Abstract
Species of pathogenic microbes are composed of an array of evolutionarily distinct chromosomal genotypes characterized by diversity in gene content and sequence (allelic variation). The occurrence of substantial genetic diversity has hindered progress in developing a comprehensive understanding of the molecular basis of virulence and new therapeutics such as vaccines. To provide new information that bears on these issues, 11 genes encoding extracellular proteins in the human bacterial pathogen group A Streptococcus identified by analysis of four genomes were studied. Eight of the 11 genes encode proteins with a LPXTG(L) motif that covalently links Gram-positive virulence factors to the bacterial cell surface. Sequence analysis of the 11 genes in 37 geographically and phylogenetically diverse group A Streptococcus strains cultured from patients with different infection types found that recent horizontal gene transfer has contributed substantially to chromosomal diversity. Regions of the inferred proteins likely to interact with the host were identified by molecular population genetic analysis, and Western immunoblot analysis with sera from infected patients confirmed that they were antigenic. Real-time reverse transcriptase–PCR (TaqMan) assays found that transcription of six of the 11 genes was substantially up-regulated in the stationary phase. In addition, transcription of many genes was influenced by the covR and mga trans-acting gene regulatory loci. Multilocus investigation of putative virulence genes by the integrated approach described herein provides an important strategy to aid microbial pathogenesis research and rapidly identify new targets for therapeutics research.
Traditionally, the study of pathogen-host interactions has been conducted one gene or protein at a time, in an inefficient linear fashion. Genome sequencing and other high-throughput analytic techniques provide far more rapid and efficient methods to identify genes and proteins that may participate in pathogenesis. As a consequence, it is now feasible to investigate simultaneously multiple putative virulence factors and screen these molecules for characteristics of interest.
In contrast to humans that have a single nucleotide polymorphism per 1,000 bp (1), virtually all pathogenic bacteria have much higher levels of allelic variation. Molecular population genetic analysis has found that some bacterial genes can differ by more than 20% at the nucleotide level, and allelic variation in regions of some genes can exceed 50% because of horizontal gene transfer events (2, 3). Strains of bacterial pathogens also can differ substantially in gene content. For example, genomes of Escherichia coli strains recovered in nature can differ in size by more than 1 Mb (4, 5). Similarly, considerable diversity in gene sequence and content has been described in Helicobacter pylori strains (6, 7). Extensive genetic variation complicates efforts to define the molecular basis of pathogenesis and indicates that a comprehensive understanding of virulence depends on knowledge of chromosomal and allelic diversity present in natural populations. This is especially true for genes encoding proteins that may confer a selective advantage, such as antibiotic resistance markers, cell-surface antigens, and regulatory molecules.
Group A Streptococcus (GAS) causes many types of human infections, including pharyngitis, cellulitis, sepsis, toxic shock syndrome, necrotizing fasciitis, rheumatic fever, and glomerulonephritis (8, 9). The pathogen expresses a large array of extracellular proteins that contribute to disease (9). Abundant chromosomal, allelic, and serologic diversity present in GAS (10) has hindered vaccine development and understanding of pathogen-host interactions and differences in strain behavior. In addition, although it has been known for decades that patients with GAS infections produce antibodies to a large number of extracellular proteins (11), the great majority of the proteins have not yet been characterized.
Here we use a combined molecular population genetic, immunologic, and genetic strategy to analyze in parallel 11 previously uncharacterized extracellular putative virulence proteins identified by comparative study of four GAS genomes. Our data provide extensive information about GAS biology and pathogenesis and have relevance to development of new therapeutics such as vaccines. The integrated approach used in this analysis is generally applicable to pathogenic microbes.
Materials and Methods
Identification of Genes Encoding Putative Virulence Factors.
To identify putative GAS virulence factor genes, we searched databases from four GAS genome sequencing projects (http://www.genome.ou.edu/strep.html, http://www.sanger.ac.uk/Projects/S_pyogenes/, and unpublished data) for ORFs encoding proteins with an LPXTG motif, or substantial homology with a virulence factor made by another bacterial pathogen. Twelve loci selected for study were present in the genome of all four GAS strains (serotype M1, M3, M5, and M18 organisms) and were distributed around the genome (Table 1). Eleven genes encode proteins presumed to be extracellular because they contain an apparent secretion signal sequence (12). Eight of the proteins have a LPXTG(L) motif that covalently links Gram-positive virulence factors to the bacterial cell surface (13). The putative function of the inferred protein encoded by each of the 12 genes was assigned by the WIT2 analysis (available at http://www.genome.ou.edu/strep.html) and blast analysis. For ease of description, each gene will be referred to by the SPy number assigned in GenBank, e.g., spy1986, spy1857, and so forth. One gene was selected for study because it encodes a presumed intracellular protein homologous to the principal Listeria monocytogenes virulence gene regulator PrfA (14). Hence, it differs in cellular location and function with the other 11 proteins and serves as a comparison gene. None of the 12 genes encodes a GAS protein that has been previously characterized.
