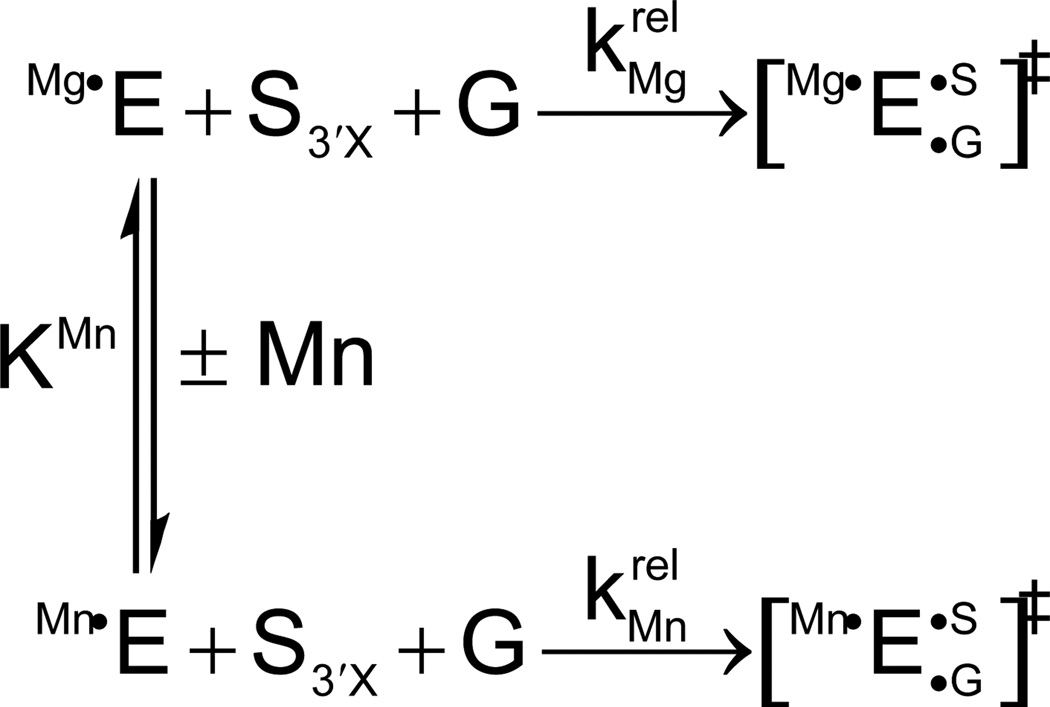Abstract
Site-bound metal ions participate in the catalytic mechanisms of many ribozymes. Understanding these mechanisms therefore requires knowledge of the specific ligands on both substrate and ribozyme that coordinate these catalytic metal ions. A number of different structural and biochemical strategies have been developed and refined for identifying metal ion binding sites within ribozymes, and for assessing the catalytic contributions of the metal ions bound at those sites. We review these approaches and provide examples of their application, focusing in particular on metal ion rescue experiments and their roles in the construction of the transition state models for the Tetrahymena group I and RNase P ribozymes.
Introduction: Metal ions and RNA catalysis
The ability of ribozymes to perform catalysis depends critically on metal ions. Their impact on catalysis is threefold. On the one hand, they screen the negative charges of the phosphodiester backbone, allowing ribozymes to fold into their active three-dimensional structures. Metal ions also coordinate functional groups on both substrates and ribozymes to position reactants and stabilize developing transition states. In addition, hydrated metal ions may donate or accept protons to facilitate general acid-base catalysis. In these ways, metal ions help ribozymes to achieve rate accelerations of as much as 1011-fold (1).
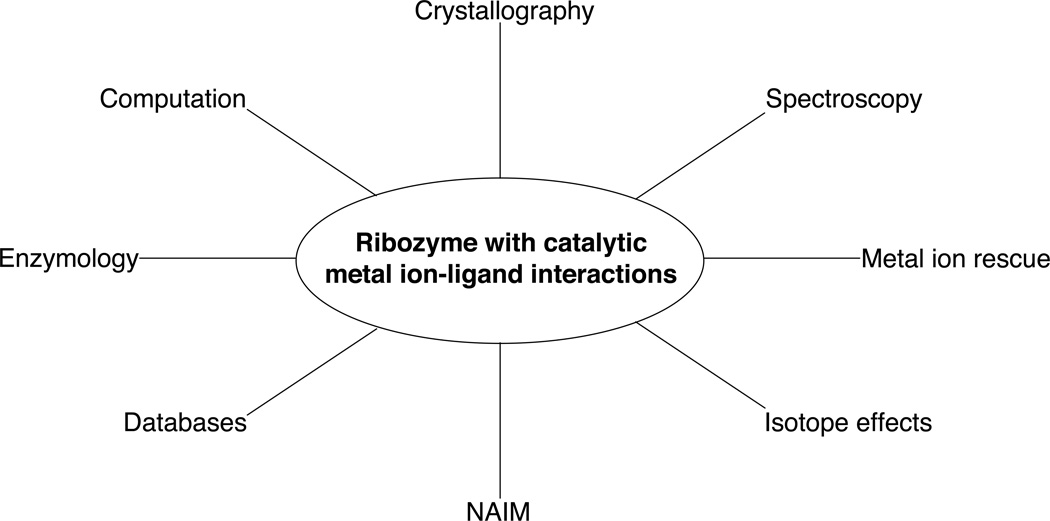
A complete understanding of the mechanisms underlying ribozyme catalysis requires a detailed description of where metal ions bind within the catalytic site. This includes the identification of specific ligands on both substrate and ribozyme that coordinate the catalytic metal ions, enabling the construction of models of the transiently-formed transition state. RNA provides a number of different ligands that can potentially bind metal ions, including both bridging and nonbridging phosphate oxygens, ribose 2’-OH groups, and nucleobase functional groups (purine N7 and O6, for example). The diffuse ion atmosphere that surrounds RNA makes the isolation of any single metal ion-ligand interaction a formidable challenge. Nevertheless, a number of experimental strategies have been developed that make possible the assignment of specific atoms as ligands for catalytic metal ions within ribozymes (Figure 1). This paper will describe several of these techniques, from the detection of metal ion binding sites to experiments that link metal ions to individual atoms in the transition state.
Figure 1.
Summary of techniques used to identify catalytic metal ion-ligand interactions within ribozymes.
Enzymologic considerations
Identifying ligands that bind the catalytic metal ions within a given ribozyme should include a thorough exploration of ribozyme enzymology. This includes determining the dependence of the reaction rate on substrate and enzyme concentrations, monovalent and divalent metal ion concentrations, ionic strength, and pH, among other variables. Equilibrium and kinetic binding parameters of substrates, cofactors, and products should be obtained with the goal of producing a rigorous description of the pathways taken from reactants to products. Although the kinetic model for a ribozyme reaction does not identify catalytic metal ion ligands per se, it does provide the experimental framework within which a meaningful ligand search can occur.
An especially important consideration is the dependence of the reaction rate on metal ion concentration. Initially, ribozymes were thought to be absolutely dependent on divalent metal ions for catalysis (2). This is true for the so-called “large” ribozymes that catalyze the attack of an external nucleophile on phosphorous (the group I and group II introns, RNase P, and the spliceosome). However, the discovery that high concentrations of monovalent ions support catalysis by the endonucleolytic ribozymes (the hammerhead, hairpin, VS, HDV, and glmS ribozymes) (3–6) has prompted a reexamination of how metal ions function in ribozyme catalysis (7). “Catalytic” roles for metal ions now include stabilization of active tertiary structures through nonspecific electrostatic effects, participation in general acid-base catalysis, modification of functional group pKa values, and site binding. A careful analysis of the mono- and divalent metal ion dependences of the reaction can help to distinguish among these possibilities. One strategy has been to monitor the divalent metal ion concentration dependence of the reaction in a background of elevated monovalent metal ion concentration (usually in the molar range). The high concentration of monovalent metal ions excludes divalent metal ions from the diffuse ion atmosphere (8). Site-bound divalent metal ions may then be characterized with respect to a number of different quantitative parameters, including binding affinity, the pH dependence of binding, and the ability of other ions to compete for different sites.
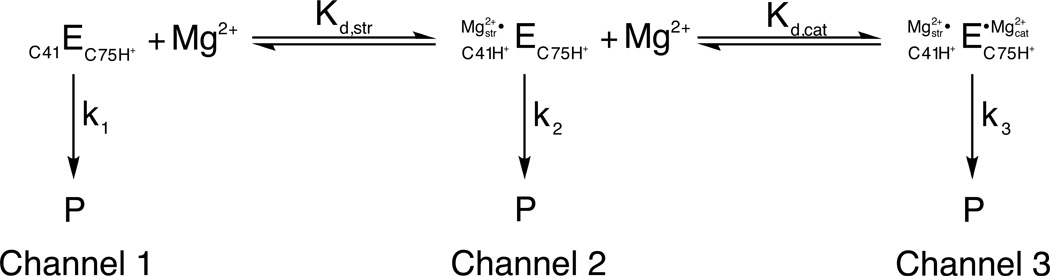
Using this approach, Bevilacqua and colleagues have performed an extensive analysis of the Mg2+- and pH-dependences of the reaction catalyzed by the hepatitis delta virus (HDV) ribozyme. Their experiments have led to a model (Figure 2) in which the ribozyme reacts via one of three separate channels depending on the concentration of Mg2+ ions (9). In the absence of Mg2+, the ribozyme reacts at a basal rate dependent only on the presence of 1 M NaCl (Figure 2, channel 1). Low concentrations of Mg2+ (10−9 to 10−7 M) do not affect this rate significantly. In contrast, intermediate concentrations of Mg2+ (10−7 to 10−4 M) accelerate the reaction rate in a log-linear fashion, consistent with a change in the predominant reaction pathway (Figure 2, channel 2). At higher Mg2+ concentrations (10−4 to 10−1 M), the reaction rate becomes independent of Mg2+ concentration, signifying a third pathway for product formation (Figure 2, channel 3).
Figure 2.
A three-channel scheme describing the Mg2+-dependent and independent pathways for HDV ribozyme catalysis. In 1 M NaCl in the absence of Mg2+, the ribozyme reacts at a basal rate with rate constant k1 (channel 1). Increasing concentrations of Mg2+ accelerate this rate in a log-linear fashion (channel 2) up to a point, after which the rate becomes independent of Mg2+ concentration (channel 3). The Mg2+ ion associated with channel 2 conditions is thought to be primarily structural, binding via inner sphere coordination to a base quadruple involving a protonated C41. Under channel 3 conditions, a second, catalytic, Mg2+ ion binds via outer sphere coordination in the vicinity of the active site. This fully hydrated Mg2+ ion is thought to participate in general acid-base catalysis. [Adapted from (10).]
Under channel 1 conditions, the reaction rate increases log-linearly with pH up to about 6, and decreases thereafter. This pH-rate profile persists at intermediate Mg2+ concentrations (channel 2). However, at high Mg2+ concentrations (channel 3) the reaction rate becomes independent of pH above 6. These distinct pH-rate profiles for channels 2 and 3 prompted the hypothesis that two different classes of Mg2+ ions accelerate the reaction rate of the HDV ribozyme. One Mg2+ ion facilitates catalysis by binding to a structural metal ion site at intermediate Mg2+ concentrations (channel 2). This interaction is thought to occur through inner sphere coordination, since the affinity decreases with increasing ionic radius and is not affected by the presence of the exchange-inert metal complex (10). In addition, the binding of the structural metal ion decreases with increasing pH (9), suggesting that binding may be linked to a protonation event. In fact, subsequent work has provided evidence that this structural metal ion binds tightly to a base quadruple whose stability depends critically on the protonation state of C41 (11).
At high Mg2+ concentrations (channel 3), a second ion contributes to catalysis. As the rate for channel 3 is independent of pH above 6, this ion must offset the inhibitory titration that occurs above pH 6 for channels 1 and 2. Consequently, the ion is thought to play a catalytic, rather than a purely structural, role in HDV ribozyme catalysis. In contrast to the structural ion, the binding affinity of the catalytic ion does not correlate with ionic radius, and inhibits the channel 3 reaction pathway (10). These observations suggest that the catalytic metal ion binds to its site via outer sphere coordination. The ion binds more tightly to its site as the pH increases (9), consistent with its proposed role as a general base in the HDV ribozyme reaction. Subsequent crystallographic (12) and spectroscopic (13) work has provided additional evidence for the existence and function of this catalytic metal ion.
X-ray crystallography
Crystal structures of ribozymes have provided a wealth of information about the local and global architectures underlying the mechanisms of catalysis. Crystals that diffract to sufficient resolution can reveal the positions, coordination geometries, and local environments of metal ions (usually Mg2+ and K+), suggesting possible ligands for further investigation. For lower resolution structures, soaking the crystals in heavier metals such as Tl+, Mn2+, Pb2+, Co3+, Os3+, Eu3+, or Ir3+ can aid in identifying metal ion binding sites and assist in phasing. This can be especially helpful in distinguishing Mg2+ ions from water molecules and other ionic species, which may yield similar patterns of electron density (14). Crystal structures of nearly all of the naturally occurring ribozymes have been solved, including the group I (15–18) and group II (19, 20) introns, RNase P (21–23), the ribosome (24–26), and the hammerhead (27–31), hairpin (32, 33), HDV (12, 34, 35), and glmS (36) ribozymes. In addition, RNA-binding proteins have improved our ability to crystallize RNA (12, 15, 32, 35, 37), whose negatively-charged backbone and relative lack of crystal contacts have been thought to impede the formation of well-ordered crystals (35). Despite their atomic-level detail, crystal structures of ribozymes have limitations with respect to their ability to predict whether apparent contacts between metal ions and ligands are relevant to the catalytic mechanism. By definition, a crystal structure is a model of a ground state conformation of the ribozyme, and not necessarily the most highly populated conformation in solution. This model may show metal ions and ribozyme functional groups in a configuration consistent with a proposed catalytic mechanism. However, the model does not prove that this mechanism actually occurs, or that the atoms involved function in the manner suggested by their arrangement. Moreover, different crystal structures of the same molecule may include different numbers of metal ions within the catalytic site. Structures of the same restriction endonuclease, for example, may show zero, one, two, or even three possible catalytic metal ions (38–40). Furthermore, crystal structures are snapshots of molecules in an instant of time under specific ionic conditions, and may not reflect the local or global dynamics that can contribute to catalysis. Instead, crystal structures are most effective at elucidating mechanism when they are interpreted in conjunction with experiments that test the functionality of the proposed catalytic players. The discrepancies between the biochemical behavior of the minimal hammerhead ribozyme and the models derived from hammerhead crystal structures (41) underscore the need to correlate both structural and functional data when investigating mechanism.
Spectroscopic methods
Several different spectroscopies have been used to locate and characterize metal ion binding sites within ribozymes. These techniques are sensitive to the coordination environment surrounding a given metal ion, and hence are especially effective at defining metal ion ligands and monitoring changes in coordination over time.
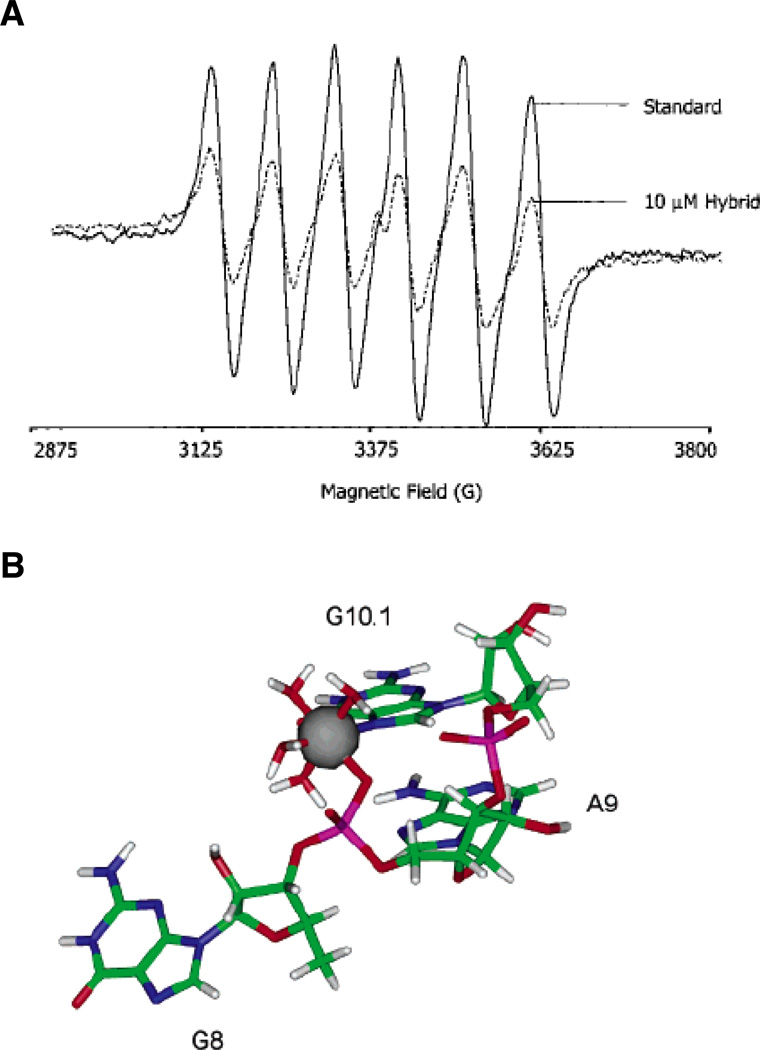
Electron paramagnetic resonance and nuclear magnetic resonance methods have found many applications in the study of interactions between metal ions and RNA. Electron paramagnetic resonance (EPR) experiments monitor signals from the spins of unpaired electrons, which may be present in either organic radicals or paramagnetic metal ions (42). As applied to metal ion binding sites in RNA, EPR relies on the ability of the paramagnetic Mn2+ ion (S = 5/2) to substitute functionally for Mg2+, which is diamagnetic. Mn2+ produces a distinctive six-peak EPR pattern that changes subtly, yet reproducibly, with RNA-induced alterations in the ion’s coordination sphere (Figure 3A). Monitoring these changes as a function of Mn2+ concentration yields binding association constants and cooperativities, as well as the number of different classes of ions that bind the RNA. Ligand identity may be inferred by using higher resolution EPR techniques such as ENDOR (electron nuclear double resonance) and ESEEM (electron spin echo envelope modulation), which report on the interactions between the unpaired Mn2+ electron and surrounding nuclei. These methods have helped to characterize a high-affinity Mn2+ binding site (Kd ≤ 10 µM in 1 M NaCl) within the minimal and full-length hammerhead ribozymes (43–46) (Figure 3B).
