inner Membrane Transporters
The critical communication between the mitochondrion and the cytosol across the outer mitochondrial membrane and the remarkably high-resistance inner membrane is dependent on numerous transporters. Thus modulation of these transporters via protein phosphorylation could dramatically affect the ability of the cytosol to influence mitochondrial reaction pathways via the exchange of metabolites and signaling molecules, as well as proteins.
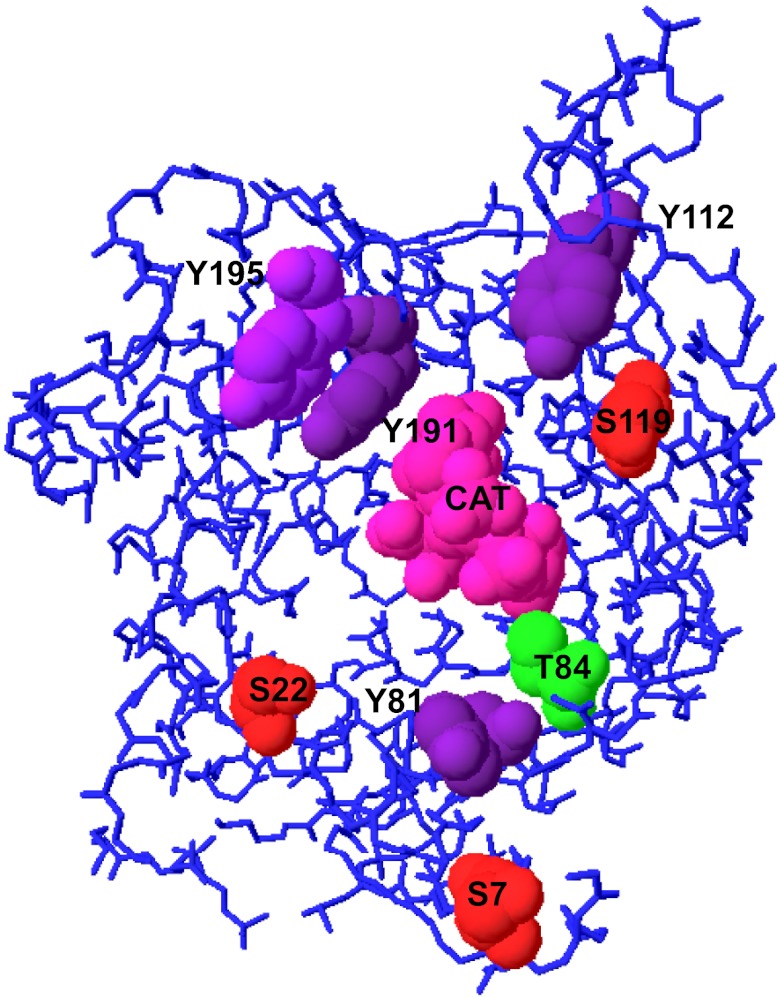
The ADP/ATP translocase (ANT) performs the exchange of cytosolic ADP with matrix ATP generated by oxidative phosphorylation (109). Phosphorylation sites have been found in all of the isoforms of ANT by numerous investigators. These sites include, in ANT-1, Ser7 (24, 236), Ser22 (24, 236), Tyr81 (24), Thr84 (24), Tyr191 (236), and Tyr195 (24, 67); in ANT-2, Thr84 (68, 236), Tyr191 (68), and Tyr195 (68); and in ANT-3 (236), Tyr112, Ser119, and Tyr195. All these phosphorylation sites are shown in Fig. 2. ANT of cardiac mitochondria from pig and rat was phosphorylated with γ-[32P]ATP in blue native gels, suggesting the possibility of autophosphorylation activity (161), although the specific sites are unknown. A series of studies by Feng et al. (67, 68) have confirmed the phosphorylation of Tyr191 and Tyr195 on the H4 helix of ANT-1 (158), showing that phosphorylation is required for maximal translocase function. In addition, Src kinases are capable of phosphorylating ANT-1 in vitro, whereas phosphorylation in intact cells was found to be blocked with 4-amino-5-(4-chloro-phenyl)-7-(t-butyl)pyrazolol[3,4,d]pyrimidine (PP2), a specific inhibitor of Src kinases. Several phenotypic changes in cells, including the inhibition of oxidative growth in yeast (68), are induced by genetically modifying these phosphorylation sites, and Tyr phosphorylation of ANT-1 has also been correlated with preconditioning in the heart (68). The Tyr phosphorylation sites on the H4 helix are exposed at the outer surface of the inner membrane (158), where intermembrane space Src kinases could phosphorylate these sites without requiring translocation of these kinases into the matrix. The one missing piece of evidence in this extensive series of studies is a measure of the mole fraction of phosphorylated ANT-1. To our knowledge, these sites have only been detected by MS after phosphopeptide enrichment or with very sensitive antibodies and radio labeling. This might imply that only a small fraction of ANT-1 is phosphorylated and thus active, which could contribute to the rate limitation of oxidative phosphorylation by this transporter, although this latter point is still the subject of controversy. A determination of the mole fraction of ANT-1 phosphorylated at Tyr191 and -195 would be extremely useful in characterizing the impact of regulation by PTMs under physiological conditions. However, Boja et al. (24) found essentially no change in Tyr196 (Tyr195 in human) phosphorylation in pig isolated cardiac mitochondria after treatment with dichloroacetate (DCA) or upon Ca2+ activation, with a statistically insignificant increase of only 20% when mitochondria were deenergized. Since these studies were conducted on isolated mitochondria, the recruitment of cytosolic kinases, such as the Src kinases, could not occur and may influence these negative results.
Fig. 2.
Reported sites of phosphorylation in the ADP/ATP translocase (ANT). The structure of the bovine ANT-1 (PDB ID 1OKC) crystallized in the presence of carboxyatractyloside (CAT) is shown looking down from the intermembrane space to the central cavity. Only the side chains of those residues reported to be sites of phosphorylation are shown space-filled on the protein Cα backbone with colors representing each type of amino acid (Ser, red; Thr, green; Tyr, purple).
Another inner membrane transport protein that is believed to play a role in the maintenance of the total adenosine pool of the matrix is the ATP-Mg/Pi carrier protein (7). This transporter is phosphorylated at Tyr324 (68), but no functional data have been reported for this site. Metabolites for intermediary metabolism also need to be transported across the inner membrane. In a very recent high-sensitivity screen by Zhao et al. (236), the aspartate-glutamate transporter Aralar 1 was found phosphorylated at Ser662 and -101, whereas the 2-oxoglutarate/malate carrier was phosphorylated at several sites (Ser6, Tyr102, Tyr202, and Ser203). Boja et al. (24) also found the Tyr102 site in addition to Thr36 on this transporter. Cui et al. (47) found several serine phosphorylation sites on two cationic amino acid transporters: Slc7a4 (Ser418, -421, and -422) and Slc7a2 (Ser463 and -645, as well as Thr466). No information on the functional significance or mole fraction of these phosphorylation sites is currently available.
The voltage-dependent anion channel (VDAC), or mitochondrial porin, is believed to play a major role in metabolite exchange across the outer membrane of the mitochondrion and is thus an important link in the energy conversion process (42). Though VDAC is not directly linked to the mitochondrial matrix, it is important to note that the phosphorylation of this transporter has been detected in most mitochondrial phosphoproteome screens and that correlations with VDAC permeability (19, 38) and enzyme complex formation (157, 197) have been reported. Thus this important solute transport process in the outer membrane may be under significant regulation by cytosolic kinases (such as GSK3b, PKA, and NEK1) and phosphatases.
It is clear that protein phosphorylation occurs on many of the transporters in the mitochondrial membranes. Of the numerous phosphorylations detected, the phosphorylation of ANT-1 and VDAC has been correlated with the function of these transporters. It should also be pointed out that the major protein transport pathway in the mitochondria, the TOM and translocase of the inner membrane (TIM) complex is also phosphorylated. Recently, Schmidt et al. (191) demonstrated that protein phosphorylation can both activate and inhibit the important protein translocation function of TOM. These data suggest a major role for protein phosphorylation in the regulation of the exchange of proteins (TOM) and metabolites (VDAC) between the cytosol and the mitochondria intermembrane space. To date, most of these regulatory sites have been found in the mitochondrial intermembrane space or in the cytosol; thus a role for a matrix kinase/phosphatase system in this control network is unclear.
Phosphorylation of the Enzymes of Intermediary Metabolism
The phosphorylation of PDH via the PDH kinase (PDHK) and PDH phosphatase (PDHP) system is the first mitochondrial enzyme shown to be regulated by protein phosphorylation as reviewed in the introduction. PDH phosphorylation and regulation are thus well documented and will not be discussed at length here. We will further discuss the PDHK and PDHP system in the kinase/phosphatases discussion below. Readers interested in this specific system are referred to the original papers (60, 102, 123, 219, 220) as well as the several reviews available on this topic (59, 133). Since quantitative measures are one of the focuses of this review, it is interesting to note that Boja et al. (24) performed one of the first quantitative MS studies on PDH phosphorylation in isolated heart mitochondria. The perturbations previously shown to activate PDH that were used in this study included additions of DCA or Ca2+, as well as substrate starved deenergization. These perturbations significantly decreased PDH serine phosphorylation, with Ser292 and Ser231 phosphorylation decreasing in the order of three- to fourfold and correlating the best with the modulation of enzyme activity. These studies demonstrate that quantitative comparisons of phosphorylation and enzyme activity are feasible using rather unambiguous MS techniques and should be useful in evaluating the functional consequences of phosphorylation sites in other mitochondrial enzymes. Another enzyme system very similar to PDH is the branched-chain α-ketoacid dehydrogenase (BCKDH) present in the heart and other tissues for the metabolism of the three essential amino acids: valine, isoleucine, and leucine (76). This enzyme has been demonstrated to have serine phosphorylation sites on its E1Bα subunit (89) that inhibit the enzyme in a similar fashion as they do in the PDH complex. A separate BCKDH kinase and phosphatase have also been described with properties that are very similar to the PDH system (49, 53, 166, 173). The phosphorylation of BCKDH has been shown to occur along with that of PDH in the intact heart using 32P (28), which correlated with the activity of the extracted enzymes. However, Boja et al. (24) actually found that phosphorylation of BCKDH E1Bα at Ser337 increased with DCA and deenergization in isolated heart mitochondria, whereas that of the E1 subunit of PDH decreased in the same study. The reason for this discrepancy is unknown and may reflect differences in the control of the BCKDH and PDHK/PDHP system.
In addition to the well characterized PDH and BCKDH systems, most of the enzymes of the citric acid cycle have been found to possess phosphorylation sites. These include in succinyl-CoA ligase [ADP forming] subunit-β, Ser257, Ser259 (236), and Ser241 (24); in citrate synthase, Ser190 (236) and Tyr80 (68); in isocitrate dehydrogenase [NADP], Ser423 (236); in malate dehydrogenase, mitochondrial Ser246, Thr235 (24, 236), Ser51, Ser31 (236), Tyr56, and Tyr80 (68); in fumarate hydratase, Tyr491 (68); and in aconitate hydratase, Ser669 (236), Tyr71, Tyr544, and Tyr655 (68). However, none of these phosphorylations have been shown to modify the activity of these enzymes, and inspection of crystal structures shows that these sites are away from the substrate binding pockets.
An interesting observation was made on succinyl-CoA ligase, where a tightly associated phosphate was found on the enzyme using 32P binding assays that correlated with enzymatic activity (162). However, it was found that this tightly associated phosphate that survived detergent, SDS, and two-dimensional electrophoresis is not a protein phosphorylation, but a noncovalent interaction that increased the maximum velocity of the succinyl-CoA ligase reaction kinetics. This tightly associated phosphate in succinyl-CoA ligase suggests that similar metabolite associations may occur in other systems and may require careful interpretation of radiolabeling experiments (5).
A major source of reducing equivalents in the heart under normal conditions is fatty acid oxidation. Phosphorylation of carnitine palmitoyltransferase-1 (CPT-1) in the outer mitochondrial membrane has been reported at Ser741 and Ser747 of the liver isoform both in vivo and in vitro using protein kinase casein kinase II, with the consequence of a slightly higher activity and a different inhibitory effect of malonyl-CoA (103). In isolated heart mitochondria, where the muscle isoform of CPT-1 is predominant, incubation with PKA and CAMK-II also generated slight differences in malonyl-CoA sensitivity of CPT-1 without altering the catalytic activity, and although changes in the phosphorylation levels of Ser and Thr residues in the muscle isoform of CPT-1 were reported using these and other kinases such as p38 MAPK, no specific sites have been found (196). Therefore, unlike the case for PDH, which regulates the entry of reducing equivalents from glycolysis, the role of phosphorylation as a regulatory mechanism for fatty acid oxidation seems to be weak. Many phosphorylation sites have been reported in the rest of the enzyme network of fatty acid oxidation. Most of the acyl dehydrogenases were detected using phosphoryl-protein sensitive dyes in heart (6, 83). In mouse heart (94), the following sites have been located in the acyl-CoA dehydrogenase family members: in ACAD9, Ser478, Thr482, and Thr488; in ACADL, Ser54, Ser55, Ser191 (not conserved in human), and Ser200; and in ACADM, Ser70 and Thr351. In enoyl-CoA hydratase (ECHS1), Thr194 was found to be phosphorylated in rat heart (68), whereas in the α-subunit of trifunctional enzyme [hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein) α-subunit], the phosphorylation sites found in mouse heart (58, 94) are Tyr283, Ser316, Thr575, Thr586, and Ser647. Phosphorylation of acetoacetyl-CoA thiolase (ACAT1), which catalyzes the last step of ketone body catabolism, has also been reported in mouse heart at Tyr90 and Ser238 (58, 94). However, no functional correlation of any of the phosphorylation sites in these enzymes with regard to fatty acid or ketone body oxidation rates has been presented.
Thus, despite the recent screening studies revealing many new phosphorylation sites in the matrix citric acid and fatty acid oxidation enzyme system, the functional significance, mole fraction, and kinase/phosphatase system remain unknown. These sites might suggest that protein phosphorylation may play a broader role in the regulation of the citric acid cycle beyond PDH as well as fatty acid oxidation; however, further investigations will be required to establish whether this regulatory system is in play.
Phosphorylation of Complex I
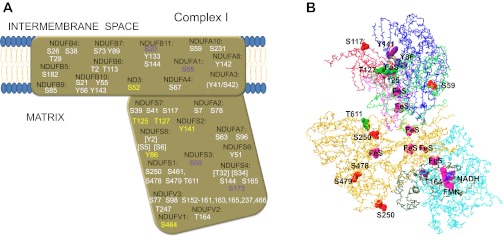
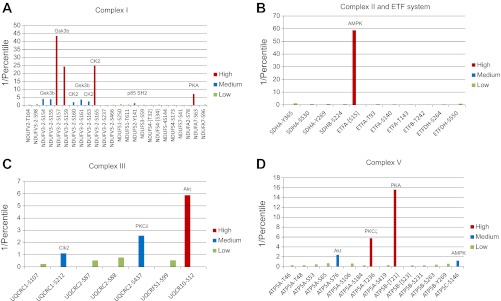
Complex I, a proton pumping NADH dehydrogenase, is the largest of the oxidative phosphorylation complexes, with a total of 46 subunits in mammals, 14 of which are the core of electron and proton transfer activities, and conserved in prokaryotes and eukaryotes (26). A total of 69 different phosphorylation sites have been reported in 26 of the complex I subunits. These are summarized in Fig. 3, A and B. To emphasize the extensive nature of protein phosphorylation detection, we are presenting two schematics for each complex: the first (A) places all of the phosphorylations roughly across the whole complex, and the second and subsequent B–D (when applicable in the corresponding figure) present a space filling model with the phosphorylation sites shown relative to active sites as discussed in the introduction. In complex I, 18 of the sites are located in the seven central proteins that comprise the membrane extrinsic peripheral arm of complex I that contains all of the redox cofactors necessary for function, and one more site is found in the hydrophobic core proteins that are essential for proton pumping. Since the only high-resolution structures available for the peripheral arm of complex I are bacterial (20, 64, 188), the relevance of those phosphorylation sites that have not been linked yet to some change in activity by empirical evidence are restricted to these eight subunits. The remaining 50 sites are distributed among 18 of the 33 accessory subunits for which structural information is not yet available. Since the role and interaction of all of the subunits of complex I is yet to be resolved, much of the extrapolation of the impact of a given protein phosphorylation on a specific site must be considered, to some extent, to be speculative.
Fig. 3.
Reported sites of phosphorylation in complex I. Residues where phosphorylation has been detected in each indicated subunit are shown schematically (A) with respect to their presumed relevance based on structural and functional evidences in a structural scheme. Sites that are unlikely to have an effect on activity based on their location in available high-resolution structures are shown in white. Sites with no known function, but that might influence activity based on their location in the structure, are shown in yellow. Sites that have been found to be phosphorylated by an identified kinase in vitro or in vivo are shown in purple. Sites that are located in the presequence and are thus absent in the assembled complex are shown in brackets. The position of each site either in the matrix space or closer to the inner mitochondrial membrane is also shown based on the structural information available. The probable location of some of these sites is shown (B) using the structure of the hydrophilic domain of complex I from Thermus thermophilus (PDB ID 3IAM) as a scaffold in which the residues that are homologous to the mammalian amino acids are colored based on the identity of the reported phosphorylation site (Ser, red; Thr, green; Tyr, purple). The Cα backbone of NDUFV1 is shown in cyan, NDUFV2 is colored dark green, NDUFS1 is shown in orange, NDUFS2 in pink, NDUFS3 in bright green, NDUFS7 in red, and NDUFS8 in blue. Redox groups (FMN and FeS centers) are space-filled and colored magenta, and NADH is represented as a blue wire model. See main text for definitions of abbreviations.