Table 1.
Putative function and expression analysis of 12 sequenced GAS genes
| SPy no.d | Putative functione | LPXTG(L)f | Expression
analysisa
|
||||
|---|---|---|---|---|---|---|---|
| Versus wild type,
exponential phaseb
|
Versus wild
type, stationary phasec
|
||||||
| Wild-type stationaryg | covR mutant exponentialh | mga mutant exponentiali | covR mutant stationaryj | mga mutant stationaryk | |||
| 2211 | Transmembrane protein | Yes | 1.78 | 1.91 | 2.32 | 0.68 | 0.37 |
| 1986 | Phosphotransferase enzyme II, ABC component | Yes | 70.71 | 1.24 | 2.81 | 0.09 | 0.89 |
| 1972 | Amylopullulanase | Yes | 31.91 | 1.42 | 0.63 | 0.07 | 1.10 |
| 1858 | XAA-PRO dipeptidyl peptidase | No | 1.74 | 1.31 | 1.33 | 0.46 | 0.48 |
| 1857 | Listeriolysin regulatory protein homolog | No | 1.78 | 1.91 | 2.32 | 0.68 | 0.37 |
| 1361 | Internalin homolog | No | 2.07 | 1.03 | 0.67 | 1.26 | 0.59 |
| 1239 | Aminopeptidase N | Yes | 0.99 | 1.59 | 1.11 | 0.98 | 1.69 |
| 0872 | 2′,3′-cyclic-nucleotide 2′-phosphodiesterase | Yes | 4.34 | 1.70 | 2.06 | 0.74 | 0.84 |
| 0843 | Cell surface proteinl | Yes | 2.97 | 2.14 | 2.67 | 0.52 | 1.23 |
| 0747 | Extracellular nuclease | Yes | 16.35 | 3.54 | 2.44 | 0.93 | 1.97 |
| 0501 | Multidrug exporter | No | 1.50 | 1.16 | 3.00 | 0.48 | 1.91 |
| 0277 | Amino acid ABC transporter permease component | Yes | 0.58 | 0.99 | 0.89 | 0.97 | 0.63 |
As assayed by RT–PCR (TaqMan). Values presented are the normalized proportional increase (>1.0) or decrease (<1.0) in transcript abundance.
Expression compared to that in wild-type M1 GAS (MGAS 5005) isolated in the exponential phase of growth.
Expression compared to that in wild-type M1 GAS (MGAS 5005) isolated in the stationary phase of growth.
SPy number refers to the corresponding numerical identifier of each ORF as listed in GenBank. The genomic position of each ORF is as follows: spy2211, 1,845,056-1,847,632; spy1986, 1,654,795-1,656,981; spy1972, 1,639,165-1,642,662; spy1858, 1,541,907-1,544,189; spy1857, 1,541,166-1,541,885; spy1361, 1,127,063-1,129,441; spy1239, 1,020,347-1,022,884; spy0872, 718,489-720,501; spy0843, 694,643-697,669; spy0747, 607,542-610,274; spy0501, 402,159-403,352; spy0277, 241,807-243,375.
As determined by WIT2 or blast analysis.
Amino acid motif used to anchor proteins to the cell wall in gram-positive organisms.
Wild-type M1 GAS (MGAS 5005) isolated in the stationary phase of growth.
CovR-negative isogenic mutant of M1 GAS (MGAS 5005) isolated in the exponential phase of growth.
Mga-negative isogenic mutant of M1 GAS (MGAS 5005) isolated in the exponential phase of growth.
CovR-negative isogenic mutant of M1 GAS (MGAS 5005) isolated in the stationary phase of growth.
Mga-negative isogenic mutant of M1 GAS (MGAS 5005) isolated in the stationary phase of growth.
Bacterial Strains and DNA Sequence Analysis.
Thirty-seven GAS strains isolated from infected humans living on two continents and representing broad GAS genomic diversity, as assessed by multilocus enzyme electrophoresis, pulsed-field gel electrophoresis of chromosomal fragments, and genome sequence data were studied (10, 15, 16) (see Table 3, which is published as supplemental material on the PNAS web site, www.pnas.org). The 37 strains include 12 M protein serotypes (M1, M2, M3, M4, M6, M12, M18, M22, M28, M49, M75, and M89) that commonly cause pharyngitis, rheumatic fever, skin infections, and invasive episodes (8, 15–18). Bacteria were grown on tryptose agar with 5% sheep blood (Becton Dickinson) overnight at 37°C, transferred to 10 ml of Todd Hewitt broth (Difco) supplemented with 0.2% yeast extract (THY medium), and incubated overnight at 37°C in an atmosphere of 5% CO2-20% O2. Chromosomal DNA was isolated with the Puregene DNA Isolation kit (Gentra Systems). DNA sequencing primers were designed on the basis of a serotype M1 genome sequence (http://www.genome.ou.edu/strep.html). Sequence data obtained from both DNA strands with an Applied Biosystems 3700 automated sequencer were analyzed by DNAstar (Madison, WI). All polymorphic sites were sequenced at least three times. Multiple-sequence alignment of the inferred amino acid sequences was performed with clustal w (version 1.8) (19). There was a rarity of insertions and deletions, and hence only subtle changes were needed for alignment of the sequence data.
