Abstract
BACKGROUND
Personalized treatment for psychopathologies, in particular alcoholism, is highly dependent upon our ability to identify patterns of genetic and environmental effects that influence a person’s risk. Unfortunately, array-based whole genome investigations into heritable factors that explain why one person becomes dependent upon alcohol and another does not, have indicated that alcohol’s genetic architecture is highly complex. That said, uncovering and interpreting the missing heritability in alcohol genetics research has become all the more important, especially since the problem may extend to our inability to model the cumulative and combinatorial relationships between common and rare genetic variants. As numerous studies begin to illustrate the dependency of alcohol pharmacotherapies on an individual’s genotype, the field is further challenged to identify new ways to transcend agnostic genomewide association approaches. We discuss insights from genetic studies of alcohol related diseases, as well as issues surrounding alcohol’s genetic complexity and etiological heterogeneity. Finally, we describe the need for innovative systems-based approaches (Systems Genetics) that can provide additional statistical power that can enhance future gene-finding strategies and help to identify heretofore-unrealized mechanisms that may provide new targets for prevention/treatments efforts. Emerging evidence from early studies suggest that Systems Genetics has the potential to organize our neurological, pharmacological, and genetic understanding of alcohol dependence into a biologically plausible framework that represents how perturbations across evolutionarily robust biological systems determine susceptibility to alcohol dependence.
Keywords: Genetics, Alcoholism, Alcohol Dependence, GWAS, Systems Genetics
1. Introduction
Alcohol Dependence (AD) is defined across all versions of the Diagnostic and Statistical Manual of Mental Disorders (DSM), and despite changes in criteria, as a disorder characterized by physiological and psychological effects in individuals who consume large amounts of alcohol (American Psychiatric Association, 1968, 1980, 1987, 2000). Individuals “addicted” to alcohol are likely to demonstrate either or all of the following: (1) a strong urge/craving for the drug, (2) an inability to limit the amount of alcohol they consume, and/or (3) a diagnosis of dependence, as defined by the DSM. Despite the many negative implications of alcohol use, AD continues to be a major public health concern in the United States of America. In fact, as of 2010, 131.3 million Americans (~52%) have been reported as current drinkers of alcohol (Substance Abuse and Mental Health Services Administration, 2011).
In our effort to understand the genetic liability to AD, research has focused on characterizing individual differences in the biological systems that regulate the breakdown of alcohol and the neuronal systems/pathways that are believed to be affected by alcohol. Research on the metabolism of alcohol suggests the involvement of several enzymes. The oxidative pathway involves aldehyde dehydrogenase (ALDH), alcohol dehydrogenase (ADH), cytochrome P450 2E1, and catalase. The non-oxidative pathway involves fatty acid ethyl ester and phospholipidase D. Differences in the functionality of the ALDH and ADH enzymes have been linked to, (1) increased risk for alcohol-induced tissue damage (cirrhosis; Chao et al., 1994), and (2) protection against developing AD (Chen et al., 2009). In the context of brain effects, the acute and chronic effects of alcohol exposure are very important, as the cycle of addiction is dependent upon how an individual responds to repeated alcohol use over time. Based on the body of literature across humans and animals, AD is likely to involve neuronal circuits involved in the Binge/Intoxication, Withdrawal/Negative Affect, and Preoccupation/Anticipation stages of the addiction cycle (Koob and Volkow, 2010); see Figure 1. In fact, neurotransmitter systems, such as dopamine, glutamate, opioid, serotonin, and γ-aminobutyric acid (GABA) systems, as well as stress response circuits (Neuropeptide-Y and Corticotropin-Releasing Factor) and appetite regulating systems are key to alcohol’s effects (Hillemacher, 2011). For instance, studies demonstrate that opioid antagonists suppress alcohol drinking (Rosner et al., 2010) and that gamma-aminobutyric acid A receptors (GABAA) mediate the rewarding effects of alcohol (Koob, 2004) and alcohol consumption (Rewal et al., 2012; Tabakoff et al., 2009). Given the role of each of these metabolic and neuronal pathways in alcohol use and addiction, they are regarded as candidate pathways for genetic studies of alcohol. It is believed that individual differences in the genetic code of these and other candidate molecules will provide insight into the risk for AD. Unfortunately, the extant body of animal and human research on alcohol has also demonstrated that alcohol, as a drug, is not specific in its effects, but rather casts a wide net in the human brain. Consequently, susceptibility to AD likely involves a network of genes across several biological systems. This has complicated the elucidation of the genetic mechanisms that drive compulsive drinking, AD, and specific AD characteristics. In the proceeding pages, we highlight the positive and negative findings from molecular genetic studies of AD and the need for analytical and interpretational approaches in the form of Systems Genetics. Systems Genetics has the potential to organize our neurological, pharmacological, and genetic understanding of AD into a biologically plausible framework that represents how perturbations across evolutionarily robust biological systems determine susceptibility to AD.
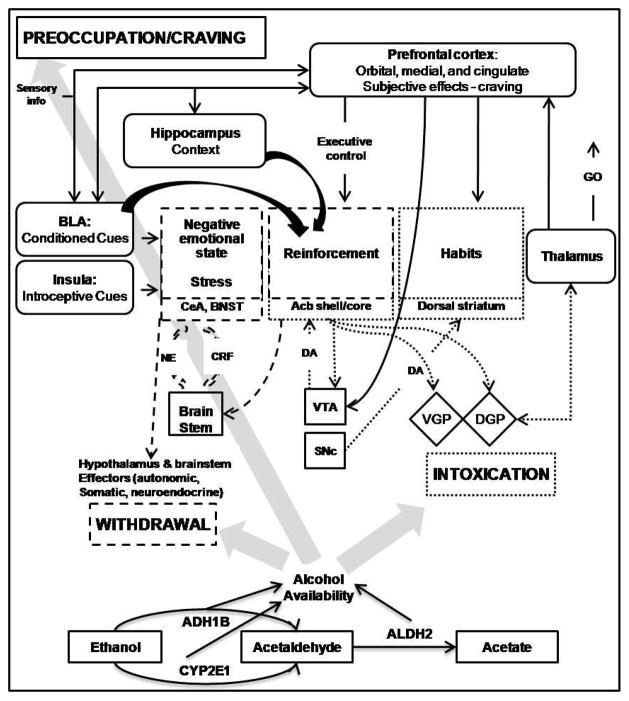
Figure 1. Example of a Theoretical Network Model for Alcohol Dependence.
This adapted figure from Koob and Volkow (2010) is used to illustrate an example of a theoretical systems model showing genes involved in alcohol metabolism and alcohol’s effect on the brain’s reward pathway. Note that for the sake of simplicity, the figure does not represent all of the pathways involved, for example, projections from the ventral tegmental area to the amygdala and hippocampus. The figure depicts each of the three components of addiction (intoxication, withdrawal, and Preoccupation), which are mediated by different neurotransmitters and systems that are compromised by alcohol; solid and dotted lines indicate glutamatergic projections, dashed arrows represent dopaminergic projections. Abbreviations. Acb - nucleus accumbens; BNST - bed nucleus of the stria terminalis; CeA - central nucleus of the amygdala; CRF - corticotropin-releasing factor; DGP - dorsal globus pallidus; NE - norepinephrine; SNc - substantia nigra pars compacta; VGP - ventral globus pallidus; VTA, ventral tegmental area. Based on the segments shown, the presence of alcohol in the system is limited by the genetic profile of the metabolic system, while alcohol’s effect on the reward pathway is limited by variation in GABAergic transmission, glutamate transmission, and dopamine transmission. Gene names shown in italics indicates possible sources of variation in the system. Adapted by permission from Macmillan Publishers Ltd: Neuropsychopharmacology (Koob, G.F., Volkow, N.D., 2010. Neurocircuitry of addiction. Neuropsychopharmacology 35, 217–238.), copyright 2010.
2. THE GENETIC EPIDEMIOLOGY OF AD
2.1 Genetic Studies of AD
Human genetic studies of alcohol are organized into two broad categories, quantitative genetic (i.e., family and twin studies) and molecular genetic studies. Quantitative genetic studies suggest that genetic differences play an important part in susceptibility to AD. Much of this evidence has been derived from early family-based studies which indicated that first-degree relatives of alcohol dependent cases were several times more likely to be later diagnosed with AD relative to first-degree relatives of control subjects (Bierut et al., 1998). Further, twin and adoption studies suggest that this familial pattern may be attributable to additive genetic factors, which account for roughly 40%–60% of the liability for AD (Agrawal and Lynskey, 2008; Knopik et al., 2004). Twin studies also suggest that a large number of genes related to AD also influence other forms of drug dependence (Palmer et al., 2012), as well as other externalizing psychopathologies (Iacono et al., 2008; Young et al., 2000).
As the genotyping technology improved, candidate gene and genomewide association methods were developed as a means to identify genetic variants that confer increased risk for AD. However, due to the etiological complexity of complex traits like AD, newer DNA sequencing methods, in particular, next generation sequencing (NGS) have become increasingly necessary as they provide a more accurate description of both common and rare variants (i.e., be they single nucleotide polymorphisms (SNPs) or structural variants). Both linkage and association studies are heavily focused on genetic variation that can be captured by genomic platforms designed to identify rare and/or common variation within a specific gene or across the entire genome, usually by relying upon linkage disequilibrium (LD). However, both methods have significant differences that affect their interpretation. Linkage studies are often regarded as being more powerful than association studies because of their ability to capture variation attributable to rare variants; however, they lack specificity as they focus on identifying stretches of DNA that either contain or are linked to the gene/genes that underlie a trait. Linkage study findings may lead to gene discovery when followed-up with targeted sequence capture; however, it is important to note that linkage study findings may be specific to the families in the pedigrees utilized. On the other hand, association studies are less powerful, but are more specific because they focus on identifying alleles (i.e., alternative forms of a gene caused by SNPs, copy number variants (CNVs), sequence repeats, etc.) that might be a contributing factor for the behavior/disease or linked to it. Notably, genomewide association studies (GWAS) thus far have utilized array-based platforms that provide good global coverage of the genome and genes within it, but primarily focus on common variants (mainly SNPs with a minor allele frequency > 10%), thereby providing very limited coverage of sequence repeats, structural variants, and rare SNPs that are more likely to be seen with NGS. So far, molecular genetic studies have linked variation across chromosomes 1, 2, 3, 4, 7, and 8 to diagnoses of AD (Foroud et al., 2000; Nurnberger et al., 2001; Reich et al., 1998; Wang et al., 2004; Williams et al., 1999), as well as chromosomes 5, 6, 9, 15, 16, and 21 using quantitative phenotypes of AD (e.g., maximum number of drinks) and neurophysiological phenotypes (e.g., event related potentials, such as P300) that are often comorbid with AD and other psychiatric disorders (Almasy et al., 2001; Begleiter et al., 1998; Foroud et al., 1998; Ghosh et al., 2003; Hill et al., 2004; Jones et al., 2004; Kuo et al., 2006; Porjesz et al., 2002; Saccone et al., 2000; Schuckit et al., 2001).
