Abstract
The National Toxicology Program is developing a high throughput screening (HTS) program to set testing priorities for compounds of interest, to identify mechanisms of action, and potentially to develop predictive models for human toxicity. This program will generate extensive data on the activity of large numbers of chemicals in a wide variety of biochemical-and cell-based assays. The first step in relating patterns of response among batteries of HTS assays to in vivo toxicity is to distinguish between positive and negative compounds in individual assays. Here, we report on a statistical approach developed to identify compounds positive or negative in a HTS cytotoxicity assay based on data collected from screening 1353 compounds for concentration-response effects in nine human and four rodent cell types. In this approach, we develop methods to normalize the data (removing bias due to the location of the compound on the 1536-well plates used in the assay) and to analyze for concentration-response relationships. Various statistical tests for identifying significant concentration-response relationships and for addressing reproducibility are developed and presented.
Keywords: high-throughput screening, dose-response, statistical modeling, viability assay
Introduction
Over the last two decades, scientists have increasingly studied critical cellular and molecular events (mechanisms) that lead to adverse responses to toxicants in animals, including humans. Mechanistic information enhances interpretation of, but does not currently replace, traditional approaches to toxicological evaluations that are the basis for most decisions related to product safety, environmental and occupational hazard assessments, and priority setting for detailed toxicity testing. Mechanistic information may also be useful in the identification of biomarkers of exposure and effect that facilitate the linkage between laboratory research and human risk. Improving the quality, quantity and utility of mechanistic knowledge is a major focus of the National Institutes of Health (NIH) and the National Toxicology Program (NTP). The use of mechanistic approaches in toxicology assessments requires a systematic and continuing evaluation of the data derived from these new approaches to determine their value in providing improved information for making public health decisions.
The development of a class designation for a group of compounds that will be handled similarly in a risk assessment can sometimes be based upon the identification of a common mechanistic target. To identify one or more mechanistic targets, approaches must be developed for screening a variety of compounds that can be used to detect members of the appropriate compound class. Data on mechanistic targets that are available for a large number of compounds through HTS assays are important in defining what types of mechanistic endpoints can truly define a class. In addition, our knowledge about key molecular targets that we know are related to chemically-induced disease (e.g. a xenobiotic in appropriately binding to the estrogen receptors) can be a guide to choosing a common target for a given disease linked to the class. Finally, high content tools that can evaluate a large number of possible mechanistic targets for a few compounds (e.g., toxicogenomics, proteomics) can help to identify single targets that work with a few compounds or even groups of targets that work in unison and link a group of chemicals to a toxicity pathway that is associated with a disease or disease process.
As part of the Molecular Libraries Initiative of the NIH Roadmap, NIH established a program to use HTS assays as a means to study the ability of a large number of small molecules to target key pathways and processes in cells as a means to advance our basic understanding of molecular biology, cellular processes and disease intervention and cure (http://mli.nih.gov/mli). As part of the NTP Roadmap8, the NTP plans to develop HTS as a means to address the development of activity clusters for compounds and to set testing priorities. The NIH Chemical Genomics Center (NCGC) leads this roadmap initiative for the NIH and has partnered with the NTP to expand these activities into HTS screening for compounds of toxicological concern.
The approach being taken by the NTP and the NCGC focuses on quantitative HTS, which uses multiple concentrations of each compound under study in order to define its concentration-response curve in biochemical- and cell-based HTS assays. The analysis of these data is novel and critical to the eventual interpretation of the results. Here, we present an approach for analyzing HTS data, including a test for significance of response, a test for quality of fit, estimates of potency and functional parameters of the concentration-response curve, and a method for categorizing the compounds into different activity classes.
Materials and Methods
Data
Data were obtained on the extent of cytotoxicity induced in 13 different cell types by 1353 compounds 11 as part of the joint HTS screening effort of the NCGC and the NTP. Assays were conducted using1536-well plates, with 18 plates per assay. The first two and last two plates served as vehicle control plates (i.e., plates inoculated with cells where dimethyl sulfoxide (DMSO), the vehicle used to solubilize the test compounds, was administered to each well). In the 1st and 17th plates, the wells used for test compounds contained DMSO at the concentration used for the compound samples in other plates, while those wells in the 2nd and 18th plates contained twice that concentration of DMSO (i.e., they were “double-dip” control plates). The 3rd through 16th plates contain increasing concentrations of the test compounds (concentration approximately doubling at each step), obtained through successive dilutions of the maximum concentration of 46 μM. The 16th plate received two dispenses from the highest concentration stock compound plate, thus doubling the DMSO concentration in that well and producing a final compound concentration of 92 μM. The compound concentrations on plates 3–16 were, therefore, 0.59 nM, 2.95 nM, 14.8 nM, 33 nM, 74 nM, 0.165 μM, 0.369 μM, 0.824 μM, 1.84 μM, 4.12 μM, 9.22 μM, 20.6 μM, 46μM, and 92 μM.