Population and Molecular Evolutionary Genetic Analysis.
The Tamura-Nei method (20) was used to calculate genetic distances with nucleotide sequence data. The strain phylogeny was constructed with the neighbor-joining algorithm in the computer program mega (21) based on a concatenated sequence consisting of all 12 genes assembled randomly in the following order: spy0277, spy1857, spy0501, spy1239, spy1972, spy0843, spy1361, spy0747, spy0872, spy1986, spy1858, and spy2211. The proportions of polymorphic synonymous (pS) and nonsynonymous (pN) sites were calculated by the method of Nei and Gojobori (22). To examine variation in the pattern of nucleotide substitution, pS and pN were calculated by sliding-window analysis of 30 codons along each gene with the program pswin (23) [pswin is available from S. D. Reid (sreid@niaid.nih.gov)]. Estimates of the sampling variance of these statistics were made by Monte Carlo simulation or bootstrapping. To investigate the role of recombination in the evolution of the 12 genes, the quality of the phylogenetic signal was analyzed by split decomposition with the program splitstree (24, 25). Unlike many other tree-building algorithms, this method does not force the data into a bifurcating tree, and, importantly, enhances the ability to detect conflicting phylogenies suggestive of recombination. In cases of high frequencies of interstrain gene transfer, split decomposition results in a reticulate network of nodes. To identify the putative endpoints of past recombination events, a computer program (maxchi) (26) that implements the maximum χ2 method was used.
Gene Fragment Cloning and Expression of Recombinant Proteins.
Cloning primers were designed on the basis of M1 genome data (http://www.genome.ou.edu/strep.html). PCR products made from strain MGAS5005 (M1) DNA were digested with NdeI and BamHI, cloned into pET-15b (Novagen), and sequenced to rule out the presence of spurious mutations. The following gene fragments were cloned (numbered on the basis of nucleotide positions in each ORF: spy2211, 1327 to 2475; spy1986, 478 to 2109; spy1972, 1 to 1215; spy1858, 1 to 1011; spy1857, 1 to 720; spy1361, 124 to 1299; spy1239, 1 to 987; spy0872, 850 to 2013; spy0843, 877 to 1752; spy0747, 1351–2436; spy0501, 1 to 1110; and spy0277, 1 to 945. To assess protein production, recombinant E. coli BL21(DE3) (Novagen) strains were grown at 37°C for 8 h in 10 ml of LB broth supplemented with 100 mg ampicillin/liter, pelleted by centrifugation, lysed, and analyzed by SDS/PAGE.
Western Immunoblot Analysis.
Proteins separated by SDS/PAGE were transferred to a nitrocellulose membrane (Millipore) and probed with sera obtained from healthy subjects with no history of severe GAS disease and acute- and convalescent-phase sera taken from patients with invasive GAS infections. Goat anti-human affinity-purified IgG (Bio-Rad) was used as the secondary antibody. Signal detection was conducted with SuperSignal West Pico chemiluminescent substrate (Pierce).
TaqMan Real-Time Reverse Transcriptase–PCR Analysis.
Bacteria were grown in 10 ml of THY medium to exponential (OD600 = 0.4) and stationary phase (OD600 = 0.8). TaqMan assays were performed with an ABI 7700 thermocycler (Perkin–Elmer) as described by Chaussee et al. (27). Specific mRNA transcript levels were expressed as fold difference between the conditions compared. Strain MGAS5005 (M1) and its isogenic nonpolar covR mutant (JRS950), and strain JRS301 (M1) and its isogenic nonpolar mga mutant (JRS403) were used for these analyses (B.L., F. R. De Leo, N. P. Hoe, M. R. Graham, S. M. Mackie, R. L. Cole, M. Liu, M. J. Federle, J. R. Scott, and J.M.M., unpublished work and ref. 29).
Results
Sequence Diversity.
The 12 genes were sequenced in 37 strains that together represent broad GAS species diversity. There were 11–15 alleles per locus except for spy1857, which had six alleles (average, 12.3 alleles per locus) (Table 2). Variation ranged between 1.2% and 6.8% at the nucleotide level and 0.4% and 7.2% at the amino acid level. Spy1857 (L. monocytogenes PrfA homolog) was the least variable protein, differing at only 0.4% of 239 aa sites. Spy1986 (glucose-specific II ABC component) and Spy1361 (related to L. monocytogenes internalin) were the most variable, with 5.9% and 7.2% polymorphic amino acid sites, respectively.