Figure 3.
EPR spectroscopy of a Mn2+ binding site within the hammerhead ribozyme. (A) EPR spectra of a 50 µM solution of Mn2+ alone (solid line) and in the presence of 10 µM of an RNA-DNA hybrid hammerhead ribozyme (dotted line). The addition of nucleic acid causes the six-line EPR spectrum of Mn2+ to diminish. (B) Model of the A9/G10.1 Mn2+ binding site in the tertiary-stabilized hammerhead ribozyme, as determined through energy minimization of crystallographic and ESEEM data. The Mn2+ ion coordinates the pro-RP nonbridging oxygen of A9, the N7 of G10.1, and four water molecules. [Taken from (43) and (46), with permission.]
In addition to its use in RNA structure determination (47–52), nuclear magnetic resonance (NMR) is also a sensitive reporter of metal ion-induced changes in secondary and tertiary structure. Metal ion binding alters the chemical shifts of NMR-active nuclei within RNA. Provided that these chemical shifts can be assigned reliably, NMR can monitor conformational changes associated with metal binding, as well as the geometries of the binding sites. Mg2+-induced conformational rearrangements have been demonstrated for a number of RNAs, including the P5abc domain from the group I ribozyme (53) and the transactivation response element (TAR) RNA from HIV-1 (54). Site-specific incorporation of phosphorothioates or spin-labeled nuclei (2H, 13C, 15N) can simplify the interpretation of NMR spectra and provide additional distance constraints within RNAs. For example, NMR was used to follow Cd2+ binding to phosphorothioate substitutions within the aforementioned high-affinity metal ion site in the hammerhead ribozyme (55). Subsequently, a combined NMR and EPR approach in the context of 2H2O and 15N-labeled guanine helped to define the coordination environment of a Mn2+ ion bound at this site (46). The paramagnetic Mn2+ ion has also found applications in pure NMR experiments that exploit the ability of bound Mn2+ ions to broaden the NMR signals from nearby ligands. This strategy helped to identify metal ion binding sites within the hairpin ribozyme (56) and an adenine-sensing riboswitch (57). In addition, direct measurements of metal ion binding are possible with NMR-active nuclei such as 25Mg (58, 59), 59Co (60), 23Na (61–63), and 105Tl (64).
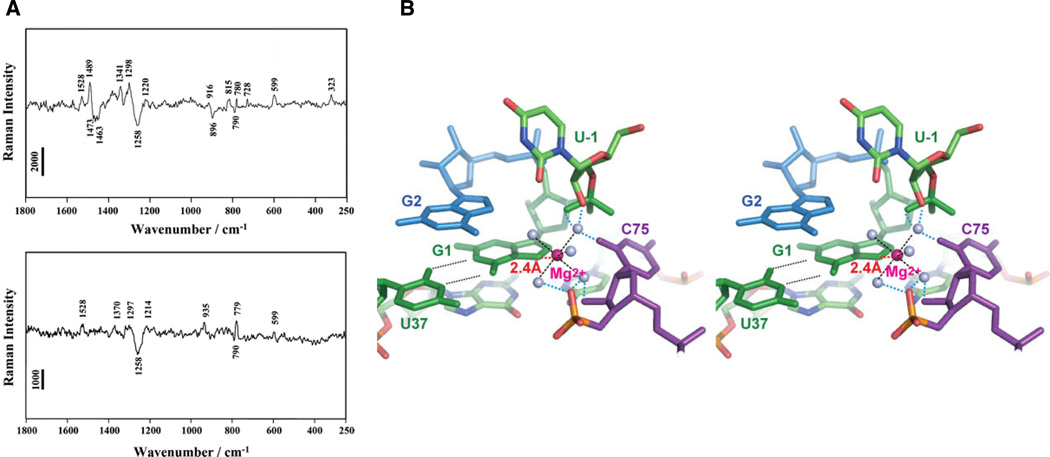
Other spectroscopic methods used to study metal ion-RNA interactions include X-ray absorption (XAS), lanthanide luminescence, and vibrational (IR, Raman) spectroscopies. In XAS, X-rays are tuned to excite selectively the s- or p-shell electrons of metal ions, usually first-row transition metals. Depending on the experiment, XAS can yield the metal ion’s oxidation state (X-ray absorption near-edge structure, or XANES) or the distances between the metal ion and atoms within 4 – 5 Å (extended X-ray absorption fine structure, or EXAFS). Nucleobase- and phosphate-bound metal ions may be distinguished readily, due to the higher electron density of phosphorous. Using EXAFS, DeRose and colleagues have obtained evidence suggesting that a Mn2+ ion binds oxygen, rather than sulfur, in a phosphorothioate-substituted hammerhead ribozyme (65). Lanthanide ions, most notably Eu3+ and Tb3+, undergo distinct f-orbital electronic transitions that are sensitive to the coordination environment of the metal ion. In particular, the luminescent lifetimes of these ions vary depending on whether the ions coordinate water molecules (which provide a pathway for luminescence quenching) or RNA ligands. Lanthanide luminescence has been used to detect and characterize metal ion binding in tRNA (66, 67), RNA hairpin loops (68), the U2–U6 spliceosomal RNAs (69), and the hammerhead (70), hairpin (71), and HDV (72) ribozymes. Vibrational spectroscopies can also detect metal ion-induced changes in RNA structure, as well as the coordination environment of specific bound metal ions. Recently, pH-dependent Raman microscopy of HDV ribozyme crystals has provided evidence that a hydrated Mg2+ ion binds a G·U wobble pair near the cleavage site (13). Previous work had established that a peak around 323 cm−1 in the HDV Raman difference spectrum arose from hydrated Mg2+ ions that coordinated at least one non-water ligand (73). By monitoring this Raman signal, the authors have shown that protonation of the catalytic cytosine C75 resulted in the loss of a single inner sphere Mg2+ hydrate per HDV molecule. In contrast, substitution of the cleavage site G1 with 7-deazaguanosine (7DG) produced a Raman difference spectrum that lacked both an N7 peak and the inner sphere Mg2+ hydrate marker at 323 cm−1 (Figure 4A). These observations suggested a model in which a partially hydrated Mg2+ ion that functions as the general base in the HDV ribozyme reaction coordinates the N7 of the cleavage site guanosine (Figure 4B).
Figure 4.
Raman spectroscopic investigation of HDV ribozyme crystals. (A) Raman difference spectra (pH 7.5 – pH 5.0) of crystals of the unmodified HDV ribozyme (top panel) and an HDV ribozyme in which 7-deazaguanosine has replaced the cleavage site G1 (bottom panel). The difference spectrum of the unmodified ribozyme shows peaks at 1489 and 323 cm−1, thought to arise from metal ion coordination of a guanosine N7 and from a single inner sphere Mg2+ hydrate, respectively. Both features are lost when 7-deazaguanosine replaces guanosine at the cleavage site. (B) Stereo model of the HDV ribozyme active site, constructed by combining the Raman spectroscopic results with pre- and post-cleavage crystallographic data. The Mg2+ ion (magenta) directly coordinates the N7 of G1, along with five water molecules (grey spheres). [Taken from (13), with permission.]
Computational and database methods
In the context of a three-dimensional structure, certain in silico modalities can predict regions within an RNA where metal ions are likely to localize. Such methods may be helpful if the structure is not of sufficient resolution to visualize metal ions, or if the patterns of electron density preclude unambiguous assignment. One approach uses the three-dimensional structure as the basis for calculating solutions to the nonlinear Poisson-Boltzmann equation (NLPB) to determine the molecule’s electrostatic potential surface. Locations of highly negative electrostatic potential that may harbor metal ion binding sites are then easily observed (74) (Figure 5). Brownian dynamics algorithms have also been applied that simulate bulk metal ion binding along an RNA’s electrostatic potential surface (75). Another program called FEATURE, developed originally to study microenvironments within proteins (76), has been adapted to study metal ion binding sites within RNAs. FEATURE starts with a training set of known metal ion microenvironments within RNA structures and applies these constraints to other RNA structures to predict likely metal ion binding sites (77). The program successfully identified site-bound metal ions observed in crystals of the P4–P6 domain derived from the Tetrahymena group I intron, and also predicted additional sites that had not been reported previously.
Figure 5.
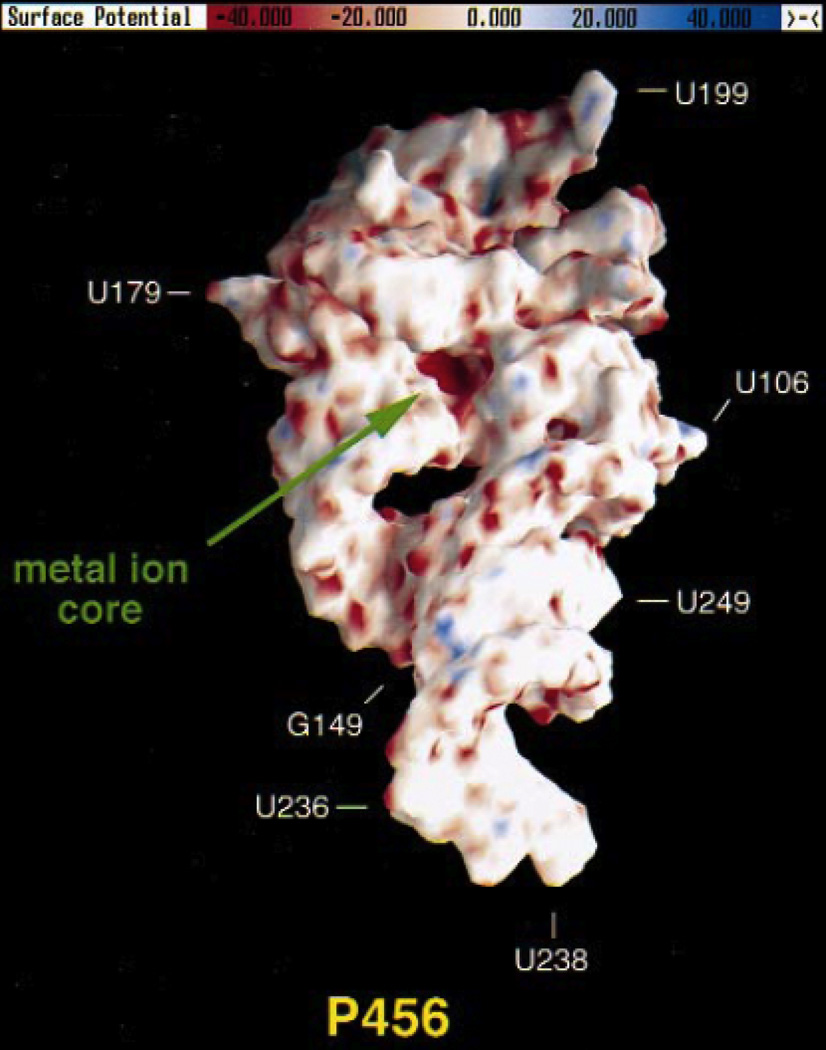
Electrostatic potential surface of the P4–P6 domain from the Tetrahymena group I ribozyme, calculated using the NLPB equation. Regions of negative electrostatic potential appear red, while electropositive regions appear blue. The A-rich bulge that coordinates two Mg2+ ions is located in a pocket of highly negative electrostatic potential (“metal ion core”) with values ranging from −80 to −100 kT/e. [Adapted from (74), with permission.]
Web-based databases are also available for studying metal ion binding sites within RNA structures. These include the Metalloprotein Database and Browser (MDB; http://metallo.scripps.edu) (78), the Metals in RNA database (MeRNA; http://merna.lbl.gov) (79), and the Metal Ions in Nucleic Acids database (MINAS; http://www.minas.uzh.ch). The MDB catalogs all examples within the Protein Data Bank (PDB) of metal ion-ligand interactions occurring within protein and nucleic acid structures. The database is searchable by author, PDB identification number, metal ion, number of ligands, and metal ion-ligand distance, among other parameters. A Java-based applet displays the metal ions and surrounding ligands within a manipulable three-dimensional environment. MeRNA lists all metal ion binding sites in RNA structures deposited in the PDB and the Nucleic Acid Database (NDB). As of 2007, the list comprises over 16,000 metal ions, representing 23 different metal ions within 389 RNA structures. In addition to the search parameters mentioned for the MDB, MeRNA is also searchable by eight different RNA metal ion-binding motifs. The database may be used in conjunction with the Structural Classification of RNA database (SCOR; http://scor.berkeley.edu) (80) to study metal ion binding sites in the context of the overall RNA structure and its tertiary interactions. MINAS was created as a tool to investigate the origin of the selectivity and specificity of nucleic acid metal ion binding sites for metal ions. To that end, the database contains the geometric parameters of every inner and outer sphere-bound metal ion present in over 1,200 nucleic acid structures derived from the PDB and the NDB. Besides inner and outer sphere coordination, users can also search the database for specific coordination environments or combinations of ligands. As stated on their website, the authors hope to classify metal ion binding sites through statistics, and to draw conclusions about the coordination properties of the metal ions involved.
Lanthanide cleavage
Lanthanide ions, notably Tb3+ and Eu3+, have been used to map RNA metal ion binding sites through regio-specific hydrolysis of the RNA backbone. These ions compete with Mg2+ for metal ion sites on RNA, binding many times more tightly than Mg2+ (66, 67). In addition, lanthanide aqua ion pKa values are close to physiologic pH, allowing micromolar concentrations to induce backbone cleavage in the vicinity of the metal ion site. Following cleavage, the locations of the sites are mapped by fractionating the products on denaturing gels. Lanthanide cleavage maps do not identify catalytic metal ion ligands per se, but as with other methods, the maps can narrow the search to specific regions of the ribozyme. Metal ion sites within the group II intron (81), RNase P (82), HDV (72, 83), and hairpin (71) ribozymes have been mapped via lanthanide cleavage. The technique can also be used to monitor RNA folding and the formation of tertiary structure, with higher concentrations of lanthanide ions (micromolar to millimolar) promoting nonspecific hydrolysis in solvent-accessible areas (83–85).
Heavy atom isotope effects
Isotopic substitution is the smallest perturbation that can be made within a catalytic system. Even so, this subtle change can affect the reaction rate and equilibrium significantly, giving rise to kinetic and equilibrium isotope effects, respectively. These effects provide information about the reaction mechanism and the structure of the transition state. Isotope effects are expressed as the ratio of the reaction rates for the lighter isotope compared to the heavier isotope (kL/kH). The effect is said to be normal if this ratio exceeds unity (favoring the lighter isotope), and inverse if it is less than unity (favoring the heavier isotope). In addition, primary isotope effects arise from substitution of atoms directly involved in making or breaking bonds, such as nucleophiles and leaving groups. Secondary isotope effects, in contrast, result from substitution of atoms that may contribute to transition state structure but do not participate directly in bond formation or cleavage. The magnitude of the effect is proportional to the relative mass difference between the two isotopes. Thus, the effect for hydrogen versus deuterium (a 100% mass difference) is significantly larger than the effect for 16O versus 18O (a 12.5% difference) or for 12C versus 13C (an 8.3% difference). As a result, detection of isotope effects for atoms other than hydrogen requires extremely precise measurements (≥ 1%) (86).
Two factors contribute to the isotope effect observed experimentally for a given reaction (86–88). A temperature-independent factor (TIF) arises from the difference between the zero point vibrational energies of the chemical bonds involving the two isotopes. Due to the increased mass, the bond to the heavier isotope vibrates at a lower energy level than the bond to the lighter isotope. As a result, formation of the transition state with the heavier isotope requires more activation energy, and hence the TIF is always normal (favoring the lighter isotope). The second contribution to the overall isotope effect reflects the change in bonding experienced by the atom of interest as it proceeds from the ground state to the transition state. This effect is influenced partly by temperature, and therefore is called the temperature-dependent factor (TDF). The TDF can be either normal or inverse, depending on the degree of bonding to the atom in question. In general, a stronger, or “stiffer”, bonding environment arising from bond formation favors the heavier isotope, while a decrease in stiffness (bond cleavage) favors the lighter isotope. The overall isotope effect therefore provides information about the extent of bonding to the reacting atoms in the transition state.
Isotope effects have been used in several instances to study reactions involving solution- or protein-mediated cleavage of model phosphate esters (89–92), as well as proton transfer in some ribozyme reactions (93, 94). The influence of metal ions on the hydrolysis of model phosphate esters has been explored (95–100), but relatively few studies have investigated isotope effects associated with metal ion binding within ribozymes. One notable example from the work of Harris and colleagues (101) extends to RNase P catalysis the results derived from water- and Mg2+-catalyzed hydrolysis of a model phosphate diester. Metal ion binding to nucleophiles generally creates a “stiffer”, more strongly bonded environment that favors heavier isotopes in a reaction (102). Hence, a metal ion-nucleophile interaction should make inverse contributions to the overall isotope effect, a prediction that was observed for Mg2+-catalyzed cleavage of 5’-p-nitrophenylphosphate (T5PNP) (101). The isotope effect for T5PNP hydrolysis in H2 18O dropped from 1.068 to 1.027 when Mg2+ was added, suggesting Mg2+ coordination to the 18O nucleophile. In the same study, the authors measured the 18O isotope effect for RNase P catalysis, which requires Mg2+ ions and in which water also acts as the nucleophile. The magnitude of the effect (1.030) was very close to that observed for Mg2+-catalyzed cleavage of T5PNP, suggesting that a metal ion coordinates the nucleophilic water in the RNase P reaction.
Aside from the requisite high precision, isotope effect experiments on ribozymes pose an additional challenge with respect to the incorporation of isotopes. The transition states of ribozyme-catalyzed phosphoryl transfer reactions can include 2’-, 3’-, and nonbridging oxygens, yet synthesis of isotopically-enriched nucleotides has been relatively limited and nucleobase-dependent. However, recent studies have reported improved methods for 18O incorporation into nucleotides and phosphoramidites at the 2’ and 3’ positions (103, 104). These molecules should facilitate the investigation of isotope effects and of ribozyme transition state structures.