Compounds for screening were placed in the 5th through 48th columns of the 32 row by 48 column plate allowing for 1408 total wells (1353 individual compounds where 1298 are tested using a single well and 55 are tested using replicate wells). Three plate formats were used for the first four columns. For most of the assays, the wells of the first two columns contain concentrations of doxorubicin and tamoxifen, respectively, while the wells of the 3rd column contain the vehicle control (DMSO or DMSO double dip as appropriate). The wells of the 4th column contain 100 μM tamoxifen as a positive control. This will be referred to as “Format 1” below. For the HepG2 assay (done in triplicate to test replicability of the assay), the wells of the 3rd column is the positive control and the wells of the 4th contain DMSO at the appropriate concentration. This will be referred to as “Format 2.” In the Jurkat cell assay, doxorubicin is in the 1st and 2nd columns and tamoxifen is in the 3rd and 4th columns of the first 24 rows of the plate. On those plates, vehicle controls are in the 1st through 4th columns of row 25 through 32 and there is no tamoxifen 100 μM positive control. This will be referred to as “Format 3.”
The CellTiter-Glo® luminescent cell viability assay (Promega Corporation, Madison, WI, USA) is a homogeneous method to measure the number of viable cells in culture and was used to screen 13 cell lines exposed to the test substances. The readout from this assay is based on quantitation of intracellular ATP, an indicator of metabolic activity, using the luciferase reaction. Luciferase catalyzes the oxidation of beetle luciferin to oxyluciferin and light in the presence of ATP. The luminescent signal is proportional to the amount of ATP present. The 13 cell lines tested include human cells (HEK 293, HepG2, SH-SY5Y, Jurkat, BJ, HUV-EC-C, MRC-5, SK-N-SH, mesenchymal cells), rat cells (primary renal proximal tubule cells, H-4-II-E) and murine cells (N2a, NIH 3T3). Details described in 11 and the normalized data are available for download from PubChem (http://pubchem.ncbi.nlm.nih.gov).
It is possible that double-dipping of the vehicle control may have an effect on the measured response. Also, there may be contamination of some amount of the study substances in the final single-dip control plate. For these reasons, this concentration-response analysis is carried out using only the first control (plate 1) and the dosed plates (including the double-dip highest concentration, plate 16). This yields a total of 15 plates in the analysis, a control plate followed by 14 plates with different concentrations of the test substances. Each plate has its own control wells and these will be used to define control response for each plate, hence the loss of the other control plates should not significantly alter the findings.
Hill function
The response for each compound is the measured luminescence, which is an indicator of ATP levels, which decreases from control if the chemical reduces cell viability and/or the rate at which cells proliferate. Many authors analyze data of this type using rescaled values (e.g., control response is 0, maximum reduction is −100%), however, we prefer an approach using the original data directly without controlling for minor variances in assay protocol like plate incubation times and detector exposure. Mean concentration-response curves for data relating to complex cellular response following chemical exposure usually follows a sigmoidal form and has been routinely analyzed using the Hill model4,7,9. Several authors have also incorporated them in the context of HTS2,3. Hill functions can take on a number of different forms; for these analyses, we used the form:
| (1) |
where i and j refer to the row and column of a plate and define the unique compound in that position, l refers to one plate, r0ijl is the control response for the chemical at (i,j), rpijl is the lowest possible activity at (i,j,l) corresponding to the high-concentration positive control, vij is the maximum fractional reduction caused by the test compound (restricted so that f is between 0 and −1 for decreasing effects, 0 and 0.1 for increasing effects, which were not as strong as the decreasing effects due to cytotoxicity of the study substances), nij is the Hill coefficient and governs the shape of the concentration response curve (e.g. a curve with a large value for n is highly sigmoidal), and kij is the concentration at which 50% of the maximum reduction occurs (the AC50). Because of the experimental design, each concentration level corresponds to a plate l but a separate symbol is used for plate vs. concentration to distinguish effects based on concentration-response from those based on plate location.
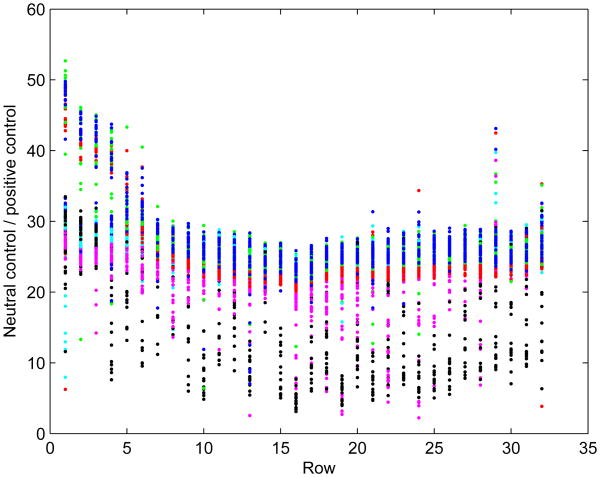
Examination of the results from control columns (columns 3 and 4) of plates in formats 1 and 2 suggests a row-dependent relationship between the values for the vehicle controls (column 3 in format 1, column 4 in format 2) and the positive controls (the other control column). This occurs in all assays, although the exact relationship varies and in some cases the difference between rows is small. In this analysis, it is assumed that the row-dependent ratio between vehicle controls and positive controls is consistent across all columns of the plate, i.e. that there is no column-dependent effect on the ratio of neutral to positive controls. If that is the case, then the positive control value rp in Eq.(1) can be treated as a function of the vehicle control value r0 and it will not need to be an optimized parameter itself. The pattern observed, with the ratio lowest in the middle, implies that the measured activity for positive controls is highest near the middle of the plate (Figure 1). In the model, this behavior was approximated with a V-shaped function, giving the following relationship for the ratio μi=r0ijl/rpijl over all j and l:
| (2) |
where m is a vector of length 32 of the form m=[16,15,14, …, 1,1,2,3, …, 16], θ1 and θ2 are parameters estimated via simple least squares and μ is a vector of length 32 containing the adjusted ratios [μ1, μ2,…, μ32]. See the supplemental material for more information on the calculation of μ. For most of the assays, the ratio between the highest and lowest values of μ is below 1.3, although it does go as high as 2.4 for one assay. With this modification, the Hill function becomes:
| (3) |
Figure 1.