Table 2.
Allelic variation present in the 12 GAS genes sequenced in 37 strains
| Gene | No. of alleles | % Polymorphic nucleotides | Protein variants* | % Polymorphic amino acids |
|---|---|---|---|---|
| spy2211 | 12 | 1.7 | 11 | 2.2 |
| spy1986 | 15 | 6.8 | 14 | 5.9 |
| spy1972 | 12 | 2.9 | 12 | 3.9 |
| spy1858 | 12 | 1.8 | 12 | 3.2 |
| spy1857 | 6 | 1.2 | 2 | 0.4 |
| spy1361 | 15 | 3.4 | 15 | 7.2 |
| spy1239 | 12 | 1.5 | 9 | 1.6 |
| spy0872 | 12 | 2.7 | 11 | 3.7 |
| spy0843 | 13 | 1.6 | 13 | 2.9 |
| spy0747 | 15 | 1.6 | 14 | 2.2 |
| spy0501 | 13 | 2.0 | 10 | 2.1 |
| spy0277 | 11 | 1.9 | 10 | 2.3 |
| Average | 12.3 | 2.4 | 11.1 | 3.1 |
Number of distinct proteins arising from amino acid replacements.
Allelic variation among strains of the same M serotype was greatly restricted compared with allelic variation per gene between strains of different M serotypes. Among strains of the same M type, allelic variation was present only in the five M28 strains (three genes), three M12 strains (three genes), three M6 strains (two genes), and two M75 strains (one gene).
Analysis of Genetic Relationships and Recombination.
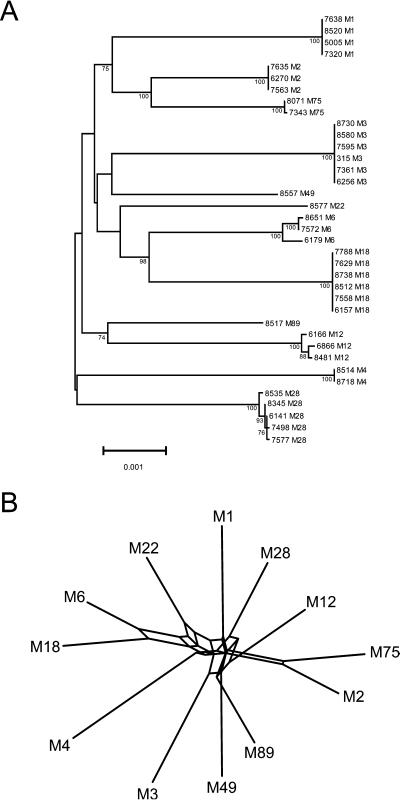
To estimate overall genetic relationships and levels of recombination, a concatenated sequence (26,946 bp) composed of all 12 genes was constructed for each of the 37 isolates. Alignment of the 37 concatenated sequences identified 658 polymorphic nucleotide sites, of which 583 were parsimony-informative; 253 of 296 polymorphic amino acid positions were parsimony-informative. The estimated mean nucleotide diversity for the 37 concatenated sequences was very low (0.0064 ± 0.0002 nt), a result consistent with the occurrence of large regions of conserved sequence among strains of the same M type identified by inspection. The average genetic distance between strains of different M types is small and very similar (Fig. 1 and data not shown). With few exceptions, bootstrap confidence limits for the ancestral nodes were <70%. In addition, the topologies of the individual gene trees were not congruent with the topology of the strain tree made from the concatenated data (data not shown). Taken together, the overall phylogeny of these strains cannot be reconstructed with confidence.
Figure 1.
(A) Genetic relationships among GAS strains based on concatenated gene sequences. The 12 genes were concatenated in the following order: spy0277, spy1857, spy0501, spy1239, spy1972, spy0843, spy1361, spy0747, spy0872, spy1986, spy1858, and spy2211. The tree was constructed by the neighbor-joining algorithm on the basis of the genetic distance determined by the Tamura-Nei method (20), which takes into consideration transitions and transversions, and GC content biases. The topologies of the trees constructed for individual genes are not cognate with the topology of the concatenated tree, suggesting the occurrence of recombination (data not shown). Bootstrap confidence levels that exceed 70% are shown. (B) Split decomposition analysis of the concatenated nucleotide sequences. A representative sequence of each of the 12 M types analyzed was used. Multiple pathways characterize each M type, a result consistent with recombination. The pairwise dissimilarity of the multilocus genotypes was estimated by using hamming (uncorrected) distances.
In light of these findings, split decomposition analysis (25) of the concatenated nucleotide sequences was used to assess the possibility that recombination had contributed to chromosomal diversification. Several alternate evolutionary pathways were identified for each strain representing the 12 M types studied (Fig. 1). Moreover, maximum χ2 analysis (26) suggested that multiple recombination events have contributed to the generation of allelic diversity in each of the 12 genes (data not shown). These results, together with the restricted allelic variation among strains of the same M type, suggest that recombination has occurred recently and insufficient time has elapsed for nucleotide polymorphisms to accumulate by other mechanisms.