Nucleotide analog interference mapping (NAIM)
Nucleotide analog interference mapping (NAIM) screens the functionality of specific atoms or groups of atoms at every position of an RNA molecule simultaneously (105). In a NAIM experiment, an RNA is transcribed in the presence of wild-type nucleotides and a nucleoside α-thiotriphosphate. The latter nucleotide contains a modification, either the α-thiophosphate by itself or combined with an atomic substitution elsewhere on the nucleotide. The concentration of nucleoside α-thiotriphosphate is adjusted such that each transcribed RNA contains one modified nucleotide on average. Following transcription and purification, the pool of RNAs is subjected to some biochemical challenge (selection), such as the ability to fold or perform catalysis. Those RNAs that perform the challenge successfully are isolated, treated with iodine to cleave the phosphorothioate linkages, and fractionated on denaturing gels alongside iodine-treated unselected RNA. Positions where the modification has interfered with the ability of the RNA to perform the challenge appear as gaps in the sequencing ladder.
Since the majority of metal ion-ligand interactions within RNAs involve nonbridging phosphate oxygens (106, 107), NAIM experiments using nucleoside α-thiotriphosphates have been especially effective at locating potential catalytic ligands. Unfortunately, these experiments survey only the RP nonbridging oxygens, since RNA polymerases incorporate only SP phosphorothioate diastereomers into the growing transcript. Despite this limitation, however, interference maps can direct the search for catalytic ligands to specific nucleotides. An early investigation of the Tetrahymena ribozyme implicated several positions where phosphorothioate substitution interfered with catalysis in the presence of Mg2+ alone (108). These positions formed part of the basis for a subsequent search for nonbridging oxygens that coordinated the catalytic metal ions. As later studies showed, the SP nonbridging oxygens of C208, C262, and A306 bound catalytic ions (109–111), even though each of these positions had been identified initially through RP phosphorothioate interference.
Metal ion rescue experiments
Metal ion rescue experiments provide a powerful strategy for identifying functional metal ion-ligand interactions within macromolecules. These assays exploit the differential affinities of two or more metal ions for two different ligands. For a given metal ion-ligand interaction critical for some function, substitution of the original ligand with one that interacts weakly with the metal ion would be expected to diminish that function. The subsequent restoration of activity upon addition of a metal ion that binds the substituted atom more strongly suggests that the metal ion-ligand interaction has been reestablished, or “rescued”. Thus, the approach isolates individual metal ion-ligand interactions, circumventing the complications of the “sea of metal ions” that surrounds macromolecules. For protein enzymes, metal ion rescue experiments have provided evidence for catalytic involvement of aspartate and glutamate residues within some DNA transposases and integrases (112–116). However, generalized substitution of individual atoms within proteins remains a challenging undertaking (117). Ribozymes, on the other hand, have been especially attractive targets for this technique since both substrates and ribozymes are more amenable to synthetic manipulation. Quantitative metal ion rescue experiments have characterized catalytic metal ion-ligand interactions within the group I (109–111, 118–124) and group II (125–128) introns, the hammerhead ribozyme (129–131), bacterial RNase P (132–139), and the spliceosome (140–142). Crystal structures of hammerhead (31) and group I (15–18) ribozymes have confirmed the validity of the approach, with the majority of the apparent metal ion-ligand interactions agreeing with the biochemical rescue data.
Mg2+ ions often facilitate ribozyme catalysis by coordinating oxygen ligands such as 2’-OH groups and bridging and nonbridging phosphate oxygens. In place of oxygen, a metal ion rescue experiment substitutes sulfur or nitrogen (usually as NH2), two ligands that do not interact particularly well with Mg2+. The rescuing metal ions are transition metals such as Mn2+, Zn2+, Cd2+, or Co2+. The higher polarizability and d-orbitals of these metal ions allow them to interact much more strongly with sulfur or nitrogen than Mg2+, which prefers to bind oxygen ligands almost exclusively (143).
To report accurately on the role of a metal ion in a ribozyme reaction, a metal ion rescue experiment must satisfy certain conditions. The assay must monitor the same transition from ground state to transition state, regardless of the presence of an atomic substitution or rescuing metal ion. The altered substrate-ribozyme combination may change the initial ground state of the reaction, or accelerate a non-chemical step unexpectedly. Identical reaction conditions may yield a log-linear pH dependence for the wild-type reaction, and a pH-independent profile for the modified reaction. Therefore, care must be taken to identify experimental conditions under which the chemical step of the reaction is rate-limiting for all modifications, substrates, and metal ions employed. This requires a rigorous thermodynamic and kinetic analysis of the reaction, including knowledge of substrate and cofactor binding parameters, relevant conformational changes, and how different modifications and conditions affect each individual step of the pathway.
In addition, any non-specific effects of the rescuing metal ion must be considered. Even under conditions in which chemistry is rate-limiting, a rescuing metal ion may confer an inherent catalytic advantage to the reaction. Alternatively, some rescuing metal ions (notably Cd2+) may inhibit a reaction at sufficiently high concentrations (124). To control for these effects, separate experiments must monitor both the wild-type and modified reactions in the presence of Mg2+ alone and with the rescuing metal ion. The rates of these reactions are then used to calculate a relative rate, krel, that controls for the non-specific effects of the rescuing metal ion. For a ribozyme system in which sulfur or nitrogen (S/N) substitutes for an oxygen ligand (O), krel is given by the expression
| (1) |
where M2+ is the rescuing metal ion and each k is the rate of the reaction in the presence of either Mg2+ alone or Mg2+ plus M2+. The magnitude of the rescue is therefore proportional to the M2+ rate acceleration of the modified reaction divided by the M2+ acceleration inherent to the wild-type reaction.
An important point regarding the interpretation of these experiments is that a lack of rescue does not imply the absence of metal ion coordination by the original ligand. The physicochemical properties of the substitution or the rescuing metal ion may preclude restoration of a disrupted metal ion-ligand interaction. Sulfur, for example, is bulkier than oxygen, and a phosphorothioate P-S bond is significantly longer than the corresponding P-O bond (1.9 Å vs. 1.5 Å, respectively). Phosphorothioate and all-oxygen linkages also differ with respect to their charge distribution (144), electronegativity, and ability to accept hydrogen bonds (145). Thus, depending on the structural and chemical context, phosphorothioate substitution may introduce a perturbation that softer metal ions cannot overcome, even though the original ligand coordinated a metal ion. In some cases, the sulfur atom may actually displace catalytic metal ions from the active site (146). In addition, the rescuing metal ions differ from Mg2+ with respect to size and coordination geometry. While Mg2+ and Mn2+ have similar ionic radii (0.72 Å vs. 0.67 Å, respectively), the more thiophilic Cd2+ ion is significantly larger (0.95 Å). Zn2+ is more thiophilic than Mn2+ and at 0.74 Å is closer in ionic radius to Mg2+, but may adopt a tetrahedral rather than an octahedral coordination geometry depending on the structural context. Each of these caveats regarding the substitution and the rescuing metal ion must be considered when a metal ion rescue experiment yields a negative result.
Metal ion rescue in practice: The Tetrahymena GROUP I intron catalytic site
The protozoan Tetrahymena thermophila contains a self-splicing group I intron within the gene encoding the 26S rRNA (147). A shortened form of this intron (Figure 6) catalyzes a phosphotransesterification reaction between an exogenous guanosine molecule and an oligonucleotide substrate that forms base pairs with the ribozyme (148):
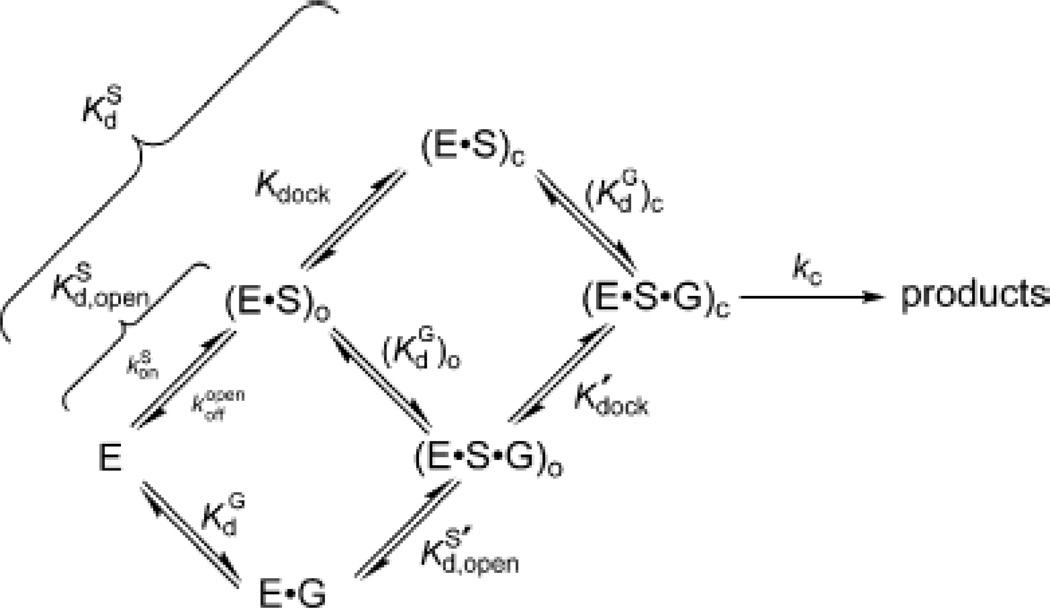
In vitro, the ribozyme is active at millimolar concentrations of Mg2+ or Mn2+, whereas neither Ca2+ nor monovalent ions alone support catalysis (149). Kinetic analyses of the reaction (1, 150–152) support the scheme shown in Figure 7, in which substrate binding occurs via the formation of two distinct intermediates. In the open complex, the substrate base pairs with the ribozyme to form the P1 helix. This species then undergoes a conformational change (“docking”) to form the closed complex, in which the P1 helix forms tertiary contacts with the ribozyme core. Metal ion rescue experiments based upon this kinetic framework have led to the development of a detailed model of the interactions between divalent metal ions and atoms of the substrate and ribozyme in the transition state (Figure 6C). The following sections describe some of the methods and strategies used to obtain the data that support this model.
Figure 6.
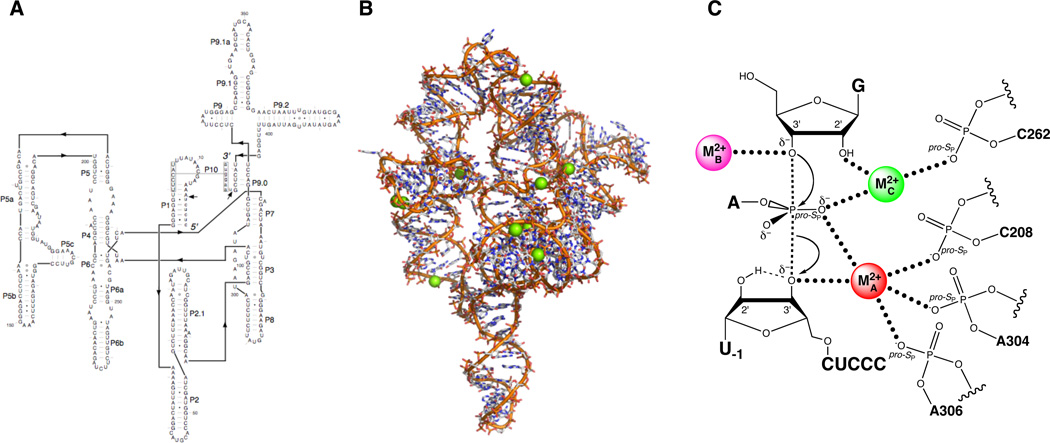
The Tetrahymena group I ribozyme. (A) Secondary structure. (B) Three-dimensional structure at 3.8 Å resolution (PDB file 1X8W). Mg2+ ions are represented as green spheres. (C) Transition state catalytic metal ion-ligand interactions within the Tetrahymena ribozyme active site, as deduced from metal ion rescue experiments.
Figure 7.
Kinetic model of the Tetrahymena ribozyme reaction. The free enzyme (E) can bind the oligonucleotide substrate (S) to form the open complex, in the presence [(E·S·G)O] or absence [(E·S)O] of guanosine. The closed complex [(E·S·G)C or (E·S)C] forms when the P1 helix carrying the substrate “docks” into the active site of the ribozyme via the formation of tertiary interactions. [Taken from (111).]
Metal A coordinates the 3’ leaving group of the substrate
Piccirilli et al. identified the first metal ion-ligand interaction within the Tetrahymena ribozyme that participates directly in the catalytic mechanism. Using a metal ion specificity switch experiment, they showed that a divalent metal ion (MA) coordinates the 3’ oxygen leaving group of the oligonucleotide substrate (118) (Figure 8A). This discovery depended in part upon two key experimental results. One was the ability to synthesize deoxyribonucleotides containing a 3’ phosphorothiolate linkage. This was accomplished through the efforts of Vyle and Cosstick, who prepared the 3’-S-phosphorothioamidite of thymidine and used it to produce a dithymidine 3’-S dinucleotide (153). Subsequent work described the incorporation of the phosphorothioamidite into longer deoxyribonucleotides via automated solid-phase synthesis (154). The other relevant result was the observation that the Tetrahymena ribozyme could cleave an all-DNA oligonucleotide substrate, albeit with weaker substrate binding and decreased rate constants compared to the all-RNA substrate (155). Importantly, while substrate binding and product release limited the reaction of the all-RNA substrate, the all-DNA substrate reacted such that the chemical step was rate-limiting at all concentrations of guanosine and substrate.
Figure 8.
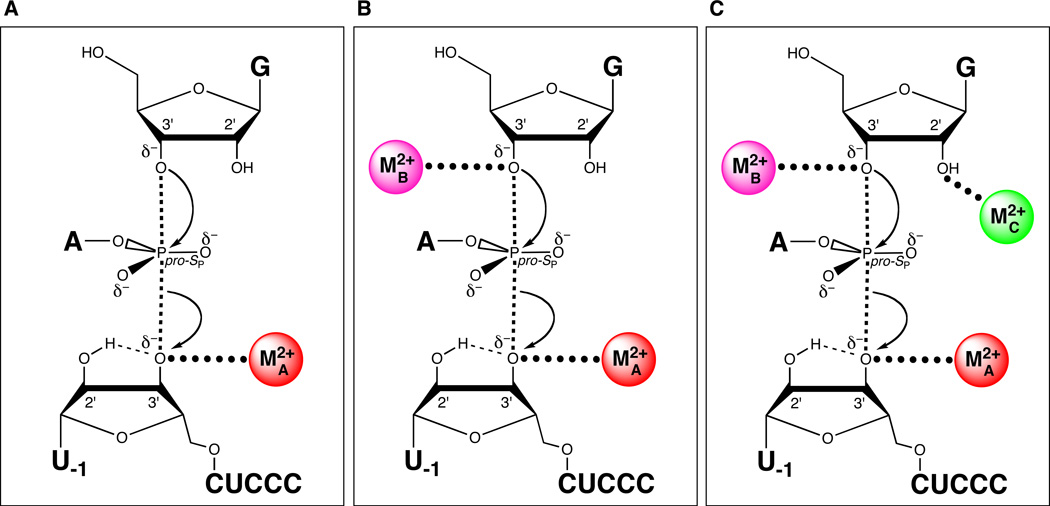
Sequential discovery of three different catalytic metal ion-ligand interactions in the transition state of the Tetrahymena group I ribozyme reaction. (A) MA coordinates the 3’ oxygen of the leaving group. (B) MB coordinates the 3’-OH of the guanosine nucleophile. (C) MC coordinates the 2’-OH of the guanosine nucleophile.
To conduct the experiment, Piccirilli et al. first synthesized the DNA substrate d(CCCUCU3’XA), where X is either oxygen or sulfur. In the presence of 12 mM Mg2+, the reactivity of the sulfur substrate in the Tetrahymena ribozyme reaction was about 1000-fold lower than the reactivity of the oxygen substrate. Because both substrates reacted from the same ground state under conditions in which chemistry was rate-limiting, this differential reactivity arose from a defect in the chemical step, rather than from a conformational change. Also, the sulfur substitution did not affect substrate binding to the ribozyme, as was shown by direct Kd measurements using native gels. Increasing the Mg2+ concentration accelerated the reaction of both substrates, but did not affect the 1000-fold relative catalytic defect. In contrast, in the presence of 10 mM Mg2+ and 2 mM Mn2+, the ribozyme cleaved the sulfur substrate only 8-fold more slowly than the oxygen substrate. The presence of Mn2+ did increase the reaction rate for the oxygen substrate by about 3-fold, likely due to enhancement of substrate docking into the closed complex (156). However, the 400-fold increase in the cleavage rate of the sulfur substrate in the presence of Mn2+ suggested strongly that a process other than nonspecific Mn2+ stimulation was at work. The additional observation that higher Mg2+ concentrations could suppress the Mn2+-stimulated reaction of the sulfur substrate strengthened the conclusion that a divalent metal ion coordinates the 3’ leaving group.
Metal B coordinates the 3’-OH nucleophile of guanosine
In a seminal 1993 paper, Steitz and Steitz proposed a generalized two-metal ion mechanism for RNA-catalyzed phosphotransesterification reactions (157). In their scheme, one metal ion activates an incoming oxygen nucleophile, a second metal ion stabilizes the leaving group oxygen, and both metal ions coordinate one of the nonbridging phosphate oxygens. These interactions are suggested to stabilize the developing negative charges on the atoms within the pentavalent transition state, analogous to the role metal ions play in catalysis by protein enzymes like DNA polymerase (158, 159) and alkaline phosphatase (160). For the Tetrahymena ribozyme, MA had been identified as the metal ion that stabilized the 3’-oxygen leaving group. Whether a second metal ion activated the 3’-OH of the guanosine nucleophile to complete a two-metal ion mechanism was unknown.