NADH binds to the NDUFV1 subunit, which contains FMN and one iron sulfur center as electron acceptors (20, 26). This protein was labeled with a phosphospecific dye (ProQ) in pig heart (83). Only one phosphorylation site has been found in this subunit in cancer cells at the COOH-terminal residue Ser464 (151). The location of this residue is unknown given the lack of conservation in this region between mammalian NDUFV1 and the Thermus thermophilus structure but is probably located far away from the redox centers and the NADH binding site. The only phosphorylation site reported in the essential subunit NDUFV2, also in HeLa cells (151), is Thr164. The corresponding residue in the bacterial structure is Thr112, which sits in a relatively well-conserved loop highly exposed to the solvent but far away from its iron sulfur center or subunit contact regions (20). Nevertheless, this subunit is labeled by ProQ and PhosTag dyes in pig heart, but not in liver (6), suggesting that other more physiologically relevant sites may exist that have escaped detection. The chain of iron sulfur centers continues in the NDUFS1 subunit, the largest of complex I, which contains three of these redox cofactors. Interestingly, this subunit is not only labeled with ProQ and PhosTag in a tissue specific manner but also with 32P in energized mitochondria, suggesting a higher turnover rate of phosphorylation (6). Of the five phosphorylation sites detected in NDUFS1, only Ser250 has been recently found in heart tissue (58). However, the corresponding residue in the bacterial structure (20) is not in contact with any other subunit interface (see Fig. 3B) and is close to an iron sulfur cluster that does not even exist in eukaryotes (26), casting doubt on its functional relevance.
The metazoan specific subunit NDUFV3 is located somewhere close to the NDUFV1 and NDUFV2 subunits, given that the three subunits can be dissociated as a subcomplex from the rest of complex I (69). NDUFV3 is the most heavily phosphorylated protein of NADH dehydrogenase, with 17 different sites in non-muscle tissues (40, 58, 61, 74, 80, 84), mostly in a string of Ser residues from position 152 to 161 and the nearby Ser163 and Ser165 of isoform 2, which exists in human and rodents. Some of these Ser phosphorylations seem to have been detected in human skeletal muscle (236). Another isoform 2 site is the Ser located four residues from the COOH-terminus (62), which exists in both isoforms and has also been detected in mouse brown fat, heart, and kidney (94). Further studies on the effect of NDUFV3 isoform expression on complex I activity are needed to understand the role of these phosphorylation sites, especially in the heart.
Subunit NDUFS8, also known as TYKY, contains two more iron sulfur clusters and has also been found to bind phosphospecific dyes in pig heart mitochondria (6, 83). Of the four phosphorylation sites reported for this protein, only Tyr86, found in rat inner medulla cells (81), exists in the mature protein. Tyr86 is close to the interface with NDUFS7 and to the membrane (14, 20), but away from redox centers (see Fig. 3B), so its functional effects, if any, remain obscure. Another central subunit of the peripheral arm labeled with PhosTag in heart but not in liver (6) is NDUFS2, which lacks redox cofactors but is phosphorylated in lung cancer cells at Tyr141 (176), located at the interface with subunit NDUFS7, also known as the PSST protein, which contains the last iron sulfur center in the chain (Fig. 3B). This interface has been proposed to form the quinone reduction site (26) and is therefore an ideal target of regulation if phosphorylation were to occur under physiological conditions and at a significant mole fraction, which is not documented yet. Phosphorylation of NDUFS3, another core protein that lacks redox centers and is labeled with PhosTag in both heart and liver mitochondria (6), has been reported at Ser59 in rat uterus homogenates upon addition of exogenous PKA after heating to inactivate endogenous kinase and phosphatase activities (88). Even though this residue lies in a potentially relevant region at the interface with NDUFS2 (Fig. 3B), the experimental manipulations applied to detect it are not applicable to physiological conditions. Also, the presence of PKA within the matrix space is still in question (see Mitochondrial Matrix Protein Kinases).
NDUFS7 has five reported phosphorylation sites, two of them found in mouse heart and kidney at Ser39 and Ser41 (94) in a poorly conserved and undefined region of the crystal structure that seems to interact with subunits of the membrane domain of complex I (20, 64). Thr125, discovered in HeLa cells upon treatment with rapamycin (37), is in contact with the iron sulfur center of NDUFS7 if the orientation of the residue in metazoans is similar to the corresponding residue (Arg) in the bacterial crystal structure (see Fig. 3B). Unfortunately, no functional information exists on the effect of phosphorylation at any of these sites on enzyme activity.
No structural information is available for the accessory subunits of the peripheral arm, so the role of the phosphorylation sites described in five of these proteins is even less clear. In the case of Ser173 of NDUFS4, at least a correlation has been observed between the amount of imported protein and the phosphorylation of this residue by PKA (54, 55). Ser96 of NDUFA7 was found to be phosphorylated in bovine heart (165), although phosphopeptide enrichment was needed to detect it, as was the case with Ser78 of NDUFA2, found in mouse brown fat, heart, kidney, and brain (94, 222), suggesting a very low mole fraction of phosphorylation.
Accessory subunits in the membrane domain that are not found in prokaryotes have been the focus of more intensive studies with respect to phosphorylation and its effects, in particular NDUFA10, NDUFA1 (also called MWFE), and NDUFB11 (known as ESSS). Ser59 (not well conserved) and Ser231 of NDUFA10 have been reported to be phosphorylated in bovine and mouse heart, respectively (58, 190), although no correlation to function or quantification of mole fraction has been established. This subunit has also been found to bind the PhosTag dye in pig heart but not in liver (6). Ser55 of NDUFA1 was phosphorylated in bovine heart complex I when incubated in vitro with PKA and cAMP (36, 225). Phosphomimetic mutations of this residue in hamster cells abolish complex I assembly, suggesting a function for phosphorylation at this site in regulating the steady-state levels of the enzyme (225). In the case of NDUFB11, Ser20 was also found to be phosphorylated in the presence of exogenous PKA (36), and phosphomimetic substitutions in nearby Ser residues in hamsters show a slight decrease in complex I activity (225), although in this case the evidence is more indirect because of the lack of residue conservation. Phosphorylation of a 18-kDa complex I protein, later shown to correspond to NDUFB11 (36), was reported to increase activity of complex I (210), although the fraction of total protein that underwent this modification was not quantified. Tyr133 and Ser144 have also been reported as phosphorylation sites in human muscle, but only using highly sensitive methods (236).
Seventeen more sites have been found in other 10 accessory subunits of the membrane domain. Sites found in heart (94) or muscle (236) are Tyr41 (or Ser42) of NDUFA3, Ser67 of NDUFA4, Ser26 and Ser38 of NDUFB4, Ser21 of NDUFB10 [which is labeled by ProQ in pig heart (83)], and Ser73 and Tyr89 in NDUFB7. NDUFA13, also known as GRIM19, an apoptosis-inducing factor that is essential for complex I assembly, was found to have a Ca2+-insensitive phosphorylation site at Thr96, detected in pig heart (24). Another accessory subunit, NDUFS5, has no reported phosphorylation sites, although it was found to be labeled both by ProQ and PhosTag dyes in pig heart (but not liver) mitochondria (6, 83). However, any functional effect of phosphorylation in these accessory subunits is probably restricted to protein assembly, given that most are either cysteine-rich chaperones likely involved in iron sulfur cluster synthesis and insertion or short single transmembrane domain polypeptides proposed to aid in the assembly and stability of the membrane domain (26), but not likely to be involved directly in proton pumping, given their absence from prokaryotic complex I.
Phosphorylation of Complex II and the ETF System
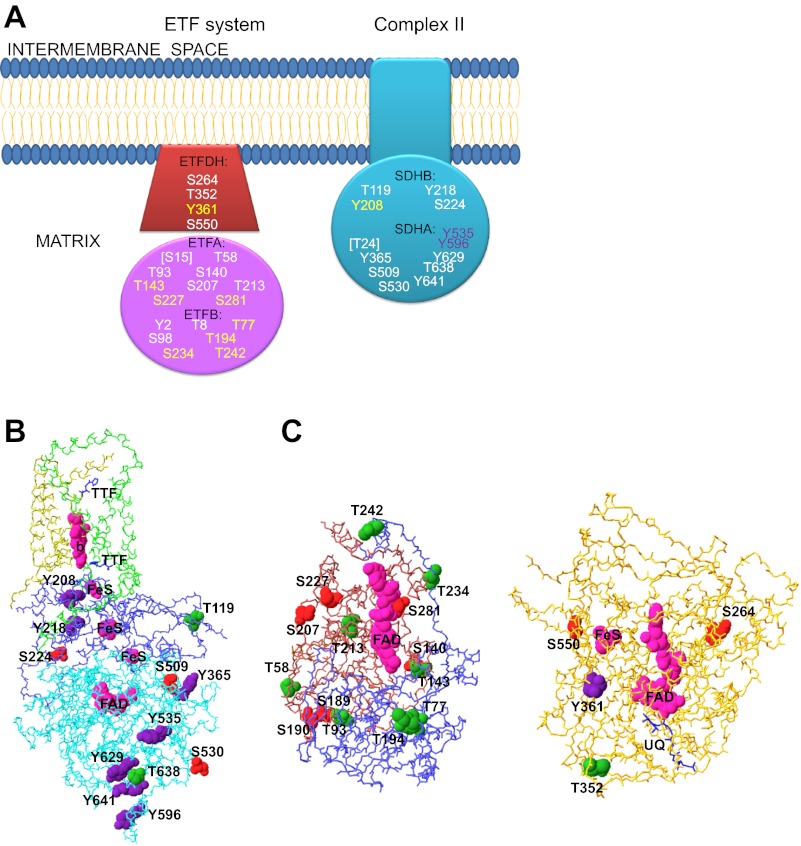
Succinate dehydrogenase (complex II) and the enzymes of the ETF system share ubiquinone as final electron acceptor in the mitochondrial inner membrane. Complex II is anchored to the membrane and oxidizes succinate to fumarate, whereas the ETF system is composed of the heterodimeric soluble ETF that shuttles electrons from a variety of dehydrogenases, most notably the acyl dehydrogenases of fatty acid oxidation, to the membrane-associated ETF: ubiquinone oxidoreductase (ETFDH). A summary of the identified phosphorylation sites in these enzymes are schematically presented in Fig. 4, A–C. Phosphorylation of the two hydrophilic subunits of complex II, SDHA, and succinate dehydrogenase subunit B (iron-sulfer protein) (SDHB) has been detected in heart and liver mitochondria using phosphosensitive dyes (6, 83). Phosphorylated subunit-β of ETF (ETFB) has also been identified using dyes in mitochondria from heart and more prominently in liver, where ETFB is also labeled by 32P (6, 83).
Fig. 4.
Reported phosphorylation sites in complex II and the ETF system. Subunits where phosphorylated residues have been found are represented schematically along with the respective residues (A) using the same color coding as in Fig. 2A. All the sites shown are exposed to the mitochondrial matrix. The location of the reported phosphorylation sites in complex II (B) and the ETF system (C) is shown according to their amino acid identities (Ser, red; Thr, green; Tyr, purple). The structure shown for complex II is that of the pig heart mitochondrial complex II (PDB ID 1ZP0), with the Cα backbone SDHA colored in cyan, SDHB in blue, SDHC in green, and SDHD in dark yellow. The two molecules of thenoyltrifluoroacetone (TTFA) shown in blue close to the b heme serve to identify the ubiquinone binding sites. The structure used for ETF and ETFDH are those from human (PDB ID 1EFV) and pig (PDB ID 2GMH), respectively. ETFA is colored dark pink, ETFB is shown in blue, and ETFDH is orange. Redox cofactors (FAD, FeS centers and the b heme) are colored magenta. Ubiquinone (UQ) in ETFDH is shown as a blue wire model. See main text for definitions of abbreviations.
The flavoprotein subunit of complex II (SDHA) is where succinate binding and oxidation takes place. Nine different phosphorylation sites have been reported in this flavin adenine dinucleotide (FAD)-containing protein, five of which are tyrosines. The crystal structure of complex II (205) shows that all the reported sites are located on the surface of SDHA away from the succinate binding site, the FAD binding pocket, and the interface with SDHB (see Fig. 4B), in line with the lack of evidence for a functional role at any of these sites. Two tyrosine phosphorylation sites (Tyr535 and Tyr596 in rat brain mitochondria) were detected after in vitro incubation of solubilized mitochondrial fractions with the tyrosine kinase Fgr followed by enrichment of phosphotyrosine peptides (180), but it is still unknown what kinases might be responsible for complex II phosphorylation in vivo. Interestingly, Phillips et al. (161) found SDHA as one of the 32P labeled proteins obtained after incubation of blue native gels with γ-[32P]ATP, suggesting an intrinsic kinase activity in complex II or in one of the proteins present in the corresponding region of the gel, where no known kinases were detected.
The iron sulfur subunit of complex II (SDHB) contains three of these redox centers and links SDHA to the two smaller hydrophobic subunits embedded in the membrane where ubiquinone binds and undergoes reduction (205). Four phosphorylation sites have been reported in SDHB, one of which, Tyr208, identified in mouse brown fat and testis (94), stands out as potentially relevant in view of its position in contact with the iron sulfur center of SDHB that is closer to the membrane, in the vicinity of one of the ubiquinone binding sites (see Fig. 4B).
Nine phosphorylation sites comprising only Ser and Thr residues have been found in ETFA, six of which have been found differentially expressed in mouse tissues, including Thr93 in heart (94, 222). Despite the lack of studies establishing a correlation with function, three phosphorylation sites could have some effect on the activity of ETF based on the crystal structures available (177, 211). Thr143, found in mouse liver (94), is close to the interface with the β-subunit (ETFB), so it could be important for the stability of the dimer, whereas Ser227 (found in brown fat) is in the FAD domain that moves during interaction with dehydrogenases (see Fig. 4C). Ser281, also detected in mouse brown fat (94), forms a hydrogen bond to one of the phosphates of FAD, so its phosphorylation could alter or even impede assembly of the FAD cofactor, although in the absence of functional data, it could simply represent an assembly intermediate (94).
The seven phosphorylation sites identified in ETFB include Thr77, detected in a human-mouse myeloid chimeric cell line (39), which is located at the surface of ETFB where acyl dehydrogenases bind (211), so it could theoretically affect electron transfer rates. Ser98, Ser234, and Thr242 were all found in mouse brown fat (94), and the last two of these residues are located, respectively, in the loop that connects to the COOH-terminal domain of ETFB that interacts with the movable FAD domain of ETFA and in the interface with ETFA close to the FAD binding pocket (Fig. 4C). Again, the lack of functional studies and the likelihood of a low fraction of phosphorylated protein at these sites preclude any conclusions on a physiological role of these sites.
ETFDH has an iron sulfur center as well as one FAD as redox cofactors, and the crystal structure has shown ubiquinone in its binding pocket (235). Only one of the four reported phosphorylation sites in EFTDH, Tyr361, found in lung cancer cells (176), is close to one of the redox cofactors, in this case, the iron sulfur center (see Fig. 4C). Ser264 and Ser550, identified respectively in mouse brown fat and in other tissues including heart and liver (94, 213), are exposed at the edge of a flat surface opposite to Thr352, which was found to be phosphorylated in pig heart, but which did not change in its phosphorylation level upon energization of mitochondria (24), suggesting a low turnover and low mole fraction.
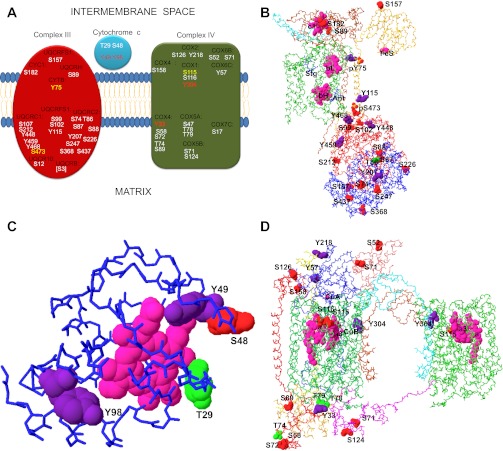
Complex III Phosphorylation
The cytochrome bc1 complex (complex III) is a dimer comprised of 11 different subunits per monomer in mammalian mitochondria (228). Phosphorylation has been detected in eight of these polypeptides, including the three subunits that contain redox prosthetic groups and are thus directly involved in electron transfer from ubiquinol to cytochrome c coupled to proton translocation, namely, cytochrome b (CYTB), the Rieske iron sulfur protein (UQCRFS1), and cytochrome c1 (CYC1). Of the 24 phosphorylation sites identified so far in complex III, 16 occur in Ser residues, 6 in Tyr, and only 1 in Thr. A summary of these sites is presented in Fig. 5, A and B.
Fig. 5.
Reported phosphorylation sites in complex III, cytochrome c, and complex IV. Residues in the indicated subunits or proteins where phosphorylation has been detected are shown schematically in different colors to indicate their presumed or demonstrated relevance based on structural and functional evidences (A). Sites that are unlikely to have an effect on activity based on their location in available high-resolution structures are shown in white. Sites with no known function, but that might influence activity based on their location in the structure, are shown in yellow. Sites that have been found to affect enzymatic activity when phosphorylated are shown in red. Sites that are located in the presequence and are thus absent in the assembled complex are shown in brackets. The position of each site relative to the intermembrane space and the matrix is also shown. The location of most of the phosphorylation sites are shown in the crystal structure of complex III (B), cytochrome c (C), and complex IV (D), colored according to the type of amino acid (Ser, red; Thr, green; Tyr, purple). Phosphate molecules are shown in contact with two of the reported sites in complex III (pY75 in CYTB and pS473 in UQCRC1), according to one of the bovine structures (PDB ID 1PPJ). Only one monomer is shown for clarity, and the Cα backbone of UQCRC1 is colored in red, UQCRC2 is shown in blue, CYTB in green, UQCRFS1 is orange, CYC1 is brown, and UQCRH is cyan. Stigmatellin (Stg) and antimycin (Ant) are shown as blue wire models bound at the quinol oxidation and reduction sites, respectively. The structures used for cytochrome c (PDB ID 2B4Z) and for complex IV (PDB ID 1V54) were both from bovine heart. COX1 is colored green in both monomers, COX2 is blue, COX4 is red, COX5A is orange, COX5B is magenta, COX5C is pink, and COX6A, which crosses from one COX1 to the other contacting Tyr304, is colored cyan in one monomer and brown in the other. Redox groups such as hemes, Cu atoms and a FeS center are colored magenta. See main text for definitions of abbreviations.