Human Immune Response to Recombinant Protein Fragments.
To identify regions of each gene that may be responding to positive selective pressure, the pattern of nucleotide substitution was analyzed by pswin (23). Eleven of the 12 genes had one or more regions in which the rate of nonsynonymous (amino acid-altering) nucleotide substitutions exceeded the rate of synonymous (silent, not resulting in amino acid replacement) substitutions (Fig. 2, which is published as supplemental material), suggesting that selection is operative. (spy1857 lacked sufficient variation to permit meaningful analysis.) These regions were cloned and except for clones derived from spy1986 and spy0501; expression of recombinant protein was confirmed by SDS/PAGE and Coomassie blue staining (data not shown).
Immune selection is one mechanism that can inflate the ratio of nonsynonymous to synonymous nucleotide substitutions. Immune selection requires that the protein is expressed in the course of pathogen-host interactions. To determine whether the proteins were made in infected patients, Western immunoblot analysis was conducted with sera obtained from 11 individuals. Seven sera taken from healthy subjects with no history of invasive GAS disease and no recent GAS infection were analyzed. Because GAS throat and skin infections are common in childhood, these individuals are likely to have been exposed to the pathogen in the past, perhaps multiple times. With the exception of Spy1857 and Spy2211, recombinant proteins obtained from the clones were reactive with one or more of the sera obtained from the healthy subjects (Table 4, which is published as supplemental material). For example, Spy1972, Spy0843, and Spy1361 were reactive with all seven sera tested. Similarly, Spy1858, Spy0747, Spy0872, Spy1239, and Spy0277 were reactive with 86%, 71%, 43%, 29%, and 14% of these seven sera, respectively. The data are consistent with the hypothesis that most of these proteins are made when GAS causes mucosal or superficial skin infections. To determine whether the proteins also were made when GAS causes severe invasive infections, paired acute and convalescent sera taken from four patients were used to screen the recombinant protein fragments. The Spy1857 and Spy2211 fragments also were unreactive with these sera. However, the four convalescent sera were reactive with the protein fragments derived from all other genes (Table 4). Although immunoreactivity with the acute sera was occasionally observed, the reactivity of the convalescent sera was far more intense, a result indicating recent exposure to the protein. The lack of serological reactivity with Spy1857 was expected because the protein does not have a secretion signal sequence and is homologous to an intracellular transcriptional regulator of L. monocytogenes. The spy2211 gene is transcribed (see below); hence, a likely explanation for the lack of reactivity of the test sera with Spy2211 is relatively poor expression of the recombinant protein (data not shown).
Analysis of Gene Transcription.
Transcription of the 12 genes was characterized with real-time reverse transcriptase–PCR (TaqMan) assays. The level of transcription induction or repression that constitutes a physiologically significant change is unknown for most genes, including the 12 studied herein. Many DNA microarray analyses have used a 2-fold change in transcription as a benchmark, although Hughes et al. (30) recently suggested that a 1.5-fold alteration in transcription can be physiologically relevant. For purposes of analysis and discussion, we will use the more conservative 2.0-fold value.
We first determined whether transcription of the 12 genes is growth phase-dependent. RNA isolated from MGAS5005 (serotype M1), a strain that is genetically representative of the M1 subclone commonly causing infections worldwide (16, 31), was used for these experiments. Transcription of spy1972 and spy1986, genes encoding proteins putatively involved in carbohydrate metabolism, was increased 32-fold and 71-fold in the stationary phase relative to exponential phase, respectively (Table 1). Up-regulation of expression of these genes may be a response to diminishing availability of carbohydrate nutrient. Transcription of the spy0747 gene encoding a putative extracellular nuclease was increased 16-fold in the stationary phase. spy0843, spy0872, and spy1361 had less substantial increases in transcription in stationary phase (4.34-, 2.97-, and 2.07-fold, respectively).
We next investigated whether transcription of these 12 genes was influenced by the CovR/CovS (control of virulence) two-component regulatory system that represses production of several critical GAS virulence factors, such as extracellular cysteine protease and capsule (32, 33). CovR represses gene transcription in exponential and stationary phase (33). Relative to wild-type strain MGAS5005, transcription of two genes (spy0843 and spy0747) was increased in exponential phase in an isogenic covR mutant strain (Table 1).