Following the logic of the experiments that found MA, the strategy to determine whether a metal ion coordinates the guanosine 3’-OH nucleophile in the transition state might have employed 3’-thioguanosine. However, as sulfur is a relatively poor nucleophile at phosphorous centers (161), the experimental assay instead monitored the reverse reaction according to
where X is either oxygen or sulfur, and I is inosine (120). In this reaction, the terminal 3’-OH of CCCUCU attacks the phosphorous atom of the dinucleotide, releasing free inosine with either a 3’ oxygen or 3’ sulfur leaving group. By the principle of microscopic reversibility, a metal ion coordinating the 3’ leaving group in the reverse reaction would coordinate the 3’ nucleophile in the forward reaction. The dinucleotide contained inosine at the 5’ position because at the time, the synthesis of the 3’-phosphorothiolate linkage was easier with inosine than with guanosine. Previous studies of this reaction found that inosine could substitute for guanosine at the 5’ position of the dinucleotide (162, 163). Although a thermodynamic and kinetic framework for the reverse reaction had not been established fully, the observation that the reactions occurred with a log-linear pH dependence from pH 5.5 to 7.0 was consistent with the chemical step being rate-limiting.
In the presence of 110 mM Mg2+ at pH 7.0, only I3’OU was a substrate in the reverse reaction, and no detectable activity was observed with I3’SU. As in the MA rescue experiments, the lack of I3’SU cleavage did not result from reduced binding to the ribozyme. Adding softer divalent metal ions stimulated the reaction of I3’SU, but only Cd2+ produced a true metal ion rescue, increasing the reaction rate of I3’SU significantly above that of I3’OU. Again, analogous to the MA experiments, increasing concentrations of Mg2+ (from 0 – 100 mM, in a background of 18.5 mM Mg2+ and 2 mM Cd2+) inhibited I3’SU cleavage, suggesting that Mg2+ competed with Cd2+ for the site of rescue. The metal ion (MB, Figure 8B) interacts with the leaving group in the transition state for the reverse reaction. Therefore, by the principle of microscopic reversability, this metal ion must interact with the nucleophile in the forward reaction.
Metal C coordinates the 2’-OH of guanosine
Around the same time that the work on MB was being performed, Strömberg and colleagues reported an interaction between a divalent metal ion and the 2’-OH of the guanosine nucleophile (119). The evidence arose from assays in which 2’-deoxy-2’-aminoguanosine (G2′NH2) replaced guanosine in the reaction of the group I intron derived from the bacteriophage T4 nrdB gene (164, 165). The experiments monitored the first step of splicing of a pre-mRNA containing 22 nucleotides of exon 1, the 598-nucleotide intron, and 200 nucleotides of exon 2. In the presence of Mg2+ alone (4 mM MgCl2, 40 mM PIPES-KOH pH 7.2, 60 mM KCl), the reaction rate with G2′NH2 was 30-fold lower than with guanosine, although the binding constants for both forms of guanosine were similar. Chemistry for this reaction appeared to be rate-limiting, since both kcat and (kcat/KM) were log-linearly dependent on pH. A mixture of 4 mM Mg2+ and 0.9 mM Mn2+ stimulated the reaction rate with G2′NH2 about 6-fold, while 0.1 mM Zn2+ increased the rate 35-fold above the Mg2+-only rate. These observations, among others, led the authors to hypothesize that the 2’-OH of guanosine positioned a metal ion so that it could activate the nearby 3’-OH nucleophile in the first step of splicing. However, the study did not control adequately for the nonspecific effects of the rescuing metal ions, since the total divalent metal ion concentration was not always held constant. The authors did note that the optimal Mn2+ concentration for the reaction involving G2′NH2 was 0.3 – 1 mM, with Mg2+ added up to a total divalent metal ion concentration of 4 mM. The corresponding reaction rate for guanosine, however, was not reported.
In subsequent work, Shan and Herschlag provided further evidence for an interaction between a metal ion and the 2’ moiety of guanosine. First, they confirmed that both Mn2+ and Zn2+ rescued the reaction of the Tetrahymena ribozyme when G2′NH2 replaced guanosine (122). In addition, they developed a thermodynamic and kinetic framework to analyze the effects of Mn2+ binding on each individual step in the reaction pathway: binding to free ribozyme (E), to the enzyme-substrate complex (E·S), and to the ternary complex (E·S·G or E·S·G2′NH2). The results showed that a single Mn2+ ion rescued the reaction involving G2′NH2. The chief effect of this ion was to increase the affinity of G2′NH2 for the E·S complex, and of substrate (S) for the E·G 2′NH2 complex. In contrast, the ion did not affect the affinity of G for E·S, or of S for E·G, nor did it alter the binding of G, G2′NH2, or S to the free ribozyme E. The ion also had little effect on the rate of the chemical step starting from either the E·S·G or the E·S·G2′NH2 ternary complex. Higher concentrations of Mg2+ decreased the affinity of the rescuing Mn2+ ion for E and E·S, indicating that Mg2+ competed with Mn2+ for binding at the site of rescue.
Taken together, these observations supported a transition state interaction between the 2’ moiety of guanosine and a divalent metal ion (MC, Figure 8C). Furthermore, the apparent cooperativity of G2′NH2 or S binding to E·S or E·G2′NH2, respectively, in the presence of Mn2+ suggested that MC provided a structural bridge between guanosine and the substrate. This bridge required the presence of the reactive phosphate linkage, since Mn2+ had no effect on the affinity of G2′NH2 for the enzyme-product complex E·P (122). The authors noted that this interaction might help to align guanosine within the catalytic site for optimal in-line attack of the 3’-OH on the scissile phosphate. In fact, later work showed that interactions between S and MC drive a conformational change that facilitates the binding of guanosine to the ribozyme (166).
Do three separate metal ion-ligand interactions arise from three distinct metal ions?
At this point, metal ion rescue experiments had identified three different catalytic interactions between divalent metal ions and specific atoms of the oligonucleotide substrate and guanosine nucleophile. However, these experiments had not been designed to determine the number of metal ions responsible for these contacts. One possibility was that three distinct metal ions (MA, MB, and MC) bound the 3’ leaving group, the guanosine 3’-OH, and the guanosine 2’-OH, respectively, in the transition state. Or, MC might align guanosine through its 2’-OH and simultaneously activate the 3’-OH nucleophile, without the need for an additional MB. Counting the number of divalent ions within the catalytic site required a strategy that could monitor the effects of individual catalytic metal ion-ligand interactions on the reaction. The Herschlag and Piccirilli groups devised a way to discriminate among individual metal ions that bound to the three known sites A, B, and C. The method, called thermodynamic fingerprint analysis (TFA), involved measuring the affinities of rescuing metal ions for each site on the free ribozyme (E) (121). Which site was being interrogated depended upon the specific substrate modification used in the reaction. For example, to obtain the affinity of a rescuing Mn2+ ion for site A, a substrate containing oxygen or sulfur as the 3’ leaving group (S3’X, where X = O or S) was used. The experiment followed the reaction of S3’X first in the presence of 10 mM Mg2+, and then in 10 mM Mg2+ with increasing concentrations of Mn2+. The overall kinetic framework is shown in Scheme 1, where KMn is the affinity of the rescuing Mn2+ ion and krel is the relative rate constant of the oxygen and sulfur substrate reactions (krel = k3’S/k3’O) performed in Mg2+ alone or with added Mn2+.
Scheme 1.
The relative reaction rates were used to control for any nonspecific effects of Mn2+ (or any other rescuing metal ion). The observed rate constant for the reaction shown in Scheme 1 depends upon the fraction of free enzyme with a metal ion bound to site A:
| (2) |
Substituting for {fractionMgA•E} and {fractionMnA•E} from the kinetic pathway in Scheme 1 yields:
| (3) |
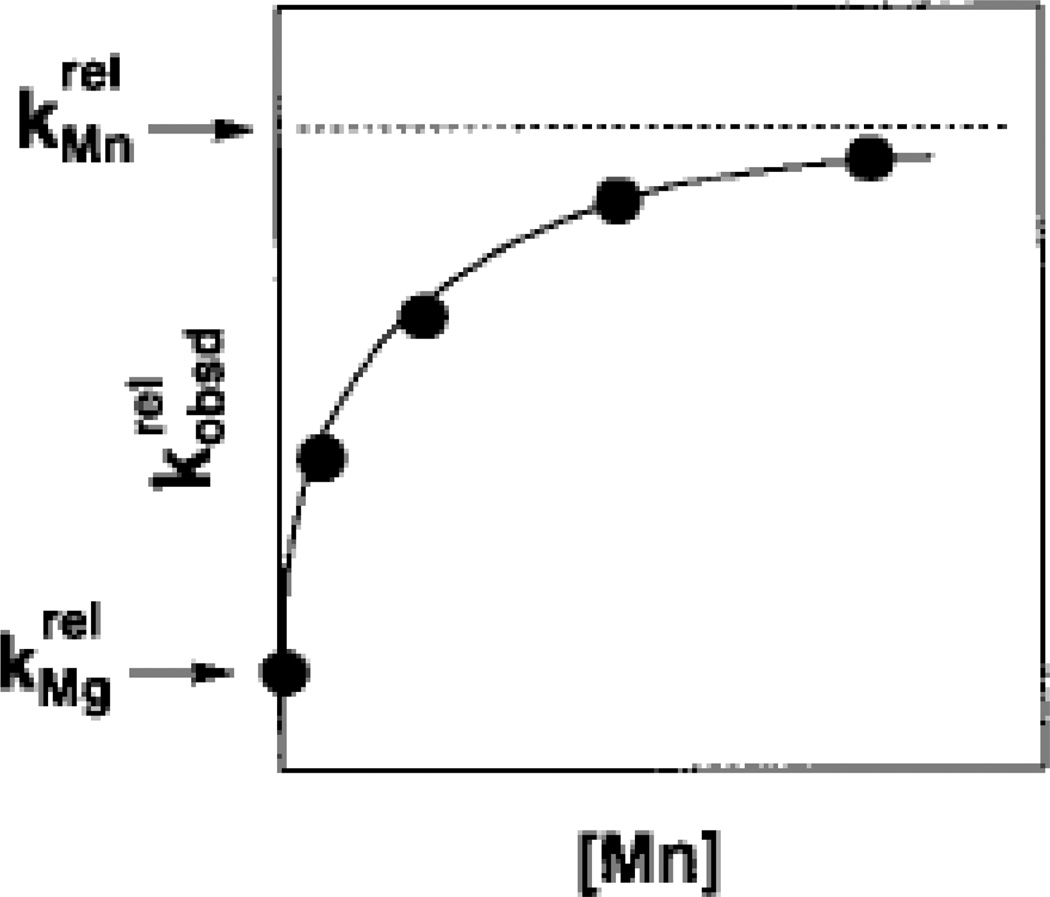
The affinity of the rescuing Mn2+ ion for site A, KMn, was then obtained by plotting versus [Mn2+] and fitting the data to Equation 3, with KMn as a fitted parameter (Figure 9). In the absence of Mn2+, , the relative rate constant for the reaction of S3’X in Mg2+ alone. The addition of Mn2+ accelerates according to Equation 3, up to a saturation value of . Thus, the plot depicts the Mn2+-dependent stimulation of , associated with an affinity KMn of the rescuing Mn2+ ion for metal site A. This affinity is a thermodynamic fingerprint, a unique signature, associated with Mn2+ rescue at site A, for the reaction shown in Scheme 1 under a specific set of conditions. Similar methods were used to obtain the affinities of rescuing metal ions for sites B and C, in the context of the appropriate substrate modifications.
Figure 9.
Mn2+ concentration dependence of the reaction of S3’S relative to S3’O () for the reaction shown in Scheme 1. The Mn2+-dependent acceleration of the reaction is associated with a specific affinity (KMn) of the rescuing Mn2+ ion for the free ribozyme. This affinity is a thermodynamic fingerprint for Mn2+ rescue of the 3’ sulfur modification. [Taken from (121).]
Importantly, these experiments were designed to monitor Mn2+ binding to the free ribozyme in the starting ground state. This ensured that the single-atom modifications of the substrate or guanosine did not affect the apparent affinities of Mn2+ ions for the ribozyme. Rescue then occurred due to the transition state interaction between Mn2+ and the modification. Because, by definition, the transition state exists only transiently, the modifications did not affect the observed Mn2+ affinities (121). For modifications involving the substrate (such as S3’O or S3’S), this was accomplished by employing substrates that reacted starting from the open complex. In the open complex, the substrate has not yet docked into the catalytic core, and therefore cannot affect the affinities of the catalytic Mn2+ ions for the free ribozyme. For modifications involving guanosine (such as G2′NH2), the experiments monitored the reaction E·S + G → products, in which the modified guanosine was not bound in the starting ground state. As a result, Mn2+ affinities obtained from experiments involving different substrate modifications could be compared directly. If a Mn2+ ion rescued one modification with the same affinity as it did a different modification, the simplest interpretation would be that the same metal ion mediates the interaction with both modifications. On the other hand, two different Mn2+ affinities would imply that distinct metal ions rescue each modification.
By using TFA, three different affinities were found for the binding of rescuing Mn2+ ions to sites A, B, and C on the Tetrahymena ribozyme (121, 122). MA rescued the reaction of S3’S with an affinity of 0.8 ± 0.2 mM (30 °C, 50 mM NaEPPS pH 7.9, 10 mM MgCl2), MB rescued the reaction of G3’SU with an affinity of 7 ± 1 mM (30 °C, 50 mM NaMES pH 6.3, 10 mM MgCl2), and MC rescued the reaction involving G2′NH2 with an affinity of 0.28 ± 0.06 mM (identical conditions as for MA). The closeness of the affinities of MA and MC prompted additional experiments to determine if these metal ions were truly distinct. Running the reaction in the presence of both S3’S and G2′NH2yielded plots of consistent with a model in which two separate ions rescue the reaction. The data therefore supported the existence of at least three separate metal ions binding within the catalytic site of the Tetrahymena ribozyme during the course of its reaction.
Interactions with the pro-SP nonbridging oxygen of the scissile phosphate
The two nonbridging oxygens at the scissile phosphate had been tested earlier as possible divalent metal ion ligands within the Tetrahymena catalytic site. Of the two, the pro-SP nonbridging oxygen seemed more likely to play this role since sulfur substitution of this oxygen reduced the reaction rate about 1000-fold (163). In comparison, sulfur substitution of the pro-RP oxygen produced only a 2.5-fold rate decrease (167). However, the addition of softer divalent metal ions did not fully rescue the reaction involving either of these substitutions (163, 167). Evidence from model compound studies suggested that the modest catalytic defect due to sulfur substitution of the pro-RP oxygen did not result from disruption of metal ion coordination (167). In contrast, the significant loss of activity upon substitution of the pro-SP oxygen with sulfur hinted at possible metal ion involvement, despite the negative rescue result.