Ratio of vehicle controls to positive controls. Each point represents one row on one plate. Data from the same assay are plotted in the same color.
If there is no row-dependent relationship in the ratio of the vehicle control to the positive control ratio, θ2 will be estimated as 0.
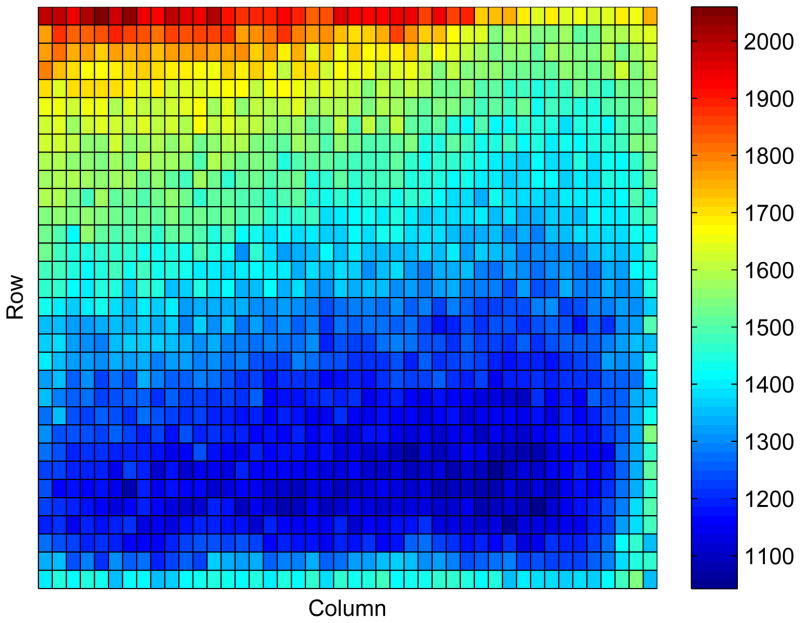
In HTS, there is a tendency to have location effects on the plates5. Figure 2 shows the location effects on one control plate. The observed data values are not randomly distributed around a constant value, but show consistent variation by row and column. Several analytical methods have been proposed to adjust for this effect6,10, but these are generally done prior to the analysis rather than as part of the formal analysis. In some cases, also, the adjustment does not completely remove location effects. Here, we adjust for plate-location effects by modeling the value for r0ijl. Preliminary analyses suggested that the control response, r0ijl, can be represented as a product of a row effect and a column effect:
| (4) |
where αil is the row effect for row i in plate l and γjl is the column effect for column j in plate l. The row-dependent effect on the control response represented by αil is distinct from the row-dependent relationship between the vehicle and positive control activity levels represented by μi. An additive rather than a multiplicative model can also fit the data on the control plates fairly well. The choice between the additive and multiplicative models is somewhat arbitrary. The use of r0ijl in this general form (4) for the test substances results in a linear dependence amongst the model parameters. Without loss of generality, we force the adjustment to be relative to appropriate control responses by setting γjl =1 for the column(s) containing vehicle controls. Given this model, a data value xijl can be converted to a normalized value yijl by the following formula:
| (5) |
Figure 2.
Raw data from columns 5 through 48 of the first control plate for BJ cells. Color corresponds to value of data, with blue the lowest and red the highest.
A normalized value of 0 corresponds to no response; a value of −1 corresponds to full suppression of activity. Because of noise and measurement error, normalized values can be greater than 0 or less than −1.
Estimation of Model Parameters
The observations are assumed to have normal error:
| (6) |
where ε ~ N (0,σ2) (A model using lognormal error instead of normal was also tested but gave significantly worse results). The parameters are constrained as follows: −0.1≤vij≤1, 0≤kij≤0.368 mM (4 times the maximum experimental concentration), 0≤nij≤5, and 0.9≤γjl≤1.1 for all i, j, and l. The limits on αik are determined by observations of the data; the lower limit for αil is 0.9 times the lowest control response on plate l and the upper limit is 1.1 times the highest control response. The limits on γjl are kept narrow so that most of the variation in the control response is contained in the αil term. The upper limit on v is 1 because the maximum possible effect is full suppression of activity (corresponding to v=1). The lower limit on v is somewhat arbitrary, but a limit of −0.1 seems in practice to be enough to account for increasing effects. See below for discussion of how often the optimized parameter values are at the limits of their ranges.