More than half of the phosphorylation sites reported in complex III are located in the two largest subunits, the ubiquinol cytochrome c reductase core 1 (UQCRC1) and core 2 (UQCRC2) proteins, which protrude toward the matrix side of the inner mitochondrial membrane (228), which is consistent with a matrix kinase activity. These proteins, however, do not participate in electron transfer or proton movement within complex III, although they are required for the assembly of CYC1, UQCRFS1, and other smaller subunits, as shown by deletion strains in yeast (232). UQCRC1 and UQCRC2 are homologous to the dimeric matrix processing metalloproteases that cleave the presequence of a considerable number of proteins translated in the cytosol and imported into the mitochondria. Some crystal structures even show the cleaved presequence of UQCRFS1, termed subunit 9, bound to UQCRC2, which suggests that these subunits are responsible for processing of UQCRFS1 upon its assembly into complex III (96). Mild detergent treatment activates the UQCRC1-UQCRC2 protease activity in isolated mammalian complex III, although the catalytic rate is considerably lower than that of the soluble matrix protease (57). However, no phosphorylation sites have been found in the active protease site. More importantly, neither UQCRC1 nor UQCRC2 has been found to change its phosphorylation state upon manipulation of cellular mitochondrial metabolism (24), and the turnover is slow as judged by the lack of labeling when mitochondria are incubated with 32P (6), suggesting that the significance of the phosphorylation status is likely limited to their role in the assembly of complex III. An interesting phosphorylation site is Ser473, reported so far only in mouse kidney (94), but which can be seen in close proximity to a phosphate molecule (Fig. 5B) in one crystal structure from bovine heart (87), suggesting a high mole fraction of phosphorylation. The phosphate molecule is also in contact with His-221 of CYTB in a region where mutagenesis in nearby residues has revealed assembly defects or altered binding properties at the ubiquinone reduction site (27).
The only other complex III subunit where more than one phosphorylation site has been reported is UQCRFS1, which, in contrast to UQCRC1 and UQCRC2, has a central role in electron transfer (230) and would thus constitute a plausible site for regulation. UQCRFS1 has a tilted transmembrane helix that traverses and possibly stabilizes the complex III dimer, and its COOH-terminal half includes a globular domain in the intermembrane space that contains the 2Fe-2S center that accepts one of the two electrons from ubiquinol (96, 230). This extramembrane domain moves from the vicinity of CYTB to interact with CYC1, functioning as a diffusible electron shuttle (46). This movement has been proposed to be involved in the asymmetric activity of complex III that results in the oxidation of ubiquinol in only one monomer at a time (45). The UQCRFS1 subunit has been reported to exhibit a phosphatase-sensitive shift in its isoelectric point when rat liver mitochondria are exposed to calcium or agents that induce the permeability transition of the mitochondrial membrane, suggesting a change in phosphorylation state of UQCRFS1 (78). However, no specific phosphorylation site has been linked to this isoelectric shift, which has not been observed in heart mitochondria (83). Moreover, UQCRFS1 phosphorylation has been detected with dyes such as ProQ and PhosTag, but not with 32P labeling (6, 83), suggesting a low turnover rate of phosphorylation, which is inconsistent with a role in acute regulation of mitochondrial function, unless a very labile phosphorylation is assumed. Nevertheless, UQCRFS1 phosphorylation, as reported by binding of antibodies against P-Ser, was observed to decrease by up to 50% upon ischemic preconditioning in rat brain, which was accompanied by an almost twofold increase in complex III activity (52). Thus it is quite feasible that some P-Ser constitutes a regulatory site in UQCRFS1.
Two of the reported sites in UQCRFS1 correspond to Ser residues detected in resting human skeletal muscle mitochondria (236). Ser99 is located on the matrix side in a bend right before the beginning of the transmembrane helix close to the NH2-terminus of UQCRC1, and thus away from the extrinsic domain which moves to transfer electrons (see Fig. 5B). Ser157 is located at the top of the movable domain, but away from the 2Fe-2S cluster and the series of residues that control its redox potential (199), and also at a considerable distance from the contact surface that interacts with CYTB or CYC1. Although phosphorylation of Ser157 would not seem to interfere with the electron transfer function of UQCRFS1 directly, it could serve as a docking point for other proteins, which upon binding could hinder the diffusion of this extrinsic domain. However, no evidence exists yet for such an interaction.
CYC1 is key to the electron transfer function of complex III because of its binding and electron transfer to cytochrome c. Interestingly, cytochrome c binding has been found to occur only to one of the CYC1 subunits in the complex III dimer, according to crystallized structures from yeast mitochondria (116, 200). Only one phosphorylation site, Ser182, has recently been reported in mitochondria from resting human muscle (236). The equivalent residue in crystallographic structures is close to the point of contact between the two CYC1 subunits in the dimer, but far from the actual binding site of cytochrome c or the c1 heme group. Furthermore, Ser182 is only found in primates, whereas Pro is found at this position in most other vertebrates, casting doubt on a functional role for phosphorylation at this site.
Another subunit that is relevant for cytochrome c binding to complex III is the acidic “hinge” protein, or subunit 6 (UQCRH). Genetic deletion (192) or removal (108) of this subunit leads to a loss of binding of cytochrome c to CYC1 depending on the ionic strength, and at least in yeast, to a decrease in cytochrome c reductase activity of 50%. Crystal structures show that this subunit does not contribute directly to the cytochrome c binding pocket (116, 200), but other studies show a perturbation of the CYC1 heme environment when UQCRH is removed (106), and its abundance of acidic residues has been proposed to direct cytochrome c to its actual binding site (200). Phosphorylation of UQCRH has been detected with PhosTag dye (6), and Ser89, located close to the COOH-terminus, has been identified as a phosphorylation site in pig heart mitochondria (24). However, this residue is not well conserved among species, so the functional relevance of phosphorylation in this subunit remains to be established.
CYTB constitutes the functional core of complex III with its two ubiquinol/ubiquinone binding sites and its two b-type hemes, where electron recycling coupled to proton uptake and release takes place. The high hydrophobicity of this subunit makes it difficult to identify by MS (90), and even its staining in gels is poor. Nevertheless, a phosphorylation site (Tyr75) has recently been reported in CYTB from human muscle (236). Crystal structures show this residue to be in one of the few solvent accessible regions of CYTB, which is mostly embedded in the membrane and surrounded by other subunits (96, 116, 228). Interestingly, as shown in Fig. 5B, a phosphate molecule can be seen in contact with Tyr75 in at least one high-resolution structure from the bovine heart enzyme (87), suggesting a high mole fraction of phosphorylation at this site under some conditions. This residue is highly conserved among species and is only four residues away from the essential Arg79 residue, which constitutes the exit pathway for protons derived from ubiquinol oxidation (90). It would therefore be interesting to determine in the future if phosphorylation at this site has an effect on the activity of complex III.
Two other small subunits are phosphorylated in complex III. Subunit 7, also called UQCRB, has been detected in pig heart with ProQ and PhosTag (6, 83). However, the phosphorylation site at Ser3 found in this subunit (24) is actually located in the short import presequence that is cleaved from this subunit before assembly into the complex, so other as yet unidentified residues in the mature protein must be responsible for the binding of phosphorylation-sensitive dyes. The function of this subunit is more related to the early assembly steps of complex III (232) and not to the actual electron transfer activity, consistent with its low phosphorylation turnover (6). Subunit 10 is the smallest subunit of complex III and has been found to be phosphorylated at Ser12 in various mouse tissues (94). This subunit 10 is required for insertion of UQCRFS1 as the last step in complex III assembly (232), so it can be considered simply as a chaperone, and its phosphorylation as related only to that function.
It is noteworthy that no specific kinases or phosphatases have been associated to any of the reported phosphorylation events at complex III, as is the case for most mitochondrial proteins. Low-level phosphorylation at complex III was detected when blue native gels were incubated with γ-[32P]ATP, suggesting at least a limited autophosphorylation activity in this complex (161). However, it remains to be determined which of the sites reported, if any, are the targets of this intrinsic kinase activity. The fact that the few phosphorylation sites found in the catalytically relevant subunits of complex III have been detected only with increasingly sensitive phosphopeptide enrichment techniques (176, 236) suggests a very low proportion of phosphorylated complexes. Only the isoelectric shift in UQCRFS1 reported in liver mitochondria upon induction of the membrane permeability transition (78) seems to reflect a PTM of a large fraction of complex III, although no Ca2+-sensitive site has been identified yet, which could reflect a very labile phosphorylation or a completely different type of PTM.
Cytochrome c and Complex IV Phosphorylation
In contrast to the highly speculative significance of phosphorylation in complexes I, II, and III, a much more solid body of evidence has emerged concerning the role of this type of modification in residues of cytochrome c and its oxidase, complex IV. As summarized in Fig. 5, A and C, there are four identified sites of phosphorylation in cytochrome c, which include two Tyr, one Ser, and one Thr residues. Of the 20 sites in complex IV subunits, four correspond to Tyr, three to Thr, and 13 to Ser residues (Fig. 5D). Two tissue-specific Tyr phosphorylation sites have been reported after purification of bovine cytochrome c in the presence of unspecific phosphatase inhibitors: Tyr49 in liver (229) and Tyr98 in heart (119). In both cases, changes in the kinetics of complex IV were observed when using the (at least partially) phosphorylated cytochrome c, which were reverted by alkaline phosphatase treatment. With the Tyr49 modified cytochrome c purified from liver, a twofold decrease in the maximal rate of isolated complex IV was observed (229), similar to what was observed when mutating this residue to Glu to mimic phosphorylation (159). This Glu49 cytochrome c also showed a lower redox potential by 45 mV, had a 30% lower binding affinity to cardiolipin, and was unable to catalyze cardiolipin oxidation or induce caspase-3 activity, which are events linked to the proapoptotic role of cytochrome c when released from the inner mitochondrial membrane (159). The Tyr98-phosphorylated cytochrome c from heart generated sigmoidal kinetics in isolated complex IV and exhibited a marked blue shift of the 695-nm spectral peak that reflects the interaction of the c heme with surrounding Met residues (119), also suggesting that a high fraction of cytochrome c molecules in the preparation contained this modification. Both Tyr49 and Tyr98 are well conserved residues, with the first forming a hydrogen bonding to the heme propionate (see Fig. 5C), and the second one being in contact with Lys-7, which is important for apoptosome formation (119, 229). Two other phosphorylation sites, Thr29 and Ser48, have recently been reported in resting human muscle (236), but no functional effect has been linked to these sites, which probably exist phosphorylated at very low mole fraction since they were detected only after high enrichment of phosphopeptides. No studies have been reported in which phosphorylated cytochrome c at any site is assayed as a substrate of complex III, which would be relevant to determine whether electron transfer at this segment of the respiratory chain is also impaired just as has been postulated for complex IV. In addition, because of the mobility of cytochrome c between different cellular compartments, the location of the kinase/phosphatase system responsible for its phosphorylation remains unclear.
Cytochrome c oxidase (complex IV) crystallizes as a dimer of 13 subunits in mammalians (212). Subunit I [cytochrome c oxidase (COX) 1] is the functional and structural core of this enzyme, containing two a-type hemes, as well as the CuB atom that constitutes, together with heme a3, the binuclear metal center where reduction of oxygen to water occurs. Three phosphorylation sites have been identified in COX1 through MS of the isolated subunit or from purified complex IV. Ser115 and Ser116 were identified in rabbit mitochondria after ischemia and reperfusion of isolated hearts (65), with phosphorylation at these two residues nearly undetectable when an inhibitor of protein kinase A was administered during ischemia. No activity assay of complex IV was reported in this study, although the complex IV levels were found to decrease significantly because of the half-hour ischemic treatment, which suggests that phosphorylation at Ser115 and Ser116 could be more related to protein degradation or turnover than to regulation of electron flow. Nevertheless, Ser115 has been proposed to be part of the exit channel for protons pumped through COX1 during normal enzymatic activity (212), and both Ser115 and Ser116 are well conserved and exposed to the solvent in the intermembrane space of mitochondria (Fig. 5D), making them susceptible to the kinase/phosphatase system well characterized in this compartment (see Mitochondrial Matrix Protein Kinases and Matrix Protein Phosphatases).
In bovine liver, isolation of mitochondria in the presence of a phosphodiesterase inhibitor to generate high cAMP levels resulted in the identification of Tyr304 as a phosphorylation site in COX1 (120). The same residue was found to be phosphorylated when bovine or mouse liver homogenates were treated with TNFα before isolation of complex IV (183). Tyr304 phosphorylation correlated with pronounced sigmoidal kinetics and inhibition of the maximal rate of oxygen consumption by complex IV when varying the concentration of cytochrome c. This well-conserved residue could be accessible from the intermembrane space if rotated out and has been postulated to be part of an adenine nucleotide binding site based on the crystal structure that shows cholate bound at this location, which is assumed to resemble ADP (212). Tyr304 is also in contact with subunit VIa of the opposite complex IV monomer, which establishes interactions with its own COX1 subunit (Fig. 5D), thereby creating a possible communication pathway for intermonomeric conformational changes that could be responsible for the allosteric regulation by ATP (118). Interestingly, the structure and sequence of the region surrounding Tyr304 is similar to that of the Tyr98 residue of cytochrome c that has been shown to be phosphorylated in bovine heart, suggesting that the same kinase/phosphatase could target both COX1 and cytochrome c in the intermembrane space of mitochondria (119). However, it is not clear what other residues in COX1 might be phosphorylated to regulate the activity of complex IV. Because of its high hydrophobicity, protein coverage of COX1 by MS has been reported to be only of 6–33% (65, 118, 183), implying that most of the protein was not surveyed. Furthermore, antibodies against P-Ser and P-Thr residues bind to COX1 after incubating isolated complex IV with protein kinase A, cAMP, and ATP, conditions that also increase the sigmoidicity of complex IV kinetics and lower the maximal rate in a phosphatase- and Ca2+-sensitive manner (79). Radioactive labeling of COX1 under these same conditions was also observed in the presence of γ-[32P]ATP (18). However, in vivo phosphorylation of complex IV was only detected when rat heart mitochondria were rapidly isolated with phosphatase inhibitors and complex IV separated by blue native electrophoresis instead of the usual long chromatographic procedure (79). We have also recently reported that bacterial lambda phosphatase activates ATP-inhibited complex IV in blue native gels (161). These characteristics of COX1 phosphorylation suggests that several other sites might have been missed in previous studies and that the properties of its kinase/phosphatase regulatory system might resemble those found in bacteria, which involve labile sites such as P-His and P-Asp residues (110). This would agree with the bacterial origin of COX1, which is still encoded in the mitochondrial genome.
Subunit II (COX2) is the only other redox center containing subunit of complex IV. Its binuclear CuA center receives electrons from cytochrome c, which binds on the surface of the COOH-terminal globular domain that extends into the mitochondrial intermembrane space (212). Of the two sites reported, only one (Ser126) has been recently identified as a phosphorylation site in heart after titanium dioxide enrichment (92). However, Ser126 is located away from the cytochrome c binding interface, although it might contact COX4 when phosphorylated (see Fig. 5D). Nevertheless, the protein band where COX2 comigrates with subunit III (COX3) appears to be labeled by γ-[32P]-ATP when complex IV is incubated with protein kinase A (18), and is recognized by antibodies against P-Tyr, P-Ser and P-Thr (79). The tyrosine kinase c-Src has been shown to phosphorylate COX2 in vitro, and its activity in cells results in immunodetection of P-Tyr residues in COX2 (and even in COX1 in mouse cells), together with a mild stimulation of complex IV activity (139). However, no specific phosphorylation sites were reported in these studies.
Subunit IV (COX4) is the protein with the most phosphorylation sites identified so far in complex IV. This subunit traverses the membrane by a single transmembrane helix in close contact with COX1 and also interacts with COX2 through its matrix and intermembrane space domains (212). COX4 appears to mediate the allosteric effect of ATP on complex IV activity by directly binding adenine nucleotides at both its matrix and intermembrane space domain (100), and expression of a second isoform in some tissues abolishes this regulation (93). COX4 has also been identified as interacting with protein kinase Cε upon activation and translocation of this kinase to mitochondria in neonatal cardiac myocytes from rat, resulting in 32P labeling of COX4 that was correlated with an increase in complex IV activity (148, 149), although it was not identified whether phosphorylation occurred at the matrix or intermembrane domain of this subunit. Given that PKCε has been implicated in cardiac ischemic or hypoxic preconditioning (137), the proposed regulation of COX activity by phosphorylation of COX4 stands out as an example of a functionally relevant modification linked to an experimental manipulation, even if, unfortunately, the mole fraction of phosphorylation has not been determined. COX4 (and COX1) were observed to become less phosphorylated when rat liver mitochondria were treated with inhibitors of a soluble adenylyl cyclase that is claimed to be present in the mitochondrial matrix and that is activated by bicarbonate (2). However, the results of these studies have not addressed the specific sites that undergo phosphorylation or dephosphorylation. We have also observed COX4 labeling by γ-[32P]ATP in situ in blue native gels where no kinases were detected by MS (161), suggesting an intrinsic complex IV phosphorylation activity acting on this and other subunits.
Phosphorylation of Tyr33 in COX4 was recently identified after purification of complex IV from bovine liver in the presence of phosphatase inhibitors and calcium chelators (120), but its identification required enrichment of phosphopeptides, implying a low mole fraction of phosphorylated Tyr33, or, alternatively, the high lability reported for Tyr phosphorylation in general in complex IV. Tyr33 is located in the matrix side of the membrane, close to the adenine nucleotide binding site in the matrix proposed by comparative modeling of the isoforms 1 and 2, the last of which does not show ATP allosteric inhibition of complex IV when expressed (93). Inspection of the crystal structure (see Fig. 5D) also shows that Tyr33 is in contact with helix XII of COX1, not far from the entrance of the channels through which protons enter COX1 to be pumped or consumed by the reduction of oxygen to water (212). However, no studies have shown whether this site has an effect on complex IV activity.
The matrix located Ser58 was also identified recently as phosphorylation site for COX4 in bovine heart after fluoride treatment of mitochondria to inhibit proteases and phosphatases, followed by phosphopeptide enrichment (79). However, phosphorylation at this site was related to a slight inhibitory effect of added ATP in the isolated complex IV, without induction of sigmoidal kinetics observed with other phosphorylation sites. Thr74 phosphorylation was detected in rabbit heart after ischemia and reperfusion (65); however, this residue (see Fig. 5D) is located at the tip of the matrix domain of COX4, far from the interface with COX1 and somewhat distant from the proposed adenine nucleotide binding site, so its functional significance, if any, is unknown.