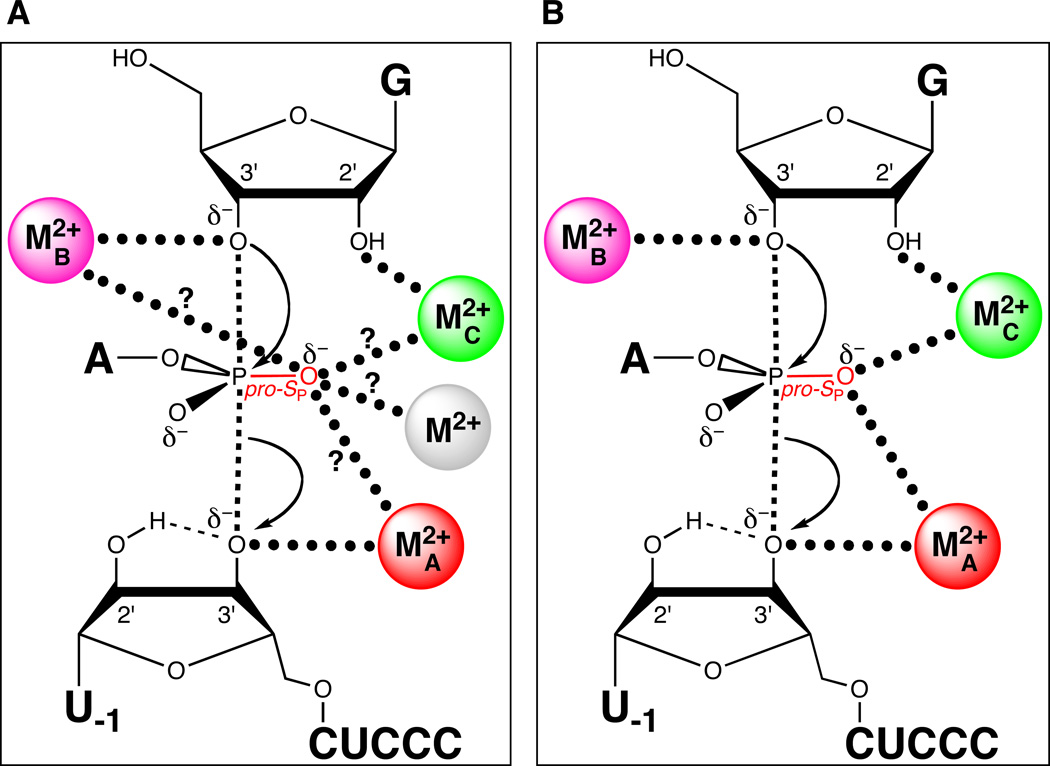
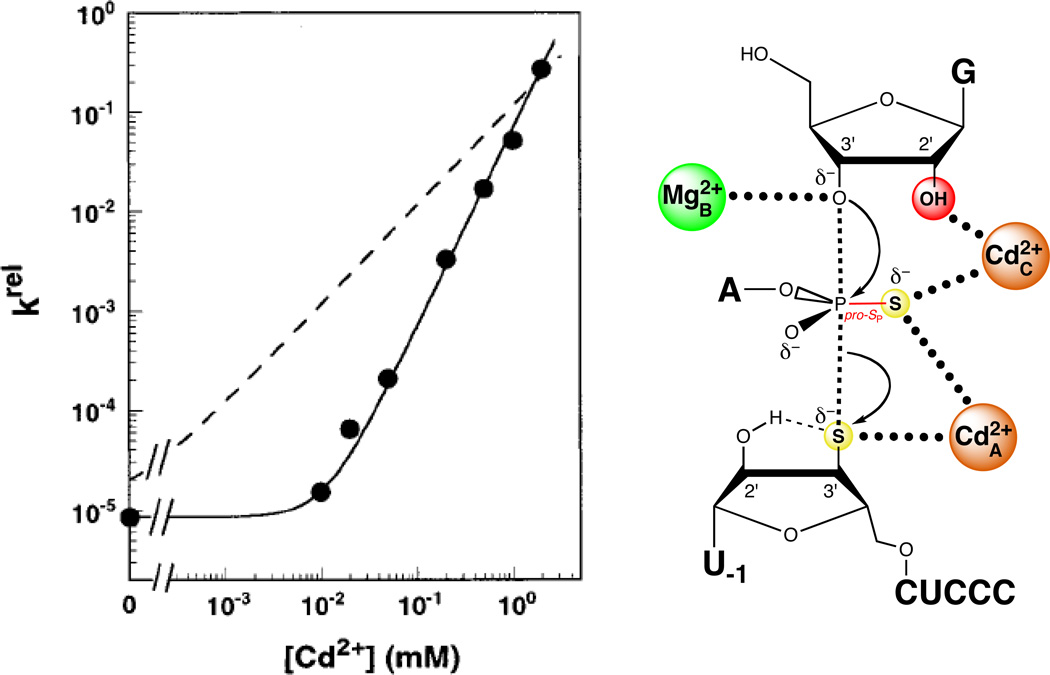
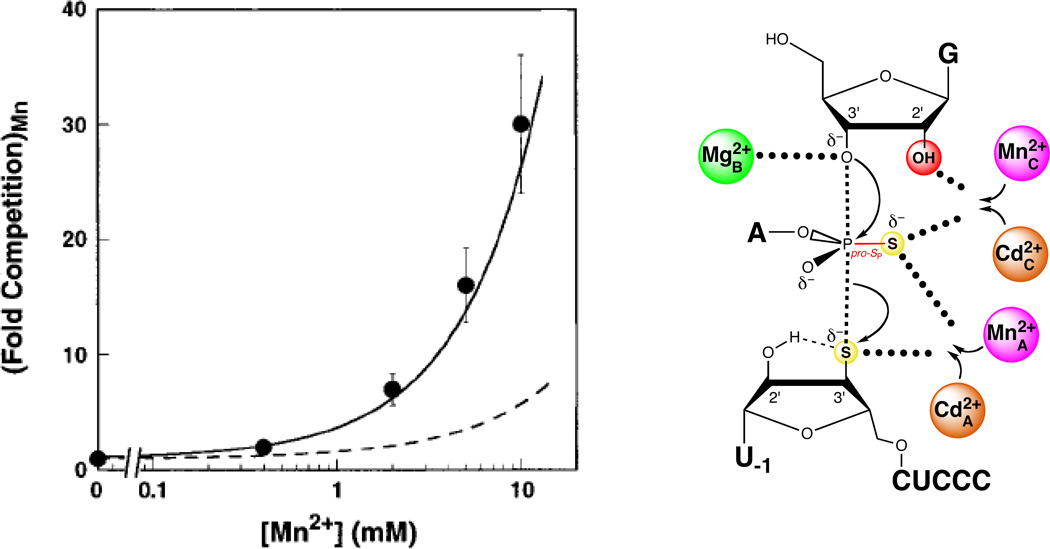
Piccirilli and colleagues resolved the role of the pro-SP nonbridging oxygen by testing the reactivity of a substrate with sulfur substitutions at both the pro-SP nonbridging and 3’ leaving group oxygens (123). In a mixture of Mg2+ and Mn2+, this phosphorodithioate substrate (S3’S,P-S) reacted more slowly than control substrates containing either a single 3’ bridging sulfur or a 3’ bridging sulfur and a pro-RP nonbridging sulfur. In contrast, the substitution of Zn2+ for Mn2+ restored the reactivity of S3’S,P-S to a level comparable to that of the single 3’ sulfur substrate. This result suggested that a metal ion or ions interacted with both the 3’ bridging and the pro-SP nonbridging oxygens in the transition state. The inability of Mn2+ or Zn2+ to rescue the reaction of the original pro-SP sulfur substrate (SP-S) was attributed to a possible geometric distortion caused by the presence of sulfur. Unlike SP-S, S3’S,P-S presented two sulfur atoms that could interact more favorably with the tetrahedral coordination geometry of Zn2+ than with the octahedral Mn2+. Although this work established that the pro-SP nonbridging oxygen interacts with a metal ion, whether this newly discovered ligand interacted with a fourth metal ion, or with one of the metal ions identified previously (MA, MB, or MC), remained unclear (Figure 10A). The lack of Mn2+ rescue in the reaction of S3’S,P-S precluded the use of standard TFA, since the MA, MB, and MC fingerprints represent apparent Mn2+ affinities. In principle, TFA might have been applied to the reactions involving S3’S,P-S by measuring the Cd2+ dependences of rescue of the MA, MB, and MC reactions. However, unlike the Mn2+ rescue profiles, the Cd2+ rescue profiles did not exhibit saturation behavior within the Cd2+ concentration range above which Cd2+ strongly inhibits the reaction. As a result, Cd2+ fingerprints analogous to those obtained with Mn2+ could not be determined. Plots of log krel versus log [Cd2+], where krel represents the observed rate constant for reaction of S3’S,P-S relative to unmodified substrate, were linear with a slope (n) of 2, implying that at least two Cd2+ ions mediate rescue of S3’S,P-S (124) (Figure 11). In contrast, Cd2+ rescue profiles for the reaction of S3’S relative to the unmodified substrate exhibited a shallower Cd2+ concentration dependence (n = 1), consistent with a single metal ion (MA) involved in the rescue. As S3’S,P-S contains a 3’ sulfur leaving group, presumably the same Cd2+ ion that mediated rescue of S3’S (MA) contributed to rescue of the dithioate by coordinating to the sulfur leaving group. The second Cd2+ ion likely interacted with the nonbridging atom in the transition state and could bind at the MB or MC site, or at a previously unidentified binding site.
Figure 10.
Both MA and MC coordinate the pro-SP nonbridging oxygen of the scissile phosphate. (A) Model showing potential metal ion-ligand interactions involving the pro-SP nonbridging oxygen (question marks). The grey sphere denotes a fourth divalent metal ion, a possibility not excluded from the results in (123). (B) Revised transition state model based on (124). Three metal ions are present in the active site, with metals A and C coordinating the pro-SP nonbridging oxygen of the scissile phosphate.
Figure 11.
Two Cd2+ ions at sites A and C rescue the reaction of S3’S,P-S by the Tetrahymena ribozyme. The plot at left shows the Cd2+ concentration dependence of krel, the reaction rate constant for S3’S,P-S relative to that of an all-oxygen substrate. The solid line fits the data according to a model in which two Cd2+ ions rescue the reaction, while the dashed line fits the data according to a model in which a single Cd2+ ion mediates rescue. [Data taken from (124).]
To assess whether the second Cd2+ ion involved in the rescue of S3’S,P-S bound in the MB or MC site, Shan et al. performed competition experiments with Mn2+ (124). Although Mn2+ does not rescue the reaction of S3’S,P-S, it does bind to the MA, MB, and MC binding sites with known affinities. Therefore, if the two Cd2+ ions involved in rescue of S3’S,P-S bound at these sites, then Mn2+ would be expected to inhibit rescue by competing with Cd2+ for binding. One of the rescuing Cd2+ ions presumably bound in the MA site (to mediate the interaction with the leaving group). If the second Cd2+ involved in the rescue of S3’S,P-S bound to the MB site, then Mn2+ would inhibit rescue by competing with Cd2+ for binding to the MA and MB sites. In contrast, if the second Cd2+ involved in the rescue of S3’S,P-S bound to the MC site, then Mn2+ would inhibit rescue by competing with Cd2+ for binding to the MA and MC sites. Because Mn2+ bound to the MC site 10-fold more strongly than to the MB site, the model involving the MA and MC sites predicted greater competition from Mn2+ than did the model involving the MA and MB sites. The competition experiments showed that Mn2+ inhibited the Cd2+-stimulated rescue of S3’S,P-S with a significantly steeper concentration dependence than predicted from the model involving the MA and MB sites. Instead, the Mn2+ competition data matched those predicted from the model in which Mn2+ competes with Cd2+ ions bound at the MA and MC sites (Figure 12). These findings led to a working model in which two Cd2+ ions rescue the reaction of S3’S,P-S. One bound at the MA site to stabilize the leaving group, while another bound at the MC site to interact with the nonbridging atom and the guanosine 2’-OH.
Figure 12.
Mn2+ ions compete with Cd2+ ions for sites A and C during rescue of the S3’S,P-S reaction. In the plot at left, the data are represented as filled spheres. The solid line shows the degree of competition expected from Mn2+ ions competing at sites A and C, while the dashed line shows the corresponding model for Mn2+ competition at sites A and B. [Data taken from (124).]
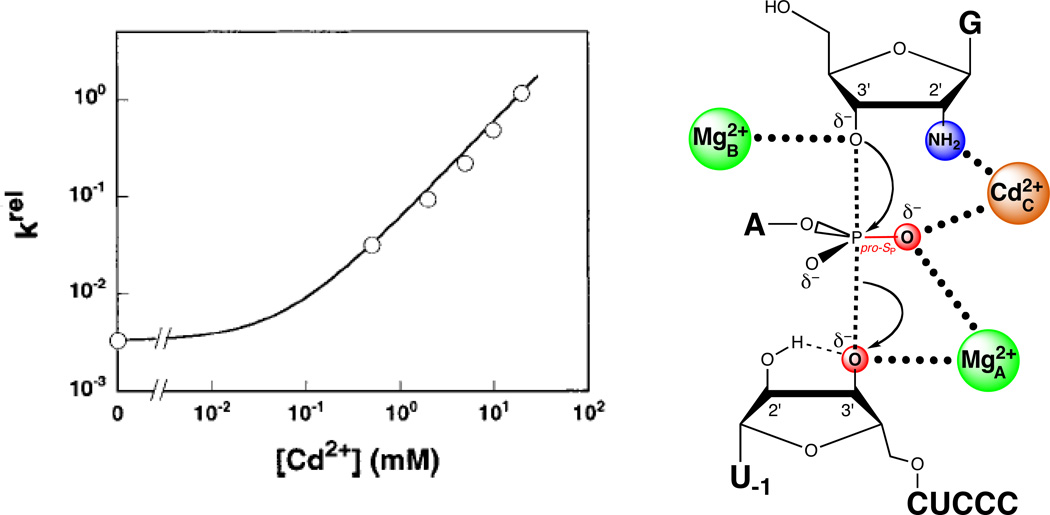
A simple prediction of this model is that the MC site can bind Cd2+. The reaction of G2′NH2 provided a means to probe Cd2+ occupancy of the MC site. Consistent with the model, Cd2+ rescued the reaction involving G2′NH2 relative to the reaction involving guanosine with a log-linear dependence of unit slope, suggesting that a single Cd2+ ion binds to the MC site to stimulate the reaction of G2′NH2 (Figure 13). As a further test of the hypothesis that MC coordinated the pro-SP nonbridging oxygen, the authors monitored the Cd2+ concentration dependence of the reaction of S3’S,P-S with G2′NH2 relative to the reaction of unmodified S with guanosine. If MC mediated interactions with both the pro-SP nonbridging oxygen and the guanosine 2’-OH, then two Cd2+ ions would have been expected to rescue the reaction of S3’S,P-S with G2′NH2 relative to the corresponding reactions involving unmodified substrates. One Cd2+ ion would populate the MA site to coordinate the 3’ sulfur leaving group, while a second would populate the MC site and interact with both the nonbridging sulfur atom of S3’S,P-S and the 2’ amino group of G2′NH2. In contrast, if a metal ion distinct from MC mediated the interaction with the nonbridging atom, then three metal ions would be involved in the rescue of S3’S,P-S in the presence of G2′NH2. The concentration dependence of both Cd2+- and Zn2+-mediated rescue of this reaction showed that only two ions were responsible for rescue, further supporting the proposed MC-pro-SP oxygen interaction (Figure 14).
Figure 13.
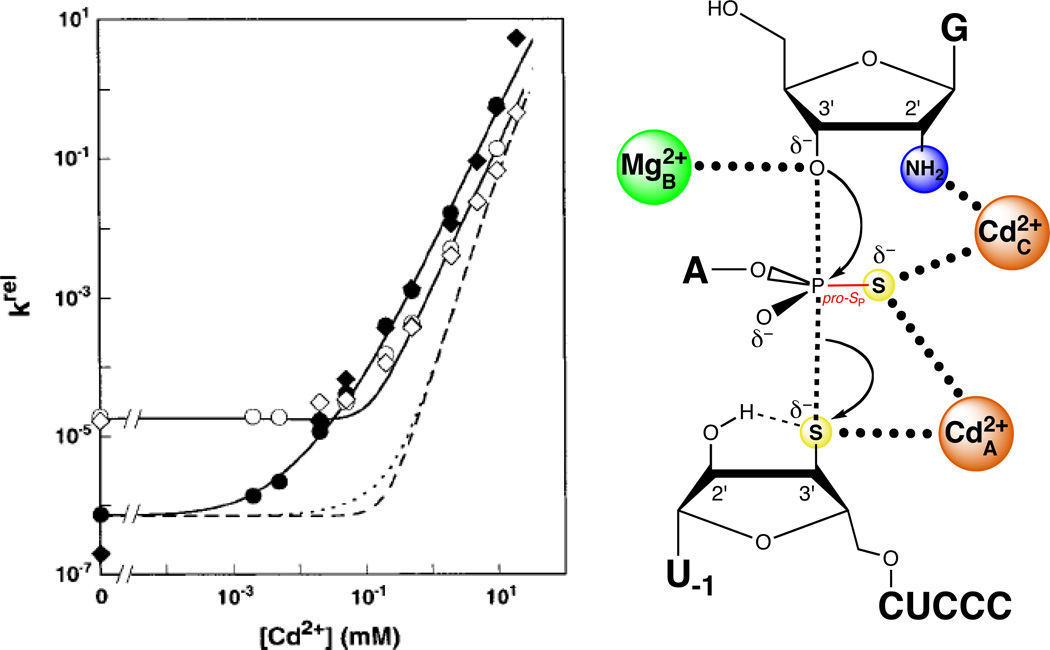
A single Cd2+ ion rescues the reaction in which G2′NH2 replaces guanosine. The plot at left shows the Cd2+ dependence of the rate constant for the reaction involving G2′NH2relative to guanosine for the all-oxygen substrate. The data are fit to a model in which a single Cd2+ ion rescues the reaction. [Data taken from (124).]
Figure 14.
Two Cd2+ ions rescue the reaction of S3’S,P-S with G2′NH2. In the plot at left, the closed symbols denote two independent determinations of the Cd2+ dependence of the reaction of S3’S,P-S with G2′NH2, relative to that of the all-oxygen substrate with guanosine. The open symbols denote two independent determinations of krel for the reaction of S3’S,P-S with guanosine, compared to that of the all-oxygen substrate with guanosine. The solid lines are fits of the data to a model in which two Cd2+ ions rescue the reaction, while the dashed lines are fits in which three Cd2+ ions rescue the reaction. [Data taken from (124).]
Additional experiments addressed the unresolved question of why softer metal ions had failed to rescue the reaction of SP-S. Although it is softer than Mg2+, Mn2+ prefers to bind oxygen rather than sulfur (143), and rescue of SP-S by Mn2+ would require formation of two Mn2+-sulfur interactions (Mn2+A and Mn2+C). On the other hand, Cd2+ rescue of the SP-S reaction would require the formation of two unfavorable Cd2+-oxygen interactions (to the 3’ leaving group oxygen and the guanosine 2’-OH). Thus, Cd2+ might not be expected to rescue the reaction of SP-S very efficiently, especially considering that Cd2+ could not saturate sites A or C even when S3’S,P-S was used as the substrate. If Cd2+ could rescue the reaction of SP-S, it should do so at high concentrations and involve the binding of two Cd2+ ions. As predicted, Cd2+ did rescue the reaction of SP-S at concentrations up to 15 mM, with a concentration dependence of two rescuing Cd2+ ions.
Functional identification of ribozyme ligands that position the catalytic metal ions
Up to this point, each of the catalytic metal ions that had been identified in the Tetrahymena ribozyme coordinated atoms of the oligonucleotide substrate. In contrast, none of the functional groups on the ribozyme itself that positioned these ions were known. As the most likely ligand candidates, the nonbridging phosphate oxygens of several conserved nucleotides within the catalytic core received special attention. An earlier NAIM study had found that phosphorothioate substitution of a number of pro-RP nonbridging oxygens inhibited splicing, and that Mn2+ could rescue the reaction of some of these sulfur-substituted ribozymes (108). This study did not explore the precise nature of this rescue, nor could it provide any information about the pro-SP nonbridging oxygens. However, coupled with phylogenetic analysis of the group I intron catalytic core (168) and results from other analog-interference experiments (169, 170), the NAIM data provided a strong starting point for a ligand search.
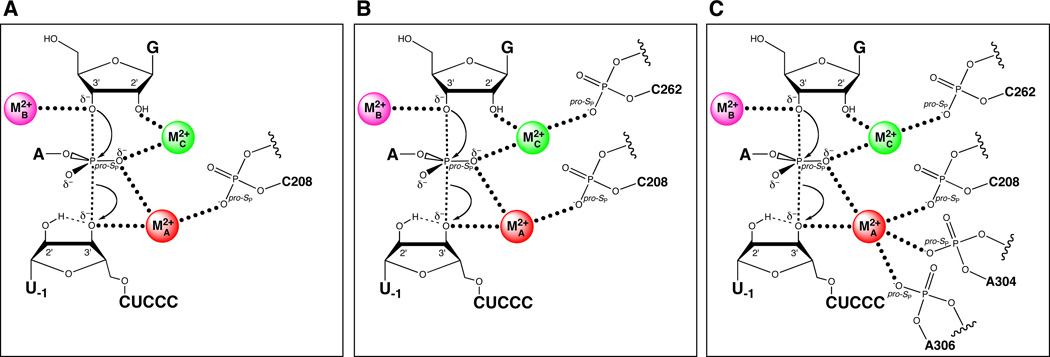
Strobel and colleagues provided evidence for one of these ligands, the pro-SP nonbridging oxygen of C208 (C208 SP) (109) (Figure 15A). Relying on a model of the Tetrahymena catalytic site (169), they singled out five positions as potential ligands, and constructed the ten corresponding phosphorothioate ribozymes (RP and SP). This required dividing the full-length ribozyme into three pieces – a synthetic oligonucleotide containing the phosphorothioate, and two transcribed flanking RNAs. The phosphorothioate oligonucleotides were first separated into RP and SP diastereomers by using reversed-phase HPLC. These were then joined to the flanking RNAs by hybridization with bridging DNA splints followed by incubation with DNA ligase (171). Of the ten constructed ribozymes, four showed substantial reductions in reactivity compared to the unmodified ribozyme, but partial Mn2+ rescue was observed only for the C208-SP ribozyme. Subsequent experiments tested the ability of this ribozyme to cleave substrates containing either 3’ phosphorothiolate or pro-SP phosphorothioate modifications. By analogy to previous work, it was thought that thiophilic metal ions like Cd2+ or Zn2+ would rescue the reaction more efficiently in the context of the double sulfur substitution (ribozyme and substrate). While neither Mn2+ nor Zn2+ rescued the reaction of either sulfur substrate in the context of the C208-SP ribozyme, Cd2+ stimulated both reactions by 70- to 100-fold. Since MA was known to coordinate both the 3’ leaving group and the scissile pro-SP nonbridging oxygen in the transition state, the authors concluded that C208 SP was a ligand for MA.