Each analysis involves 15 plates with 14 treatment levels for 1408 chemical wells and 32 untreated control wells yielding a total of 21,600 data values for each run (for the assay with format 3 plates, there are 21,360 data values). There are 32×15=480 αil parameters which will be collected into a matrix denoted by α=[αil]. Using similar notation, there are 44×15=660 γjl’s in γ, 32×44=1408 nij’s in n, 1408 vij’s and kij’s, and a single value for σ. The values of the adjustable parameters (α, γ, v, k, n) are found using a MATLAB@(The Mathworks, Inc., Natick, MA, USA 2007) least squares minimization routine (lsqnonlin) to maximize the log-likelihood1 of the data. While simultaneous optimization of the entire set of 5365 parameters is possible, it is not practical and a stratified estimation procedure is used, with repetition of the following steps:
Optimization by individual substance: α, γ, and σ are kept constant, while v, k, and n are optimized. With α and γ constant, the maximum of the likelihood can be obtained through independent estimation of vij, kij, and nij for each test substance.
Optimization by plate, control-response parameters only: parameters other than α and γ are kept constant. For each plate l, the values of αil and γjl are estimated for all i, j.
Optimization by row, control-response parameters only: parameters other than α are kept constant. For each row i, the values of αil are estimated for all l.
Optimization by column, control-response parameters only: parameters other than γ are kept constant. For each column j, the values of γjl are estimated for all l.
Optimization by row: γ and σ are held constant, while α, v, k, and n are optimized. In this case, the maximum of the likelihood can be obtained through independent estimation of vij, kij, nij, and αil for each row (i) of test substances for all j and k.
Optimization by column: α and σ are held constant, while γ, v, k, and n are optimized. In this case, the maximum of the likelihood can be obtained through independent estimation of vij, kij, nij, and γjl for each column (j) of test substances for all i and l.
Reduction of parameter space: The significance of the concentration-response for each compound is tested (see below). If the p value for compound ij is greater than a preset cutoff value (in the results below, the cutoff is 0.05/1408), then vij is set to 0.
The steps are carried out in a specified order with repetition as follows:
Perform step 5 and step 6 to find a set of starting values for the parameters.
Perform steps 1–4; if the change in the log-likelihood after all 4 steps is less than a prespecified small value, δ, then go on to the third step; if not, repeat this step.
Perform steps 5 and 6; if the change in the log-likelihood is less than a prespecified δ, then go on to the next step; if not, return to the second step.
Do a final optimization for each substance individually (step 1).
Do step 7. If no new compounds are found to have nonsignificant concentration-response, stop here; otherwise, return to 2).
The order of steps is partly arbitrary, and partly chosen to improve optimization time by having more repetitions of the steps 1–4 (which are the quickest, since they use the fewest optimizable parameters) than of steps 5 and 6. Including step 7 helps to reduce the number of compounds that have significant but weak response. Those compounds may be false positives. Consequences of changing the cutoff value in step 7 are discussed in the supplemental material. When vij is fixed at 0, then none of the Hill function parameters (v, k, or n) for (i,j) needs to be optimized. In a least squares problem such as this, the standard deviation σ does not need to be computed during the optimization; instead, it is computed from the sum of squared errors using the final model parameter estimates.
Outliers
An initial optimization revealed some data points that seemed to be outliers; for example, there were cases where all of the observed activity levels were within 10% of the control level, except for one data point at a low concentration with almost no measured luciferase activity. Identifying and removing outliers is complex and does not readily lend itself to simple statistical rules. For this reason, we propose a combination of three activities to identify outliers: a statistical/quantitative rule, visual inspection of model fits, and examination of the normalized values directly.
Computer code was written to identify outliers based on the following criteria.
The difference between the data point xijk and the model fit must be >3.5 σ AND
The absolute value of the normalized value of the data must be greater than 0.3 AND
If a downward trend is present in the data for yijk(the normalized value of xijk) (i.e., yijk−1>yijk>yijk+1), then: xijk is not a low outlier unless at least one its neighbors (xijk−1 and xijk+1) is also a low outlier; it is not a high outlier unless one of its neighbors is also a high outlier.
These criteria were intended to be conservative and not identify too many outliers. A list of possible outliers was produced by the code and these data points were inspected visually in plots of normalized data vs. model fit. Points that seemed to satisfy the outlier criterion because of overall poor quality of fit rather than because they were isolated points of abnormally low or high response were excluded from the list of possible outliers.
Finally, plots of data values for each plate (in the same format as Figure 2) were also examined to find possible outliers not revealed by the above procedure. Several outliers were also identified in this way, all of them located in the corners of plates (at or near row 1, column 48, or row 32, column 48). These points had very low measured activity compared to the nearby wells on their plates, and their normalized data values represented very low responses in relation to the responses for the next higher and next lower doses. It is possible that functional form for the plate-location effects more complicated than that given in equation (4) would be able to normalize these points without treating them as outliers. However, because the measured responses at those points are so low, the error in the normalized data would probably be large. Overall, 126 data points (of 323,760; ~0.06%) were identified as outliers. The estimation procedure above was then redone with the reduced data.
Statistical Testing
A likelihood ratio test 1 is used to test each compound for a significant concentration-response trend. For the compound in position ij, the null hypothesis is that there is no concentration-response (vij=0); the alternative hypothesis is that vij≠0. The likelihoods are calculated using only the data for substance ij, using the standard deviation σ from the overall fit. The likelihoods for the unrestricted (alternative hypothesis) and restricted (null hypothesis) models are compared using a likelihood ratio test (chi squared with 3 degrees of freedom, corresponding to the 3 removed parameters vij, kij and nij) to produce a p-value. This calculation is also carried out using only the first 14 data points (i.e., excluding the data for the highest concentration) to determine whether the significance of the response is sensitive to the high concentration data point. This procedure for testing trend is an approximation. Since all model parameters for all substances are actually related in the optimization algorithm, an exact calculation of the p-value would require a full optimization of all model parameters after setting vij=0 for a particular substance. Therefore, the calculated p-value is an underestimate of the true p-value and an overestimate of the significance of the trend.