Subunit Va of complex IV (COX5A) interacts with COX4 and to a lesser extent with COX2 on the matrix side of the membrane (212). It has been reported to abolish the allosteric effect of ATP on complex IV kinetics in the presence of thyroid hormones (8) and is readily detected by phosphospecific dyes and by γ-[32P]ATP labeling (6, 83, 161). COX5A is located in contact with the matrix domain of COX4, close to the proposed regulatory ATP binding site (93), and also interacts with the single matrix loop of COX2 (see Fig. 5D). Ser47 and Thr78 were identified as phosphorylation sites in complex IV from fresh bovine heart isolated in the presence of phosphatase inhibitors and after phosphopeptide enrichment (79), suggesting a low mole fraction (as was the case with Ser58 in COX4), which would explain the lack of significant effects on enzymatic activity.
Subunit Vb (COX5B) is also located in the matrix in contact with COX1, COX3, COX4, and COX5A. Its phosphorylation has been detected with PhosTag (6), by protein kinase A in vitro (18), in blue native gels by an intrinsic phosphorylation activity in complex IV (161), and also by P-Ser antibodies (167). Phosphorylation of Ser71 in COX5B was found after ischemia and reperfusion of rabbit hearts followed by blue native electrophoresis of mitochondria without enrichment of phosphopeptides and was sensitive to an inhibitor of protein kinase A (65). However, no attempt to quantify complex IV activity was made to evaluate the functional relevance of this phosphorylation. Ser124 phosphorylation was reported in pig heart COX5B after phosphopeptide enrichment (24), although most other mammals have a Pro residue at this position. Both Ser71 and Ser124 are located away from the contacting surfaces of COX5B with other complex IV subunits and exposed to the solvent (see Fig. 5D), so their functional effect in enzyme catalysis is probably very weak.
Three more subunits in complex IV have phosphorylation sites for which functional significance and mole fraction are unknown. Sites in subunit VIb (COX6B) have recently been reported in mouse, but not in heart or muscle (94), although it has been detected by labeling with γ-[32P]ATP in blue native gels from pig and rat heart (161). COX6B interacts with COX1 and COX2 on the intermembrane space (212) and is reported to slow the rate of complex IV and to modify the affinity for cytochrome c (218). Phosphorylation has also been reported at Ser17 in subunit VIIc (COX7C) in human resting muscle (236) after phosphopeptide enrichment, which is the very NH2-terminal residue of COX7C, and is probably located very close to f-Met 1 of COX1 in the matrix, although its position is not defined in the structure (212). However, since the role of COX6C and COX7C in complex IV activity and/or assembly is obscure, very little can be inferred about possible effects of phosphorylation at these sites or whether they are physiologically relevant at all.
In summary, the functional correlation in cytochrome c and complex IV with specific phosphorylation sites constitutes some of the strongest data in support of a role of protein phosphorylation in the regulation of the cytochrome chain. The potential involvement of cAMP-sensitive kinases, PKA, and PKCε in complex IV is also compelling; however, the precise localization of the kinase activity that interacts with complex IV remains unclear and could simply occur under certain conditions that favor translocation of kinases into the intermembrane space. Furthermore, unknown or low confidence kinase recognition motifs characterize the amino acid sequence where phosphorylation sites are located in cytochrome c and complex IV (92), which is also the case for the majority of other sites reported in the rest of the mitochondrial proteome (see Mitochondrial Matrix Protein Kinases). Thus the actual kinase and phosphatase network regulating these sites is still an open question.
Complex V Phosphorylation
ATP synthesis is accomplished by the rotational catalysis of the complex V enzyme, comprised by the extrinsic F1 region and the membrane-embedded Fo sector. A total of 67 different phosphorylation sites have been reported in 12 of the 16 subunits of mammalian complex V, in addition to five more sites in the inhibitor protein of F1 (IF1) and factor B, which are proteins that associate with complex V to modify its activity. Almost two-thirds of these sites are found in the F1 domain, mainly in the hexamer formed by the α- and β-subunits. Of the remaining sites, 16 are found in four subunits (OSCP, b, d, and F6) that, although traditionally classified as part of the Fo region, actually constitute a membrane-anchored stator that extends to the tip of F1 to hold it in place against the rotation of a central stalk composed of a ring of c-subunits in Fo and of subunits-γ, -δ, and -ε in F1. Only six phosphorylation sites have been reported in four of the eight subunits that constitute the hydrophobic core of the Fo sector. These sites are schematically presented in Fig. 6, A–D.
Fig. 6.
Reported sites of phosphorylation in complex V. In the schematic representation of phosphorylation sites (A), residues in the indicated regions and subunits are shown in white if no functional effect has been reported and their location in the structure suggests no impact on activity. Sites colored in yellow have no known function but are located in a region that might influence activity according to available high-resolution structures. Site that could influence function based on structure and that have been found to change its phosphorylation level in different physiological states are shown in orange. Sites that are located in the presequence and absent in the assembled complex are shown in brackets. The location of several of the reported sites in the F1 region are shown in the crystal structure (PDB ID 2JDI) where only the α- and β-subunits in the “closed” conformation with adenosine 5′-(β,γ-imino) triphosphate (ANP) in the active site are shown (B). Subunit-α is colored blue, subunit-β is red, and subunit-γ is cyan. Other phosphorylation sites (C) are shown in another crystal structure (PDB ID 2WSS) that shows part of the stator attached to one of the α-subunits (blue). The β-subunit closer to the stator is also shown (magenta), with subunit-γ in the center of F1 (cyan) and subunit-δ (pink) at its foot. In the stator region, OSCP is colored brown, subunit-b is orange, F6 is blue, and subunit d is yellow. The IF1 protein with its two reported phosphorylation sites is also shown attached to F1 (D, top), based on a different crystal structure (PDB ID 1OHH). Only the α (green)- and β (red)-subunits with which IF1 (blue) establishes contacts are shown, together with subunit-γ (cyan). The tetrameric (inactive) IF1 is shown (D, bottom) with Ser63 highlighted in one of the oligomerization interfaces (PDB ID 1GMJ). See main text for definitions of abbreviations.
In the F1 region, subunit-α is labeled by both PhosTag and ProQ dyes in pig heart and by PhosTag in pig liver (6, 83). It is also radioactively labeled by incubating pig heart (but not liver) mitochondria with 32P (6), although part of this can be due to metabolite association as indicated by the observation that some labeling occurs when incubating the purified complex V or blue native gel strips with α-[32P]ATP (161). Our own evaluation shows that ADP might be one of the metabolites tightly associated with complex V (R. Covian, R. L. Levine, and R. S. Balaban, unpublished observations). Subunit-α contains 17 reported phosphorylation sites that correspond only to Ser or Thr residues. A cluster of seven sites is located near the NH2-terminal region of the mature protein, with Thr46 establishing contact with the OSCP subunit that forms the tip of the stator (174). This residue was detected in isolated mouse heart mitochondria (58). Ser53 establishes contacts with residues from the adjacent β-subunit and has been found to be phosphorylated in human resting muscle (236). In pig heart, the phosphorylation level of this residue increased threefold when mitochondria were energized with substrates and ATP (24). Ser65 phosphorylation was found in mouse heart (94) and in human muscle (236), but its position in a solvent exposed loop far away from any other subunit makes it unlikely to affect catalysis (34). Ser76 is in close contact with subunit-β, and its phosphorylation has been detected in pig heart (24) and mouse heart (94, 213, 214, 222). However, the functional impact of phosphorylation even at these sites that interact with the OSCP or β-subunits is unclear, given that little conformational changes during the catalytic cycle of complex V is observed in this upper region of the F1 hexamer (84). The same can be said of phosphorylation at Ser106, found in mouse heart (58, 94), given that this residue is away from any subunit interface. Thr236 and Ser521, found to be phosphorylated in mouse heart (94, 222, 231), also fall in the category of functionally irrelevant residues because of their position and poor conservation across species.
Only four residues where phosphorylation has been reported in subunit-α are potentially relevant in a functional sense from a structural point of view (see Fig. 6B). Ser184, Ser419, and Thr432 move significantly during the closure of the catalytic site as the α-β interface cycles between the empty, loose, and tight conformations (203) and are also in the path of substrate entry and exit. Phosphorylation has been found in mouse brown fat and heart for Ser184 and Ser419 (94) and during mouse osteoblast differentiation in the case of Thr432 (105). Another phosphorylation site identified in mouse brain, Ser451, establishes a transient hydrogen bond via a water molecule with an Arg residue at the foot of the γ-subunit that is visible in crystal structures when the α-β interface is in the empty conformation (25). Given that this is a region that changes during rotation of γ, a phosphate group attached to this Ser could prevent this movement either by strengthening the bond between α and γ or by introducing a steric hindrance to the relative displacement of the two subunits. However, the lack of data on the mole fraction of phosphorylation at any of these structurally relevant residues makes any discussion on their functional significance speculative.
Subunit-β is labeled by the PhosTag dye and by incubation of mitochondria with 32P in pig heart and liver (6) and also by ProQ in heart (83). However, most radioactive labeling can be attributed to tight association of adenine nucleotides and phosphate to the binding site, which are revealed as shifts in two-dimensional gels (6). Of the 23 phosphorylation sites reported in subunit-β, those reported in mouse heart or human skeletal muscle are Ser128 (58, 151), Tyr230 (82), and the COOH-terminal residues Ser528 and Ser529 (94, 236). However, these sites seem to have little significance based on their location on the surface of the β-subunit and away from the catalytic site (Fig. 6B). Tyr269, which was found in skeletal muscle (82), is in close contact with the adjacent α-subunit, but in the upper half of F1, where little conformational changes occur during catalysis (25). Thr312 and Ser316, both detected as phosphorylation sites in mouse heart (94) and the first also in human muscle (82), are away from any subunit interface and buried inside the hexamer, which raises questions on how they can be phosphorylated in the assembled complex. Thr312 has also been reported in rabbit ventricular myocytes subjected to pharmacological preconditioning by being exposed to adenosine (9), along with four other sites not reported elsewhere: Ser106, Thr107, Thr262 or Ser263, and Thr368. Except for the last of these residues, their location in the structure does not seem to support a functional role, as shown in a phosphomimetic mutagenic study in yeast that showed primarily assembly defects with some weak direct enzymatic activity changes (101). Thr368 is in contact with the γ-subunit inside the F1 hexamer, but its poor accessibility suggests that its phosphorylation does not occur in the assembled complex V.
Only four phosphorylation sites in subunit-β stand out as potentially regulatory for enzymatic activity based on available structures because of being part of the substrate binding pocket (25). As shown in Fig. 6B, Tyr395 and Thr475 are in contact with the adenine ring, whereas Tyr361 is close to the γ-phosphate of ATP and Thr213 coordinates the Mg ion of the substrate complex. Interestingly, all four residues were found as phosphorylation sites in human skeletal muscle, and both Thr213 and Tyr361 were shown to have a 30% higher phosphorylation in biopsies from obese individuals (82). Tyr361 also showed a 50% increase in its phosphorylation level upon insulin administration in healthy individuals but not in diabetics. However, it is not stated in this study what fraction of sites was phosphorylated at basal levels; an increase of 50% from a fraction of a percent would be meaningless in terms of modifying the overall rate of oxidative phosphorylation. The fact that most sites were detected only after a targeted MS/MS approach indicates that the abundance of phosphopeptides was very low. Since the effects of obesity and diabetes were also related to a decrease in protein levels of oxidative phosphorylation enzymes, including complex V (82), it can be speculated that the phosphorylation of residues in the active site of the β-subunit could be a mechanism to prevent ATP hydrolysis by free F1 that has dissociated from Fo and is in the process of being disassembled for proteolysis. It is also troubling that none of the β-subunit sites label with 32P in intact heart mitochondria (5, 6) and the amount of phosphorylation does not change with a wide range of perturbations (24), suggesting that these sites are not dynamically regulated.
Subunit-γ constitutes the axle that transmits the rotation energy of the ring of c-subunits in Fo to the α-β-hexamer to induce the conformational changes that modify the catalytic sites where adenine nucleotides and phosphate bind (203). It is noteworthy that this subunit is labeled by PhosTag and ProQ dyes in pig heart, but not in liver (6, 83), and that it is labeled with 32P in a calcium-sensitive manner that depends on mitochondrial energization which cannot be explained by metabolite association or intrinsic kinase activity in blue native gels (6, 161), suggesting that an as yet unidentified soluble kinase is responsible for phosphorylation of γ in the mitochondrial matrix. Four phosphorylation sites have been identified in this subunit, two of which could in theory impede the rotation of γ relative to the α-β-hexamer based on the structural information available (25, 203). Thr45 is inside the F1 hexamer in close contact with one of the β-subunits (see Fig. 6, B and C); however, the inaccessibility of this residue from the solvent and its being found only in a human cancer cell line (151), suggest that it is not a physiologically relevant site, perhaps being phosphorylated abnormally when γ is disassembled from the rest of the enzyme. The other residue, Ser108, is less than 7 Å away from the interface with the hexamer (Fig. 6, B and C), so an attached phosphate in an extended conformation of the side chain could make contact with the α- or β-subunits. This site, however, has only been reported in mouse kidney (94), suggesting it is unrelated to the phosphospecific labeling observed in heart (6, 83). Another reported phosphopeptide from bovine heart mitochondria is phosphorylated on either Tyr69 or Tyr77 (63), both of which are over 30 Å away from the interface with the F1 hexamer (Fig. 6C), which in addition to the low abundance of the corresponding phosphopeptide (63) argues against its functional significance in terms of affecting oxidative phosphorylation rates. The most commonly found phosphorylation site in subunit-γ is Ser146, which has been reported in pig heart (24), where its relative abundance did not change significantly upon calcium stimulation of mitochondrial respiration. This residue is ∼ 11 Å from the interface with α and β, much closer than the Tyr residues in the phosphopeptide found in bovine heart (63), but still too far away to propose a direct interference with rotation by the simple addition of a phosphate group (see Fig. 6C).
Phosphorylation of the δ-subunit, which is associated (and rotates with) γ at the interface with the c ring of Fo (203), has been pinpointed to its only Tyr residue (112, 233). However, in a structural sense (Fig. 6C), it is not evident how modification of this residue could affect the rotational catalysis, given that there is no interaction of the δ-subunit with other regions of complex V that do not rotate. Nevertheless, phosphorylation at this site has been linked to PDGF signaling in mouse cortical neurons (233), as well as in fibroblasts and kidney cells (112), although its role, if any, remains obscure.
The stator region of complex V is highly exposed to the matrix. Its function seems to be much less dynamic that the F1 sector: simply holding the latter in place relative to the rotation of γ, although some discussion has taken place regarding the possible effects of flexibility in the stator region during the catalytic cycle (174, 215). Therefore, the phosphorylation of stator subunits does not seem to have an intuitively relevant role in modifying complex V activity, unless it is assumed that phosphorylation takes place on the stator sufficiently close to the middle or lower regions of the F1 hexamer so as to obstruct the outward displacements of α and β as rotation of γ induces the active sites to open and close sequentially. Two of the identified phosphorylation sites that are somewhat close to this region are Ser30 and Ser106 of the d-subunit, detected, respectively, in dog heart (187) and in human skeletal muscle. Some of these sites might also be close to the interface between complex V dimers that has been proposed to be important for oligomerization and subsequent formation of tubular mitochondrial cristae (43), but a role of phosphorylation in this kind of phenomenon is highly speculative at this point.
All other phosphorylation sites identified in the stator region are close to the tip of F1 (see Fig. 6C) where contact is made with the hexamer (174), including Ser155, found in human muscle (236), and Thr145, identified in pig heart, both located in OSCP. Ser226 and Thr229 of subunit-b, and Ser64 and Ser65 (which are not conserved in other mammals) of the F6 subunit, all found in skeletal muscle (236), are also close to this upper region where little functional effects can be expected. It is not known whether any of the above-mentioned sites are responsible for the labeling of the OSCP and d-subunits with PhosTag, ProQ, or 32P detected in heart mitochondria (6, 83, 161). Little can be said about the functional significance of the five phosphorylation sites located in mature Fo subunits because their structure and relation to other subunits is not known. None of these sites has been found in heart or muscle proteomic screens.
Other accessory polypeptides that affect activity of complex V and where phosphorylation sites have been found are factor B and the inhibitor protein (IF1). The first has been implicated in enhancing the coupling of proton movement to ATP synthesis by an as yet unidentified mechanism (16). Three phosphorylation sites (Thr154, Ser162, and Thr167) have been found in this protein, but only in human lymphoma cells (131), so their physiological role is unlikely. IF1 has been proposed to be important to prevent hydrolysis of ATP under ischemic conditions in which mitochondrial membrane potential collapses, binding to the lower region of F1 and preventing the conformational changes required for catalysis in the α- and β-subunits (71). IF1 inhibition takes place under slightly acidic conditions in which its inactive tetrameric form dissociates into a dimer that binds to two complex V molecules simultaneously (30, 31, 70). Two phosphorylation sites, Ser39 and Ser63, have been reported in this protein. The first site was found in human skeletal muscle (236) and is located inside F1 in the dimeric form (see Fig. 6D), in contact with subunit-β (15), so in theory it could be envisioned that phosphorylation at this site might interfere with binding. However, it is not universally conserved, being substituted by Ala in ungulates. The second site was found in cultured cells transfected with an angiotensin receptor (40) and lies on the interface of tetramer formation (32). However, this residue is even less conserved, changing to Thr in rodents and to Ala in ungulates, which precludes the assignment of a physiological role to this site.
In summary, many phosphorylation sites have been identified in complex V; however, the functional significance of these events remains obscure. Furthermore, their kinase/phosphatase system is poorly defined in either the intermembrane space or the matrix.