Figure 15.
Sequential discovery of ligands within the Tetrahymena ribozyme that position the catalytic metal ions. (A) The pro-SP nonbridging oxygen of C208 coordinates MA. (B) The pro-SP nonbridging oxygen of C262 coordinates MC. (C) Additional ligands for MA include The pro-SP nonbridging oxygens of A304 and A306.
In the absence of thermodynamic fingerprints or the dependence of krel on the rescuing metal ion concentration, the range of metal ions that confers rescue can provide a qualitative approach to link catalytic metal ions to their ligands within the ribozyme. For example, in the Tetrahymena ribozyme, incorporation of the SP phosphorothioate at residue C208 inhibited reaction of the unmodified substrate in Mg2+. Mn2+ and Zn2+ rescued this effect, but Cd2+ did not. However, when the mutant ribozyme was assayed using a substrate containing a 3’ phosphorothiolate at the cleavage site, Cd2+, but neither Mn2+ nor Zn2+, could rescue the reaction (109). Phosphorothioate substitutions at other positions in the ribozyme did not show this behavior. These results functionally linked the C208 pro-SP phosphorothioate to the 3’ phosphorothiolate in the substrate, suggesting that the C208 pro-SP oxygen within the ribozyme provides a ligand to the catalytic metal ion that stabilizes the 3’ oxygen leaving group. In-depth quantitative and structural analyses later confirmed this interaction. Thus, functional linkage between groups may be inferred when the combination of modifications leads to rescue behavior distinct from that of either modification alone.
The Herschlag and Piccirilli groups used the concentration dependence of krel on Cd2+ as a signature to identify ligands for the catalytic metal ions. Prior to the publication of the group I crystal structures, they combined substrate atomic mutagenesis with site- and stereospecific phosphorothioate substitutions at six sites within the J6/7 and P7 regions of the ribozyme to identify the ligands to MA, MB, and MC. They tested whether Cd2+, a thiophilic metal ion that, like Mg2+, can adopt octahedral coordination geometry, stimulated the catalytic activity of the phosphorothioate-containing ribozymes in the reaction of G with S3’O. Under conditions of saturating E and G, they found that several of the phosphorothioates significantly affected catalysis. However, upon addition of 0.1 – 1.0 mM Cd2+, only one of the variant ribozymes, the C262-SP variant, experienced significant stimulation, suggesting that Cd2+ may form a functionally important interaction with the sulfur at position C262. They used the modified substrates that report on the MA, MB, and MC sites (described above) to ascertain whether the C262 phosphorothioate affected the rescue behavior. If the C262 SP phosphorothioate directly interacted with a Cd2+ ion bound in the MA, MB, or MC sites, they expected that the stronger Cd2+-sulfur interaction (compared to the Cd2+-oxygen interaction in the unmodified ribozyme) would shift the metal rescue profile toward lower Cd2+ concentrations compared to the corresponding rescue profile using the unmodified ribozyme. The MA and MB rescue profiles remained essentially unaffected by the C262-SP phosphorothioate. In contrast, the MC rescue profile (using G2′NH2 as the modified substrate) shifted such that 16-fold lower Cd2+ concentrations were required to achieve the same level of rescue as with the unmodified ribozyme. The observation that the C262-SP phosphorothioate shifted the MC rescue profile, but not the MA and MB profiles, to lower Cd2+ concentrations suggested that the sulfur atom interacted directly with the Cd2+ ion binding at the MC site (Figure 15B). Further independent tests supported this model, as did the available crystal structures (see below). Recently, Forconi et al. extended this approach to identify two additional ligands for MA (the pro-SP nonbridging oxygens of A304 and A306) (111) (Figure 15C), verifying interactions inferred from the Azoarcus crystal structures. As of this writing, no ligands for MB have been identified.
Comparison with structural data
Most of the preceding experiments were conducted in the absence of high-resolution structural information. During the transition of the investigation from defining substrate ligands to defining ribozyme ligands, crystal structures of three group I introns emerged, providing for the first time a structural framework with which to evaluate more than a decade of functional experiments. The agreement between the structural and biochemical models is striking. The structures show that nearly all of the predicted ligands are located in close proximity to metal ions residing within the catalytic site. While the resolution of the structures is not sufficient to view Mg2+ ions directly, their positions have been inferred based on anomalous scattering maps of heavy metal ion derivatives.
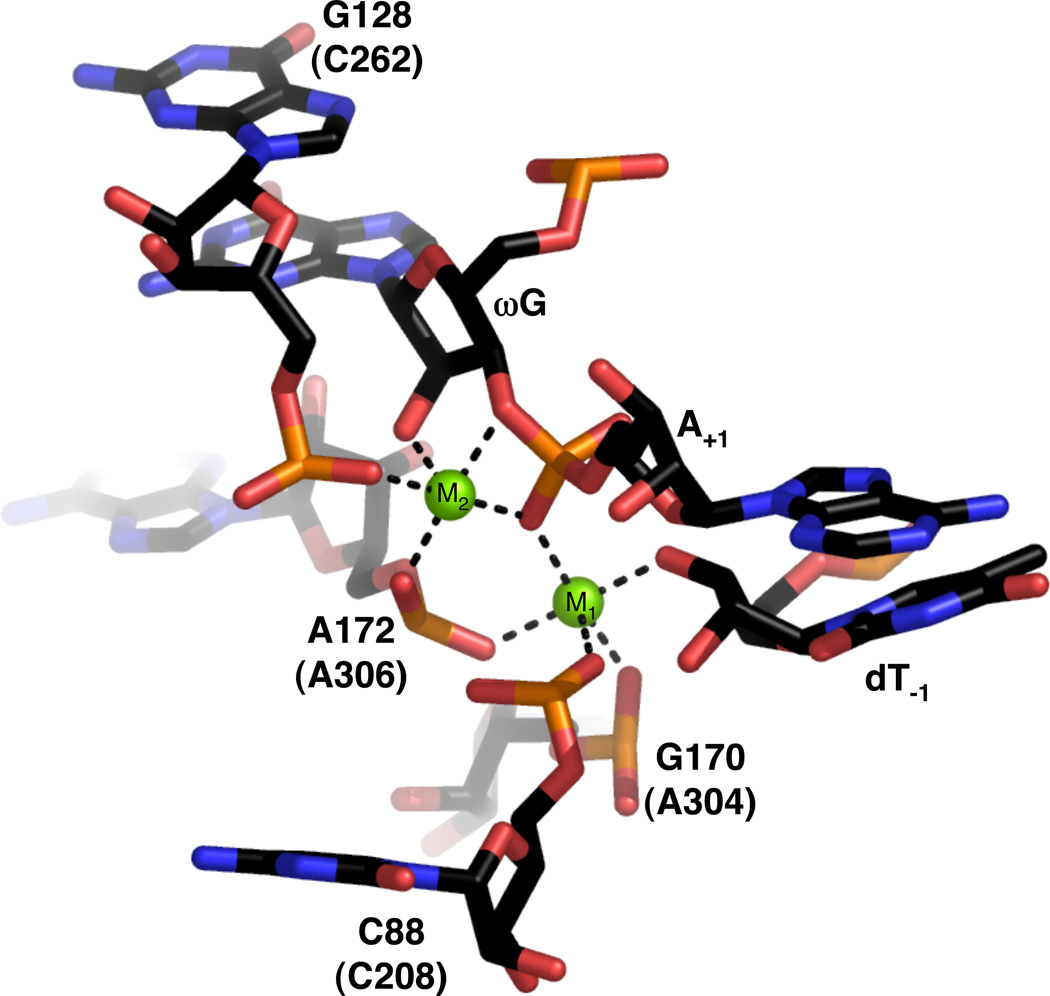
In terms of inferring catalytic interactions, the most recent structure of the Azoarcus group I ribozyme (18) likely provides the information relevant to the catalytic mechanism. The 3.4-Å structure captures the ribozyme just after the first step of splicing, with both exons poised for ligation and riboguanosine bound at the 3’ splice site. Two Mg2+ ions, designated M1 and M2, are modeled into the catalytic site based on Yb3+ and Mn2+ soaks (Figure 16). M1 makes apparent inner sphere contacts with the 3’ oxygen of dT−1 (the leaving group in the forward reaction), and the pro-RP nonbridging oxygen of the scissile phosphate (corresponding to the pro-SP oxygen in the forward reaction). In addition, the pro-SP nonbridging oxygens of C88, G170, and A172 (Tetrahymena nucleotides C208, A304, and A306, respectively) all lie within inner sphere coordination distance of M1. A possible outer sphere sixth ligand, the pro-SP oxygen of U173 (Tetrahymena U307), is also present, although the functional significance of this interaction is unclear (111). This set of ligands suggests that M1 corresponds to MA. M2, on the other hand, appears to correspond to MC. It lies within 2.2 Å of the 2’-OH of ωG (corresponding to the 2’-OH of the guanosine nucleophile in the forward reaction), the pro-RP nonbridging oxygen of the scissile phosphate, and the pro-SP nonbridging oxygen of G128 (Tetrahymena C262). In addition, the pro-RP nonbridging oxygen of A172 appears to contact M2 via inner sphere coordination, and initial rescue data support the existence of this interaction in Tetrahymena (172). Interestingly, M2 also resides 2.1 Å from the 3’-OH of ωG, suggesting a possible role in leaving group stabilization in the second splicing step (or nucleophile activation in the first step).
Figure 16.
View of the active site of the Azoarcus group I ribozyme (PDB file 1ZZN). Nucleotide numbers in parentheses refer to homologous positions within the Tetrahymena ribozyme. Mg2+ ions are represented as green spheres. Black lines denote putative inner sphere interactions between ribozyme ligands and the metal ions.
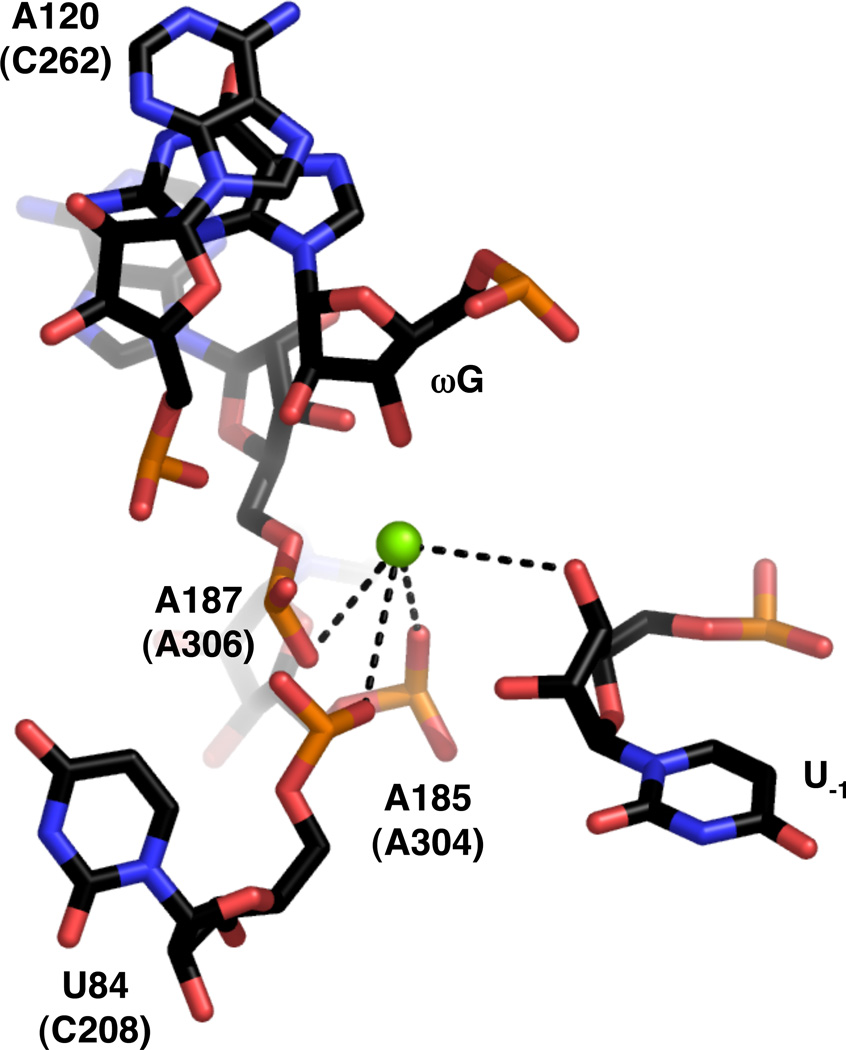
The crystal structure of the group I intron derived from bacteriophage Twort was solved to 3.6 Å resolution as a complex between the ribozyme and a four-nucleotide product analog (17). Following co-crystallization with Mn2+, a single metal ion within the catalytic site was observed that appeared to correspond to MA. This metal ion resides within 4 Å of the 3’-OH of U−1 and the pro-SP nonbridging oxygens of residues U84, A185, and A187 (Tetrahymena nucleotides C208, A304, and A306, respectively) (Figure 17). Although the scissile phosphate is absent in this structure, its approximate position could be modeled plausibly so that the pro-SP nonbridging oxygen lies within inner sphere coordination distance of the metal ion. A candidate for MC was not found, but a possible location for such an ion was modeled within 2.5 Å of the ωG 2’-OH and the pro-SP nonbridging oxygen of A120 (Tetrahymena C262). The pro-RP nonbridging oxygen of A187 was also proposed as a ligand for MC, although subsequent biochemical work has not confirmed this interaction in the Tetrahymena ribozyme (111).
Figure 17.
View of the active site of the group I ribozyme derived from bacteriophage Twort (PDB file 1Y0Q). Numbering and colors are as in Figure 16.
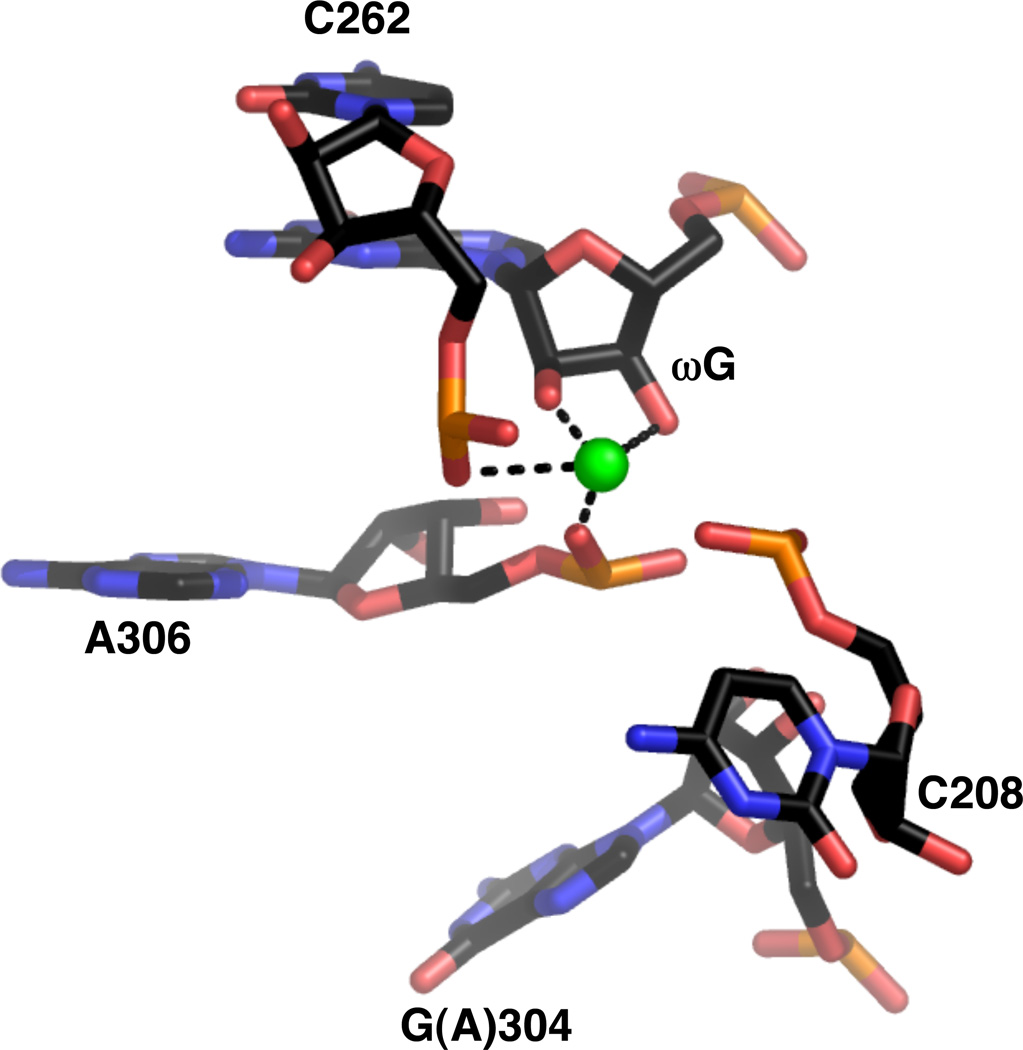
The Tetrahymena ribozyme itself was solved to 3.8 Å resolution in the absence of substrate (16), after mutations were introduced that improved the thermostability of the ribozyme without significantly affecting catalytic activity (173). Eu3+ and Ir3+ derivatives revealed a number of metal ion binding sites that were assumed to represent Mg2+ sites. Within the catalytic site, one metal ion was found in proximity to both the 2’- and 3’-OH groups of ωG, as well as the nonbridging phosphate oxygens of C262, U305, and A306 (Figure 18). Given this ligand environment, the metal ion was thought to be MC rather than MB, despite its apparent contact with the ωG 3’-OH group. The authors noted, however, that the position of this ion would likely shift significantly prior to catalysis due to conformational changes associated with substrate binding. Thus, in the presence of substrate, the ion potentially could become any one of the three catalytic metal ions identified biochemically.