It is important to know whether replication in this HTS assay yields reproducible results. There are two types of replication in the data; 55 test substances are replicated within plates and the HepG2 assay was conducted in triplicate. A likelihood ratio test was used to compare the triplicate data. Under the null hypothesis, replicates were assumed to share equal values for common parameters (v, k, and n). The alternative hypothesis was that the parameter values are different among replicates. For duplicates, the values of k, the AC50 parameter, were compared.
When using p-values to evaluate results in a large group of tests such as this set of high throughput screening data, the issue of multiple comparisons becomes important. In most of the results discussed below, a “significant” p-value will be taken to be one with p<0.05/1408 (3.5511 × 10−5).
A simulation study was performed to test the performance of the algorithm on simulated data with known parameters. Details of the simulation are given in the supplemental material. Results from the simulation will be compared to those from the actual data when relevant.
Results
Normalization and quality of fit
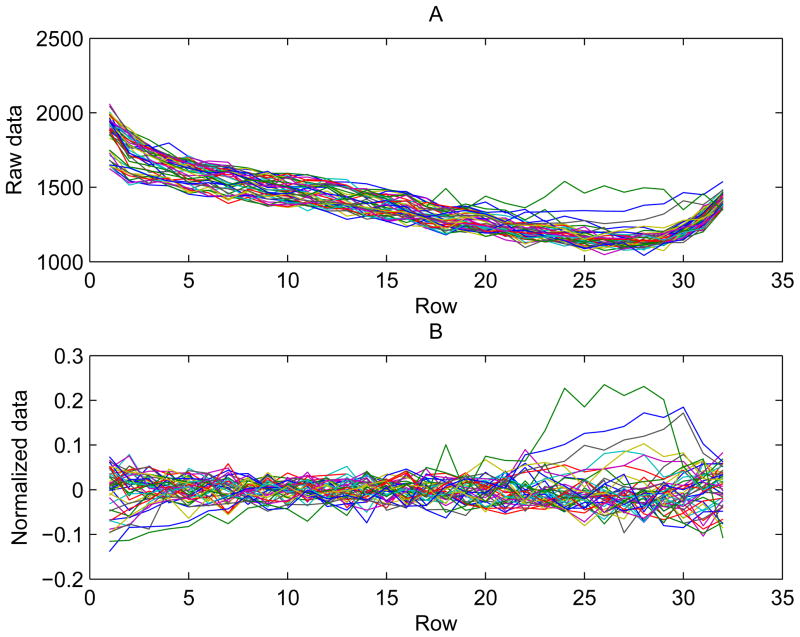
The normalized data range in value from −1.07 to 0.63, with one high outlier at a value of1. 35. Figure 3 illustrates how the normalization removes general trend across the plate for the same data used in Figure 2. The raw luminescence data (3-A) vary over a range of nearly twofold, with a very strong row effect. After normalization (3-B), the row effect has been almost completely removed. A measure of the variability of the data is the ratio of the fitted value σ to the mean of the raw data on the control plate. That ratio ranges from 0.028 to 0.0574 over all 15 assays, with a mean of 0.0389. The normalization by removal of row and column effects greatly reduces the variability of the data. The ratio of the standard deviation of the raw data on the control plate to its mean ranges from 0.043 to 0.143, with mean 0.081 over all assays. Figure 4 shows the raw and fitted values for columns 5 to 48 on the control plates. The fitted values for the control plates, with zero concentration of the study chemicals, are given by equation (4). For most points, the fit is good. Examination of the residuals by plate location (not shown) reveals that the residuals mostly show little to no trend with respect to row or column. However, there is a tendency for the absolute value of the residuals to be higher near the edges of the plate. This could be due to higher measurement error at the edges or to non-error effects that are not fully modeled by equation (4).
Figure 3.
(A) Raw and (B) normalized data from columns 5–48 of the control plate for BJ cells. Each colored line is one column of data.
Figure 4.
Raw data values from columns 5:48 of the control plates and the fitted values from the model (from equation (4)), for all plates.
The frequency at which fitted values of parameters were at the limits of their ranges from the optimization algorithm depended on the p-cutoff used in the parameter reduction step, decreasing as the cutoff became stricter. The parameter values most likely to reach their limits were v and n, which reached their upper limits 7.5% and 6.6% of the time, respectively. There is more discussion in the supplemental material.