To date, at least one variant in roughly 602 genes has been linked to Alcoholism and/or AD (visit: http://www.hugenavigator.net/HuGENavigator/startPagePhenoPedia.do; (Yu et al., 2010). Amongst these genes, there are several systems that have received special attention due to their neurological and pharmacological relevance to indentifying (1) molecular targets of alcohol, and (2) brain systems that are altered by the presence of alcohol. Unlike other substances (e.g., cocaine) alcohol has global neuronal effects. Alcohol alters the membranes, ion channels, enzymes, and receptors of neurons (Valenzuela, 1997). Alcohol has also been shown to alter the binding of receptors for acetylcholine, serotonin, GABA, and the NMDA receptors for glutamate (Mukherjee et al., 2008; Nevo and Hamon, 1995). Indisputably, the wide range of symptoms seen as part of AD are likely the result of individual differences in alcohol metabolism and alcohol-induced neuroplastic changes.
2.2 Genetics in the Context of Pharmacology and Neuroscience
Understanding how individual differences in genetic risk factors influences the risk for AD across individuals requires interpreting gene effects across different systems because no single gene determines the overall risk for AD. While a complete review of the pharmacology and neurobiology of AD is beyond the scope of this paper, the moderate success of addiction pharmacotherapies highlights the importance of the opioid, GABA, serotonin, dopamine, and corticotropin-releasing factor systems in the liability to alcoholism. We refer readers to recent a review by Vengeliene et al. (2008), which describes the pharmacodynamic and pharmacokinetics of AD, a review by Heilig et al. (2011), which highlights brain systems that have been targeted by pharmacological agents in the treatment of alcoholism and a review by Koob and Volkow (2010) that describes different systems that facilitate preoccupation with addictive substances. Based on research in these areas, a key component to understanding alcohol’s effect on the brain can be found in variation both within and across biological mechanisms that regulate ethanol concentration and intercellular communication among neurons. Studies have demonstrated that the liver enzymes, alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) regulate the degradation of alcohol into acetate. Pharmacogenetic research has also shown that variation within ADH and ALDH genes alters a person’s risk for developing alcohol problems (Alcohol_Alert, 2007). For instance, of the seven genes that code for different forms of ADH (clustered on chromosome 4q), variants within the genes encoding the hepatic forms, ADH1B and ADH1C, have been related to alcohol dependence. The ADH1B*2 alleles has also been shown to protect against alcoholism in males and females of different ethnic origins (Lorenzo et al., 2006; Toth et al., 2010). Koob and Volkow’s review of the neurobiology of drug addiction highlights several circuits that mediate the ‘binge/intoxication’, ‘withdrawal/negative affect’, and ‘preoccupation/anticipation (craving)’ stages of the addiction cycle. These stages have been linked to specific neural networks. For example, in regards to the Binge/Intoxication Stage, the acute reinforcing effects of alcohol are hypothesized to invoke the release of dopamine in the nucleus accumbens through its actions in the ventral tegmental area or nucleus accumbens). Candidate gene studies focused on neurotransmitters whose levels or end function are altered by the acute or chronic presence of alcohol have supported neuroimaging studies. As one example, dopamine (a key component of the brain’s reward circuitry) and serotonin (the primary contributor to motivation behaviors and mood) are considered to be among alcohol’s major liability factors. Alcohol’s ability to modulate dopamine levels results in neuronal adaptation that perpetuates further alcohol/other drug use. Although the different mechanisms by which alcohol evokes its acute reinforcing effects on the brain remain poorly understood, its effects appear to be mediated by the actions of the dopamine, serotonin, opioid, and GABA systems in the basal forebrain. For instance, alcohol use leads to the release of endogenous opioids, which activate mu-opioid receptors on GABAergic interneurons in the ventral tegmental area, resulting in attenuation of inhibitory tone from these onto mesolimbic dopamine-neurons, and ultimately increased dopamine release in the nucleus accumbens. Over time, chronic alcohol use eventually leads to a hypodopaminergic state that becomes the driving force behind continued alcohol and/or other drug seeking behaviors (Volkow et al., 2007). To date, genetic studies of AD have highlighted polymorphisms in the genes for GABA (e.g., GABRA2; Dick et al., 2006a; 2006b; Edenberg et al., 2004; Fehr et al., 2006; Ittiwut et al., 2011; Lind et al., 2008a; Lydall et al., 2011; Matthews et al., 2007; Philibert et al., 2009), dopamine (e.g., DRD4; Connor et al., 2007; Dick et al., 2007; Park et al., 2011; Pinto et al., 2009; Yang et al., 2008), serotonin (e.g., SLC6A4; McHugh et al., 2010; Philibert et al., 2008), and opioid receptors (e.g., OPRM1; Chen et al., 2011a), and the dopamine enzyme, catechol-O-methyl-transferase (COMT; He et al., 2008) among others.
2.3 GWAS of AD
We are aware of several GWAS of AD (Bierut et al., 2010; Edenberg et al., 2010; Heath et al., 2011; Johnson et al., 2006; Kendler et al., 2011; Kerner et al., 2011; Lind et al., 2010; Treutlein et al., 2009; Wang et al., 2011b, 2012; Zuo et al., 2011a, 2011b) and AD related endophenotypes, such as theta-band oscillations (Hodgkinson et al., 2010; Kang et al., 2012; Zlojutro et al., 2011), and drinking phenotypes (Baik et al., 2011; Heath et al., 2011; Pei et al., 2012; Schumann et al., 2011). These studies suggest that (1) AD is genetically heterogeneous, and (2) the complete statistical characterization of the genetic susceptibility to AD requires novel analytic techniques that can utilize all of the molecular data from both array and NGS technologies. Although a complete review is beyond the scope of this paper, we suggest recent review papers that cover GWAS of AD (Kimura and Higuchi, 2011; Treutlein and Rietschel, 2011). Notably, several GWAS have validated some candidate genes. For instance, the ADH1C gene, which has over 50 publications linking it to alcohol use/alcoholism/AD, has also been evidenced in two recent GWAS of AD (Kendler et al., 2011; Treutlein et al., 2009). Similarly, the muscarinic receptor (CHRM2) has been replicated in two GWAS (Dick et al., 2008; Kendler et al., 2011), and several other well replicated candidate genes (i.e., >10 studies), such as GABRA2 (Bierut et al., 2010), MAOA (Wang et al., 2011a), GRIN2B (Joslyn et al., 2010), and ANKK1 (Kendler et al., 2011) have emerged in recent GWAS.
3. Insights and Limitations from GWAS of AD
GWASs of AD and related phenotypes have identified numerous loci, however, these loci alone have limited utility (i.e., they each account for less than 1% of the variance in liability of the disease/trait). Despite this limitation, alcohol GWAS continue to be studied because they (1) support pathways that were previously hypothesized from linkage study findings, and (2) highlight pathways that were not initially considered (Heath et al., 2011); for example, genes involved in the specification and maintenance of neuronal connections. The major success and challenge of alcohol GWAS is that hundreds of genetic variants, each with a modest effect size, contribute to its liability. This observation presents several new challenges to modeling the relationships between different genes and alcohol phenotypes, such as (1) the identification and selection of polymorphisms, (2) reducing the heterogeneity of alcohol phenotypes, (3) the design and implementation of mathematical approaches that provide the necessary power, and (4) the development of a conceptual framework that will provide a meaningful interpretation of the findings. For the remainder of the paper we discuss factors as they relate to the missing heritability in alcohol GWAS.
3.1 Genetic Variation: Common and Rare Variants
Missing heritability (i.e., the disparity between genetic effects identified in family/twin studies and molecular genetic studies) in GWAS has been attributed to the emphasis on common genetic variants that have low penetrance (i.e., the proportion of individuals carrying a particular allele/genotype that also express a particular behavior). As the number of variants tested on GWAS platforms has evolved from testing thousands of variants to more than 1 million, the likelihood of capturing variants that are in LD with rare variants has increased. Current 1M chip platforms have identified hundreds of possible candidate variants for AD, but only a few of these have replicated across independent samples and have functional implications. Altogether, these observations suggest that when treated individually, common variants, such as SNPs, and rare variants, such as copy number variants, account for a small fraction of the missing heritability of diseases (Orozco et al., 2010). The most likely solution to this problem would be the incorporation of both common and rare genetic variants in genetic studies of alcohol using whole genome sequencing platforms. Unfortunately, a noteworthy drawback to the inclusion of rare-moderately-penetrant and common-weakly-penetrant alleles in the genetic model of any disease is that it decreases the power to detect true associations. As a result, larger studies of AD would be necessary (Wray et al., 2007, 2008); alternatively, family-based association studies, such as the Collaborative Studies on the Genetics of Alcoholism, would continue to have great utility as they are better powered to detect rare variants. Capitalizing on the idea that common variants can capture variance attributable to low-frequency functional variants, whole genome prediction models (WGPMs), such as genome-wide complex trait analysis (Yang et al., 2011b), have been able to capture more of the variability in complex traits (e.g., height and body mass index) using current genotyping platforms (Lee et al., 2011), thus demonstrating that cumulative/aggregate genetic risk scores should prove useful in capturing more of the missing heritability (De Jager et al., 2009; Kohli et al., 2010; Purcell et al., 2009). However, a notable limitation of these models is that they lack the degree of specificity needed to inform the development of prevention/treatment tools.