Figure 18.
Active site metal ion within the crystal structure of the Tetrahymena ribozyme (PDB file 1X8W). The guanosine at position 304 is one of several mutations introduced based on experiments that selected for thermostable variants of the ribozyme.
In spite of the overall agreement between the structures and the biochemical data, some discrepancies remain unresolved. The most significant is the lack of any electron density in the structures that might confirm convincingly the presence of MB. Several possibilities could explain this observation, with the simplest being that MB does not occupy its binding site in these crystals. This may result from the crystallization conditions themselves. Of the three catalytic metal ions, MB binds an order of magnitude more weakly than MA or MC, and its binding might therefore be more sensitive to variables like ionic strength or the presence of other salts or organic compounds. Alternatively, the ribozymes within the crystals could exist in conformations incompatible with the binding of MB. Or, MB could be present in the crystals, but disordered and therefore not visible at the current resolutions. Other possibilities involve the precise role of MB in the reaction mechanism. The evidence for the presence of MB comes from metal ion rescue experiments using a sulfur-modified oligonucleotide substrate in the reverse reaction. Rescue of this particular reaction may require an ion acting as MB, even though the unmodified forward reaction does not. The ribozymes themselves may even have slightly different reaction mechanisms. The three-metal ion model relies on data obtained from the Tetrahymena ribozyme only. This ribozyme may indeed use MB to effect catalysis, while the Azoarcus and Twort ribozymes may not. Applying metal ion rescue and TFA techniques to other group I ribozymes should help to address whether a three-metal ion mechanism is unique to Tetrahymena.
Metal ion rescue in RNase P
RNase P is an essential endonuclease found in nearly all bacterial, archaeal, and eukaryal organisms (174, 175) (Figure 19). It removes the 5’ leader sequence of precursor tRNA molecules (pre-tRNAs) via hydrolysis to produce tRNAs with mature 5’ ends. With few exceptions (176), all RNases P are ribonucleoprotein complexes consisting of one or more protein subunits ranging from 120 to 1000 amino acids, combined with a 200- to 400-nucleotide RNA (P RNA). This RNA component contains the catalytic apparatus, as evidenced by the ability of bacterial (177) and some archaeal (178) P RNAs to cleave pre-tRNA in vitro in the absence of protein.
Figure 19.
The RNA component of RNase P. (A) Secondary structure of E. coli P RNA. (B) Transition state model for the reaction catalyzed by RNase P. Question marks denote putative metal ion-ligand interactions awaiting experimental verification. [Adapted from (175), with permission.] (C) Crystal structure of the P RNA from B. stearothermophilus at 3.3 Å (PDB file 2A64). (D) Location (in red) of the P4 helix within the B. stearothermophilus P RNA.
The hydrolysis reaction produces a mature tRNA with a 5’ phosphate, along with the leader sequence. Catalysis is absolutely dependent on divalent metal ions (179), with a Hill analysis suggesting the involvement of at least three Mg2+ ions (180). Site-specific atomic substitutions coupled with metal ion rescue experiments have implicated several ligands on both pre-tRNA and RNase P as important for metal ion binding during catalysis. On the pre-tRNA substrate, phosphorothioate substitution of the pro-RP nonbridging oxygen at the cleavage site reduces the reaction rate by 103- to 104-fold (132–135, 181). The addition of either Mn2+ or Cd2+ restores much of this activity (132–135, 182), indicating that the pro-RP nonbridging oxygen coordinates a divalent metal ion during catalysis. This same ion has also been proposed to activate water to generate the hydroxyl nucleophile, based partly on the 6.4-fold rate increase associated with Mn2+ rescue of the RP phosphorothioate reaction (133). In contrast, sulfur substitution of either the pro-SP or the 3’ bridging oxygen at the cleavage site shifts cleavage to the next unmodified phosphodiester linkage in the 5’ direction (132, 134, 135, 183). The rate of miscleavage is greatly reduced compared with the rate of canonical cleavage in the unmodified reaction. However, softer metal ions have only a minimal effect on the reaction rate and do not change the position of cleavage. This observation suggests that the pro-SP and 3’ oxygens at the cleavage site play important roles in catalysis, but whether they coordinate metal ions is unclear. These atoms could be involved in tertiary interactions that the bulkier sulfur atom cannot accommodate. Alternatively, the sulfur atom might exclude catalytic metal ions from the active site, as has been observed for a phosphorothioate substrate bound to the 3’→5’ exonuclease site of E. coli DNA polymerase I (146, 183).
The 2’-OH group at the cleavage site also appears to coordinate one of the catalytic metal ions. Deoxy substitution of the 2’-OH reduces the reaction rate by 150- to 3400-fold (138, 180, 184). In addition, the Mg2+ affinity decreases by four orders of magnitude, with a concomitant reduction in cooperativity from 3 ions to 2 (180). Experiments in which an amino group was installed at the 2’ position also suggested a role for the 2’-OH as a metal ion ligand (138). At low pH, substrates bearing a 2’-NH2 group at the cleavage site showed significant miscleavage in the presence of either Mg2+ or Mn2+. Since the 2’-NH2 group would be protonated at low pH, it would be unable to donate a lone pair of electrons for metal ion coordination. Moreover, the protonated amino group would also repulse any positively-charged Mg2+ ions, further contributing to the loss of cleavage fidelity. At higher pH in the presence of 10 mM Mg2+, the 2’-NH2 modification resulted in a more than 2500-fold decrease in the reaction rate. While 10 mM Mn2+ accelerated the reaction by almost 100-fold, it had a similar effect in the presence of the 2’-H substitution. This result led the authors to propose that the 2’-OH group coordinated a metal ion indirectly through a bound water molecule (outer sphere).
The active site picture that emerges from these experiments is shown in Figure 19B, in which water molecules and functional groups on the pre-tRNA substrate coordinate three metal ions (175). One metal ion binds both the pro-RP nonbridging oxygen and the attacking hydroxyl nucleophile. A second metal ion binds both the pro-RP nonbridging and the 3’ bridging oxygens, while a third interacts with the pro-SP nonbridging oxygen and indirectly with the cleavage site 2’-OH group. However, most of these interactions have not yet been confirmed, through either functional experiments or crystallography, to the same extent as those within the Tetrahymena active site.
On the P RNA itself, much effort has focused on the P4 helix within the catalytic domain as a candidate site for metal ion coordination (Figure 19D). Phylogenetic analysis has shown that several residues within P4 are highly conserved among bacterial, archaeal, and eukaryal RNase P RNAs (185). In addition, NAIM studies have implicated specific nonbridging oxygens and purine N7 groups within helix P4 as potential ligands (136, 186, 187). In particular, phosphorothioate substitution of either nonbridging oxygen of A67 (E. coli numbering) reduces the catalytic rate constant by 103- to 104-fold (136). Both Mn2+ and Cd2+ rescue the reactions of the A67-RP and A67-SP P RNAs, supporting a role for these nonbridging oxygens in metal ion coordination during catalysis. Similar results were obtained for the homologous position in B. subtilis P RNA, although the extent of rescue varied depending on the presence or absence of the P protein (188). Quantitative analysis of Mn2+ rescue of the A67-RP and SP ribozyme reactions revealed two different affinities for the rescuing Mn2+ ion (137). As with the Tetrahymena ribozyme (121), this result suggested that two distinct metal ions bound each of the nonbridging oxygens of A67 during catalysis. Experiments using the A67-SP ribozyme combined with 7-deaza-adenosine substitution of A66 supported a model in which one of these metal ions bound the N7 of A66 and the pro-SP oxygen of A67 simultaneously (137). Evidence for a possible third metal ion binding within the P4 helix came form analagous experiments with 7-deaza-adenosine substitution of A65 (137).
Although these results suggest that the P4 helix binds metal ions important for catalysis, the specific atoms within P4 that coordinate metal ions at the cleavage site are not yet known. A cross-linking study that substituted 4-thiouridine for the conserved bulged uridine U69 demonstrated that the P4 helix does lie in the vicinity of the 5’ leader sequence (139). However, the bulge itself, along with other residues postulated to position catalytic metal ions, is situated about 5 nucleotides downstream from the cleavage site, too far to interact with it directly. This result was consistent with models derived from two different crystal structures of bacterial P RNAs (22, 23), which indicated that the P4 metal binding sites were distant from the proposed catalytic center. To determine whether P4 was linked to the catalytic metal ions at the cleavage site, Christian et al. mutated the bulged U69 and determined the Cd2+ dependence for rescue of an RP phosphorothioate at the cleavage site (139). Replacing U69 with adenine did not affect the affinity of rescuing Cd2+ ions. In contrast, deletion or repositioning of the bulged uridine within the helix resulted in a significant decrease in Cd2+ affinity and cooperativity. Substitution of 4-thiouridine for a bulged uridine repositioned at U68 resulted in a cross-link shifted further into the 5’ leader sequence relative to the cross-link obtained with unmodified P RNA. Taken together, these observations suggested that the P4 helix positions both the catalytic metal ions and the pre-tRNA substrate within the active site. As of this writing, however, an atomic model of the RNase P transition state comparable to that deduced for the Tetrahymena ribozyme has not been described.
Conclusions
Identifying ligands that bind and position the catalytic metal ions within ribozymes remains an essential prerequisite for the elucidation of ribozyme mechanisms. A plethora of both low and high resolution techniques – biochemical, crystallographic, spectroscopic – have delineated many of the qualitative and quantitative contributions of these interactions to catalysis. As the preceding discussions illustrate, no individual strategy by itself can locate unequivocally all of the metal ions that activate the reacting species and facilitate catalysis. Rather, the most complete picture of metal ion interactions within ribozyme transition states emerges when observations derived from many different approaches are combined and reconciled with each other.
Acknowledgements
The authors would like to thank Mr. Raghuvir Sengupta for his thoughtful advice and comments on the manuscript. J.K.F. is supported by the Medical Scientist Training Program of the University of Chicago Pritzker School of Medicine. J.A.P. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Herschlag D, Cech TR. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 2.Pyle AM. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- 3.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. Chem Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 4.Nakano S, Chadalavada DM, Bevilacqua PC. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 5.Perrotta AT, Been MD. Biochemistry. 2006;45:11357–11365. doi: 10.1021/bi061215+. [DOI] [PubMed] [Google Scholar]
- 6.Roth A, Nahvi A, Lee M, Jona I, Breaker RR. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigel RK, Pyle AM. Chem Rev. 2007;107:97–113. doi: 10.1021/cr0502605. [DOI] [PubMed] [Google Scholar]
- 8.Bukhman YV, Draper DE. J Mol Biol. 1997;273:1020–1031. doi: 10.1006/jmbi.1997.1383. [DOI] [PubMed] [Google Scholar]
- 9.Nakano S, Proctor DJ, Bevilacqua PC. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- 10.Nakano S, Cerrone AL, Bevilacqua PC. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- 11.Nakano S, Bevilacqua PC. Biochemistry. 2007;46:3001–3012. doi: 10.1021/bi061732s. [DOI] [PubMed] [Google Scholar]
- 12.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 13.Chen JH, Gong B, Bevilacqua PC, Carey PR, Golden BL. Biochemistry. 2009;48:1498–1507. doi: 10.1021/bi8020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams LD. Top Curr Chem. 2005;253:77–88. [Google Scholar]
- 15.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 16.Guo F, Gooding AR, Cech TR. Mol Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Golden BL, Kim H, Chase E. Nat Struct Mol Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 18.Stahley MR, Strobel SA. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 19.Toor N, Keating KS, Taylor SD, Pyle AM. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toor N, Rajashankar K, Keating KS, Pyle AM. Nat Struct Mol Biol. 2008;15:1221–1222. doi: 10.1038/nsmb.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krasilnikov AS, Yang X, Pan T, Mondragon A. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 22.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Proc Natl Acad Sci USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 24.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 25.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 26.Clemons WM, Jr, Brodersen DE, McCutcheon JP, May JL, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. J Mol Biol. 2001;310:827–843. doi: 10.1006/jmbi.2001.4778. [DOI] [PubMed] [Google Scholar]
- 27.Pley HW, Lindes DS, DeLuca-Flaherty C, McKay DB. J Biol Chem. 1993;268:19656–19658. [PubMed] [Google Scholar]
- 28.Pley HW, Flaherty KM, McKay DB. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 29.Scott WG, Finch JT, Klug A. Nucleic Acids Symp Ser; 1995. pp. 214–216. [DOI] [PubMed] [Google Scholar]
- 30.Scott WG, Murray JB, Arnold JR, Stoddard BL, Klug A. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- 31.Martick M, Scott WG. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rupert PB, Ferre-D'Amare AR. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 33.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 34.Ferre-D'Amare AR, Zhou K, Doudna JA. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 35.Ferre-D'Amare AR, Doudna JA. J Mol Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 36.Klein DJ, Ferre-D'Amare AR. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 37.Ye JD, Tereshko V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Koide S, Kossiakoff AA, Piccirilli JA. Proc Natl Acad Sci USA. 2008;105:82–87. doi: 10.1073/pnas.0709082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovall RA, Matthews BW. Curr Opin Chem Biol. 1999;3:578–583. doi: 10.1016/s1367-5931(99)00012-5. [DOI] [PubMed] [Google Scholar]
- 39.Pingoud A, Jeltsch A. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galburt EA, Stoddard BL. Biochemistry. 2002;41:13851–13860. doi: 10.1021/bi020467h. [DOI] [PubMed] [Google Scholar]
- 41.Nelson JA, Uhlenbeck OC. Mol Cell. 2006;23:447–450. doi: 10.1016/j.molcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 42.DeRose VJ. In: Nucleic Acid-Metal Ion Interactions. Hud NV, editor. Cambridge, UK: Royal Society of Chemistry; 2009. pp. 154–179. [Google Scholar]
- 43.Horton TE, Clardy DR, DeRose VJ. Biochemistry. 1998;37:18094–18101. doi: 10.1021/bi981425p. [DOI] [PubMed] [Google Scholar]
- 44.Morrissey SR, Horton TE, DeRose VJ. J Am Chem Soc. 2000;122:3473–3481. [Google Scholar]
- 45.Kisseleva N, Khvorova A, Westhof E, Schiemann O. RNA. 2005;11:1–6. doi: 10.1261/rna.7127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogt M, Lahiri S, Hoogstraten CG, Britt RD, DeRose VJ. J Am Chem Soc. 2006;128:16764–16770. doi: 10.1021/ja057035p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salemink PJ, Swarthof T, Hilbers CW. Biochemistry. 1979;18:3477–3485. doi: 10.1021/bi00583a007. [DOI] [PubMed] [Google Scholar]
- 48.Hilbers CW, Heerschap A, Haasnoot CA, Walters JA. J Biomol Struct Dyn. 1983;1:183–207. doi: 10.1080/07391102.1983.10507434. [DOI] [PubMed] [Google Scholar]
- 49.Cheong C, Varani G, Tinoco I., Jr Nature. 1990;346:680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- 50.Dieckmann T, Suzuki E, Nakamura GK, Feigon J. RNA. 1996;2:628–640. [PMC free article] [PubMed] [Google Scholar]