Mitochondrial Matrix Protein Kinases
From the discussion above it is clear that phosphorylated proteins, at some level, are widespread within the mitochondrial matrix. A few years ago, protein phosphorylation was believed to occur only on PDH or BCKDH; however, the presence of this potential regulatory PTM is much more widespread. The first issue to address is where did the protein phosphorylation occur? Since most of the proteins of the mitochondria come from nuclear DNA, the majority of proteins must traverse the cytosol as well as be processed through a complex protein trafficking pathway (the TOM-TIM complex) across the outer membrane, the intermembrane space, and the remarkably “tight” inner membrane (145). Thus protein phosphorylations could occur along this transport pathway, and if appropriate phosphatases are not present in the matrix, these phosphorylations could persist indefinitely. A screen of the protein phosphorylations and the theoretical kinases interactions was performed by Cui et al. (47), who found that over 52 kinases could be responsible for this profile. However, the vast majority of these kinases lack a mitochondrial targeting sequence (MTS), implying they are not located in the matrix, unless novel protein transport mechanisms are in play. This is also the case in yeast as reviewed by Lubomir (127) where it was suggested that “mitochondria, like obligatory intracellular bacterial parasites, are no longer dependent on signaling mechanisms mediated by protein kinases residing within the mitochondria. Instead, the nucleo-mitochondrial communication system requiring protein phosphorylation may be predominantly regulated by protein kinases, which are cytosolic and/or anchored to the outer mitochondrial membrane.” We would modify this quote to include the intermembrane space and outer regions of the inner membrane based on the observations presented below. Since many of these kinases have not been found in the matrix or possess a MTS, many of the phosphorylation sites in mitochondrial proteins might be the result of cytosolic kinase reactions that take place before being imported, as suggested by Lubomir. Cytosolic or intermembrane space protein phosphorylation has been demonstrated to influence the import of some proteins located both in the cytosol and matrix such as mitochondrial-targeted cytochrome P-450, family 1, subfamily A, polypeptide 1 (51); glutathione S-transferase (172); and complex 1 NDUFS4 (54), supporting this view. We also found many examples of protein phosphorylation in targeting (MTS) portions of the protein, which is consistent with a possible role in protein translocation. However, the extent of proteins that are transported into the matrix with phosphorylations from the cytosol is still poorly defined.
Besides the well-characterized PDH and BCKDH kinase phosphatase system, the best evidence for the existence of matrix kinase activity is protein phosphorylation with 32P in isolated mitochondria. As originally shown by Sale and Randle (179), the addition of 32P to energized mitochondria can be demonstrated to directly phosphorylate PDH in the steady state, suggesting a rapid turnover of these regulatory sites. This high turnover confirmed the existence of an active kinase and phosphatase in the matrix. Subsequently, this approach has been used extensively in intact potato mitochondria (201), isolated potato mitochondria membranes (164, 204), porcine heart mitochondria (5, 6, 83, 161), as well as in blue native gel resolved mitochondrial complexes (161). These studies demonstrate extensive 32P incorporation into matrix-exclusive proteins once proper controls are run for weak and tight metabolite associations (5). Since 32P incorporation requires these sites to turn over, that is, to dephosphorylate and rephosphorylate, these data imply that there is a wide range of matrix kinase and phosphatase reactions occurring that do not require cytosolic elements, and this phosphorylation process is widely conserved phylogenetically.
What are these matrix kinase reactions? There is no question that cytosolic kinases are associated with purified mitochondrial preparations and that these kinases can be recruited to this subcellular fraction under different experimental conditions (23, 104, 115, 129, 139, 182). These later observations have been supported by the association of several kinase anchoring proteins including AKAP (86), SKIP (134), PICK1 (216), Grb10 (143), Sab (221), OMP25 (44), and Rab32 (4). Thus, at least on the outer membrane and outer surface of the inner membrane, there is a growing body of evidence that many kinases are sequestered on the mitochondrion at different times to accomplish a variety of intriguing signaling chores. These intermembrane space kinases include PINK (41), PKA (4, 134), Src kinase (182), and PKCδ (3). However, how these kinases can influence matrix phosphorylation events remains elusive. One of the difficulties in this process is differentiating kinase activities on the surface of the mitochondrion from those occurring in the matrix space. Thus the difference between mitochondrial association and true matrix localization has been difficult to prove.
One of the most extensively studied systems associated with the mitochondria is a cAMP-dependent PKA system that was found in isolated membrane fractions by Papa's group (185, 209) well before the recent developments in screening methodology. Subsequently, it has been demonstrated that PKA can phosphorylate many mitochondrial proteins when combined in vitro (17, 36, 136); however, the evidence that PKA is within the matrix space is still controversial. For example, one of the early studies on the topology of PKA evaluated cAMP-sensitive protein phosphorylation in intact deenergized oligomycin-treated mitochondria using γ[32P]ATP (186). The authors found that the addition of the purified catalytic subunit of PKA was more effective than the endogenous PKA activity at phosphorylating the same protein fractions, suggesting that PKA outside of the matrix could be influencing these results. The authors suggested that this was due to the same molecular weight proteins being fortuitously phosphorylated on the outer surface of the inner membrane; however, the specific sites were not determined in these early studies. An inhibitor of the adenylate translocase blocked much of the native phosphorylation, but not the exogenously added PKA. This latter result suggested that a cAMP-sensitive matrix pool of protein phosphorylation existed; however, the specific activity of the ATP was not determined in these complicated experiments with compromised mitochondria.
Electron microscopy immune-localization of gold particles has been used to confirm the location of PKA on the mitochondrion inner membrane (184, 193); however, whether PKA actually has access to the matrix is still controversial. Indeed, recent reviews have models locating this enzyme in the intermembrane space (66), whereas others propose a matrix-PKA (155). Most recently, Means et al. (134) provided convincing evidence that one of prominent substrates for PKA is the intermembrane helix protein ChChd3, supporting the notion of its localization outside the matrix. Again, the evidence that PKA plays a role in mitochondrial intermembrane space phosphorylation reactions and that a complex “transduceosome” seems to exist in this space is compelling [reviewed in Feliciello et al. (66)]. However, the evidence that PKA is translocated into the matrix is still not as well developed. One of the major issues is how this translocation can occur for this and other kinases that lack a MTS. Sardanelli et al. (184) suggested that dissociated PKA may diffuse through cellular membranes just as it does across the nucleus (75); however, the nuclear membrane is permeable to proteins up to ∼40 kDa, making it very different from the remarkably tight inner membrane of the mitochondria.
To date, the prediction of Lubomir (127) that the cytoplasmic kinases effects seem to be limited to the region of the outer membrane and intermembrane space is clearly correct for the large majority of conventional protein kinases. How these reactions influence the matrix function is a continuing area of investigation. However, it is still unclear how 32P protein phosphorylation is occurring in isolated mitochondria at numerous sites with very few demonstrated matrix kinases. To illustrate this point, a computational search of classical kinase recognition motifs in MOPC subunits that face to the matrix, except complex IV, where a search yielding similar results has already been reported elsewhere (92), is shown in Fig. 7. Out of the >140 reported sites in these subunits, only 19 were identified in silico with at least medium confidence as substrates of known kinases, and almost half of these were in the string of Ser residues located in the NDUFV3 (isoform 2) subunit of complex I (see Fig. 7A). Interestingly, two of the sites with high similarity to consensus recognition sequences are located in the import presequence of ETFA (Fig. 7B) and the β-subunit of complex V (Fig. 7D), clearly indicating that phosphorylation of these residues occurred in the cytosol. Only a handful of sites with medium to high confidence kinase motifs appear in the mature proteins, some of which are thought to be important only for assembly of the complexes such as the accessory subunits UQCRC1, UQCRC2, and UQCR10 of complex III (Fig. 7C) and NDUFA7 of complex I (Fig. 7A). The few sites that are located in catalytically relevant subunits are either in structurally uninteresting regions of the protein, such as Tyr365 of SDHA in complex II, Ser76 and Thr236 of subunit-α, and Ser146 of -γ in complex V, or have only been found in cancer cells, such as Tyr141 of NDUFS2 in complex I and Ser89 of COX4 in complex IV (92, 151). No common kinase activation pathway in the matrix can therefore be discerned in mitochondrial matrix proteins that could support the assumption that oxidative phosphorylation is regulated by cytosolic kinases that are translocated into the matrix.
Fig. 7.
Kinase recognition motifs in selected complex I (A), II (B), III (C), and V (D) subunits located in the mitochondrial matrix. The Scansite 2 program (147) was used to analyze the subunits that face the matrix in the indicated MOPC to identify kinase consensus motif sequences. The confidence level is related to the reciprocal of the percentile score provided by the Scansite program in the following manner: high (red), >5; medium (blue), and 1–5; low (green), 0.2–1. The kinase with the highest score for each site is indicated above the medium and high confidence sites. Residues in brackets are located in the import presequence and are thus not present in the assembled complex. Reported sites that were not recognized at the lowest confidence threshold are not shown. See main text for definitions of abbreviations.
Kinase dependent protein phosphorylation relies on the main product of oxidative phosphorylation, ATP, thus the possibility that [ATP] could regulate protein phosphorylation is feasible and an intriguing feedback mechanism. However, the fact that a metabolic homeostasis exists in the heart, maintaining the [ATP] essentially constant over wide work ranges (11), suggests that [ATP] is not regulatory under normal physiological conditions. However, [ATP] regulation of kinase activity during metabolic compromises could occur if the ATP drops sufficiently. Most classical protein kinases have affinities of 10 to 100 μM for ATP, depending on the [Mg2+], well below the resting millimolar concentration of ATP (195, 206). Based on these classical protein kinase ATP affinity values, the [ATP] would need to drop roughly an order of magnitude to influence protein kinase reaction rates.
Matrix Protein Phosphatases
As is the case for the kinase system, the number of documented phosphatases in the matrix is much lower than those found associated with a mitochondrial enriched fraction from tissues [for example, see Hopper et al. (83)]. Again, the best characterized are the PDHPs where the Ca2+-sensitive PDHP 1 is the major isoform in heart (85), generating the sensitivity of PDH activity to Ca2+. The BCKDH phosphatase protein has also been characterized (50). Recently, a serine/threonine phosphatase was identified in the matrix (PP2Cm), with high-sequence homology with yeast Ptc4, which was shown to be critical in cell survival, possibly responsible for the dephosphorylation of BCKDH (98, 126). Since the phosphorylation of BCKDH was found to change opposite to that of PDH under a number of conditions (24), the differences between these process might be the regulation of PP2Cm. Outside of these phosphatases, the only other confirmed phosphatase is PTMT1, which was identified as a tyrosine phosphatase (152); however, subsequent studies demonstrated it was a phosphatidylglycerophosphate phosphatase that plays a role in cardiolipin biosynthesis (234).
Other phosphatases have been localized to the outer membrane or intermembrane space, such as Aup1p (207), PPL2a (178), Shp-2 (181), PHLPP-1 (138), and DSP-18 (171), consistent with the dominant localization of cytosolic protein kinases in the same space. Thus, even though extensive phosphatase activity is present in the matrix based on 32P turnover and quantitative MS studies (24), the phosphatases of the matrix remain obscure. Added evidence that these phosphatases exist in the aqueous phase in the matrix was the demonstration of phosphatase activity on the inner membrane by a matrix extract in potato mitochondria (204). Clearly, the protein phosphatase system within the mitochondria matrix is as ill defined as are the matrix kinase components, and a clear mechanism to explain the larger turnover of protein phosphorylation in the matrix is yet to be resolved.
Nonconventional Protein Phosphorylation
Nonconventional protein phosphorylations are events not associated with conventional serine, threonine, and tyrosine sites. Of particular interest are the protein phosphorylation events associated with the bacterial system that includes histidine, arginine, aspartate, cysteine, and lysine. Many of these phospho-amino acids have high energy phosphate bonds and permit a phosphotransfer relay between sites without a specific kinase. Recently, it has become appreciated that “remnants” of this signaling systems are present in mammalian cells (99, 110, 117). These are remarkably different from the conventional protein phosphorylation systems in numerous ways. The first is that these sites are highly labile at acid pH, making many conventional proteomic procedures miss them entirely. In addition, the actual phosphorylation reactions are often done via autophosphorylation mechanisms not requiring a specialized secondary kinase. Finally, due to the labile nature of these events, no antibodies are available for detection.
The reasons for suspecting these reactions in the mitochondrion are severalfold. First, many of the bacterial systems involved in protein synthesis, DNA structure, and processing are very similar to the mitochondrial system (153), supporting the notion that the mitochondria originated from some type of symbiotic relationship early in the development of the eukaryotic cell (130). Thus it is not unreasonable to assume that the bacterial signaling systems might also be conserved in the mitochondrion. Interestingly, the bacterial two-component system is particularly suited to generate cellular signals to adapt to extracellular events, much like the mitochondrion samples and “serves” the cytosol. Beyond this rationalization, what is the evidence that these events are occurring in the mitochondria? First, it has been known since the 1960s that most of the 32P associated with mitochondrial proteins is labile at acid pH (122, 160, 168, 190). One of these acid labile sites was identified to be phosphohistidine by several investigators (160, 168, 198). This acid labile phosphorylation pool is also modulated by Ca2+ (146), which is interesting given the proposed role of Ca2+ in regulating mitochondrial energy conversion (10). Estimates by Pressman (168) suggests that up to 99% of the initial 32P protein association is acid labile, implying not only a widespread distribution but also a significant fraction of the total protein phosphorylation in the matrix. This observation was recently confirmed by Aponte et al. (5) in blue native electrophoresis gels of mitochondrial proteins labeled in the intact mitochondrion after 20 min, where most of the 32P labeling was removed by warm acid washing. Nonconventional protein phosphorylation sites are usually initially detected based on their acid sensitivity, since other methods of detection are difficult using conventional proteomic methodologies. Finally, Phillips et al. (161) demonstrated protein phosphorylation by γ-[32P]ATP in oxidative phosphorylation complexes resolved with blue native gel electrophoresis with no detectable kinases associated within the protein complexes. These later results are consistent with the autocatalytic processes in the bacterial phosphorylation pathways.
From a kinase sequence or motif searching approach, the two protein kinases that are unequivocally shown to be in the matrix, PDHK and BCKDH kinase (BCKDHK), have more sequence similarity to bacterial histidine kinases motifs than to cytosolic serine protein kinases (77). However, no evidence exists that these kinases phosphorylate anything other than the E1α serines of their target proteins. We and others have screened the mammalian mitochondrial nuclear and mitochondrial genome for the limited consensus eukaryotic phospho-histidine kinases (154) and other nonconventional kinases motifs with little success. Specifically, we have used the bacterial histidine kinase motifs (PF02895) from PFAM (169) to BLAST against the mouse genome. A brief overview of these searches with regard to mitochondrial proteins include 1) IPR003660 HAMP linker domain; no proteins; 2) IPR005467 signal transduction histidine kinase, core; no proteins; 3) IPR003594 ATPase-like, ATP-binding domain; the previously identified BCKDHK and PDH isoforms, as well as heat shock protein 75 (TNF receptor-associated protein 1), and topoisomerase; 4) IPR010559 signal transduction histidine kinase, internal region; no protein; and 5) IPR004358 signal transduction histidine kinase-related protein, COOH-terminal; apparent isoforms of BCKDHK. This search confirms the sequence similarity of PDHK and BCKDHK with bacterial kinases, whereas any protein kinase activity associated with the mitochondrial heat shock proteins and topoisomerase ATPase activity has to our knowledge not been reported but might be an interesting area of investigation. However, despite the considerable evidence for labile phosphorylation events, the limited detection of bacterial histidine kinase motifs, generated from bacteria or simple eukaryotic systems, suggests that either new motifs have evolved in mammalia or completely different mechanisms for these labile phosphorylation sites are in play. The evolution of new motifs is not surprising given the different environmental pressures and nuclear genomic recombination occurring in the mitochondria versus bacterial systems.
Though definitive evidence is not available for the existence of these nonconventional protein phosphorylations in the mitochondrial matrix, the circumstantial evidences discussed, as well as the apparent origins of many mitochondrial matrix reactions, suggest that a careful examination of this process should be conducted. Recent developments in MS methods suitable for the identification of these sites should provide useful tools in this effort (21, 117, 238).
Conclusions and Future Directions
It is clear that mitochondrial matrix protein phosphorylation is extensive when detected at high sensitivity. However, the functional impact of these phosphorylations and their kinase/phosphatase system remain generally obscure for most of the sites identified. We take the general position that most of the sites detected with these high-sensitivity approaches are not impacting enzyme function but are low level phosphorylation “noise” from numerous protein kinase reactions. The issue in the field is separating the noise from the regulatory sites. One of the major limitations for the metabolic enzymes is the lack of information on the fraction of the enzyme that is phosphorylated. The development of quantitative protein phosphorylation methods is underway around the world and, we predict, will greatly reduce the number candidate sites for regulatory activity in the energy metabolism enzyme network in two ways: first, establishing the fraction of the enzymatic pool phosphorylated and second, quantitatively relating the amount of phosphorylation with the enzyme activity [for example, see Boja et al. (24)]. It is important to note that cataloging the phosphorylation sites is still very important for several reasons. First, the detection of the phosphorylation event confirms that the site can be modified by existing kinases. Second, the limited functional analysis we perform and the conditions we can create may miss the functional consequences of these phosphorylations on processes such as assembly, transport, and unknown functions that we cannot or know not to assay.
Many protein kinases and phosphatases are selectively targeted to the outer compartments of the mitochondria through very sophisticated cell signaling processes; however, the functional significance of this localization is less defined. With regard to the matrix, the currently established matrix kinase and phosphatase system is inadequate to explain the extent of protein phosphorylation detected. One possibility was that most of the phosphorylation events occur in the cytosol or kinase rich intermembrane space and outer mitochondrial membrane before, or during, protein transport into the mitochondria. However, 32P studies on intact mitochondria clearly establish extensive matrix protein kinase activity. In silico surveys of the phosphorylation sites suggest no specific conventional protein kinases responsible for the phosphorylation patterns observed, with some exceptions. Thus the mechanisms for much of the matrix protein phosphorylation detected, especially with 32P labeling in intact mitochondria, remain unknown. The mitochondrion has an early bacterial origin, and based on several lines of evidence including the pH and temporal sensitivity of protein phosphorylation and the presence of autophosphorylation events within the complexes, it can be suggested that bacterial nonconventional protein phosphorylation transfer events may be playing a role in protein phosphorylation. The test of this hypothesis will require the application of nonconventional methods to monitor these putative high-energy protein phosphorylation events. It is important to note that very little genetic evidence exists for conserved nonconventional kinase activity in the mitochondrial proteome. These later results suggest that if these bacterial signaling pathways exist, they have likely evolved new functional motifs.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author (s).