3.2 Gene-Gene Interactions (Epistasis)
Another aspect of missing heritability in alcohol GWAS is the layers of interactions between genes within the context of the rest of the genome and the environment in which they exist. This non-linear combination of genes/gene-products is referred to as epistasis. Because AD is a complex developmental disease that involves impaired neural development and function, it is undoubtedly fraught with molecular interactions (i.e., DNA and proteins) that are difficult to model statistically. For example, Palmer et al. (2003) suggested that the background of a knockout mouse might be important when studying response to ethanol. In their study of two different DRD2 knockout mouse strains, the authors showed that the effects of the null allele on ethanol’s stimulant and sensitizing effects differed based on the background used to develop the knockout strain (Palmer et al., 2003). So far, there has been modest evidence of statistical epistatic effects on AD in the human literature, possibly because of a lack of power in most studies. In a recent study, Kumar et al. discovered epistatic effects between the mu and kappa opioid system, with respect to alcohol (Kumar et al., 2012). The researchers showed that although individual markers were not associated with alcoholism, locus-locus interactions between OPRM1 and OPRK1 led to a two-fold increase [2.318 (1.025 to 5.24)] in the risk for alcoholism. Coupled with findings from complex diseases, these results suggest that genetic interactions may be far more important than initially thought. Furthermore, it has been suggested that the failure to model epistatic effects in biometrical studies may have inflated the heritability of complex traits (Zuk et al., 2012). The inherent problem with capturing epistatic effects using variants with modest effect sizes arises from the vast number of possible combinations that can exist within the human genome. For example, a model containing two SNPs, each with three genotypes (i.e., AA, Aa, and aa), would have nine possible genotypes, while a model containing four SNPs would have 81 possible genotypes (i.e., 3# of SNPs). Capturing epistatic effects is further complicated by the fact that the power to detect a genetic effect is dependent upon the minor allele frequency (MAF) of the risk allele being studied. Specifically, the power to detect relatively modest genotypic risk ratios observed in AD GWAS increases as the MAF of the risk allele being tested increases. However, in the case of epistatic studies we would be dealing with the MAF of different combinations of risk alleles, which further increases the likelihood of obtaining a false positive result. Recently, it has been suggested that the inclusion of epistatic effects, as well as gene-environment interaction effects (discussed in section 4.2) in association studies provides a slight improvement in predictive power; notably, predictive power increases as (1) the number of risk factors increases and (2) the effect size of causal variants increases (Aschard et al., 2012).
Non-parametric methods (e.g., data mining, machine learning, and neural network modeling) have been proposed as a discovery tool for exploring these highly dimensional spaces without the need of a priori hypotheses. Of particular interest are non-parametric data-mining methods, such as multifactor dimensionality reduction (MDR; Chen et al., 2011b; Hahn et al., 2003; Ritchie et al., 2003a), because they are easier to interpret than neural network models (Lucek and Ott, 1997; Motsinger-Reif et al., 2008; Ritchie et al., 2003b). Notably, despite the utility of data-mining approaches, the possibility of obtaining false-positive results that can result from chance patterns in the data still exists. MDR has been applied to several complex phenotypes, such as multiple sclerosis, coronary artery disease, and cancer (Agirbasli et al., 2011; Brassat et al., 2006; Gui et al., 2011), however, applications to addiction phenotypes are lacking.
3.3 The Phenotypic and Genetic Complexity of AD
Missing heritability in AD GWAS is also attributable to the fact that AD’s liability is genetically heterogeneous (i.e., different individuals posses different combinations of susceptibility alleles within/across genes) and phenotypically heterogeneous (i.e., individuals might arrive at a diagnosis of AD with a combination of different characteristics/symptoms). This largely reflects the fact that people become addicted or remain addicted to alcohol for different reasons. Hence, the lack of power in alcohol GWAS AD can also be attributed to the use of phenotypes that fail to capture the biological underpinnings of AD, which would ultimately led to the classification of groups/types of alcoholics that may be more genetically homogeneous. This is primarily obvious in summary phenotypes, such as AD, which combine physiological characteristics of the disorder with psychosocial aspects. This, in turn, results in different combinations of individuals with different problems. Consequently, every GWAS of AD has had to average the score across individuals with different aspects of an underlying inability to regulate their alcohol consumption. For example, a recent GWAS by Kendler et al. (2011) indicated that while symptoms of AD formed a single factor there were no SNPs that approached the 5×10−8 threshold for genome-wide significance for the AD factor score.
3.3.1 Using Comorbidity to Understand Heterogeneity
One approach to understanding the phenotypic/etiological complexity of alcohol is to understand the nature of its relationship with other traits and to account for them in studies. Past studies show that individuals who use/misuse alcohol are also likely to use/misuse tobacco and other drugs (Palmer et al., 2009), thus pointing to a general liability for dependence across multiple substances (Palmer et al., 2012). Alcoholics are also more likely to be diagnosed with other psychiatric disorders, such as major depression or antisocial personality disorder (Edwards et al., 2012; Iacono et al., 2008; Sher et al., 2005). In fact, many psychiatric disorders are often comorbid with alcohol use and AD and in some instances, precede it (Elkins et al., 2006, 2007). For instance, we recently examined a sample of older adolescents and determined that those who exhibited high levels of DSM-IV CD symptoms and novelty seeking tendencies were more likely to exhibit high levels of alcohol, tobacco, and cannabis DSM-IV dependence symptoms during young adulthood (Palmer et al., 2011). To further complicate matters, AD is one component of the latent Externalizing (EXT) or Behavioral Disinhibition (BD) dimension that represents an inability to controls one’s own impulsive thoughts and actions (Krueger et al., 2002; Young et al., 2000). In addition, one of the largest studies of comorbid psychopathologies (i.e., internalizing and externalizing disorders) by Kendler et al. found separate genetic factors that predispose to internalizing disorders (i.e., major depression, generalized anxiety, and phobia) and externalizing disorders (i.e., AD, other drug dependence, adult antisocial behavior, and conduct disorder; Kendler et al., 2003), suggesting that most of the genetic variance associated with AD is shared with adult antisocial behavior, other drug dependence, and childhood conduct disorder. Consequently, it is important to consider whether genetic studies of AD, truly indicate susceptibility factors specifically related to AD.
3.3.2 The Endophenotype Approach
The primary approach to overcoming etiological heterogeneity has been the use of endophenotypes (i.e., a heritable biological and/or psychological characteristic of a disease that (1) has a strong biological basis, (2) manifests whether or not the illness is active within the individual, (3) relates to the disease in the population, and (4) co-segregates with the disease in families; Gottesman and Gould, 2003), which should improve the power to identify alcohol susceptibility genes because they reduce the complexity of both the phenotype and the genetic analysis (i.e., a less complex genetic architecture). Several of the most studied endophenotypes of AD have been electrophysiological measures, such as electroencephalography and event-related potentials (e.g., alpha and beta waves and alpha power, as well as the P300 amplitude; Carlson and Iacono, 2006; Chorlian et al., 2007), sensitivity to alcohol (i.e., level of alcohol response; Schuckit, 1994; Schuckit et al., 2004)), alcohol metabolism (Lind et al., 2008b; Martin et al., 1985a; Martin et al., 1985b), and alcohol craving (Anton, 1999; Mackillop et al., 2007, 2010; Monti et al., 2000; Sinha and O’Malley, 1999; Verheul et al., 1999). A detailed review of alcohol endophenotypes is presented elsewhere (Hines et al., 2005). Another approach to limiting heterogeneity has been the utilization of animal models and post-gene studies (i.e., transcriptome and proteome studies) to identify genetic factors related to specific components of alcoholism, such as alcohol consumption. For example, researchers have used whole-brain gene expression data of several mouse models of alcohol consumption to identify candidate genes and functional pathways related to voluntary alcohol consumption (Mulligan et al., 2006). By characterizing differences between mice that were never exposed to alcohol, but were characteristically known to differ in their levels of alcohol consumption, Mulligan et al. demonstrated that there are multitudes of neuronal pathways that differ between mice that are inherently destined to consume low/high amounts of alcohol. Likewise, there are many genes located within these pathways that are differentially expressed between these groups of mice because of variation within and among them. Notably, several of the 3800 unique genes identified in Mulligan’s study (2006) were present in gene loci that had previously been linked to AD in humans. Overall, these approaches point to a physiological domain (linked to metabolism) and a neurological domain (i.e., neuronal profiles susceptibility to alcohol’s effects) that support the pharmacology and neuroscience literature. However, they also indicate complexity in accounting for genetic heterogeneity in association studies as it would require large datasets with extensive phenotyping or alternatively experimental studies that are (1) ethically challenging, (2) experimentally challenging, and (3) fiscally infeasible in humans alone.
4. ACHIEVING A SYSTEMS-BASED APPROACH TO STUDYING AD
4.1 The Need for Genomewide Systems-based Studies of AD
Alcohol’s genetic complexity highlights the need for comprehensive models that account for the cumulative, pleiotropic, and epistatic effects of genes in the context of the rest of the genome and the environment. System-based genetic studies (i.e., Systems Genetics) of AD have become increasingly possible because of the major advances in genomics, proteomics, gene x environment interaction and correlation studies, and epigenetics. Systems-based approaches that conceptualize and model the susceptibility to AD as combinatorial effects of genetic, epigenetic, transcriptomic, and proteomic variation are likely to prove useful in overcoming these challenges. The advantage of a systems-based framework over agnostic testing procedures is that it organizes the distribution of “relatively modestly effective” variants into profiles that might better inform our understanding of specific aspects of the development of alcohol use disorders. We propose Systems Genetics (a combination of Systems Biology and Genetic Association Studies) over individual pathway or gene-set enrichment approaches because it conceptualizes and explores vulnerability to AD as a function of the joint and multiplicative distribution of gene, epigene, and proteomic effects that constitute evolutionarily robust biological systems disrupted by alcohol and other drugs. Such models [which include biological (e.g., genetic/transcriptomic variants) and environmental (e.g., presence/absence of alcohol cues or alcohol using relatives/peers) variables] describe how any one perturbation in one aspect of the system (e.g., “Motivation” in Figure 1) affects other components of the system and the manifestation of the disease/trait. Given the lack of large NGS databases, a starting point for Systems Genetics would be the application of sophisticated network-based models to existing GWAS data in order to identify/confirm candidate systems (i.e., network of variation across different biological pathways) that are likely to be more stable and reproducible across independent samples. Notably, the lack of comparable environmental assessments/arrays across studies will make it difficult to replicate environmental effects. In addition to GWAS data network-based models can be made to incorporate micro-array/RNA-Seq (i.e., whole genome sequencing of mRNA transcripts) and Chip-Seq (i.e., whole genome sequencing of immune-precipitation-enriched genomic DNA) data. For instance, although microarray studies have suffered from multiple testing issues resulting from the agnostic interrogation of the expression of thousands of genes, they have demonstrated that alcohol induces changes in the expression level of DNA binding and cell signaling genes within the prefrontal cortex (Flatscher-Bader et al., 2006). Alcohol has also been shown to influence the expression of genes involved in matrix remodeling, proliferation, and cell morphogenesis in the nucleus accumbens and ventral tegmental area (Flatscher-Bader et al., 2010). The combination of DNA whole genome genetic variation with epigenetic, transcriptomic, and proteomic profiles taken from select neural tissues involved in different stages of addiction (e.g., hippocampus) would be the ideal approach to achieving Systems-based models of AD. By accounting for the relationship between the genome as a whole and the transcriptome of select tissue as a whole, we would obtain effects that are more robust. For instance, recent studies that have examined the covariation between variation in the genome and the transcriptome, and the proteome suggest modest correlations between them (Colantuoni et al., 2011; Ghazalpour et al., 2011). For example, Colantuoni et al’s (2011) examination of how genomewide sequence variation affects gene expression in the prefrontal cortex (PFC) showed that across different racial/ethnic groups, individual SNPs can alter the expression of individual genes in the PFC; furthermore, although the level of gene expression in the PFC varies across the lifespan, it is a consistent set of genes that is expressed. Thus, for future studies, the joint analysis of the genome, transcriptome, and proteome will be essential to understanding the structural and functional changes in our brain and metabolism. A recent example of this approach in alcohol research involved the use of RNA-Seq, ChIP-Seq, and histone H3 lysine 4 trimethylation (H3K4me3) data to identify expression differences in post-mortem hippocampus tissue collected from alcohol and cocaine dependent cases and matching controls (Zhou et al., 2011). Similarly, Schumann et al. (2011) followed up on their GWAS findings that pointed to AUTS2 as a regulator of alcohol consumption, by demonstrating significant expression-level differences in human prefrontal cortex, and whole-brain extracts from mice, as well as, reduced consumption in drosophila insertion mutants.