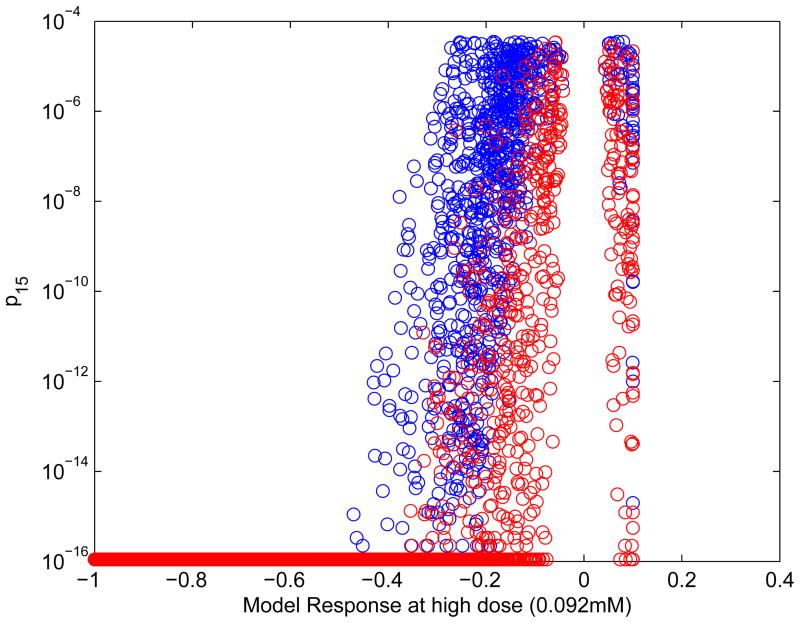
The significance of cytotoxic effects was evaluated using two statistical tests. In the first, all of the experimental concentrations were used; the second removed the highest concentration when trying to determine whether there was a significant concentration-response. Doing the test without the high-concentration data point helps to show whether a response is significant only because of a single data point. Figure 5 shows the p-values for the two tests (p15 and p14 for the 15 and 14 point tests respectively). As the modeled response at the highest concentration drops (−1 is complete loss of viability), the chances of seeing a significant response increase until finally every evaluation is positive for both 14 and 15 concentrations. Some of the results are significant when all concentration points are taken into account but not when the highest concentration is excluded (blue points). Over all 15 assays (including the triplicate HepG2 assays), 14% of concentration-response curves have simultaneously significant (with positive response) values of both p15 and p14; 6% have significant values for p15 but not p14; and 80.0% have nonsignificant values of both p15 and p14. When the value of p15 is below the significance level but p14 is not, the significance of the result depends on the one data point at the highest concentration; that data point has a small but real chance of being an outlier. If it is not, the p values suggest that response data at concentration levels between the two highest levels may be needed to better characterize the concentration-response curve.
Figure 5.
Plot of p-value (p15) based on all concentrations, with color coding based on both p15 and p14. Results plotted are from all assays, including triplicate HepG2 assays. Red: both p15 and p14 are significant. Blue: only p15 is significant. Model response is on the normalized scale. P values less than 10−16are plotted as 10−16. Values with model response 0 (i.e. v=0) and p15=1 are omitted.
As mentioned above, the likelihood ratio test used here overestimates the significance of the response. A better measure of concentration-response strength might be one similar to that used in Inglese et al. 3. In this article, the classification will be into three categories: active, inactive, and marginal. An active concentration-response curve is one satisfying these criteria: 1) the response is significant (p<0.05/1408) with or without the high-concentration data point; 2) the response is cytotoxic, = (i.e., the Hill parameter v>0); 3) the AC50 is below the highest concentration (i.e., the Hill function parameter k<0.092 mM); 4) the normalized response at the highest concentration is less than −0.1 (i.e., there is at least a 10% loss of viability). An inactive curve is one with v≤0 or p>0.05/1408. All other concentration-responses are marginal. With this classification, 11.4% of all the concentration-response curves are active, 80.0% are inactive, and 8.6% are marginal.
In any testing method whose, there is a tradeoff between sensitivity (the ability of the test to detect actual effects) and specificity (the ability of the test to detect lack of effect). Changing the criterion which the test classifies substances as having or lacking an effect will affect both the sensitivity and the specificity. The classification criterion for this method can be adjusted at two stages in the calculation. The p-value cutoff in the parameter reduction step can be changed, or the nominal significance level in the final test of significance can be changed. In this model, the cutoff for the parameter reduction step and the nominal significance level for the final test of significance are both set at p=0.05/1408. Using higher p-values at either stage (or omitting the parameter reduction step entirely by setting the p-cutoff to 1) will increase the number of substances classified as having a significant concentration-response. The supplemental material gives more detail on this effect. The main effect of increasing the p-cutoff for the parameter reduction step was to change the classification of substances from “no loss of viability” to “weak response, marginal or inactive”. The number of substances classified as active changes less when the p-cutoff was changed, probably because the criteria for an active classification are strict enough that they include mostly stronger responses which will be detected under any reasonable analysis method. Under the assumptions of the simulation study, the criteria used here were found to give a very high specificity (0.98) but a much lower sensitivity (0.53). The simulation found that using triplicated study data would give a great improvement in sensitivity (or, if less strict selection criteria were used, an improvement in specificity and a smaller improvement in sensitivity). Details are in the supplemental material.
The plate effects in this model are represented as the product of a row effect and a column effect, with the row and column effects varying by plate. Another possibility for removing the plate effects would be to use the control plates at the beginning and end of the run to generate corrections, as was done, for example, in a previous analysis of these data11. One way to do such a correction would be to calculate the plate parameters α and γ for the initial and final plates and then interpolate to find the values for intermediate plates. This approach was also tried and the model was found to give a significantly worse fit for all but one of the assays, although examination of the values of α and γ from the current analysis shows that there is a rough trend in their values across plates. Nevertheless, fitting the Hill function model to the normalized data used in the previous analysis gives very similar results to those from the algorithm used in this paper. This is discussed further in the supplemental material.