AUTHOR CONTRIBUTIONS
R.C. and R.S.B. conception and design of research; R.C. and R.S.B. analyzed data; R.C. and R.S.B. interpreted results of experiments; R.C. and R.S.B. prepared figures; R.C. and R.S.B. drafted manuscript; R.C. and R.S.B. edited and revised manuscript; R.C. and R.S.B. approved final version of manuscript; R.S.B. performed experiments.
REFERENCES
- 1. Acin-Perez R, Gatti DL, Bai Y, Manfredi G. Protein phosphorylation and prevention of cytochrome oxidase inhibition by ATP: coupled mechanisms of energy metabolism regulation. Cell Metab 13: 712–719, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acin-Perez R, Salazar E, Kamenetsky M, Buck J, Levin LR, Manfredi G. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab 9: 265–276, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Acin-Perez R, Hoyos B, Gong J, Vinogradov V, Fischman DA, Leitges M, Borhan B, Starkov A, Manfredi G, Hammerling U. Regulation of intermediary metabolism by the PKCdelta signalosome in mitochondria. FASEB J 24: 5033–5042, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alto NM, Soderling J, Scott JD. Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J Cell Biol 158: 659–668, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aponte AM, Phillips D, Harris RA, Blinova K, French S, Johnson DT, Balaban RS. 32P labeling of protein phosphorylation and metabolite association in the mitochondria matrix. Methods Enzymol 457: 63–80, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aponte AM, Phillips D, Hopper RK, Johnson DT, Harris RA, Blinova K, Boja ES, French S, Balaban RS. Use of (32)P to study dynamics of the mitochondrial phosphoproteome. J Proteome Res 8: 2679–2695, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aprille JR. Mechanism and regulation of the mitochondrial ATP-Mg/P(i) carrier. J Bioenerg Biomembr 25: 473–481, 1993 [DOI] [PubMed] [Google Scholar]
- 8. Arnold S, Goglia F, Kadenbach B. 3,5-Diiodothyronine binds to subunit Va of cytochrome-c oxidase and abolishes the allosteric inhibition of respiration by ATP. Eur J Biochem 252: 325–330, 1998 [DOI] [PubMed] [Google Scholar]
- 9. Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, Robinson J, Murata M, Murphy AM, Marbán E, Van Eyk JE. Proteomic analysis of pharmacological preconditioning: novel protein targets converge to mitochondrial metabolism pathways. Circ Res 99: 706–714, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Balaban RS. Cardiac energy metabolism homeostasis: role of cytosolic calcium. J Mol Cell Cardiol 34: 1259–1271, 2002 [DOI] [PubMed] [Google Scholar]
- 11. Balaban RS. Maintenance of the metabolic homeostasis of the heart: developing a systems analysis approach. Ann NY Acad Sci 1080: 140–153, 2006 [DOI] [PubMed] [Google Scholar]
- 12. Balaban RS. Domestication of the cardiac mitochondrion for energy conversion. J Mol Cell Cardiol 46: 832–841, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balaban RS. The role of Ca2+ signaling in the coordination of mitochondrial ATP production with cardiac work. Biochim Biophys Acta 1787: 1334–1341, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baranova EA, Holt PJ, Sazanov LA. Projection structure of the membrane domain of Escherichia coli respiratory complex I at 8 A resolution. J Mol Biol 366: 140–154, 2007 [DOI] [PubMed] [Google Scholar]
- 15. Bason JV, Runswick MJ, Fearnley IM, Walker JE. Binding of the inhibitor protein IF(1) to bovine F(1)-ATPase. J Mol Biol 406: 443–453, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Belogrudov GI. Recent advances in structure-functional studies of mitochondrial factor B. J Bioenerg Biomembr 41: 137–143, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bender A, Hajieva P, Moosmann B. Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc Natl Acad Sci USA 105: 16496–16501, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bender E, Kadenbach B. The allosteric ATP-inhibition of cytochrome c oxidase activity is reversibly switched on by cAMP-dependent phosphorylation. FEBS Lett 466: 130–134, 2000 [DOI] [PubMed] [Google Scholar]
- 19. Bera AK, Ghosh S. Dual mode of gating of voltage-dependent anion channel as revealed by phosphorylation. J Struct Biol 135: 67–72, 2001 [DOI] [PubMed] [Google Scholar]
- 20. Berrisford JM, Sazanov LA. Structural basis for the mechanism of respiratory complex I. J Biol Chem 284: 29773–29783, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Besant PG, Attwood PV. Detection and analysis of protein histidine phosphorylation. Mol Cell Biochem 329: 93–106, 2009 [DOI] [PubMed] [Google Scholar]
- 22. Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4: 1633–1649, 2004 [DOI] [PubMed] [Google Scholar]
- 23. Boerner JL, Demory ML, Silva C, Parsons SJ. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol Cell Biol 24: 7059–7071, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boja ES, Phillips D, French SA, Harris RA, Balaban RS. Quantitative mitochondrial phosphoproteomics using iTRAQ on an LTQ-Orbitrap with high energy collision dissociation. J Proteome Res 8: 4665–4675, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bowler MW, Montgomery MG, Leslie AG, Walker JE. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 A resolution. J Biol Chem 282: 14238–14242, 2007 [DOI] [PubMed] [Google Scholar]
- 26. Brandt U. Energy converting NADH: quinone oxidoreductase (complex I). Annu Rev Biochem 75: 69–92, 2006 [DOI] [PubMed] [Google Scholar]
- 27. Brasseur G, Saribas AS, Daldal F. A compilation of mutations located in the cytochrome b subunit of the bacterial and mitochondrial bc1 complex. Biochim Biophys Acta 1275: 61–69, 1996 [DOI] [PubMed] [Google Scholar]
- 28. Buxton DB, Olson MS. Regulation of the branched chain alpha-keto acid and pyruvate dehydrogenases in the perfused rat heart. J Biol Chem 257: 15026–15029, 1982 [PubMed] [Google Scholar]
- 29. Bykova NV, Egsgaard H, Moller IM. Identification of 14 new phosphoproteins involved in important plant mitochondrial processes. FEBS Lett 540: 141–146, 2003 [DOI] [PubMed] [Google Scholar]
- 30. Cabezon E, Arechaga I, Jonathan P, Butler G, Walker JE. Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1. J Biol Chem 275: 28353–28355, 2000 [DOI] [PubMed] [Google Scholar]
- 31. Cabezon E, Montgomery MG, Leslie AG, Walker JE. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat Struct Biol 10: 744–750, 2003 [DOI] [PubMed] [Google Scholar]
- 32. Cabezon E, Runswick MJ, Leslie AG, Walker JE. The structure of bovine IF(1), the regulatory subunit of mitochondrial F-ATPase. EMBO J 20: 6990–6996, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates JR., 3rd Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res 7: 1346–1351, 2008 [DOI] [PubMed] [Google Scholar]
- 34. Carbajo RJ, Kellas FA, Yang JC, Runswick MJ, Montgomery MG, Walker JE, Neuhaus D. How the N-terminal domain of the OSCP subunit of bovine F1Fo-ATP synthase interacts with the N-terminal region of an alpha subunit. J Mol Biol 368: 310–318, 2007 [DOI] [PubMed] [Google Scholar]
- 35. Castro L, Demicheli V, Tortora V, Radi R. Mitochondrial protein tyrosine nitration. Free Radic Res 45: 37–52, 2011 [DOI] [PubMed] [Google Scholar]
- 36. Chen R, Fearnley IM, Peak-Chew SY, Walker JE. The phosphorylation of subunits of complex I from bovine heart mitochondria. J Biol Chem 279: 26036–26045, 2004 [DOI] [PubMed] [Google Scholar]
- 37. Chen Y, Lu B, Yang Q, Fearns C, Yates JR, III, Lee JD. Combined integrin phosphoproteomic analyses and small interfering RNA-based functional screening identify key regulators for cancer cell adhesion and migration. Cancer Res 69: 3713–3720, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y, Gaczynska M, Osmulski P, Polci R, Riley DJ. Phosphorylation by Nek1 regulates opening and closing of voltage dependent anion channel 1. Biochem Biophys Res Commun 394: 798–803, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PN, Bohmer FD, Gerke V, Schmidt-Arras DE, Berdel WE, Muller-Tidow C, Mann M, Serve H. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol Cell 36: 326–339, 2009 [DOI] [PubMed] [Google Scholar]
- 40. Christensen GL, Kelstrup CD, Lyngso C, Sarwar U, Bogebo R, Sheikh SP, Gammeltoft S, Olsen JV, Hansen JL. Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol Cell Proteomics 9: 1540–1553, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chu CT. Tickled PINK1: mitochondrial homeostasis and autophagy in recessive Parkinsonism. Biochim Biophys Acta 1802: 20–28, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Colombini M. VDAC: the channel at the interface between mitochondria and the cytosol. Mol Cell Biochem 256–257: 107–115, 2004 [DOI] [PubMed] [Google Scholar]
- 43. Couoh-Cardel SJ, Uribe-Carvajal S, Wilkens S, Garcia-Trejo JJ. Structure of dimeric F1F0-ATP synthase. J Biol Chem 285: 36447–36455, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Court N, Ingley E, Klinken SP, Bogoyevitch MA. Outer membrane protein 25-a mitochondrial anchor and inhibitor of stress-activated protein kinase-3. Biochim Biophys Acta 1744: 68–75, 2005 [DOI] [PubMed] [Google Scholar]
- 45. Covian R, Trumpower BL. Regulatory interactions between ubiquinol oxidation and ubiquinone reduction sites in the dimeric cytochrome bc1 complex. J Biol Chem 281: 30925–30932, 2006 [DOI] [PubMed] [Google Scholar]
- 46. Crofts AR, Guergova-Kuras M, Huang L, Kuras R, Zhang Z, Berry EA. Mechanism of ubiquinol oxidation by the bc(1) complex: role of the iron sulfur protein and its mobility. Biochemistry 38: 15791–15806, 1999 [DOI] [PubMed] [Google Scholar]
- 47. Cui Z, Hou J, Chen X, Li J, Xie Z, Xue P, Cai T, Wu P, Xu T, Yang F. The profile of mitochondrial proteins and their phosphorylation signaling network in INS-1 beta cells. J Proteome Res 9: 2898–2908, 2010 [DOI] [PubMed] [Google Scholar]
- 48. Dai J, Jin WH, Sheng QH, Shieh CH, Wu JR, Zeng R. Protein phosphorylation and expression profiling by Yin-yang multidimensional liquid chromatography (Yin-yang MDLC) mass spectrometry. J Proteome Res 6: 250–262, 2007 [DOI] [PubMed] [Google Scholar]
- 49. Damuni Z, Merryfield ML, Humphreys JS, Reed LJ. Purification and properties of branched-chain alpha-keto acid dehydrogenase phosphatase from bovine kidney. Proc Natl Acad Sci USA 81: 4335–4338, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Damuni Z, Reed LJ. Purification and properties of the catalytic subunit of the branched-chain alpha-keto acid dehydrogenase phosphatase from bovine kidney mitochondria. J Biol Chem 262: 5129–5132, 1987 [PubMed] [Google Scholar]
- 51. Dasari VR, Anandatheerthavarada HK, Robin MA, Boopathi E, Biswas G, Fang JK, Nebert DW, Avadhani NG. Role of protein kinase C-mediated protein phosphorylation in mitochondrial translocation of mouse CYP1A1, which contains a non-canonical targeting signal. J Biol Chem 281: 30834–30847, 2006 [DOI] [PubMed] [Google Scholar]
- 52. Dave KR, Defazio RA, Raval AP, Torraco A, Saul I, Barrientos A, Perez-Pinzon MA. Ischemic preconditioning targets the respiration of synaptic mitochondria via protein kinase C epsilon. J Neurosci 28: 4172–4182, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Davie JR, Wynn RM, Meng M, Huang YS, Aalund G, Chuang DT, Lau KS. Expression and characterization of branched-chain alpha-ketoacid dehydrogenase kinase from the rat. Is it a histidine-protein kinase? J Biol Chem 270: 19861–19867, 1995 [DOI] [PubMed] [Google Scholar]
- 54. De Rasmo D, Panelli D, Sardanelli AM, Papa S. cAMP-dependent protein kinase regulates the mitochondrial import of the nuclear encoded NDUFS4 subunit of complex I. Cell Signal 20: 989–997, 2008 [DOI] [PubMed] [Google Scholar]
- 55. De Rasmo D, Palmisano G, Scacco S, Technikova-Dobrova Z, Panelli D, Cocco T, Sardanelli AM, Gnoni A, Micelli L, Trani A, Di Luccia A, Papa S. Phosphorylation pattern of the NDUFS4 subunit of complex I of the mammalian respiratory chain. Mitochondrion 10: 464–471, 2010 [DOI] [PubMed] [Google Scholar]
- 57. Deng K, Shenoy SK, Tso SC, Yu L, Yu CA. Reconstitution of mitochondrial processing peptidase from the core proteins (subunits I and II) of bovine heart mitochondrial cytochrome bc(1) complex. J Biol Chem 276: 6499–6505, 2001 [DOI] [PubMed] [Google Scholar]
- 58. Deng N, Zhang J, Zong C, Wang Y, Lu H, Yang P, Wang W, Young GW, Wang Y, Korge P, Lotz C, Doran P, Liem DA, Apweiler R, Weiss JN, Duan H, Ping P. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Mol Cell Proteomics. 2011. February; 10(2):M110.000117. Epub 2010 May 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Denton RM. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim Biophys Acta 1787: 1309–1316, 2009 [DOI] [PubMed] [Google Scholar]
- 60. Denton RM, Randle P, Martin BR. Stimulation by calciums ions of pyruvate dehydrogenase phosphate phosphatase. Biochem J 128: 161–163, 1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA 105: 10762–10767, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Depontieu FR, Qian J, Zarling AL, McMiller TL, Salay TM, Norris A, English AM, Shabanowitz J, Engelhard VH, Hunt DF, Topalian SL. Identification of tumor-associated, MHC class II-restricted phosphopeptides as targets for immunotherapy. Proc Natl Acad Sci USA 106: 12073–12078, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Di PF, Bisetto E, Alverdi V, Mavelli I, Esposito G, Lippe G. Differential steady-state tyrosine phosphorylation of two oligomeric forms of mitochondrial F0F1ATPsynthase: a structural proteomic analysis. Proteomics 6: 921–926, 2006 [DOI] [PubMed] [Google Scholar]
- 64. Efremov RG, Sazanov LA. Structure of the membrane domain of respiratory complex I. Nature 476: 414–420, 2011 [DOI] [PubMed] [Google Scholar]
- 65. Fang JK, Prabu SK, Sepuri NB, Raza H, Anandatheerthavarada HK, Galati D, Spear J, Avadhani NG. Site specific phosphorylation of cytochrome c oxidase subunits I, IVi1 and Vb in rabbit hearts subjected to ischemia/reperfusion. FEBS Lett 581: 1302–1310, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Feliciello A, Gottesman ME, Avvedimento EV. cAMP-PKA signaling to the mitochondria: protein scaffolds, mRNA and phosphatases. Cell Signal 17: 279–287, 2005 [DOI] [PubMed] [Google Scholar]
- 67. Feng J, Lucchinetti E, Enkavi G, Wang Y, Gehrig P, Roschitzki B, Schaub MC, Tajkhorshid E, Zaugg K, Zaugg M. Tyrosine phosphorylation by Src within the cavity of the adenine nucleotide translocase 1 regulates ADP/ATP exchange in mitochondria. Am J Physiol Cell Physiol 298: C740–C748, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Feng J, Zhu M, Schaub MC, Gehrig P, Roschitzki B, Lucchinetti E, Zaugg M. Phosphoproteome analysis of isoflurane-protected heart mitochondria: phosphorylation of adenine nucleotide translocator-1 on Tyr194 regulates mitochondrial function. Cardiovasc Res 80: 20–29, 2008 [DOI] [PubMed] [Google Scholar]
- 69. Galante YM, Hatefi Y. Resolution of complex I and isolation of NADH dehydrogenase and an iron-sulfur protein. Methods Enzymol 53: 15–21, 1978 [DOI] [PubMed] [Google Scholar]
- 70. Garcia JJ, Morales-Rios E, Cortes-Hernandez P, Rodriguez-Zavala JS. The inhibitor protein (IF1) promotes dimerization of the mitochondrial F1F0-ATP synthase. Biochemistry 45: 12695–12703, 2006 [DOI] [PubMed] [Google Scholar]
- 71. Gledhill JR, Montgomery MG, Leslie AG, Walker JE. How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc Natl Acad Sci USA 104: 15671–15676, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Goodwin GW, Taylor CS, Taegtmeyer H. Regulation of energy metabolism of the heart during acute increase in heart work. J Biol Chem 273: 29530–29539, 1998 [DOI] [PubMed] [Google Scholar]
- 73. Gu TL, Deng X, Huang F, Tucker M, Crosby K, Rimkunas V, Wang Y, Deng G, Zhu L, Tan Z, Hu Y, Wu C, Nardone J, Macneill J, Ren J, Reeves C, Innocenti G, Norris B, Yuan J, Yu J, Haack H, Shen B, Peng C, Li H, Zhou X, Liu X, Rush J, Comb MJ. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One 6: e15640, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Han G, Ye M, Liu H, Song C, Sun D, Wu Y, Jiang X, Chen R, Wang C, Wang L, Zou H. Phosphoproteome analysis of human liver tissue by long-gradient nanoflow LC coupled with multiple stage MS analysis. Electrophoresis 31: 1080–1089, 2010 [DOI] [PubMed] [Google Scholar]
- 75. Harootunian AT, Adams SR, Wen W, Meinkoth JL, Taylor SS, Tsien RY. Movement of the free catalytic subunit of cAMP-dependent protein kinase into and out of the nucleus can be explained by diffusion. Mol Biol Cell 4: 993–1002, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annu Rev Nutr 4: 409–454, 1984 [DOI] [PubMed] [Google Scholar]
- 77. Harris RA, Popov KM, Zhao Y, Kedishvili NY, Shimomura Y, Crabb DW. A new family of protein kinases—the mitochondrial protein kinases. Adv Enzyme Regul 35: 147–162, 1995 [DOI] [PubMed] [Google Scholar]