4.2 Including the Environment as a Part of the System
Although not the focus of this paper, it is important that we mention the environment as a key factor that provides the context in which biological systems operate. Environmental exposure plays a critical role in the liability to AD; according to twin/family studies, roughly one-half of the liability to AD is attributable to environmental factors. Environments that influence the risk for AD differ in both proximity to the disorder and mechanism of action. The risk for AD is elevated among (1) children that were prenatally exposed to alcohol, (2) children that grow up in a home with an alcoholic parent, and (3) children that are poorly monitored by their parents, to name a few (Sher et al., 2005). Many of these environments are thought to interact or correlate with the individual’s biological/genetic background resulting in an increased/decreased risk for the development of AD. For instance, genetic effects on drinking has been shown to be greater in urban versus rural residential settings (Dick et al., 2001; Rose et al., 2001), possibly because of differences in the level of social control between rural and urban environments (i.e., social control or structural constraints may be greater in rural environments, limiting the manifestation of genetically determined behaviors). Findings from the COGA study also suggest that variation within an AD susceptibility gene (GABRA2; rs279871) is related to a person’s marital status. Individuals with the high risk GABRA2 variant were less likely to be married partly because of their elevated risk for antisocial personality disorder; marital status also moderated the effect of other variants within GABRA2 on AD (Dick et al., 2006a). In addition to the evidence for gene x environment interaction, gene-environmental correlations are also relevant to alcohol. For example, several studies (Cleveland et al., 2005; Fowler et al., 2007; Harden et al., 2008) have shown that a person’s genes influence their exposure to 1) alcohol, and 2) their exposure to peers who use alcohol. In their study of 862 twin pairs, Fowler et al. (2007) found significant correlations (>0.60) between genetic influences on friends alcohol use and problem use and a person’s own use and problem use. Studies such as these indicate that individuals from high-risk backgrounds (i.e., alcohol abusing parents or relatives) may be more likely to place themselves in high-risk environments for AD (as seen above, interacting with substance abusing peers). If we are to obtain robust estimates of the contribution of genetic factors to the liability of alcohol dependence, the synergy between environmental factors and genetic factors must be acknowledged in molecular genetics approaches. In a previous report, Heath and Nelson (2002) highlighted the need for well-designed prospective studies and family-based association studies to identify important environmental risk factors and account for intergenerational processes that can confound the genetic risk for AD with other psychiatric disorders (Heath and Nelson, 2002). While there have been several advances in the environmental literature, our understanding of how genes and environments interact is still limited (Duncan and Keller, 2011). More importantly, environmental measures are not consistent across studies, highlighting the need for high-throughput techniques, such as an environment-array that can be used to broadly survey environmental measures related to AD.
4.3 The Integration of Biology into Novel Statistical and Computational Approaches
Current approaches to capturing the missing heritability of complex diseases involve the application of gene-set analyses, whole genome prediction analyses, the use of biological data in the form of pathway-enrichment approaches, and genomewide modeling of gene-gene interactions (i.e., epistasis). Gene-set methods, such as the set-based method in PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/anal.shtml#set; Purcell et al., 2007) are suitable to large-scale candidate gene studies, and more recently, sets of genes identified after applying strict r2 (i.e., correlation coefficient between a pair of alleles) thresholds to GWAS data (Kendler et al., 2011). Gene-set approaches provide the benefit of large-scale permutation testing on a specified number of SNPs that enable the identification of genes that achieve gene-wise significance. Whole genome prediction models complement the gene-set approach by examining the total amount of phenotypic variance attributable to variants present on a selected platform. Although this approach has not yet been applied to AD, a recent study on height (Yang et al., 2011a) indicated that if all the SNPs on a particular genotyping platform are considered simultaneously, approximately 45% of the variation in height can be captured. Consequently, the modeling of cumulative variant/gene effects and the capture of rare causal variants are crucial to the examination of complex traits. Unlike gene-set and whole genome prediction, pathway enrichment approaches capitalize on the wealth of data stored in bioinformatics databases, such as the Gene Ontology database (Ashburner et al., 2000), the Mouse Genome Informatics database (Blake et al., 2011), the Kyoto Encyclopedia of Genes and Genomes (Kanehisa, 2002), the Human Protein Reference Database (Mishra et al., 2006), HumanCyc (Romero et al., 2005), and Panther Pathways (Mi and Thomas, 2009). For example, a recent GWAS by Kendler et al. (2011) used ALIGATOR (Holmans et al., 2009), a method useful for testing the overrepresentation of Gene Ontology terms in gene lists identified from a GWAS study, to examine the genetic etiology of AD in the Molecular Genetics of Schizophrenia Control Sample. Although no SNPs survived multiple testing corrections, Kendler et al. identified six genes in the European American sample and five genes in the African American sample that met the criteria for gene-wise significance using Plink, as well as a large number of enriched categories/pathways in Europeans (up to 347) and African Americans (up to 254). The advantage of pathway-enriched approaches is that they overcome the issue of genetic heterogeneity, which greatly reduces the power to detect an association. Thus, by shifting attention to the frequency of occurrence of variants related to a pathway, pathway-enriched approaches improve the power to detect an association. Similar pathway/enrichment approaches include: GenGen (http://www.openbioinformatics.org/gengen/; Wang et al., 2007), GSEA (for RNA expression analysis; http://www.broadinstitute.org/gsea/index.jsp; Subramanian et al., 2007, 2005), the SNP-ratio test (http://sourceforge.net/projects/snpratiotest/; O’Dushlaine et al., 2009), and INRICH (can be used for combinations of SNPs, CNVs, genes; http://atgu.mgh.harvard.edu/inrich/; Lee et al., 2012), to name a few. It is important to note however that pathway approaches are limited to existing knowledge about a gene and the biological pathways relevant to the disease. Furthermore, study results may not be generalizable because the approach assumes that genes influence the disease/trait through a common biological pathway, which in the case of addiction phenotypes is only partially true (Palmer et al., 2012). Genomewide epistatic modeling (GEM) approaches provide a means to model biological interactions while also utilizing the clustering approach used in pathway/gene-set analyses.
Unfortunately, this form of research is still under development with most of the nonparametric tools being applied to cancer phenotypes. Given its nature, parametric logistic regression cannot be employed in GEM studies because large sample sizes would be required. Alternatively, researchers may opt to limit the 1 million markers on a DNA micro array to only those belonging to genes in candidate pathways (given prior knowledge to select these markers; Grady et al., 2011). The likely alternative approach is the application of nonparametric methods that are currently being adapted to GWAS, such as MDR and HotNet (Vandin et al., 2012, 2011). Many of these approaches provide flexibility in model specification providing a way to reduce the burden of genetic heterogeneity and multiple testing. For instance, MDR is a machine-learning alternative to logistic regression that assumes no particular genetic model while identifying combinations of SNPs that influence the likelihood of a disease state. As part of its method, MDR combines attribute selection and construction (i.e., the creation of a single attribute by pooling data across SNPs) with permutation testing. Like MDR, HotNet, identifies groups of genes related to a disease but does so differently. HotNet uses a diffusion model and a two-stage statistical test to identify groups of mutated/perturbed genes related to a disease. HotNet first formulates an influence measure between pairs of genes using a diffusion process, which is a type of flow problem that has been implemented in protein function prediction on protein interaction networks with significant success (Vandin et al., 2011). Each measure of influence considers a gene to influence another gene if (1) they are both close in distance on the network, and (2) there are relatively few paths between them in the network. Second, subnetworks are identified using an enhanced influence model, in which the number of mutations (e.g., alleles leading to altered protein function) in a gene weights the influence between pairs of genes. Overall, these combinatorial approaches, coupled with the increasing accessibility of GWAS and NGS data across multiple domains provide the means to the joint effect of genetic, epigenetic, transcriptomic, and proteomic factors on biochemical pathways related to AD susceptibility.