Duplicate substances
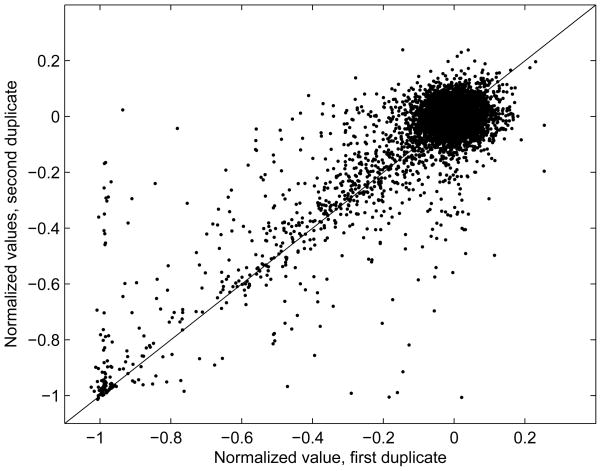
Of the 1353 substances tested, 55 were tested in duplicate. Comparing the data and the model results for the duplicated substances provides an indication of the reproducibility of the test results. Figure 6 compares the two normalized data values from a pair of duplicates. 91% of the duplicate values differ by less than 0.1, but there are some values for which the two duplicates are very different. Examination of individual concentration-response profiles showed varying patterns of difference between duplicates. In many cases, the responses at the highest concentration are roughly similar for the two duplicates, both being near full loss of viability, while the responses at lower concentrations vary. In many cases the duplicate compounds were drawn from two lots from different suppliers, which may be reflected in the results. It is difficult to tell from the current data what the sources of variability in the duplicated measurements are. The current model allows only two sources of variability: plate location effects and normally-distributed noise. It would be possible to use a model with a more complicated error model. For example, the random error could be dependent on the slope of the concentration-response curve, which would correspond to uncertainty in the concentration of the administered substances. However, the optimization for such a model would be considerably more difficult.
Figure 6.
Normalized data values for duplicate substances on all plates used in the model, using all 15 concentration levels. X axis is the normalized data value for first occurrence of the duplicate, Y axis is the value for the second occurrence.
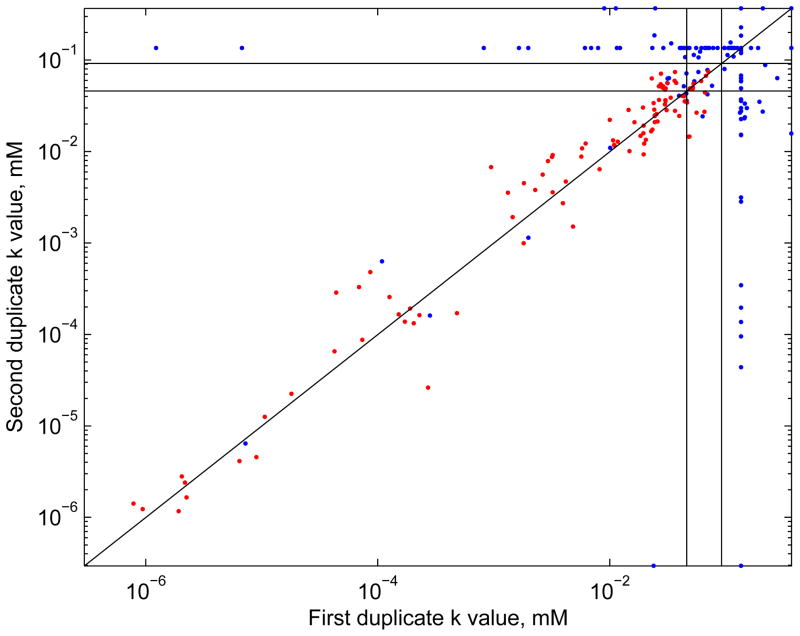
A major factor describing the potency of a compound in these assays using the Hill model for analysis is the AC50 (i.e., k). Concentration-response curves with identical maximum responses (v) can have very different AC50 values if their responses at low concentrations are different. In general there is considerable variation in the values for k between duplicates (Figure 7). Luckily, much of this disagreement is for compounds with non-significant concentration-response curves, since compounds for which both duplicates show significant concentration-response have more consistent values of k (red data points in Figure 7). In those cases, the k values seem to match more closely for compounds with low k values, possibly because of the closer spacing of exposures in the low-concentration part of the exposure range. Table 1 shows Pearson correlation coefficients for several variables between duplicates for the duplicate pairs for which both members of the pair were active. Of the 825 (55×15) duplicate pairs, 612 had neither duplicate significant and positive, 67 had only one significant, and 146 had both significant, so 92% of the pairs agreed in significance between duplicates. Using the activity classification, 677/825 had neither duplicate active, 103 had both active, and 45 had only one active, so 94.5% agreed in classification of active vs. marginal or inactive.
Figure 7.
Values of the Hill parameter k by duplicate. Red: both duplicate concentration-response curves for the duplicated substance in the given assay are active. Blue: neither, or only one, is active. The vertical and horizontal lines are at the location of the two highest concentrations (0.046 and 0.092 mM). Cluster of k values along vertical or horizontal lines correspond to k values set to an arbitrary constant when v is fixed at 0 in step 7 of the algorithm; in that case, the values of k are not meaningful.
Table 1.