The influence of Mga (multiple gene activator) on transcription of the 12 genes was studied next. Mga is a trans-acting positive transcriptional regulator of genes encoding the virulence factors M protein, C5a peptidase, streptococcal inhibitor of complement, and streptococcal collagen-like protein 1 (29, 34–36). Mga influences gene transcription in the exponential phase, but not in the stationary phase, and its action is linked to Mga-binding sites located upstream of the regulated genes (35). None of the 12 genes studied has a consensus Mga-binding site in presumed regulatory regions (data not shown), and consistent with this observation, none of the genes was down-regulated in the isogenic mutant (Table 1). However, transcription of seven of the 12 genes (spy0747, spy1986, spy1857, spy0843, spy2211, spy0501, and spy0872) was up-regulated modestly (2- to 3-fold) in exponential phase, a paradoxical effect that could be due to the absence of Mga-influenced transcripts in the mutant.
Discussion
Recombination and Genetic Diversity.
We identified evidence that horizontal gene transfer has contributed to allelic diversity at all 12 loci. GAS lacks plasmids and is not known to be naturally transformable. However, the species has many bacteriophages (37), suggesting that transduction is the primary mechanism mediating recombination. The 12 genes studied are located in chromosomal regions that do not contain obvious phage-related genes, suggesting that if recombination is phage-mediated, generalized transduction is operative. Data obtained from comparative sequencing of many proven and putative virulence genes (e.g., streptokinase; M protein; M-like proteins; streptococcal pyrogenic exotoxin A, B, and C; streptococcal superantigen; hyaluronidase; and streptococcal collagen-like protein-1 and -2) (36, 38–43) has suggested that recombination has been a major contributor to allelic variation and chromosomal heterogeneity in GAS. The general lack of congruence between the individual gene trees and the genetic relationships inferred by analysis of the concatenated sequences strongly confirms the importance of recombination in the evolution of GAS. Lateral transfer permits the pathogen to sample many new gene and allele combinations, perhaps increasing the likelihood of generating a more fit organism or enhancing other biomedically relevant traits such as antimicrobial resistance, niche expansion, and virulence.
Characterization of Gene Transcription.
TaqMan assays proved to be a very rapid method to obtain quantitative information about transcription of all 12 genes. Our analysis found that transcription of six genes was up-regulated in stationary phase. In addition, transcription of several genes was influenced by CovR and Mga, two major regulators of GAS virulence factor production. These data provide information about gene regulatory circuits in GAS. Additional regulatory genes have been described in GAS (27, 44–46), and other probable regulators are present in the four genomes analyzed (data not shown). Hence, high-throughput TaqMan assays will permit rapid assessment of the influence of each regulatory gene on target genes such as those encoding virulence factors, extracellular molecules, and other transcriptional regulators.
Implications for Pathogenesis and Development of Therapeutics.
Eight of the 12 genes were selected for analysis because they encode a protein with an LPXTG(L) cell-wall anchor motif. More than 50 extracellular proteins with this motif have been described in Gram-positive bacteria, and many of them are virulence factors (38). For example, in GAS this motif is present in M protein, M-like proteins, C5a peptidase, GRAB protein, serum opacity factor, a fibronectin-binding protein, streptococcal protective antigen, and two collagen-like proteins (13, 43, 47–49). All of these proteins are expressed on the GAS cell surface. With the exception of the streptococcal collagen-like protein-2, which was described very recently but not yet characterized in pathogenesis studies, all of these GAS proteins are virulence determinants in one or more model systems (13, 43, 47–49). Involvement in pathogenesis was not studied, but two lines of evidence suggest that some of the 12 proteins we characterized participate in host-pathogen interactions, broadly defined. First, the Western immunoblot data show that the proteins are made in human infections, including severe invasive episodes. Second, several of the proteins are homologous to virulence factors produced by other Gram-positive pathogens. For example, Spy1361 is homologous to internalins made by L. monocytogenes that are required for entry into host cells (50, 51). Moreover, Spy1858 and Spy1239 are homologous to virulence proteins made by the related pathogen group B Streptococcus that were identified recently by use of signature-tagged mutagenesis and a rat sepsis model (52). Spy0843 has regions of homology with a choline-binding protein made by Streptococcus pneumoniae (53) and Listeria ivanovii internalin B. Spy0843 has a leucine-rich repeat motif that has been implicated in diverse protein–protein interactions (54).
GAS causes human morbidity and mortality worldwide. Estimates of the annual direct health-care costs in the United States for pharyngitis alone exceed $1 billion, and GAS is responsible for 10,000–15,000 cases of serious invasive disease with high morbidity and mortality each year in the U.S. (8, 9). The pathogen also is the most common cause of preventable pediatric heart disease globally due to rheumatic fever and subsequent rheumatic heart disease. There is no vaccine available for prevention of GAS infections. All five GAS cell-surface proteins with an LPXTG anchor motif or closely related amino acid sequence that have been extensively studied confer protective immunity in mouse models, including M protein, C5a peptidase, serum opacity factor, a fibronectin-binding protein, and streptococcal protective antigen (48, 49, 55, 56). Two other secreted proteins found free in culture supernatants also confer protective immunity in mouse or rabbit disease models (57, 58). Although speculative, one or more of the 11 extracellular proteins we studied also may contribute to protective immunity. Three observations lead us to suggest that further analysis of the suitability of these proteins as candidate antigens for conferring protective immunity is warranted. First, none of the proteins was hypervariable (mean percent amino acid polymorphism = 3.1), and few had long regions of greatly increased amino acid variation. Second, all of the genes were transcribed and the Western immunoblot results indicated that the proteins are made in infected hosts. Third, as noted, several of the proteins are homologous to virulence factors made by other bacteria, suggesting that immune-mediated inhibition of these molecules may be therapeutic.