4.4 Interpreting Systems-based Analysis of AD: The Need for Alcohol-focused Ontologies
Taken together, the different –omics’ illustrate the complexity of the genetic and environmental mechanisms involved in the liability to AD. Genomic studies highlight quantitative trait loci that confer risk or protect against AD. Transcriptome studies demonstrate that alcohol changes the expression level of genes in several brain regions (Flatscher-Bader et al., 2006; Mulligan et al., 2006), such as the nucleus accumbens (Bell et al., 2009; Flatscher-Bader et al., 2010; Obara et al., 2009), and the extended amygdala (McBride et al., 2010) to name a few. Protein expression studies of alcohol use/dependence also indicate protein-level changes that may account for individual differences in drinking behaviors (McBride et al., 2009). Gene-environment studies highlight the strong role of proximal and distal environmental factors that might correlate with genetic factors and/or moderate their effects (Enoch, 2006). Moving forward, the largest obstacles to designing systematic approaches to AD and other complex diseases appear to be data integration, analysis, and interpretation. Integrating genomics, transcriptomics, proteomics, and most eventually environmental effects into a format that will be applicable to AD requires extensive mining of bioinformatic databases with the intent to build a framework upon evidence from model organisms like drosophila, mouse, and simple organisms. This knowledge base will be essential to modeling specific aspects of AD because functional experiments in these animals will be the key to unlocking epistatic processes in different environmental situations (as best studied under controlled conditions using pre-clinical models). Notably, data sharing across genetic, proteomic, transcriptomic, and epigenetic databases are already underway, leaving the interpretation of systems-based studies as the latest challenge in genetics research; unfortunately, environmental databases that connect environmental factors to AD using replicable high-throughput techniques are still in their infancy. Since graph-based models will be particularly useful in modeling complex biological systems, systems-genetics models of AD will most likely resemble a collection of nodes, which would represent genes and/or proteins, and edges that represent the relationship between nodes and the means by which susceptibility to disease is transmitted through the system (Figure 2). Given the biology of AD, formal ontologies (i.e., formal models of a domain of knowledge and the relationship between entities that are being modeled) will be needed to interpret the findings of network models. An ever-evolving AD ontology could describe the cascade of events involved in the metabolic clearance of alcohol and the neuronal circuitry that regulates (1) alcohol’s rewarding and reinforcing effects (i.e., how alcohol activates the mesolimbic reward system (i.e., the nucleus accumbens neurons, ventral tegmental area, amygdala, and hippocampus), (2) executive control (i.e., the dorsolateral prefrontal cortex and its connections to the mesolimbic system), (3) the development of an alcohol habit (i.e., the cerebellum, the amygdala, the basal ganglia (primarily the striatum), and the hippocampus) (Zahr and Sullivan, 2008), and (4) behavioral response to stressors (i.e., the extra-hypothalamic corticotrophin releasing factor system; Merlo Pich et al., 1995). Formalizing an ontology of AD (including all of its different domains) will help to organize and communicate all of the important risk and protective factors and phenotypes into a structured representation. In addition, ontologies of AD (global or focused on a particular domain of AD (stress circuits and AD)) can serve as a base for exploration in future research. More specifically, researchers will be able to design computational models that explore variation across multiple domains (genomics, transcriptomics, proteomics, etc.) and levels (e.g., cell, tissue, behavior) of the system.
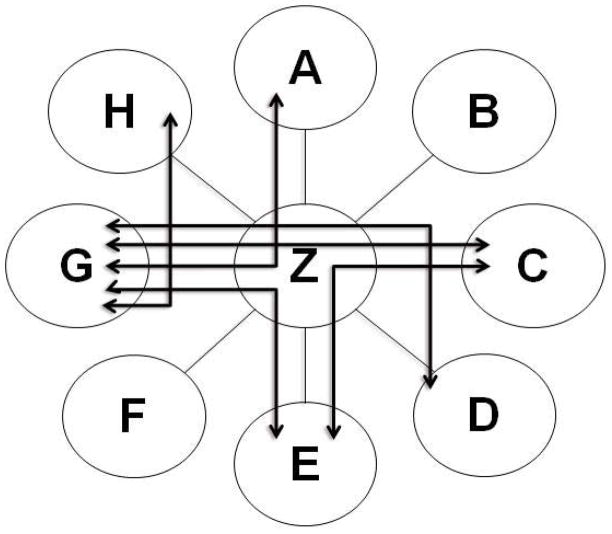
Figure 2. Example of a Generic Network Model.
Visualization of the results of a typical network model analysis. Nodes (with the letters A thru H) represent variables. Edges represent pathways though which perturbations within each variable propagate through the system.
5. IMPLICATIONS FOR TREATMENT AND PREVENTION
The use of a combined systems biology and GEM approach will ideally account for more genetic variance in a particular phenotype than that which can be attributed to any single genetic variant. In some cases, this may complicate the clinical translation of these findings as single variant findings are more readily translatable into pharmacologically-based interventions whereas the likely group of variants implicated by a systems biology approach may span multiple neurotransmitter systems in a fashion not readily amenable to monotherapies or medication development. Systems biology may highlight a particular biological pathway that could be targeted pharmacologically at various levels (e.g., presynaptically, synaptically or postsynaptically). However, this information offers a distinct advantage over single variant methods by providing an increased understanding of epistasis, or interactions of genes relevant to potentially different neurotransmitter systems. Accordingly, it is possible that these systems based approaches may define the genetic contributions to larger scale polygenic phenomena that could be targeted behaviorally. For example if variation in multiple genes were implicated in explaining differential urge for alcohol (in a fashion that was not readily addressed through pharmacologic monotherapies), use of a genetically-defined “urge sensitivity” profile might be used to personalize treatment for such individuals to focus on urge management strategies behaviorally. This is not to suggest that there is no place for pharmacogenetic approaches in the treatment of alcohol dependence (in fact, the evidence suggests otherwise). Rather we highlight the possibility for combination pharmacotherapies and also targeting behavioral interventions using genetic information until such a time as targeted drug delivery and pharmacological target specificity is obtained (Pajer et al., 2012).
6. COMMENT
Numerous technologies have demonstrated that AD is a function of genetic differences, gene expression differences, protein-level differences, and differences in environmental exposure. Genetic association studies have uncovered genetic variants and environments that explain individual differences in susceptibility to AD. Genomewide association studies of alcohol indicate that its genetic etiology is highly complex. Transcriptome and proteome studies have shown differences in gene and protein expression in candidate tissues affected by alcohol and other drugs. Collectively, variability across these different systems may contribute a greater understanding of alcohol dependence; however, future strategies will require modeling techniques that capture ubiquitous principles that underlies all biological processes and complex diseases, epistasis and pleiotropy. Challenges for future genetic studies of alcohol, will be (1) the identification of phenotypes that assist in the assessment of how genetic variation in different neural systems influence and relate to different developmental stages of alcohol dependence, (2) the development and application of computational techniques that model the cumulative and combinatorial effects of genes and environments in a systematic manner, (3) the incorporation of a consistent set of proximal (e.g., peers) and distal (e.g., residency location) environmental measures across genetic studies of alcohol and other drugs, and (4) the development of ontologies of alcohol dependencies that can facilitate the development of future studies and the interpretation of their findings. In the future, family-based whole-genome studies and animal studies may be the best approaches to understanding all of these mechanisms; family studies, because they are likely to include extreme cases of alcoholics with an ancestral history of alcohol problems and associated neuronal and genetic susceptibilities; animal studies, because selection and congenic experiments provide more than adequate control over genetic background effects, as well as epigenetic and environmental effects. The application of systems biology and GEM to neuronal and alcohol metabolism processes involved in alcohol related diseases will help to provide a better understanding of how individual parts of the liability to alcohol-related diseases work as a whole. Further, incorporating environmental measures will be a powerful method for better understanding the nature of gene-environment interaction and its contribution to the etiology of behavioral variation.
Acknowledgments
Role of funding source
This paper was supported by MH019927 (Rohan Palmer), L30DA032090 (Rohan Palmer), DA023134 (Valerie Knopik (PI) and Sarah Francazio), HG005690 (Benjamin Raphael), AA011998 (Andrew Heath), and GM076516 and 24480 from the Templeton Foundation (Lander).
Footnotes
Contributors
Authors Rohan Palmer and Sarah Francazio managed the literature searches and summaries of previous work. Author Rohan Palmer wrote the first draft of the manuscript under John McGeary, Andrew Heath, Arthur Lander, and Valerie Knopik’s mentorship. All authors contributed to and have approved the final manuscript.
Conflict of interest
All of the listed authors declare that they have no conflicts of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agirbasli M, Guney A, Ozturhan H, Agirbasli D, Ulucan K, Sevinc D, Kirac D, Ryckman K, Williams S. Multifactor dimensionality reduction analysis of MTHFR, PAI-1, ACE, PON1, and eNOS gene polymorphisms in patients with early onset coronary artery disease. Eur J Cardiovasc Prev Rehabil. 2011;18:803–809. doi: 10.1177/1741826711398806. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT. Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction. 2008;103:1069–1081. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- Alcohol_Alert. Alcohol Metabolism: An Update. National Institute on Alcohol Abuse and Alcoholism Publications Distribution Center; Rockville, MD: 2007. [Google Scholar]
- Almasy L, Porjesz B, Blangero J, Goate A, Edenberg HJ, Chorlian DB, Kuperman S, O’Connor SJ, Rohrbaugh J, Bauer LO, Foroud T, Rice JP, Reich T, Begleiter H. Genetics of event-related brain potentials in response to a semantic priming paradigm in families with a history of alcoholism. Am J Hum Genet. 2001;68:128–135. doi: 10.1086/316936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 2. Washington DC: 1968. [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 3. Washington, DC: 1980. [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 3. Washington DC: 1987. Revised. [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4. Washington, DC: 2000. text rev. [Google Scholar]