- 78. He L, Lemasters JJ. Dephosphorylation of the Rieske iron-sulfur protein after induction of the mitochondrial permeability transition. Biochem Biophys Res Commun 334: 829–837, 2005 [DOI] [PubMed] [Google Scholar]
- 79. Helling S, Vogt S, Rhiel A, Ramzan R, Wen L, Marcus K, Kadenbach B. Phosphorylation and kinetics of mammalian cytochrome c oxidase. Mol Cell Proteomics 7: 1714–1724, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hoffert JD, Nielsen J, Yu MJ, Pisitkun T, Schleicher SM, Nielsen S, Knepper MA. Dynamics of aquaporin-2 serine-261 phosphorylation in response to short-term vasopressin treatment in collecting duct. Am J Physiol Renal Physiol 292: F691–F700, 2007 [DOI] [PubMed] [Google Scholar]
- 81. Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA 103: 7159–7164, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hojlund K, Yi Z, Lefort N, Langlais P, Bowen B, Levin K, Beck-Nielsen H, Mandarino LJ. Human ATP synthase beta is phosphorylated at multiple sites and shows abnormal phosphorylation at specific sites in insulin-resistant muscle. Diabetologia 53: 541–551, 2010 [DOI] [PubMed] [Google Scholar]
- 83. Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry 45: 2524–2536, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332: 1317–1322, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huang B, Gudi R, Wu P, Harris RA, Hamilton J, Popov KM. Isoenzymes of pyruvate dehydrogenase phosphatase. J Biol Chem 273: 17680–17688, 1998 [DOI] [PubMed] [Google Scholar]
- 86. Huang LJ, Wang L, Ma Y, Durick K, Perkins G, Deerinck TJ, Ellisman MH, Taylor SS. NH2-terminal targeting motifs direct dual specificity A-kinase-anchoring protein 1 (D-AKAP1) to either mitochondria or endoplasmic reticulum. J Cell Biol 145: 951–959, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Huang LS, Cobessi D, Tung EY, Berry EA. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J Mol Biol 351: 573–597, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang SY, Tsai ML, Chen GY, Wu CJ, Chen SH. A systematic MS-based approach for identifying in vitro substrates of PKA and PKG in rat uteri. J Proteome Res 6: 2674–2684, 2007 [DOI] [PubMed] [Google Scholar]
- 89. Hughes WA, Halestrap AP. The regulation of branched-chain 2-oxo acid dehydrogenase of liver, kidney and heart by phosphorylation. Biochem J 196: 459–469, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hunte C, Palsdottir H, Trumpower BL. Protonmotive pathways and mechanisms in the cytochrome bc1 complex. FEBS Lett 545: 39–46, 2003 [DOI] [PubMed] [Google Scholar]
- 91. Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Glutathionylation of mitochondrial proteins. Antioxid Redox Signal 7: 999–1010, 2005 [DOI] [PubMed] [Google Scholar]
- 92. Huttemann M, Helling S, Sanderson TH, Sinkler C, Samavati L, Mahapatra G, Varughese A, Lu G, Liu J, Ramzan R, Vogt S, Grossman LI, Doan JW, Marcus K, Lee I. Regulation of mitochondrial respiration and apoptosis through cell signaling: cytochrome c oxidase and cytochrome c in ischemia/reperfusion injury and inflammation. Biochim Biophys Acta 1817: 598–609, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Huttemann M, Kadenbach B, Grossman LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene 267: 111–123, 2001 [DOI] [PubMed] [Google Scholar]
- 94. Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Imami K, Sugiyama N, Kyono Y, Tomita M, Ishihama Y. Automated phosphoproteome analysis for cultured cancer cells by two-dimensional nanoLC-MS using a calcined titania/C18 biphasic column. Anal Sci 24: 161–166, 2008 [DOI] [PubMed] [Google Scholar]
- 96. Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281: 64–71, 1998 [DOI] [PubMed] [Google Scholar]
- 97. Johnson ET, Parson WW. Electrostatic interactions in an integral membrane protein. Biochemistry 41: 6483–6494, 2002 [DOI] [PubMed] [Google Scholar]
- 98. Joshi M, Jeoung NH, Popov KM, Harris RA. Identification of a novel PP2C-type mitochondrial phosphatase. Biochem Biophys Res Commun 356: 38–44, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jung K, Jung H. A new mechanism of phosphoregulation in signal transduction pathways. Sci Signal 2: e71, 2009 [DOI] [PubMed] [Google Scholar]
- 100. Kadenbach B, Napiwotzki J, Frank V, Arnold S, Exner S, Huttemann M. Regulation of energy transduction and electron transfer in cytochrome c oxidase by adenine nucleotides. J Bioenerg Biomembr 30: 25–33, 1998 [DOI] [PubMed] [Google Scholar]
- 101. Kane LA, Youngman MJ, Jensen RE, Van Eyk JE. Phosphorylation of the F(1)F(o) ATP synthase beta subunit: functional and structural consequences assessed in a model system. Circ Res 106: 504–513, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kerbey AL, Randle PJ, Cooper RH, Whitehouse S, Pask HT, Denton RM. Regulation of pyruvate dehydrogenase in rat heart. Mechanism of regulation of proportions of dephosphorylated and phosphorylated enzyme by oxidation of fatty acids and ketone bodies and of effects of diabetes: role of coenzyme A, acetyl-coenzyme A and reduced and oxidized nicotinamide-adenine dinucleotide. Biochem J 154: 327–348, 1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kerner J, Distler AM, Minkler P, Parland W, Peterman SM, Hoppel CL. Phosphorylation of rat liver mitochondrial carnitine palmitoyltransferase-I: effect on the kinetic properties of the enzyme. J Biol Chem 279: 41104–41113, 2004 [DOI] [PubMed] [Google Scholar]
- 104. Kharbanda S, Saxena S, Yoshida K, Pandey P, Kaneki M, Wang Q, Cheng K, Chen YN, Campbell A, Sudha T, Yuan ZM, Narula J, Weichselbaum R, Nalin C, Kufe D. Translocation of SAPK/JNK to mitochondria and interaction with Bcl-xL in response to DNA damage. J Biol Chem 275: 322–327, 2000 [DOI] [PubMed] [Google Scholar]
- 105. Kim BG, Lee JH, Ahn JM, Park SK, Cho JH, Hwang D, Yoo JS, Yates JR, 3rd, Ryoo HM, Cho JY. ‘Two-stage double-technique hybrid (TSDTH)’ identification strategy for the analysis of BMP2-induced transdifferentiation of premyoblast C2C12 cells to osteoblast. J Proteome Res 8: 4441–4454, 2009 [DOI] [PubMed] [Google Scholar]
- 106. Kim CH, Yencha AJ, Bunker G, Zhang G, Chance B, King TE. Effect of the hinge protein on the heme iron site of cytochrome c1. Biochemistry 28: 1439–1441, 1989 [DOI] [PubMed] [Google Scholar]
- 107. Kim SC, Chen Y, Mirza S, Xu Y, Lee J, Liu P, Zhao Y. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J Proteome Res 5: 3446–3452, 2006 [DOI] [PubMed] [Google Scholar]
- 108. King TE, Kim CH. Preparation of hinge protein and its requirement for interaction of cytochrome c with cytochrome c1. Methods Enzymol 126: 238–253, 1986 [DOI] [PubMed] [Google Scholar]
- 109. Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta 1778: 1978–2021, 2008 [DOI] [PubMed] [Google Scholar]
- 110. Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in proteins from vertebrates. Sci Signal 2: e13, 2009 [DOI] [PubMed] [Google Scholar]
- 111. Knebel A, Morrice N, Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. EMBO J 20: 4360–4369, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ko YH, Pan W, Inoue C, Pedersen PL. Signal transduction to mitochondrial ATP synthase: evidence that PDGF-dependent phosphorylation of the delta-subunit occurs in several cell lines, involves tyrosine, and is modulated by lysophosphatidic acid. Mitochondrion 1: 339–348, 2002 [DOI] [PubMed] [Google Scholar]
- 113. Kobayashi K, Neely JR. Mechanism of pyruvate dehydrogenase activation by increased cardiac work. J Mol Cell Cardiol 15: 369–382, 1983 [DOI] [PubMed] [Google Scholar]
- 114. Krebs EG, Fischer EH. The phosphorylase b to a converting enzyme of rabbit skeletal muscle. Biochim Biophys Acta 20: 150–157, 1956 [DOI] [PubMed] [Google Scholar]
- 115. Kumar S, Bharti A, Mishra NC, Raina D, Kharbanda S, Saxena S, Kufe D. Targeting of the c-Abl tyrosine kinase to mitochondria in the necrotic cell death response to oxidative stress. J Biol Chem 276: 17281–17285, 2001 [DOI] [PubMed] [Google Scholar]
- 116. Lange C, Hunte C. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc Natl Acad Sci USA 99: 2800–2805, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Lapek JD, Jr, Tombline G, Friedman AE. Mass spectrometry detection of histidine phosphorylation on NM23-H1. J Proteome Res 10: 751–755, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lee I, Salomon AR, Ficarro S, Mathes I, Lottspeich F, Grossman LI, Huttemann M. cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity. J Biol Chem 280: 6094–6100, 2005 [DOI] [PubMed] [Google Scholar]
- 119. Lee I, Salomon AR, Yu K, Doan JW, Grossman LI, Huttemann M. New prospects for an old enzyme: mammalian cytochrome c is tyrosine-phosphorylated in vivo. Biochemistry 45: 9121–9128, 2006 [DOI] [PubMed] [Google Scholar]
- 120. Lee I, Salomon AR, Yu K, Samavati L, Pecina P, Pecinova A, Huttemann M. Isolation of regulatory-competent, phosphorylated cytochrome C oxidase. Methods Enzymol 457: 193–210, 2009 [DOI] [PubMed] [Google Scholar]
- 121. Lee J, Xu Y, Chen Y, Sprung R, Kim SC, Xie S, Zhao Y. Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Mol Cell Proteomics 6: 669–676, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Linberg O, Duffy JJ, Norman AW, Boyer PD. Characteristics of bound phosphohistidine labeling in mitochondria. J Biol Chem 240: 2850–2854, 1965 [PubMed] [Google Scholar]
- 123. Linn TC, Pettit FH, Reed LJ. Alpha-keto acid dehydrogenase complexes. X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc Natl Acad Sci USA 62: 234–241, 1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Louro RO, Catarino T, Paquete CM, Turner DL. Distance dependence of interactions between charged centres in proteins with common structural features. FEBS Lett 576: 77–80, 2004 [DOI] [PubMed] [Google Scholar]
- 125. Lu G, Sun H, Korge P, Koehler CM, Weiss JN, Wang Y. Functional characterization of a mitochondrial Ser/Thr protein phosphatase in cell death regulation. Methods Enzymol 457: 255–273, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lu G, Sun H, She P, Youn JY, Warburton S, Ping P, Vondriska TM, Cai H, Lynch CJ, Wang Y. Protein phosphatase 2Cm is a critical regulator of branched-chain amino acid catabolism in mice and cultured cells. J Clin Invest 119: 1678–1687, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lubomir T. Mitochondrial protein phosphorylation: lessons from yeasts. Gene 255: 59–64, 2000 [DOI] [PubMed] [Google Scholar]
- 128. Lund AA, Rhoads DM, Lund AL, Cerny RL, Elthon TE. In vivo modifications of the maize mitochondrial small heat stress protein, HSP22. J Biol Chem 276: 29924–29929, 2001 [DOI] [PubMed] [Google Scholar]
- 129. Majumder PK, Mishra NC, Sun X, Bharti A, Kharbanda S, Saxena S, Kufe D. Targeting of protein kinase C delta to mitochondria in the oxidative stress response. Cell Growth Differ 12: 465–470, 2001 [PubMed] [Google Scholar]
- 130. Margulis L, Sagan D. Acquiring Genomes: A Yheory of the Origins of Species. New York: Basic Books, 2002 [Google Scholar]
- 131. Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal 2: ra46, 2009 [DOI] [PubMed] [Google Scholar]
- 132. McCormack JG, Denton RM. The activation of pyruvate dehydrogenase in the perfused rat heart by adrenaline and other inotropic agents. Biochem J 194: 639–643, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. McCormack JG, Halestrap AP, Denton RM. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol Rev 70: 391–425, 1990 [DOI] [PubMed] [Google Scholar]
- 134. Means CK, Lygren B, Langeberg LK, Jain A, Dixon RE, Vega AL, Gold MG, Petrosyan S, Taylor SS, Murphy AN, Ha T, Santana LF, Tasken K, Scott JD. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc Natl Acad Sci USA 108: E1227–E1235, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Mehler EL, Guarnieri F. A self-consistent, microenvironment modulated screened coulomb potential approximation to calculate pH-dependent electrostatic effects in proteins. Biophys J 77: 3–22, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Miller JL, Cimen H, Koc H, Koc EC. Phosphorylated proteins of the mammalian mitochondrial ribosome: implications in protein synthesis. J Proteome Res 8: 4789–4798, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Miura T, Tanno M, Sato T. Mitochondrial kinase signalling pathways in myocardial protection from ischaemia/reperfusion-induced necrosis. Cardiovasc Res 88: 7–15, 2010 [DOI] [PubMed] [Google Scholar]
- 138. Miyamoto S, Purcell NH, Smith JM, Gao T, Whittaker R, Huang K, Castillo R, Glembotski CC, Sussman MA, Newton AC, Brown JH. PHLPP-1 negatively regulates akt activity and survival in the heart/novelty and significance. Circ Res 107: 476–484, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Miyazaki T, Neff L, Tanaka S, Horne WC, Baron R. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J Cell Biol 160: 709–718, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Mootha VK, Arai AE, Balaban RS. Maximum oxidative phosphorylation capacity of the mammalian heart. Am J Physiol Heart Circ Physiol 272: H769–H775, 1997 [DOI] [PubMed] [Google Scholar]
- 141. Moritz A, Li Y, Guo A, Villen J, Wang Y, Macneill J, Kornhauser J, Sprott K, Zhou J, Possemato A, Ren JM, Hornbeck P, Cantley LC, Gygi SP, Rush J, Comb MJ. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal 3: ra64, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Murphy E, Kohr M, Sun J, Nguyen T, Steenbergen C. S-nitrosylation: a radical way to protect the heart. J Mol Cell Cardiol 52: 568–577, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Nantel A, Huber M, Thomas DY. Localization of endogenous Grb10 to the mitochondria and its interaction with the mitochondrial-associated Raf-1 pool. J Biol Chem 274: 35719–35724, 1999 [DOI] [PubMed] [Google Scholar]
- 144. Neubauer S. The failing heart—an engine out of fuel. N Engl J Med 356: 1140–1151, 2007 [DOI] [PubMed] [Google Scholar]
- 145. Neupert W, Brunner M. The protein import motor of mitochondria. Nat Rev Mol Cell Biol 3: 555–565, 2002 [DOI] [PubMed] [Google Scholar]
- 146. Norman AW, Bieber LL, Lindber GO, Boyer PD. Relationships of Ca++ to “protein-bound” phosphate fractions of mitochondria. J Biol Chem 240: 2855–2862, 1965 [PubMed] [Google Scholar]
- 147. Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31: 3635–3641, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Ogbi M, Chew CS, Pohl J, Stuchlik O, Ogbi S, Johnson JA. Cytochrome c oxidase subunit IV as a marker of protein kinase Cepsilon function in neonatal cardiac myocytes: implications for cytochrome c oxidase activity. Biochem J 382: 923–932, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Ogbi M, Johnson JA. Protein kinase Cepsilon interacts with cytochrome c oxidase subunit IV and enhances cytochrome c oxidase activity in neonatal cardiac myocyte preconditioning. Biochem J 393: 191–199, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Old WM, Shabb JB, Houel S, Wang H, Couts KL, Yen CY, Litman ES, Croy CH, Meyer-Arendt K, Miranda JG, Brown RA, Witze ES, Schweppe RE, Resing KA, Ahn NG. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol Cell 34: 115–131, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3: ra3, 2010 [DOI] [PubMed] [Google Scholar]
- 152. Pagliarini DJ, Wiley SE, Kimple ME, Dixon JR, Kelly P, Worby CA, Casey PJ, Dixon JE. Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic beta cells. Mol Cell 19: 197–207, 2005 [DOI] [PubMed] [Google Scholar]
- 153. Pallen MJ. Time to recognise that mitochondria are bacteria? Trends Microbiol 19: 58–64, 2011 [DOI] [PubMed] [Google Scholar]
- 154. Pao GM, Saier MH., Jr Nonplastid eukaryotic response regulators have a monophyletic origin and evolved from their bacterial precursors in parallel with their cognate sensor kinases. J Mol Evol 44: 605–613, 1997 [DOI] [PubMed] [Google Scholar]
- 155. Papa S, Rasmo DD, Technikova-Dobrova Z, Panelli D, Signorile A, Scacco S, Petruzzella V, Papa F, Palmisano G, Gnoni A, Micelli L, Sardanelli AM. Respiratory chain complex I, a main regulatory target of the cAMP/PKA pathway is defective in different human diseases. FEBS Lett 586: 568–577, 2012 [DOI] [PubMed] [Google Scholar]
- 156. Paradela A, Albar JP. Advances in the analysis of protein phosphorylation. J Proteome Res 7: 1809–1818, 2008 [DOI] [PubMed] [Google Scholar]
- 157. Pastorino JG, Hoek JB, Shulga N. Activation of glycogen synthase kinase 3beta disrupts the binding of hexokinase II to mitochondria by phosphorylating voltage-dependent anion channel and potentiates chemotherapy-induced cytotoxicity. Cancer Res 65: 10545–10554, 2005 [DOI] [PubMed] [Google Scholar]
- 158. Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJM, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426: 39–44, 2003 [DOI] [PubMed] [Google Scholar]
- 159. Pecina P, Borisenko GG, Belikova NA, Tyurina YY, Pecinova A, Lee I, Samhan-Arias AK, Przyklenk K, Kagan VE, Huttemann M. Phosphomimetic substitution of cytochrome C tyrosine 48 decreases respiration and binding to cardiolipin and abolishes ability to trigger downstream caspase activation. Biochemistry 49: 6705–6714, 2010 [DOI] [PubMed] [Google Scholar]