Our studies add to the emerging concept that potential virulence factors and therapeutics candidates can be identified very rapidly by whole-genome investigations. This notion was made very clear recently by Pizza et al. (28), who sequenced a Neisseria meningitidis genome and identified seven new surface proteins that may confer protective immunity. The importance of delineating potential antigenic variability by comparative sequencing of the genes encoding candidate proteins in strains representing known natural population diversity, widespread localities, and the most common serotypes responsible for human infections is also shown by our study. In addition, this study combines TaqMan transcription analysis of isogenic mutants with protein overexpression and Western blot results to rapidly identify and characterize potential new vaccine candidates. Analysis of the role of these proteins in host-pathogen interactions, and their ability to stimulate protective immunity in animal models, may provide much-needed new avenues for control of GAS disease.
Supplementary Material
Acknowledgments
We thank Robert L. Cole, Stacey M. Mackie, Gail L. Sylva, and Cristi Migliaccio for technical assistance; D. Low, H. Hill, and L. G. Veasy for generously providing sera; J. R. Scott for providing strains, and N. P. Hoe for critical reading of the manuscript.
Abbreviation
- GAS
group A Streptococcus
References
- 1.Wang D G, Fan J-B, Siao C-J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 2.Maynard Smith J, Feil E J, Smith N H. BioEssays. 2000;22:1115–1122. doi: 10.1002/1521-1878(200012)22:12<1115::AID-BIES9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 3.Dowson C G, Hutchison A, Woodford N, Johnson A P, George R C, Spratt B G. Proc Natl Acad Sci USA. 1990;87:5858–5862. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergthorsson U, Ochman H. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 5.Ochman H, Jones I B. EMBO J. 2000;19:6637–6643. doi: 10.1093/emboj/19.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alm R A, Ling L-S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, Dejonge B L, et al. Nature (London) 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 7.Salama N, Guillemin K, McDaniel T K, Sherlock G, Tompkins L, Falkow S. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musser J M, Krause R M. In: Emerging Infections. Krause R M, editor. New York: Academic; 1998. pp. 185–218. [Google Scholar]
- 9.Cunningham M W. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reid S D, Hoe N P, Smoot L, Musser J M. J Clin Invest. 2001;107:393–399. doi: 10.1172/JCI11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halbert S P, Keatinge S L. J Exp Med. 1961;113:1013–1028. doi: 10.1084/jem.113.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tjalsma H, Bolhuis A, Jongbloed J D, Bron S, van Dijl J M. Microbiol Mol Biol Rev. 2000;64:515–547. doi: 10.1128/mmbr.64.3.515-547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischetti V A. In: Gram-Positive Pathogens. Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 11–24. [Google Scholar]
- 14.Leimeister-Wachter M, Haffner C, Domann E, Goebel W, Chakraborty T. Proc Natl Acad Sci USA. 1990;87:8336–8340. doi: 10.1073/pnas.87.21.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musser J M, Kapur V, Szeto J, Pan X, Swanson D, Martin D. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson D R, Stevens D L, Kaplan E L. J Infect Dis. 1992;166:374–382. doi: 10.1093/infdis/166.2.374. [DOI] [PubMed] [Google Scholar]
- 18.Beall B, Facklam R, Hoenes T, Schwartz B. J Clin Microbiol. 1997;35:1231–1235. doi: 10.1128/jcm.35.5.1231-1235.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Tamura K, Nei M. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 22.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 23.Reid S D, Selander R K, Whittam T S. J Bacteriol. 1999;181:153–160. doi: 10.1128/jb.181.1.153-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huson D H. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 25.Bandelt H-J, Dress A W M. Mol Phylogenet Evol. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- 26.Maynard Smith J. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 27.Chaussee M S, Watson R O, Smoot J C, Musser J M. Infect Immun. 2001;69:822–831. doi: 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pizza M, Scarlato V, Masignani V, Giuliani M M, Arico B, Comanducci M, Jennings G T, Baldi L, Bartolini E, Cappechi B, et al. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Casal J F, Dillon H F, Husmann L K, Graham B, Scott J R. Infect Immun. 1993;61:5426–5430. doi: 10.1128/iai.61.12.5426-5430.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 31.Hoe N P, Nakashima K, Lukomski S, Grigsby D, Liu M, Kordari P, Dou S J, Pan X, Vuopio-Varkila J, Salmelinna S, et al. Nat Med. 1999;5:924–929. doi: 10.1038/11369. [DOI] [PubMed] [Google Scholar]