- Anton RF. What is craving? Models and implications for treatment. Alcohol Res Health. 1999;23:165–173. [PMC free article] [PubMed] [Google Scholar]
- Aschard H, Chen J, Cornelis MC, Chibnik LB, Karlson EW, Kraft P. Inclusion of gene-gene and gene-environment interactions unlikely to dramatically improve risk prediction for complex diseases. Am J Hum Genet. 2012;90:962–972. doi: 10.1016/j.ajhg.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik I, Cho NH, Kim SH, Han BG, Shin C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr. 2011;93:809–816. doi: 10.3945/ajcn.110.001776. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Reich T, Edenberg HJ, Goate A, Blangero J, Almasy L, Foroud T, Van Eerdewegh P, Polich J, Rohrbaugh J, Kuperman S, Bauer LO, O’Connor SJ, Chorlian DB, Li TK, Conneally PM, Hesselbrock V, Rice JP, Schuckit MA, Cloninger R, Nurnberger J, Jr, Crowe R, Bloom FE. Quantitative trait loci analysis of human event-related brain potentials: p3 voltage. Electroencephalogr Clin Neurophysiol. 1998;108:244–250. doi: 10.1016/s0168-5597(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Bell RL, Kimpel MW, McClintick JN, Strother WN, Carr LG, Liang T, Rodd ZA, Mayfield RD, Edenberg HJ, McBride WJ. Gene expression changes in the nucleus accumbens of alcohol-preferring rats following chronic ethanol consumption. Pharmacol Biochem Behav. 2009;94:131–147. doi: 10.1016/j.pbb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Dinwiddie SH, Begleiter H, Crowe RR, Hesselbrock V, Nurnberger JI, Jr, Porjesz B, Schuckit MA, Reich T. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism. Arch Gen Psychiatry. 1998;55:982–988. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39:D842–848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassat D, Motsinger AA, Caillier SJ, Erlich HA, Walker K, Steiner LL, Cree BA, Barcellos LF, Pericak-Vance MA, Schmidt S, Gregory S, Hauser SL, Haines JL, Oksenberg JR, Ritchie MD. Multifactor dimensionality reduction reveals gene-gene interactions associated with multiple sclerosis susceptibility in African Americans. Genes Immun. 2006;7:310–315. doi: 10.1038/sj.gene.6364299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SR, Iacono WG. Heritability of P300 amplitude development from adolescence to adulthood. Psychophysiology. 2006;43:470–480. doi: 10.1111/j.1469-8986.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- Chao YC, Liou SR, Chung YY, Tang HS, Hsu CT, Li TK, Yin SJ. Polymorphism of alcohol and aldehyde dehydrogenase genes and alcoholic cirrhosis in Chinese patients. Hepatology. 1994;19:360–366. [PubMed] [Google Scholar]
- Chen D, Liu L, Xiao Y, Peng Y, Yang C, Wang Z. Ethnic-specific meta-analyses of association between the OPRM1 A118G polymorphism and alcohol dependence among Asians and Caucasians. Drug Alcohol Depend. 2011a;123:1–6. doi: 10.1016/j.drugalcdep.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Chen GB, Zhu J, Lou XY. A faster pedigree-based generalized multifactor dimensionality reduction method for detecting gene-gene interactions. Stat Interface. 2011b;4:295–304. doi: 10.4310/sii.2011.v4.n3.a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Peng GS, Wang MF, Tsao TP, Yin SJ. Polymorphism of ethanol-metabolism genes and alcoholism: correlation of allelic variations with the pharmacokinetic and pharmacodynamic consequences. Chem Biol Interact. 2009;178:2–7. doi: 10.1016/j.cbi.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Chorlian DB, Tang Y, Rangaswamy M, O’Connor S, Rohrbaugh J, Taylor R, Porjesz B. Heritability of EEG coherence in a large sib-pair population. Biol Psychol. 2007;75:260–266. doi: 10.1016/j.biopsycho.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland HH, Wiebe RP, Rowe DC. Sources of exposure to smoking and drinking friends among adolescents: a behavioral-genetic evaluation. J Genet Psychol. 2005;166:153–169. [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JP, Young RM, Lawford BR, Saunders JB, Ritchie TL, Noble EP. Heavy nicotine and alcohol use in alcohol dependence is associated with D2 dopamine receptor (DRD2) polymorphism. Addict Behav. 2007;32:310–319. doi: 10.1016/j.addbeh.2006.04.006. [DOI] [PubMed] [Google Scholar]
- De Jager PL, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC, Aubin C, Bauer D, Heubach JF, Sandbrink R, Tyblova M, Lelkova P, Havrdova E, Pohl C, Horakova D, Ascherio A, Hafler DA, Karlson EW. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–1119. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Agrawal A, Schuckit MA, Bierut L, Hinrichs A, Fox L, Mullaney J, Cloninger CR, Hesselbrock V, Nurnberger JI, Jr, Almasy L, Foroud T, Porjesz B, Edenberg H, Begleiter H. Marital status, alcohol dependence, and GABRA2: evidence for gene-environment correlation and interaction. J Stud Alcohol. 2006a;67:185–194. doi: 10.15288/jsa.2006.67.185. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Wang JC, Grucza RA, Schuckit M, Kuperman S, Kramer J, Hinrichs A, Bertelsen S, Budde JP, Hesselbrock V, Porjesz B, Edenberg HJ, Bierut LJ, Goate A. Using dimensional models of externalizing psychopathology to aid in gene identification. Arch Gen Psychiatry. 2008;65:310–318. doi: 10.1001/archpsyc.65.3.310. [DOI] [PubMed] [Google Scholar]
- Dick DM, Bierut L, Hinrichs A, Fox L, Bucholz KK, Kramer J, Kuperman S, Hesselbrock V, Schuckit M, Almasy L, Tischfield J, Porjesz B, Begleiter H, Nurnberger J, Jr, Xuei X, Edenberg HJ, Foroud T. The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behav Genet. 2006b;36:577–590. doi: 10.1007/s10519-005-9041-8. [DOI] [PubMed] [Google Scholar]
- Dick DM, Rose RJ, Viken RJ, Kaprio J, Koskenvuo M. Exploring gene-environment interactions: socioregional moderation of alcohol use. J Abnorm Psychol. 2001;110:625–632. doi: 10.1037//0021-843x.110.4.625. [DOI] [PubMed] [Google Scholar]
- Dick DM, Wang JC, Plunkett J, Aliev F, Hinrichs A, Bertelsen S, Budde JP, Goldstein EL, Kaplan D, Edenberg HJ, Nurnberger J, Jr, Hesselbrock V, Schuckit M, Kuperman S, Tischfield J, Porjesz B, Begleiter H, Bierut LJ, Goate A. Family-based association analyses of alcohol dependence phenotypes across DRD2 and neighboring gene ANKK1. Alcohol Clin Exp Res. 2007;31:1645–1653. doi: 10.1111/j.1530-0277.2007.00470.x. [DOI] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr, O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Jr, Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, Kramer J, Kuperman S, Nurnberger JI, Jr, Schuckit MA, Porjesz B, Dick DM. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr Genet. 2012;22:31–41. doi: 10.1097/YPG.0b013e32834acd07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins IJ, King SM, McGue M, Iacono WG. Personality traits and the development of nicotine, alcohol, and illicit drug disorders: prospective links from adolescence to young adulthood. J Abnorm Psychology. 2006;115:26–39. doi: 10.1037/0021-843X.115.1.26. [DOI] [PubMed] [Google Scholar]
- Elkins IJ, McGue M, Iacono WG. Prospective effects of attention-deficit/hyperactivity disorder, conduct disorder, and sex on adolescent substance use and abuse. Arch Gen Psychiatry. 2007;64:1145–1152. doi: 10.1001/archpsyc.64.10.1145. [DOI] [PubMed] [Google Scholar]
- Enoch MA. Genetic and environmental influences on the development of alcoholism: resilience vs. risk. Ann NY Acad Sci. 2006;1094:193–201. doi: 10.1196/annals.1376.019. [DOI] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Dahmen N, Schmidt LG, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, Harrison E, Matsumoto I, Wilce PA. Genes associated with alcohol abuse and tobacco smoking in the human nucleus accumbens and ventral tegmental area. Alcohol Clin Exp Res. 2010;34:1291–1302. doi: 10.1111/j.1530-0277.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug MP, Landis N, Hwang JW, Harrison E, Wilce PA. Comparative gene expression in brain regions of human alcoholics. Genes Brain Behav. 2006;5(Suppl 1):78–84. doi: 10.1111/j.1601-183X.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- Foroud T, Bucholz KK, Edenberg HJ, Goate A, Neuman RJ, Porjesz B, Koller DL, Rice J, Reich T, Bierut LJ, Cloninger CR, Nurnberger JI, Jr, Li TK, Conneally PM, Tischfield JA, Crowe R, Hesselbrock V, Schuckit M, Begleiter H. Linkage of an alcoholism-related severity phenotype to chromosome 16. Alcohol Clin Exp Res. 1998;22:2035–2042. [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- Fowler T, Shelton K, Lifford K, Rice F, McBride A, Nikolov I, Neale MC, Harold G, Thapar A, van den Bree MB. Genetic and environmental influences on the relationship between peer alcohol use and own alcohol use in adolescents. Addiction. 2007;102:894–903. doi: 10.1111/j.1360-0443.2007.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, Park CC, Wen PZ, Brewer H, Weitz K, Camp DG, 2nd, Pan C, Yordanova R, Neuhaus I, Tilford C, Siemers N, Gargalovic P, Eskin E, Kirchgessner T, Smith DJ, Smith RD, Lusis AJ. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Begleiter H, Porjesz B, Chorlian DB, Edenberg HJ, Foroud T, Goate A, Reich T. Linkage mapping of beta 2 EEG waves via non-parametric regression. Am J Med Genet B Neuropsychiatr Genet. 2003;118B:66–71. doi: 10.1002/ajmg.b.10057. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Grady BJ, Torstenson ES, McLaren PJ, PI DEB, Haas DW, Robbins GK, Gulick RM, Haubrich R, Ribaudo H, Ritchie MD. Use of biological knowledge to inform the analysis of gene-gene interactions involved in modulating virologic failure with efavirenz-containing treatment regimens in art-naive actg clinical trials participants. Pac Symp Biocomput. 2011:253–264. doi: 10.1142/9789814335058_0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J, Moore JH, Kelsey KT, Marsit CJ, Karagas MR, Andrew AS. A novel survival multifactor dimensionality reduction method for detecting gene-gene interactions with application to bladder cancer prognosis. Hum Genet. 2011;129:101–110. doi: 10.1007/s00439-010-0905-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- Harden KP, Hill JE, Turkheimer E, Emery RE. Gene-environment correlation and interaction in peer effects on adolescent alcohol and tobacco use. Behav Genet. 2008;38:339–347. doi: 10.1007/s10519-008-9202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He GF, Zhong SR, Jing Q. [Genetic polymorphism for genes of alcohol dependence] Yi Chuan. 2008;30:413–418. doi: 10.3724/sp.j.1005.2008.00413. [DOI] [PubMed] [Google Scholar]
- Heath AC, Nelson EC. Effects of the interaction between genotype and environment. Research into the genetic epidemiology of alcohol dependence. Alcohol Res Health. 2002;26:193–201. [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PAF, Montgomery GW. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatr. 2011;70:513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Goldman D, Berrettini W, O’Brien CP. Pharmacogenetic approaches to the treatment of alcohol addiction. Nat Rev Neurosci. 2011;12:670–684. doi: 10.1038/nrn3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am J Med Genet B Neuropsychiatr Genet. 2004;128B:102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillemacher T. Biological mechanisms in alcohol dependence--new perspectives. Alcohol Alcohol. 2011;46:224–230. doi: 10.1093/alcalc/agr026. [DOI] [PubMed] [Google Scholar]