- 51.Butcher SE, Dieckmann T, Feigon J. EMBO J. 1997;16:7490–7499. doi: 10.1093/emboj/16.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sashital DG, Venditti V, Angers CG, Cornilescu G, Butcher SE. RNA. 2007;13:328–338. doi: 10.1261/rna.418407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu M, Tinoco I., Jr Proc Natl Acad Sci USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Hashimi HM, Pitt SW, Majumdar A, Xu W, Patel DJ. J Mol Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maderia M, Hunsicker LM, DeRose VJ. Biochemistry. 2000;39:12113–12120. doi: 10.1021/bi001249w. [DOI] [PubMed] [Google Scholar]
- 56.Butcher SE, Allain FH, Feigon J. Biochemistry. 2000;39:2174–2182. doi: 10.1021/bi9923454. [DOI] [PubMed] [Google Scholar]
- 57.Noeske J, Schwalbe H, Wohnert J. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reid SS, Cowan JA. Biochemistry. 1990;29:6025–6032. doi: 10.1021/bi00477a021. [DOI] [PubMed] [Google Scholar]
- 59.Grant CV, Frydman V, Frydman L. J Am Chem Soc. 2000;122:11743–11744. [Google Scholar]
- 60.Grant CV, Frydman V, Harwood JS, Frydman L. J Am Chem Soc. 2002;124:4458–4462. doi: 10.1021/ja012353j. [DOI] [PubMed] [Google Scholar]
- 61.Marincola FC, Casu M, Saba G, Manetti C, Lai A. Phys Chem Chem Phys. 2000;2:2425–2428. [Google Scholar]
- 62.Denisov VP, Halle B. Proc Natl Acad Sci USA. 2000;97:629–633. doi: 10.1073/pnas.97.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marincola FC, Denisov VP, Halle B. J Am Chem Soc. 2004;126:6739–6750. doi: 10.1021/ja049930z. [DOI] [PubMed] [Google Scholar]
- 64.Gill ML, Strobel SA, Loria JP. J Am Chem Soc. 2005;127:16723–16732. doi: 10.1021/ja055358f. [DOI] [PubMed] [Google Scholar]
- 65.Hunsicker-Wang LM. Ph.D. Thesis. Texas A&M University; 2001. [Google Scholar]
- 66.Kayne MS, Cohn M. Biochemistry. 1974;13:4159–4165. doi: 10.1021/bi00717a014. [DOI] [PubMed] [Google Scholar]
- 67.Draper DE. Biophys Chem. 1985;21:91–101. doi: 10.1016/0301-4622(85)85011-0. [DOI] [PubMed] [Google Scholar]
- 68.Greenbaum NL, Mundoma C, Peterman DR. Biochemistry. 2001;40:1124–1134. doi: 10.1021/bi002210u. [DOI] [PubMed] [Google Scholar]
- 69.Yuan F, Griffin L, Phelps L, Buschmann V, Weston K, Greenbaum NL. Nucleic Acids Res. 2007;35:2833–2845. doi: 10.1093/nar/gkm134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feig AL, Panek M, Horrocks WD, Jr, Uhlenbeck OC. Chem Biol. 1999;6:801–810. doi: 10.1016/s1074-5521(99)80127-6. [DOI] [PubMed] [Google Scholar]
- 71.Walter NG, Yang N, Burke JM. J Mol Biol. 2000;298:539–555. doi: 10.1006/jmbi.2000.3691. [DOI] [PubMed] [Google Scholar]
- 72.Tinsley RA, Walter NG. Biol Chem. 2007;388:705–715. doi: 10.1515/BC.2007.088. [DOI] [PubMed] [Google Scholar]
- 73.Gong B, Chen Y, Christian EL, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. J Am Chem Soc. 2008;130:9670–9672. doi: 10.1021/ja801861s. [DOI] [PubMed] [Google Scholar]
- 74.Chin K, Sharp KA, Honig B, Pyle AM. Nat Struct Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- 75.Hermann T, Westhof E. Structure. 1998;6:1303–1314. doi: 10.1016/s0969-2126(98)00130-0. [DOI] [PubMed] [Google Scholar]
- 76.Bagley SC, Altman RB. Protein Sci. 1995;4:622–635. doi: 10.1002/pro.5560040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banatao DR, Altman RB, Klein TE. Nucleic Acids Res. 2003;31:4450–4460. doi: 10.1093/nar/gkg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Castagnetto JM, Hennessy SW, Roberts VA, Getzoff ED, Tainer JA, Pique ME. Nucleic Acids Res. 2002;30:379–382. doi: 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stefan LR, Zhang R, Levitan AG, Hendrix DK, Brenner SE, Holbrook SR. Nucleic Acids Res. 2006;34:D131–D134. doi: 10.1093/nar/gkj058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klosterman PS, Tamura M, Holbrook SR, Brenner SE. Nucleic Acids Res. 2002;30:392–394. doi: 10.1093/nar/30.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sigel RK, Vaidya A, Pyle AM. Nat Struct Biol. 2000;7:1111–1116. doi: 10.1038/81958. [DOI] [PubMed] [Google Scholar]
- 82.Kaye NM, Zahler NH, Christian EL, Harris ME. J Mol Biol. 2002;324:429–442. doi: 10.1016/s0022-2836(02)01094-x. [DOI] [PubMed] [Google Scholar]
- 83.Harris DA, Tinsley RA, Walter NG. J Mol Biol. 2004;341:389–403. doi: 10.1016/j.jmb.2004.05.074. [DOI] [PubMed] [Google Scholar]
- 84.Hargittai MR, Musier-Forsyth K. RNA. 2000;6:1672–1680. doi: 10.1017/s135583820000128x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harris DA, Walter NG. Curr Protoc Nucleic Acid Chem. 2003;Chapter 6(Unit 6.8) doi: 10.1002/0471142700.nc0608s13. [DOI] [PubMed] [Google Scholar]
- 86.Harris ME, Cassano AG. Curr Opin Chem Biol. 2008;12:626–639. doi: 10.1016/j.cbpa.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buddenbaum WE, Shiner VJ. Can J Chem. 1976;54:1146–1161. [Google Scholar]
- 88.Paneth P, Oleary MH. J Am Chem Soc. 1991;113:1691–1693. [Google Scholar]
- 89.Hengge AC, Sowa GA, Wu L, Zhang ZY. Biochemistry. 1995;34:13982–13987. doi: 10.1021/bi00043a003. [DOI] [PubMed] [Google Scholar]
- 90.Hengge AC, Bruzik KS, Tobin AE, Cleland WW, Tsai MD. Bioorg Chem. 2000;28:119–133. doi: 10.1006/bioo.2000.1170. [DOI] [PubMed] [Google Scholar]
- 91.Gerratana B, Sowa GA, Cleland WW. J Am Chem Soc. 2000;122:12615–12621. [Google Scholar]
- 92.Cassano AG, Anderson VE, Harris ME. J Am Chem Soc. 2002;124:10964–10965. doi: 10.1021/ja020823j. [DOI] [PubMed] [Google Scholar]
- 93.Shih IH, Been MD. Proc Natl Acad Sci USA. 2001;98:1489–1494. doi: 10.1073/pnas.98.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith MD, Collins RA. Proc Natl Acad Sci USA. 2007;104:5818–5823. doi: 10.1073/pnas.0608864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rishavy MA, Hengge AC, Cleland WW. Bioorg Chem. 2000;28:283–292. doi: 10.1006/bioo.2000.1179. [DOI] [PubMed] [Google Scholar]
- 96.Humphry T, Forconi M, Williams NH, Hengge AC. J Am Chem Soc. 2002;124:14860–14861. doi: 10.1021/ja027671c. [DOI] [PubMed] [Google Scholar]
- 97.Rawlings J, Cleland WW, Hengge AC. J Inorg Biochem. 2003;93:61–65. doi: 10.1016/s0162-0134(02)00435-x. [DOI] [PubMed] [Google Scholar]
- 98.Humphry T, Forconi M, Williams NH, Hengge AC. J Am Chem Soc. 2004;126:11864–11869. doi: 10.1021/ja047110g. [DOI] [PubMed] [Google Scholar]
- 99.Yang MY, Iranzo O, Richard JP, Morrow JR. J Am Chem Soc. 2005;127:1064–1065. doi: 10.1021/ja044617i. [DOI] [PubMed] [Google Scholar]
- 100.Rawlings J, Cleland WW, Hengge AC. J Am Chem Soc. 2006;128:17120–17125. doi: 10.1021/ja065931a. [DOI] [PubMed] [Google Scholar]
- 101.Cassano AG, Anderson VE, Harris ME. Biochemistry. 2004;43:10547–10559. doi: 10.1021/bi049188f. [DOI] [PubMed] [Google Scholar]
- 102.Cassano AG, Anderson VE, Harris ME. Biopolymers. 2004;73:110–129. doi: 10.1002/bip.10517. [DOI] [PubMed] [Google Scholar]
- 103.Dai Q, Frederiksen JK, Anderson VE, Harris ME, Piccirilli JA. J Org Chem. 2008;73:309–311. doi: 10.1021/jo701727h. [DOI] [PubMed] [Google Scholar]
- 104.Zhong M, Strobel SA. J Org Chem. 2008;73:603–611. doi: 10.1021/jo702070m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ryder SP, Strobel SA. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- 106.Freisinger E, Sigel RKO. Coord Chem Rev. 2007;251:1834–1851. [Google Scholar]
- 107.Klein DJ, Moore PB, Steitz TA. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Christian EL, Yarus M. Biochemistry. 1993;32:4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- 109.Szewczak AA, Kosek AB, Piccirilli JA, Strobel SA. Biochemistry. 2002;41:2516–2525. doi: 10.1021/bi011973u. [DOI] [PubMed] [Google Scholar]
- 110.Hougland JL, Kravchuk AV, Herschlag D, Piccirilli JA. PLoS Biol. 2005;3:e277. doi: 10.1371/journal.pbio.0030277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Forconi M, Lee J, Lee JK, Piccirilli JA, Herschlag D. Biochemistry. 2008;47:6883–6894. doi: 10.1021/bi800519a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sarnovsky RJ, May EW, Craig NL. Embo J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 113.Allingham JS, Pribil PA, Haniford DB. J Mol Biol. 1999;289:1195–1206. doi: 10.1006/jmbi.1999.2837. [DOI] [PubMed] [Google Scholar]
- 114.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Genes Dev. 1999;13:3070–3380. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Landree MA, Wibbenmeyer JA, Roth DB. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gao K, Wong S, Bushman F. J Virol. 2004;78:6715–6722. doi: 10.1128/JVI.78.13.6715-6722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schwarzer D, Cole PA. Curr Opin Chem Biol. 2005;9:561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 118.Piccirilli JA, Vyle JS, Caruthers MH, Cech TR. Nature. 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 119.Sjögren AS, Pettersson E, Sjöberg BM, Strömberg R. Nucleic Acids Res. 1997;25:648–653. doi: 10.1093/nar/25.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weinstein LB, Jones BC, Cosstick R, Cech TR. Nature. 1997;388:805–808. doi: 10.1038/42076. [DOI] [PubMed] [Google Scholar]
- 121.Shan S, Yoshida A, Sun S, Piccirilli JA, Herschlag D. Proc Natl Acad Sci USA. 1999;96:12299–12304. doi: 10.1073/pnas.96.22.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shan SO, Herschlag D. Biochemistry. 1999;38:10958–10975. doi: 10.1021/bi990388e. [DOI] [PubMed] [Google Scholar]
- 123.Yoshida A, Sun S, Piccirilli JA. Nat Struct Biol. 1999;6:318–321. doi: 10.1038/7551. [DOI] [PubMed] [Google Scholar]
- 124.Shan S, Kravchuk AV, Piccirilli JA, Herschlag D. Biochemistry. 2001;40:5161–5171. doi: 10.1021/bi002887h. [DOI] [PubMed] [Google Scholar]
- 125.Sontheimer EJ, Gordon PM, Piccirilli JA. Genes Dev. 1999;13:1729–1741. doi: 10.1101/gad.13.13.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gordon PM, Sontheimer EJ, Piccirilli JA. Biochemistry. 2000;39:12939–12952. doi: 10.1021/bi001089o. [DOI] [PubMed] [Google Scholar]
- 127.Gordon PM, Piccirilli JA. Nat Struct Biol. 2001;8:893–898. doi: 10.1038/nsb1001-893. [DOI] [PubMed] [Google Scholar]
- 128.Gordon PM, Fong R, Piccirilli JA. Chem Biol. 2007;14:607–612. doi: 10.1016/j.chembiol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 129.Peracchi A, Beigelman L, Scott EC, Uhlenbeck OC, Herschlag D. J Biol Chem. 1997;272:26822–26826. doi: 10.1074/jbc.272.43.26822. [DOI] [PubMed] [Google Scholar]
- 130.Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- 131.Osborne EM, Schaak JE, DeRose VJ. RNA. 2005;11:187–196. doi: 10.1261/rna.7950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Warnecke JM, Furste JP, Hardt WD, Erdmann VA, Hartmann RK. Proc Natl Acad Sci USA. 1996;93:8924–8928. doi: 10.1073/pnas.93.17.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen Y, Li X, Gegenheimer P. Biochemistry. 1997;36:2425–2438. doi: 10.1021/bi9620464. [DOI] [PubMed] [Google Scholar]
- 134.Warnecke JM, Held R, Busch S, Hartmann RK. J Mol Biol. 1999;290:433–445. doi: 10.1006/jmbi.1999.2890. [DOI] [PubMed] [Google Scholar]
- 135.Pfeiffer T, Tekos A, Warnecke JM, Drainas D, Engelke DR, Seraphin B, Hartmann RK. J Mol Biol. 2000;298:559–565. doi: 10.1006/jmbi.2000.3655. [DOI] [PubMed] [Google Scholar]
- 136.Christian EL, Kaye NM, Harris ME. RNA. 2000;6:511–519. doi: 10.1017/s1355838200000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Christian EL, Kaye NM, Harris ME. EMBO J. 2002;21:2253–2262. doi: 10.1093/emboj/21.9.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Persson T, Cuzic S, Hartmann RK. J Biol Chem. 2003;278:43394–43401. doi: 10.1074/jbc.M305939200. [DOI] [PubMed] [Google Scholar]
- 139.Christian EL, Smith KM, Perera N, Harris ME. RNA. 2006;12:1463–1467. doi: 10.1261/rna.158606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sontheimer EJ, Sun S, Piccirilli JA. Nature. 1997;388:801–805. doi: 10.1038/42068. [DOI] [PubMed] [Google Scholar]
- 141.Gordon PM, Sontheimer EJ, Piccirilli JA. RNA. 2000;6:199–205. doi: 10.1017/s1355838200992069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yean SL, Wuenschell G, Termini J, Lin RJ. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pecoraro VL, Hermes JD, Cleland WW. Biochemistry. 1984;23:5262–5271. doi: 10.1021/bi00317a026. [DOI] [PubMed] [Google Scholar]
- 144.Frey PA, Sammons RD. Science. 1985;228:541–545. doi: 10.1126/science.2984773. [DOI] [PubMed] [Google Scholar]
- 145.Rablen PR, Lockman JW, Jorgensen WL. J Phys Chem A. 1998;102:3782–3797. [Google Scholar]
- 146.Brautigam CA, Steitz TA. J Mol Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- 147.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 148.Zaug AJ, Been MD, Cech TR. Nature. 1986;324:429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- 149.Grosshans CA, Cech TR. Biochemistry. 1989;28:6888–6894. doi: 10.1021/bi00443a017. [DOI] [PubMed] [Google Scholar]
- 150.Bevilacqua PC, Kierzek R, Johnson KA, Turner DH. Science. 1992;258:1355–1358. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- 151.Herschlag D. Biochemistry. 1992;31:1386–1399. doi: 10.1021/bi00120a015. [DOI] [PubMed] [Google Scholar]
- 152.Herschlag D, Khosla M. Biochemistry. 1994;33:5291–5297. doi: 10.1021/bi00183a036. [DOI] [PubMed] [Google Scholar]
- 153.Cosstick R, Vyle JS. Nucleic Acids Res. 1990;18:829–835. doi: 10.1093/nar/18.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vyle JS, Connolly BA, Kemp D, Cosstick R. Biochemistry. 1992;31:3012–3018. doi: 10.1021/bi00126a024. [DOI] [PubMed] [Google Scholar]
- 155.Herschlag D, Cech TR. Nature. 1990;344:405–409. doi: 10.1038/344405a0. [DOI] [PubMed] [Google Scholar]
- 156.Shan SO, Herschlag D. RNA. 2000;6:795–813. doi: 10.1017/s1355838200000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Steitz TA, Steitz JA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Freemont PS, Friedman JM, Beese LS, Sanderson MR, Steitz TA. Proc Natl Acad Sci USA. 1988;85:8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Beese LS, Steitz TA. Embo J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kim EE, Wyckoff HW. J Mol Biol. 1991;218:449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- 161.Dantzman CL, Kiessling LL. J Am Chem Soc. 1996;118:11715–11719. [Google Scholar]
- 162.Bass BL, Cech TR. Nature. 1984;308:820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- 163.McConnell TS, Cech TR. Biochemistry. 1995;34:4056–4067. doi: 10.1021/bi00012a024. [DOI] [PubMed] [Google Scholar]
- 164.Gott JM, Shub DA, Belfort M. Cell. 1986;47:81–87. doi: 10.1016/0092-8674(86)90368-5. [DOI] [PubMed] [Google Scholar]
- 165.Stromberg R, Hahne S, Sjogren AS, Sjoberg BM. Biochem Biophys Res Commun. 1992;183:842–848. doi: 10.1016/0006-291x(92)90560-8. [DOI] [PubMed] [Google Scholar]
- 166.Shan SO, Herschlag D. RNA. 2002;8:861–872. doi: 10.1017/s1355838202020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Herschlag D, Piccirilli JA, Cech TR. Biochemistry. 1991;30:4844–4854. doi: 10.1021/bi00234a003. [DOI] [PubMed] [Google Scholar]
- 168.Michel F, Westhof E. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 169.Ortoleva-Donnelly L, Szewczak AA, Gutell RR, Strobel SA. RNA. 1998;4:498–519. doi: 10.1017/s1355838298980086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Strauss-Soukup JK, Strobel SA. J Mol Biol. 2000;302:339–358. doi: 10.1006/jmbi.2000.4056. [DOI] [PubMed] [Google Scholar]
- 171.Moore MJ, Sharp PA. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 172.Forconi M, Lee J, Hougland JL, Piccirilli JA, Herschlag D. unpublished. [Google Scholar]
- 173.Guo F, Cech TR. Nat Struct Biol. 2002;9:855–861. doi: 10.1038/nsb850. [DOI] [PubMed] [Google Scholar]
- 174.Evans D, Marquez SM, Pace NR. Trends Biochem Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 175.Kazantsev AV, Pace NR. Nat Rev Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- 176.Wang MJ, Davis NW, Gegenheimer P. EMBO J. 1988;7:1567–1574. doi: 10.1002/j.1460-2075.1988.tb02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 178.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. Proc Natl Acad Sci USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Beebe JA, Kurz JC, Fierke CA. Biochemistry. 1996;35:10493–10505. doi: 10.1021/bi960870m. [DOI] [PubMed] [Google Scholar]
- 180.Smith D, Pace NR. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- 181.Kahle D, Kust B, Krupp G. Biochimie. 1993;75:955–962. doi: 10.1016/0300-9084(93)90145-i. [DOI] [PubMed] [Google Scholar]
- 182.Warnecke JM, Green CJ, Hartmann RK. Nucleosides & Nucleotides. 1997;16:721–725. [Google Scholar]
- 183.Warnecke JM, Sontheimer EJ, Piccirilli JA, Hartmann RK. Nucleic Acids Res. 2000;28:720–727. doi: 10.1093/nar/28.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Loria A, Pan T. Biochemistry. 1998;37:10126–10133. doi: 10.1021/bi980220d. [DOI] [PubMed] [Google Scholar]
- 185.Chen JL, Pace NR. RNA. 1997;3:557–560. [PMC free article] [PubMed] [Google Scholar]
- 186.Harris ME, Pace NR. RNA. 1995;1:210–218. [PMC free article] [PubMed] [Google Scholar]
- 187.Kaye NM, Christian EL, Harris ME. Biochemistry. 2002;41:4533–4545. doi: 10.1021/bi012158h. [DOI] [PubMed] [Google Scholar]
- 188.Crary SM, Kurz JC, Fierke CA. RNA. 2002;8:933–947. doi: 10.1017/s1355838202025025. [DOI] [PMC free article] [PubMed] [Google Scholar]