- 32.Levin J C, Wessels M R. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 33.Federle M J, McIver K S, Scott J R. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McIver K S, Scott J R. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McIver K S, Thurman A S, Scott J R. J Bacteriol. 1999;181:5373–5383. doi: 10.1128/jb.181.17.5373-5383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukomski S, Nakashima K, Abdi I, Cipriano V, Shelvin B J, Graviss E A, Musser J M. Infect Immun. 2001;69:1729–1738. doi: 10.1128/IAI.69.3.1729-1738.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McShan W M. In: Gram-Positive Pathogens. Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 105–116. [Google Scholar]
- 38.Kehoe M A, Kapur V, Whatmore A, Musser J M. Trends Microbiol. 1996;4:436–443. doi: 10.1016/0966-842x(96)10058-5. [DOI] [PubMed] [Google Scholar]
- 39.Kapur V, Kanjilal S, Hamrick M R, Li L-L, Whittam T S, Sawyer S A, Musser J M. Mol Microbiol. 1995;16:509–519. doi: 10.1111/j.1365-2958.1995.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 40.Marciel A M, Kapur V, Musser J M. Microb Pathog. 1997;22:209–217. doi: 10.1006/mpat.1996.9999. [DOI] [PubMed] [Google Scholar]
- 41.Reda K B, Kapur V, Goela D, Lamphear J G, Musser J M, Rich R R. Infect Immun. 1994;64:1161–1165. doi: 10.1128/iai.64.4.1161-1165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Proft T, Moffatt S L, Weller K D, Paterson A, Martin D, Fraser J D. J Exp Med. 2000;191:1765–1776. doi: 10.1084/jem.191.10.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukomski S, Nakashima K, Abdi I, Cipriano V, Ireland R M, Reid S D, Adams G G, Musser J M. Infect Immun. 2000;68:6542–6553. doi: 10.1128/iai.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podbielski A, Woischnik M, Leonard B A, Schmidt K H. Mol Microbiol. 1999;31:1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- 45.Granok A B, Parsonage D, Ross R P, Caparon M G. J Bacteriol. 2000;182:1529–1540. doi: 10.1128/jb.182.6.1529-1540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kreikemeyer B, Boyle M D P, Buttaro B A, Heinemann M, Podbielski A. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 47.Rasmussen M, Muller H-P, Bjorck L. J Biol Chem. 1999;274:15336–15344. doi: 10.1074/jbc.274.22.15336. [DOI] [PubMed] [Google Scholar]
- 48.Courtney H S, Hasty D L, Li Y, Chiang H C, Thacker J L, Dale J B. Mol Microbiol. 1999;32:89–98. doi: 10.1046/j.1365-2958.1999.01328.x. [DOI] [PubMed] [Google Scholar]
- 49.Dale J B, Chiang E Y, Liu S, Courtney H S, Hasty D L. J Clin Invest. 1999;103:1261–1268. doi: 10.1172/JCI5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaillard J L, Berche P, Frehel C, Gouin E, Cossart P. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- 51.Lingnau A, Domann E, Hudel M, Bock M, Nichterlein T, Wehland J, Chakraborty T. Infect Immun. 1995;63:3896–3903. doi: 10.1128/iai.63.10.3896-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones A L, Knoll K M, Rubens C E. Mol Microbiol. 2000;37:1444–1455. doi: 10.1046/j.1365-2958.2000.02099.x. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez-Beato A R, Lopez R, Garcia J L. FEMS Microbiol Lett. 1998;164:207–214. doi: 10.1111/j.1574-6968.1998.tb13087.x. [DOI] [PubMed] [Google Scholar]
- 54.Marino M, Braun L, Cossart C, Ghosh P. Proc Natl Acad Sci USA. 2000;97:8784–8788. doi: 10.1073/pnas.97.16.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dale J B. Infect Dis Clin North Am. 1999;13:227–243. doi: 10.1016/s0891-5520(05)70052-0. [DOI] [PubMed] [Google Scholar]
- 56.Guzman C A, Talay S R, Molinari G, Medina E, Chhatwal G S. J Infect Dis. 1999;179:901–906. doi: 10.1086/314655. [DOI] [PubMed] [Google Scholar]
- 57.Kapur V, Maffei J T, Greer R S, Li L-L, Adams G J, Musser J M. Microb Pathog. 1994;16:443–450. doi: 10.1006/mpat.1994.1044. [DOI] [PubMed] [Google Scholar]
- 58.McCormick J K, Tripp T J, Olmsted S B, Matsuka Y V, Gahr P J, Ohlendorf D H, Schlievert P. J Immunol. 2000;165:2306–2312. doi: 10.4049/jimmunol.165.4.2306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.