- Hines LM, Ray L, Hutchison K, Tabakoff B. Alcoholism: the dissection for endophenotypes. Dialogues Clin Neurosci. 2005;7:153–163. doi: 10.31887/DCNS.2005.7.2/lhines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Enoch MA, Srivastava V, Cummins-Oman JS, Ferrier C, Iarikova P, Sankararaman S, Yamini G, Yuan Q, Zhou Z, Albaugh B, White KV, Shen PH, Goldman D. Genome-wide association identifies candidate genes that influence the human electroencephalogram. Proc Natl Acad Sci USA. 2010;107:8695–8700. doi: 10.1073/pnas.0908134107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Owen MJ, O’Donovan MC, Craddock N. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, McGue M. Behavioral disinhibition and the development of early-onset addiction: common and specific influences. Ann Rev Clin Psychol. 2008;4:325–348. doi: 10.1146/annurev.clinpsy.4.022007.141157. [DOI] [PubMed] [Google Scholar]
- Ittiwut C, Yang BZ, Kranzler HR, Anton RF, Hirunsatit R, Weiss RD, Covault J, Farrer LA, Gelernter J. GABRG1 and GABRA2 Variation Associated with Alcohol Dependence in African Americans. Alcohol Clin Exp Res. 2011;36:588–593. doi: 10.1111/j.1530-0277.2011.01637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Porjesz B, Almasy L, Bierut L, Goate A, Wang JC, Dick DM, Hinrichs A, Kwon J, Rice JP, Rohrbaugh J, Stock H, Wu W, Bauer LO, Chorlian DB, Crowe RR, Edenberg HJ, Foroud T, Hesselbrock V, Kuperman S, Nurnberger J, Jr, O’Connor SJ, Schuckit MA, Stimus AT, Tischfield JA, Reich T, Begleiter H. Linkage and linkage disequilibrium of evoked EEG oscillations with CHRM2 receptor gene polymorphisms: implications for human brain dynamics and cognition. Int J Psychophysiol. 2004;53:75–90. doi: 10.1016/j.ijpsycho.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Joslyn G, Ravindranathan A, Brush G, Schuckit M, White RL. Human variation in alcohol response is influenced by variation in neuronal signaling genes. Alcohol Clin Exp Res. 2010;34:800–812. doi: 10.1111/j.1530-0277.2010.01152.x. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. discussion 101–103, 119–128, 244–152. [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Almasy L, Brooks A, Chorlian DB, Dick D, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Rice J, Schuckit M, Tischfield J, Bierut LJ, Edenberg HJ, Goate A, Foroud T, Porjesz B. Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes Brain Behav. 2012 doi: 10.1111/j.1601-183X.2012.00803.x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin Exp Res. 2011;35:963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 2003;60:929–937. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- Kerner B, Lambert CG, Muthen BO. Genome-wide association study in bipolar patients stratified by co-morbidity. PLoS One. 2011;6:e28477. doi: 10.1371/journal.pone.0028477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Higuchi S. Genetics of alcohol dependence. Psychiatr Clin Neurosci. 2011;65:213–225. doi: 10.1111/j.1440-1819.2011.02190.x. [DOI] [PubMed] [Google Scholar]
- Knopik VS, Heath AC, Madden PA, Bucholz KK, Slutske WS, Nelson EC, Statham D, Whitfield JB, Martin NG. Genetic effects on alcohol dependence risk: reevaluating the importance of psychiatric and other heritable risk factors. Psychol Med. 2004;34:1519–1530. doi: 10.1017/s0033291704002922. [DOI] [PubMed] [Google Scholar]
- Kohli MA, Salyakina D, Pfennig A, Lucae S, Horstmann S, Menke A, Kloiber S, Hennings J, Bradley BB, Ressler KJ, Uhr M, Muller-Myhsok B, Holsboer F, Binder EB. Association of genetic variants in the neurotrophic receptor-encoding gene NTRK2 and a lifetime history of suicide attempts in depressed patients. Arch Gen Psychiatry. 2010;67:348–359. doi: 10.1001/archgenpsychiatry.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF. A role for GABA mechanisms in the motivational effects of alcohol. Biochem Pharmacol. 2004;68:1515–1525. doi: 10.1016/j.bcp.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. Etiologic connections among substance dependence, antisocial behavior, and personality: modeling the externalizing spectrum. J Abnorm Psychol. 2002;111:411–424. [PubMed] [Google Scholar]
- Kumar D, Chakraborty J, Das S. Epistatic effects between variants of kappa-opioid receptor gene and A118G of mu-opioid receptor gene increase susceptibility to addiction in Indian population. Prog Neuropsychopharmacol Biol Psychiatry. 2012;36:225–230. doi: 10.1016/j.pnpbp.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, Patterson DG, Thiselton DL, van den Oord EJ, Walsh D, Kendler KS, Prescott CA. Identification of susceptibility loci for alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence. Alcohol Clin Exp Res. 2006;30:1807–1816. doi: 10.1111/j.1530-0277.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: Interval-based Enrichment Analysis for Genome Wide Association Studies. Bioinformatics. 2012;28:1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Agrawal A, Montgomery GW, Heath AC, Martin NG, Whitfield JB. The role of GABRA2 in alcohol dependence, smoking, and illicit drug use in an Australian population sample. Alcohol Clin Exp Res. 2008a;32:1721–1731. doi: 10.1111/j.1530-0277.2008.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, MacGregor S, Montgomery GW, Heath AC, Martin NG, Whitfield JB. Effects of GABRA2 variation on physiological, psychomotor and subjective responses in the alcohol challenge twin study. Twin Res Hum Genet. 2008b;11:174–182. doi: 10.1375/twin.11.2.174. [DOI] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, de Moor MH, Smit AB, Hottenga JJ, Richter MM, Heath AC, Martin NG, Willemsen G, de Geus EJ, Vogelzangs N, Penninx BW, Whitfield JB, Montgomery GW, Boomsma DI, Madden PA. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res Hum Genet. 2010;13:10–29. doi: 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo A, Auguet T, Vidal F, Broch M, Olona M, Gutierrez C, Lopez-Dupla M, Sirvent JJ, Quer JC, Santos M, Richart C. Polymorphisms of alcohol-metabolizing enzymes and the risk for alcoholism and alcoholic liver disease in Caucasian Spanish women. Drug Alcohol Depend. 2006;84:195–200. doi: 10.1016/j.drugalcdep.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Lucek PR, Ott J. Neural network analysis of complex traits. Genet Epidemiol. 1997;14:1101–1106. doi: 10.1002/(SICI)1098-2272(1997)14:6<1101::AID-GEPI90>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Lydall GJ, Saini J, Ruparelia K, Montagnese S, McQuillin A, Guerrini I, Rao H, Reynolds G, Ball D, Smith I, Thomson AD, Morgan MY, Gurling HM. Genetic association study of GABRA2 single nucleotide polymorphisms and electroencephalography in alcohol dependence. Neurosci Lett. 2011;500:162–166. doi: 10.1016/j.neulet.2011.05.240. [DOI] [PubMed] [Google Scholar]
- Mackillop J, Menges DP, McGeary JE, Lisman SA. Effects of craving and DRD4 VNTR genotype on the relative value of alcohol: an initial human laboratory study. Behav Brain Funct. 2007;3:11. doi: 10.1186/1744-9081-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKillop J, Miranda R, Jr, Monti PM, Ray LA, Murphy JG, Rohsenow DJ, McGeary JE, Swift RM, Tidey JW, Gwaltney CJ. Alcohol demand, delayed reward discounting, and craving in relation to drinking and alcohol use disorders. J Abnorm Psychol. 2010;119:106–114. doi: 10.1037/a0017513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NG, Oakeshott JG, Gibson JB, Starmer GA, Perl J, Wilks AV. A twin study of psychomotor and physiological responses to an acute dose of alcohol. Behav Genet. 1985a;15:305–347. doi: 10.1007/BF01070893. [DOI] [PubMed] [Google Scholar]
- Martin NG, Perl J, Oakeshott JG, Gibson JB, Starmer GA, Wilks AV. A twin study of ethanol metabolism. Behav Genet. 1985b;15:93–109. doi: 10.1007/BF01065891. [DOI] [PubMed] [Google Scholar]
- Matthews AG, Hoffman EK, Zezza N, Stiffler S, Hill SY. The role of the GABRA2 polymorphism in multiplex alcohol dependence families with minimal comorbidity: within-family association and linkage analyses. J Stud Alcohol Drugs. 2007;68:625–633. doi: 10.15288/jsad.2007.68.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, Schultz JA, McClintick JN, Edenberg HJ, Bell RL. Changes in gene expression in regions of the extended amygdala of alcohol-preferring rats after binge-like alcohol drinking. Alcohol. 2010;44:171–183. doi: 10.1016/j.alcohol.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Schultz JA, Kimpel MW, McClintick JN, Wang M, You J, Rodd ZA. Differential effects of ethanol in the nucleus accumbens shell of alcohol-preferring (P), alcohol-non-preferring (NP) and Wistar rats: a proteomics study. Pharmacol Biochem Behav. 2009;92:304–313. doi: 10.1016/j.pbb.2008.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh RK, Hofmann SG, Asnaani A, Sawyer AT, Otto MW. The serotonin transporter gene and risk for alcohol dependence: a meta-analytic review. Drug Alcohol Depend. 2010;108:1–6. doi: 10.1016/j.drugalcdep.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo Pich E, Lorang M, Yeganeh M, Rodriguez de Fonseca F, Raber J, Koob GF, Weiss F. Increase of extracellular corticotropin-releasing factor-like immunoreactivity levels in the amygdala of awake rats during restraint stress and ethanol withdrawal as measured by microdialysis. J Neurosci. 1995;15:5439–5447. doi: 10.1523/JNEUROSCI.15-08-05439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, Menon S, Hanumanthu G, Gupta M, Upendran S, Gupta S, Mahesh M, Jacob B, Mathew P, Chatterjee P, Arun KS, Sharma S, Chandrika KN, Deshpande N, Palvankar K, Raghavnath R, Krishnakanth R, Karathia H, Rekha B, Nayak R, Vishnupriya G, Kumar HG, Nagini M, Kumar GS, Jose R, Deepthi P, Mohan SS, Gandhi TK, Harsha HC, Deshpande KS, Sarker M, Prasad TS, Pandey A. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti PM, Rohsenow DJ, Hutchison KE. Toward bridging the gap between biological, psychobiological and psychosocial models of alcohol craving. Addiction. 2000;95(Suppl 2):S229–236. doi: 10.1080/09652140050111799. [DOI] [PubMed] [Google Scholar]
- Motsinger-Reif AA, Fanelli TJ, Davis AC, Ritchie MD. Power of grammatical evolution neural networks to detect gene-gene interactions in the presence of error. BMC Res Notes. 2008;1:65. doi: 10.1186/1756-0500-1-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Das SK, Vaidyanathan K, Vasudevan DM. Consequences of alcohol consumption on neurotransmitters-an overview. Curr Neurovasc Res. 2008;5:266–272. doi: 10.2174/156720208786413415. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo I, Hamon M. Neurotransmitter and neuromodulatory mechanisms involved in alcohol abuse and alcoholism. Neurochem Int. 1995;26:305–336. doi: 10.1016/0197-0186(94)00139-l. discussion 337–342. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L, Reich T, Schuckit M, Reich W. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158:718–724. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- O’Dushlaine C, Kenny E, Heron EA, Segurado R, Gill M, Morris DW, Corvin A. The SNP ratio test: pathway analysis of genome-wide association datasets. Bioinformatics. 2009;25:2762–2763. doi: 10.1093/bioinformatics/btp448. [DOI] [PubMed] [Google Scholar]