- 160. Peter JB, Boyer PD. The formation of bound phosphohistidine from adenosine triphosphate-P32 in mitochondria. J Biol Chem 238: 1180–1182, 1963 [PubMed] [Google Scholar]
- 161. Phillips D, Aponte AM, Covian RG, Balaban RS. Intrinsic protein kinase activity of mitochondrial oxidative phosphorylation complexes. Biochemistry 50: 2515–2529, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Phillips D, Aponte AM, French SA, Chess DJ, Balaban RS. Succinyl-CoA synthetase is a phosphate target for the activation of mitochondrial metabolism. Biochemistry 48: 7140–7149, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Phillips D, Reilley MJ, Aponte AM, Wang G, Boja E, Gucek M, Balaban RS. Stoichiometry of STAT3 and mitochondrial proteins: implications for the regulation of oxidative phosphorylation by protein-protein interactions. J Biol Chem 285: 23532–23536, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Pical C, Fredlund KM, Petit PX, Sommarin M, Moller IM. The outer membrane of plant mitochondria contains a calcium-dependent protein kinase and multiple phosphoproteins. FEBS Lett 336: 347–351, 1993 [DOI] [PubMed] [Google Scholar]
- 165. Pocsfalvi G, Cuccurullo M, Schlosser G, Scacco S, Papa S, Malorni A. Phosphorylation of B14.5a subunit from bovine heart complex I identified by titanium dioxide selective enrichment and shotgun proteomics. Mol Cell Proteomics 6: 231–237, 2007 [DOI] [PubMed] [Google Scholar]
- 166. Popov KM, Zhao Y, Shimomura Y, Kuntz MJ, Harris RA. Branched-chain alpha-ketoacid dehydrogenase kinase. Molecular cloning, expression, and sequence similarity with histidine protein kinases. J Biol Chem 267: 13127–13130, 1992 [PubMed] [Google Scholar]
- 167. Prabu SK, Anandatheerthavarada HK, Raza H, Srinivasan S, Spear JF, Avadhani NG. Protein kinase A-mediated phosphorylation modulates cytochrome c oxidase function and augments hypoxia and myocardial ischemia-related injury. J Biol Chem 281: 2061–2070, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Pressman BC. Metabolic function of phosphohistidine. Biochem Biophys Res Commun 15: 556–561, 1964 [Google Scholar]
- 169. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res 40: D290–D301, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Rao S, Schmidt O, Harbauer AB, Schonfisch B, Guiard B, Pfanner N, Meisinger C. Biogenesis of the preprotein translocase of the outer mitochondrial membrane: protein kinase A phosphorylates the precursor of Tom40 and impairs its import. Mol Biol Cell 23: 1618–1627, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Rardin MJ, Wiley SE, Murphy AN, Pagliarini DJ, Dixon JE. Dual specificity phosphatases 18 and 21 target to opposing sides of the mitochondrial inner membrane. J Biol Chem 283: 15440–15450, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Raza H. Dual localization of glutathione S-transferase in the cytosol and mitochondria: implications in oxidative stress, toxicity and disease. FEBS J 278: 4243–4251, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Reed LJ, Damuni Z, Merryfield ML. Regulation of mammalian pyruvate and branched-chain alpha-keto acid dehydrogenase complexes by phosphorylation-dephosphorylation. Curr Top Cell Regul 27: 41–49, 1985 [DOI] [PubMed] [Google Scholar]
- 174. Rees DM, Leslie AG, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci USA 106: 21597–21601, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Reinders J, Sickmann A. State-of-the-art in phosphoproteomics. Proteomics 5: 4052–4061, 2005 [DOI] [PubMed] [Google Scholar]
- 176. Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131: 1190–1203, 2007 [DOI] [PubMed] [Google Scholar]
- 177. Roberts DL, Frerman FE, Kim JJ. Three-dimensional structure of human electron transfer flavoprotein to 2.1-A resolution. Proc Natl Acad Sci USA 93: 14355–14360, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Ruvolo PP, Deng X, Ito T, Carr BK, May WS. Ceramide induces Bcl2 dephosphorylation via a mechanism involving mitochondrial PP2A. J Biol Chem 274: 20296–20300, 1999 [DOI] [PubMed] [Google Scholar]
- 179. Sale GJ, Randle PJ. Analysis of site occupancies in [32P]phosphorylated pyruvate dehydrogenase complexes by aspartyl-prolyl cleavage of tryptic phosphopeptides. Eur J Biochem 120: 535–540, 1981 [DOI] [PubMed] [Google Scholar]
- 180. Salvi M, Morrice NA, Brunati AM, Toninello A. Identification of the flavoprotein of succinate dehydrogenase and aconitase as in vitro mitochondrial substrates of Fgr tyrosine kinase. FEBS Lett 581: 5579–5585, 2007 [DOI] [PubMed] [Google Scholar]
- 181. Salvi M, Stringaro A, Brunati AM, Agostinelli E, Arancia G, Clari G, Toninello A. Tyrosine phosphatase activity in mitochondria: presence of Shp-2 phosphatase in mitochondria. Cell Mol Life Sci 61: 2393–2404, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Salvi M, Brunati AM, Bordin L, La Rocca N, Clari G, Toninello A. Characterization and location of Src-dependent tyrosine phosphorylation in rat brain mitochondria. Biochim Biophys Acta 1589: 181–195, 2002 [DOI] [PubMed] [Google Scholar]
- 183. Samavati L, Lee I, Mathes I, Lottspeich F, Huttemann M. Tumor necrosis factor alpha inhibits oxidative phosphorylation through tyrosine phosphorylation at subunit I of cytochrome c oxidase. J Biol Chem 283: 21134–21144, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Sardanelli AM, Signorile A, Nuzzi R, Rasmo DD, Technikova-Dobrova Z, Drahota Z, Occhiello A, Pica A, Papa S. Occurrence of A-kinase anchor protein and associated cAMP-dependent protein kinase in the inner compartment of mammalian mitochondria. FEBS Lett 580: 5690–5696, 2006 [DOI] [PubMed] [Google Scholar]
- 185. Sardanelli AM, Technikova-Dobrova Z, Scacco SC, Speranza F, Papa S. Characterization of proteins phosphorylated by the cAMP-dependent protein kinase of bovine heart mitochondria. FEBS Lett 377: 470–474, 1995 [DOI] [PubMed] [Google Scholar]
- 186. Sardanelli AM, Technikova-Dobrova Z, Speranza F, Mazzocca A, Scacco S, Papa S. Topology of the mitochondrial cAMP-dependent protein kinase and its substrates. FEBS Lett 396: 276–278, 1996 [DOI] [PubMed] [Google Scholar]
- 187. Sawicki G, Jugdutt BI. Valsartan reverses post-translational modifications of the delta-subunit of ATP synthase during in vivo canine reperfused myocardial infarction. Proteomics 7: 2100–2110, 2007 [DOI] [PubMed] [Google Scholar]
- 188. Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311: 1430–1436, 2006 [DOI] [PubMed] [Google Scholar]
- 189. Schieven G, Martin GS. Nonenzymatic phosphorylation of tyrosine and serine by ATP is catalyzed by manganese but not magnesium. J Biol Chem 263: 15590–15593, 1988 [PubMed] [Google Scholar]
- 190. Schilling B, Aggeler R, Schulenberg B, Murray J, Row RH, Capaldi RA, Gibson BW. Mass spectrometric identification of a novel phosphorylation site in subunit NDUFA10 of bovine mitochondrial complex I. FEBS Lett 579: 2485–2490, 2005 [DOI] [PubMed] [Google Scholar]
- 191. Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, Meisinger C. Regulation of mitochondrial protein import by cytosolic kinases. Cell 144: 227–239, 2011 [DOI] [PubMed] [Google Scholar]
- 192. Schmitt ME, Trumpower BL. Subunit 6 regulates half-of-the-sites reactivity of the dimeric cytochrome bc1 complex in Saccharomyces cerevisiae. J Biol Chem 265: 17005–17011, 1990 [PubMed] [Google Scholar]
- 193. Schwoch G, Trinczek B, Bode C. Localization of catalytic and regulatory subunits of cyclic AMP-dependent protein kinases in mitochondria from various rat tissues. Biochem J 270: 181–188, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Segal HL. Enzymatic interconversion of active and inactive forms of enzymes. Science 180: 25–32, 1973 [DOI] [PubMed] [Google Scholar]
- 195. Shaffer J, Adams JA. Detection of conformational changes along the kinetic pathway of protein kinase A using a catalytic trapping technique. Biochemistry 38: 12072–12079, 1999 [DOI] [PubMed] [Google Scholar]
- 196. Sharma V, Abraham T, So A, Allard MF, McNeill JH. Functional effects of protein kinases and peroxynitrite on cardiac carnitine palmitoyltransferase-1 in isolated mitochondria. Mol Cell Biochem 337: 223–237, 2010 [DOI] [PubMed] [Google Scholar]
- 197. Sheldon KL, Maldonado EN, Lemasters JJ, Rostovtseva TK, Bezrukov SM. Phosphorylation of voltage-dependent anion channel by serine/threonine kinases governs its interaction with tubulin. PLoS One 6: e25539, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Slater E, Kemp A. Rate of labelling of mitochondrial phosphohistidine by radioactive inorganic phosphate. Nature 204: 1268–1271, 1964 [DOI] [PubMed] [Google Scholar]
- 199. Snyder CH, Merbitz-Zahradnik T, Link TA, Trumpower BL. Role of the Rieske iron-sulfur protein midpoint potential in the protonmotive Q-cycle mechanism of the cytochrome bc1 complex. J Bioenerg Biomembr 31: 235–242, 1999 [DOI] [PubMed] [Google Scholar]
- 200. Solmaz SR, Hunte C. Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J Biol Chem 283: 17542–17549, 2008 [DOI] [PubMed] [Google Scholar]
- 201. Sommarin M, Petit PX, Moller IM. Endogenous protein phosphorylation in purified plant mitochondria. Biochim Biophys Acta 1052: 195–203, 1990 [DOI] [PubMed] [Google Scholar]
- 202. Steinberg RA. Cyclic AMP-dependent phosphorylation of the precursor to beta subunit of mitochondrial F1-ATPase: a physiological mistake? J Cell Biol 98: 2174–2178, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Stock D, Gibbons C, Arechaga I, Leslie AG, Walker JE. The rotary mechanism of ATP synthase. Curr Opin Struct Biol 10: 672–679, 2000 [DOI] [PubMed] [Google Scholar]
- 204. Struglics A, Fredlund KM, Konstantinov YM, Allen JF, Moller IM. Protein phosphorylation/dephosphorylation in the inner membrane of potato tuber mitochondria. FEBS Lett 475: 213–217, 2000 [DOI] [PubMed] [Google Scholar]
- 205. Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121: 1043–1057, 2005 [DOI] [PubMed] [Google Scholar]
- 206. Sun G, Budde RJ. Requirement for an additional divalent metal cation to activate protein tyrosine kinases. Biochemistry 36: 2139–2146, 1997 [DOI] [PubMed] [Google Scholar]
- 207. Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem 282: 5617–5624, 2007 [DOI] [PubMed] [Google Scholar]
- 208. Tao WA, Wollscheid B, O'Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat Methods 2: 591–598, 2005 [DOI] [PubMed] [Google Scholar]
- 209. Technikova-Dobrova Z, Sardanelli AM, Papa S. Phosphorylation of mitochondrial proteins in bovine heart. Characterization of kinases and substrates. FEBS Lett 322: 51–55, 1993 [DOI] [PubMed] [Google Scholar]
- 210. Technikova-Dobrova Z, Sardanelli AM, Speranza F, Scacco S, Signorile A, Lorusso V, Papa S. Cyclic adenosine monophosphate-dependent phosphorylation of mammalian mitochondrial proteins: enzyme and substrate characterization and functional role. Biochemistry 40: 13941–13947, 2001 [DOI] [PubMed] [Google Scholar]
- 211. Toogood HS, van Thiel A, Basran J, Sutcliffe MJ, Scrutton NS, Leys D. Extensive domain motion and electron transfer in the human electron transferring flavoprotein.medium chain Acyl-CoA dehydrogenase complex. J Biol Chem 279: 32904–32912, 2004 [DOI] [PubMed] [Google Scholar]
- 212. Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272: 1136–1144, 1996 [DOI] [PubMed] [Google Scholar]
- 213. Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA 104: 1488–1493, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Vosseller K, Hansen KC, Chalkley RJ, Trinidad JC, Wells L, Hart GW, Burlingame AL. Quantitative analysis of both protein expression and serine / threonine post-translational modifications through stable isotope labeling with dithiothreitol. Proteomics 5: 388–398, 2005 [DOI] [PubMed] [Google Scholar]
- 215. Walker JE, Dickson VK. The peripheral stalk of the mitochondrial ATP synthase. Biochim Biophys Acta 1757: 286–296, 2006 [DOI] [PubMed] [Google Scholar]
- 216. Wang WL, Yeh SF, Chang YI, Hsiao SF, Lian WN, Lin CH, Huang CY, Lin WJ. PICK1, an anchoring protein that specifically targets protein kinase Calpha to mitochondria selectively upon serum stimulation in NIH 3T3 cells. J Biol Chem 278: 37705–37712, 2003 [DOI] [PubMed] [Google Scholar]
- 217. Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci USA 105: 13793–13798, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Weishaupt A, Kadenbach B. Selective removal of subunit VIb increases the activity of cytochrome c oxidase. Biochemistry 31: 11477–11481, 1992 [DOI] [PubMed] [Google Scholar]
- 219. Wieland O, Jagow-Westermann B. ATP-dependent inactivation of heart muscle pyruvate dehydrogenase and reactivation by Mg++. FEBS Lett 3: 271–274, 1969 [DOI] [PubMed] [Google Scholar]
- 220. Wieland O, Siess E. Interconversion of phospho- and dephospho- forms of pig heart pyruvate dehydrogenase. Proc Natl Acad Sci USA 65: 947–954, 1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Wiltshire C, Matsushita M, Tsukada S, Gillespie DA, May GH. A new c-Jun N-terminal kinase (JNK)-interacting protein, Sab (SH3BP5), associates with mitochondria. Biochem J 367: 577–585, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Wisniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J Proteome Res 9: 3280–3289, 2010 [DOI] [PubMed] [Google Scholar]
- 223. Wu QZ, Li DM, Kuang WH, Zhang TJ, Lui S, Huang XQ, Chan RC, Kemp GJ, Gong QY. Abnormal regional spontaneous neural activity in treatment-refractory depression revealed by resting-state fMRI. Hum Brain Mapp 32: 1290–1299, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0: a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics 7: 1598–1608, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Yadava N, Potluri P, Scheffler IE. Investigations of the potential effects of phosphorylation of the MWFE and ESSS subunits on complex I activity and assembly. Int J Biochem Cell Biol 40: 447–460, 2008 [DOI] [PubMed] [Google Scholar]
- 226. Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol 19: 348–353, 2001 [DOI] [PubMed] [Google Scholar]
- 227. Yang F, Stenoien DL, Strittmatter EF, Wang J, Ding L, Lipton MS, Monroe ME, Nicora CD, Gristenko MA, Tang K, Fang R, Adkins JN, Camp DG, Chen DJ, Smith RD. Phosphoproteome profiling of human skin fibroblast cells in response to low- and high-dose irradiation. J Proteome Res 5: 1252–1260, 2006 [DOI] [PubMed] [Google Scholar]
- 228. Yu CA, Xia D, Kim H, Deisenhofer J, Zhang L, Kachurin AM, Yu L. Structural basis of functions of the mitochondrial cytochrome bc1 complex. Biochim Biophys Acta 1365: 151–158, 1998 [DOI] [PubMed] [Google Scholar]
- 229. Yu H, Lee I, Salomon AR, Yu K, Huttemann M. Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochim Biophys Acta 1777: 1066–1071, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Yu PH, Deng YL. Endogenous formaldehyde as a potential factor of vulnerability of atherosclerosis: involvement of semicarbazide-sensitive amine oxidase-mediated methylamine turnover. Atherosclerosis 140: 357–363, 1998 [DOI] [PubMed] [Google Scholar]
- 231. Zanivan S, Gnad F, Wickstrom SA, Geiger T, Macek B, Cox J, Fassler R, Mann M. Solid tumor proteome and phosphoproteome analysis by high resolution mass spectrometry. J Proteome Res 7: 5314–5326, 2008 [DOI] [PubMed] [Google Scholar]
- 232. Zara V, Conte L, Trumpower BL. Biogenesis of the yeast cytochrome bc1 complex. Biochim Biophys Acta 1793: 89–96, 2009 [DOI] [PubMed] [Google Scholar]
- 233. Zhang J, Duncker DJ, Ya X, Zhang Y, Pavek T, Wei H, Merkle H, Ugurbil K, From AH, Bache RJ. Effect of left ventricular hypertrophy secondary to chronicX pressure overload on transmural myocardial 2-deoxyglucose uptake. X A 31P NMR spectroscopic study. Circulation 92: 1274–1283, 1995 [DOI] [PubMed] [Google Scholar]
- 234. Zhang J, Murphy AN, Wiley SE, Perkins G, Worby CA, Engel JL, Heacock P, Nguyen OK, Wang JH, Raetz CRH, Dowhan W, Dixon JE. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab 13: 690–700, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Zhang J, Frerman FE, Kim JJ. Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proc Natl Acad Sci USA 103: 16212–16217, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Zhao X, León IR, Bak S, Mogensen M, Wrzesinski K, Højlund K, Jensen ON. Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol Cell Proteomics 10: 1–14, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Zhou L, Cabrera ME, Huang H, Yuan CL, Monika DK, Sharma N, Bian F, Stanley WC. Parallel activation of mitochondrial oxidative metabolism with increased cardiac energy expenditure is not dependent on fatty acid oxidation in pigs. J Physiol 579: 811–821, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Zu XL, Besant PG, Imhof A, Attwood PV. Mass spectrometric analysis of protein histidine phosphorylation. Amino Acids 32: 347–357, 2007 [DOI] [PubMed] [Google Scholar]