- Obara I, Bell RL, Goulding SP, Reyes CM, Larson LA, Ary AW, Truitt WA, Szumlinski KK. Differential effects of chronic ethanol consumption and withdrawal on homer/glutamate receptor expression in subregions of the accumbens and amygdala of P rats. Alcohol Clin Exp Res. 2009;33:1924–1934. doi: 10.1111/j.1530-0277.2009.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco G, Barrett JC, Zeggini E. Synthetic associations in the context of genome-wide association scan signals. Hum Mol Genet. 2010;19:R137–144. doi: 10.1093/hmg/ddq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajer K, Andrus BM, Gardner W, Lourie A, Strange B, Campo J, Bridge J, Blizinsky K, Dennis K, Vedell P, Churchill GA, Redei EE. Discovery of blood transcriptomic markers for depression in animal models and pilot validation in subjects with early-onset major depression. Translational Psychiatry. 2012;2:e101. doi: 10.1038/tp.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AA, Low MJ, Grandy DK, Phillips TJ. Effects of a Drd2 deletion mutation on ethanol-induced locomotor stimulation and sensitization suggest a role for epistasis. Behav Genet. 2003;33:311–324. doi: 10.1023/a:1023450625826. [DOI] [PubMed] [Google Scholar]
- Palmer RH, Button TM, Rhee SH, Corley RP, Young SE, Stallings MC, Hopfer CJ, Hewitt JK. Genetic etiology of the common liability to drug dependence: evidence of common and specific mechanisms for DSM-IV dependence symptoms. Drug Alcohol Depend. 2012;123(Suppl 1):S24–S32. doi: 10.1016/j.drugalcdep.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RH, Young SE, Hopfer CJ, Corley RP, Stallings MC, Crowley TJ, Hewitt JK. Developmental epidemiology of drug use and abuse in adolescence and young adulthood: evidence of generalized risk. Drug Alcohol Depend. 2009;102:78–87. doi: 10.1016/j.drugalcdep.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RH, Young SE, Stallings MC, Corley RP, Hopfer CJ, Knopik VS, Hewitt JK. Genetics of the associations between adolescent indicators of behavioral disinhibition and young adult measures of alcohol, tobacco and other substance use disorders. Behav Genet. 2011;41:1. [Google Scholar]
- Park A, Sher KJ, Todorov AA, Heath AC. Interaction between the DRD4 VNTR polymorphism and proximal and distal environments in alcohol dependence during emerging and young adulthood. J Abnorm Psychol. 2011;120:585–595. doi: 10.1037/a0022648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei YF, Zhang L, Yang TL, Han Y, Hai R, Ran S, Tian Q, Shen H, Li J, Zhu XZ, Luo X, Deng HW. Genome-wide association study of copy number variants suggests LTBP1 and FGD4 are important for alcohol drinking. PLoS One. 2012;7:e30860. doi: 10.1371/journal.pone.0030860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Gunter TD, Beach SR, Brody GH, Hollenbeck N, Andersen A, Adams W. Role of GABRA2 on risk for alcohol, nicotine, and cannabis dependence in the Iowa Adoption Studies. Psychiatr Genet. 2009;19:91–98. doi: 10.1097/YPG.0b013e3283208026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:543–549. doi: 10.1002/ajmg.b.30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto E, Reggers J, Gorwood P, Boni C, Scantamburlo G, Pitchot W, Ansseau M. The TaqI A DRD2 polymorphism in type II alcohol dependence: a marker of age at onset or of a familial disease? Alcohol. 2009;43:271–275. doi: 10.1016/j.alcohol.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Porjesz B, Begleiter H, Wang K, Almasy L, Chorlian DB, Stimus AT, Kuperman S, O’Connor SJ, Rohrbaugh J, Bauer LO, Edenberg HJ, Goate A, Rice JP, Reich T. Linkage and linkage disequilibrium mapping of ERP and EEG phenotypes. Biol Psychol. 2002;61:229–248. doi: 10.1016/s0301-0511(02)00060-1. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Rewal M, Donahue R, Gill TM, Nie H, Ron D, Janak PH. Alpha4 subunit-containing GABAA receptors in the accumbens shell contribute to the reinforcing effects of alcohol. Addict Biol. 2012;17:309–321. doi: 10.1111/j.1369-1600.2011.00333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003a;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, White BC, Parker JS, Hahn LW, Moore JH. Optimization of neural network architecture using genetic programming improves detection and modeling of gene-gene interactions in studies of human diseases. BMC Bioinformatics. 2003b;4:28. doi: 10.1186/1471-2105-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RJ, Dick DM, Viken Rj, Kaprio J. Gene-environment interaction in patterns of adolescent drinking: regional residency moderates longitudinal influences on alcohol use. Alcohol Clin Exp Res. 2001;25:637–643. [PubMed] [Google Scholar]
- Rosner S, Hackl-Herrwerth A, Leucht S, Vecchi S, Srisurapanont M, Soyka M. Opioid antagonists for alcohol dependence. Cochrane Database Syst Rev. 2010:CD001867. doi: 10.1002/14651858.CD001867.pub3. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Edenberg HJ, Kalmijn J, Flury L, Smith TL, Reich T, Bierut L, Goate A, Foroud T. A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol Clin Exp Res. 2001;25:323–329. [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Kalmijn J. The search for genes contributing to the low level of response to alcohol: patterns of findings across studies. Alcohol Clin Exp Res. 2004;28:1449–1458. doi: 10.1097/01.alc.0000141637.01925.f6. [DOI] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivieres S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proenca C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernandez-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Nunez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tonjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V, Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin MR, Elliott P. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA. 2011;108:7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher KJ, Grekin ER, Williams NA. The development of alcohol use disorders. Annu Rev Clin Psychol. 2005;1:493–523. doi: 10.1146/annurev.clinpsy.1.102803.144107. [DOI] [PubMed] [Google Scholar]
- Sinha R, O’Malley SS. Craving for alcohol: findings from the clinic and the laboratory. Alcohol Alcohol. 1999;34:223–230. doi: 10.1093/alcalc/34.2.223. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration. Results from the 2010 National Survey on Drug Use and Health: Summary of National Findings. Rockville, MD: 2011. [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL, Hubner N, Heinig M, Pravenec M, Mangion J, Legault L, Dongier M, Conigrave KM, Whitfield JB, Saunders J, Grant B, Hoffman PL. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth R, Pocsai Z, Fiatal S, Szeles G, Kardos L, Petrovski B, McKee M, Adany R. ADH1B*2 allele is protective against alcoholism but not chronic liver disease in the Hungarian population. Addiction. 2010;105:891–896. doi: 10.1111/j.1360-0443.2009.02876.x. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Rietschel M. Genome-wide association studies of alcohol dependence and substance use disorders. Curr Psychiatr Rep. 2011;13:147–155. doi: 10.1007/s11920-011-0176-4. [DOI] [PubMed] [Google Scholar]
- Valenzuela CF. Alcohol and neurotransmitter interactions. Alcohol Health Res World. 1997;21:144–148. [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Clay P, Upfal E, Raphael BJ. Discovery of mutated subnetworks associated with clinical data in cancer. Pac Symp Biocomput. 2012:55–66. [PubMed] [Google Scholar]
- Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol. 2011;18:507–522. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- Vengeliene V, Bilbao A, Molander A, Spanagel R. Neuropharmacology of alcohol addiction. Br J Pharmacol. 2008;154:299–315. doi: 10.1038/bjp.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheul R, van den Brink W, Geerlings P. A three-pathway psychobiological model of craving for alcohol. Alcohol Alcohol. 1999;34:197–222. doi: 10.1093/alcalc/34.2.197. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Telang F, Fowler JS, Logan J, Jayne M, Ma Y, Pradhan K, Wong C. Profound decreases in dopamine release in striatum in detoxified alcoholics: possible orbitofrontal involvement. J Neurosci. 2007;27:12700–12706. doi: 10.1523/JNEUROSCI.3371-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, Kwon JM, Wu W, Dick DM, Rice J, Jones K, Nurnberger JI, Jr, Tischfield J, Porjesz B, Edenberg HJ, Hesselbrock V, Crowe R, Schuckit M, Begleiter H, Reich T, Goate AM, Bierut LJ. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum Mol Genet. 2004;13:1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu X, Aragam N, Jian X, Mullersman JE, Liu Y, Pan Y. Family-based association analysis of alcohol dependence in the COGA sample and replication in the Australian twin-family study. J Neural Transm. 2011a;118:1293–1299. doi: 10.1007/s00702-011-0628-3. [DOI] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. J Psychiatr Res. 2011b;45:1419–1425. doi: 10.1016/j.jpsychires.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Wu LY, Zeng M. Genome-wide association study identifies 5q21 and 9p24.1 (KDM4C) loci associated with alcohol withdrawal symptoms. J Neural Transm. 2012;119:425–433. doi: 10.1007/s00702-011-0729-z. [DOI] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk of complex disease. Curr Opin Genet Dev. 2008;18:257–263. doi: 10.1016/j.gde.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol Clin Exp Res. 2008;32:2117–2127. doi: 10.1111/j.1530-0277.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011a;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011b;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SE, Stallings MC, Corley RP, Krauter KS, Hewitt JK. Genetic and environmental influences on behavioral disinhibition. Am J Med Genet. 2000;96:684–695. [PubMed] [Google Scholar]
- Yu W, Clyne M, Khoury MJ, Gwinn M. Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26:145–146. doi: 10.1093/bioinformatics/btp618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr NM, Sullivan EV. Translational Studies of Alcoholism: Bridging the Gap. Alcohol Res Health. 2008;31:215–230. [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci USA. 2011;108:6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Bierut LJ, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Jr, Rice JP, Schuckit MA, Foroud T, Edenberg HJ, Porjesz B, Almasy L. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109:193–198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CS, Wang F, Zhang XY, Deng HW, Krystal JH, Zhang F, Luo X. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology. 2011a;37:557–566. doi: 10.1038/npp.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang CK, Wang F, Li CS, Zhao H, Lu L, Zhang XY, Zhang H, Zhang F, Krystal JH, Luo X. A novel, functional and replicable risk gene region for alcohol dependence identified by genome-wide association study. PLoS One. 2011b;6:e26726. doi: 10.1371/journal.pone.0026726. [DOI] [PMC free article] [PubMed] [Google Scholar]