Pearson correlation coefficients between parameters in sets of duplicates for all duplicate pairs in which both duplicates are active (103 pairs) or in which both duplicates are active and have at least 50% loss of viability (68 pairs)
| Parameter | Correlation, both active | Correlation, both active, >50% |
|---|---|---|
| k | 0.79 | 0.76 |
| v | 0.91 | 0.83 |
| n | 0.73 | 0.84 |
| Normalized model response at highest concentration (0.092 mM) | 0.93 | 0.79 |
When the concentration-response data are nearly linear, there is considerable dependence among the parameters, because the noise and measurement error in the data produce uncertainty as to the true shape of the concentration-response curve. This results in uncertainty in the value of the parameters, which may partly explain why the correlation between high-concentration responses is greater than that between actual parameters in Table 1 when looking at pairs with both substances active. When the criterion for choosing the pairs is stricter, requiring both activity and a >50% response, the correlations for the variables are more similar. These results resemble those seen in a simulation study (see the supplemental material). A concordance was computed to compare values of the AC50 parameter k between duplicates. Values of k were compared for either the duplicate pairs with at least one active or for pairs with both active. The parameter values k were considered to match if the ratio of the larger k to the smaller was less than a cutoff value, either 2 or 10. They were considered to mismatch if the ratio was larger than the cutoff value or if only one duplicate was active. The concordance equals the fraction of matching k values in the whole set of k values. For a cutoff of 2, the concordance is 0.54 for pairs with at least one active, and 0.78 for pairs with both active; for a cutoff of 10, the concordance is 0.69 for pairs with at least one active and 0.99 for pairs with both active.
Triplicate HepG2 data
The assay for HepG2 cells was done in triplicate to test the reproducibility of the assay. The p values varied among the replicates. Out of all 1408 substances (counting duplicates separately), 105 had active concentration-response curves in all 3 triplicates, 1187 did not have an active concentration-response in any of the triplicates, 69 were significant only for one replicate, and the other 47 were significant for two replicates. This means that 92% of the triplicates had the same active or inactive/marginal classification for all triplicates. The correlation coefficient of various parameters between each of 3 possible pairs of the triplicates for the cases in which all 3 triplicates were active was calculated as a measure of similarity. The correlation of v between triplicates ranged from 0.89 to 0.91. The AC50 (k) had lower correlation (0.73 to 0.85), as did the shape parameter n (0.59–0.81). The normalized data had higher correlations, especially at the highest concentrations. The correlations ranged between 0.93 and 0.95 for the data at the highest concentration and between 0.92 and 0.96 for the second highest concentration.
The results of the likelihood ratio test for parameter equivalence were examined on a pooled basis by considering all 1408 results as a common data set. Duplicated substances were considered as separate data points. The triplicate data were fitted to the Hill function model with all parameters constrained to be equal and with no constraints on the parameters. A likelihood ratio test was used to evaluate the null hypothesis that the triplicates could be described using the same parameters. Of the 105 triplicates where all three responses were active, 26% showed a significant difference in the parameter values (p<0.05/1408). It appears that the main difference between the triplicates is in the value of k. When the likelihood ratio test was carried out for a null hypothesis of all parameters constrained to be equal vs. an alternative hypothesis of only 2 constrained to be equal (and thus with 1 parameter allowed to vary), the null hypothesis was rejected 11% of the time when n was allowed to vary, 17% of time when v was allowed to vary, and 29% of the time when k was allowed to vary.
Substance behavior across all assays
Most compounds were inactive or marginal for most of the assays. 936 of the compounds were inactive or marginal in all 15 assays. 36 of the compounds were active in all 15 assays.
The concentration-response curves can also be classified by strength of response. For the following results, a “strong” curve is one with normalized modeled response at the highest concentration which is below −0.8. A “medium” curve is one with response between −0.3 and −0.8, and a “weak” curve is one with modeled response at the high concentration between 0 and −0.3. A “negative” curve is one which shows no loss of viability (i.e., the response is 0 or more [equivalently, the v parameter is 0 or less]). The classification of “negative” is stricter than the “inactive” classification above, which does not require that there be a complete lack of concentration-response. Of all concentration-response curves (15×1408=21,120) evaluated, 80.0% (16,898) are negative. 7.4% (1561) of the responses are weak. The majority of the weak responses (1161 of them) are inactive. The medium responses make up 7.2% (1520) of the results, with most of them (944) being active. 5.4% (1141) of the results are strong, with the vast majority, 1065 of them, being active. Because all of the strong-response curves were significant according to the likelihood ratio test, the only way a strong response could be not active was to be classified as marginal because the response was nonsignificant when tested without the high-concentration data point (i.e., strong non-actives are curves whose significance depends on the strength of the response at the highest concentration). Of the 576 medium-strength substances that are inactive, 376 are so because they are nonsignificant without the highest-concentration data point. Table 2 shows a breakdown of this classification by assay. Twelve of the 1408 compounds had strong active response for all 15 assays.
Table 2.
Classification of concentration-response curves by strength and activity classification for each assay and for all assays combined
| Assay | No loss of viability | Weak, not active | Weak, active | Medium, not active | Medium, active | Strong, not active | Strong, active |
|---|---|---|---|---|---|---|---|
| N2a | 1131 | 68 | 6 | 55 | 45 | 14 | 89 |
| HUV-EC-C | 1250 | 38 | 10 | 30 | 34 | 5 | 41 |
| NIH 3T3 | 1034 | 108 | 51 | 51 | 68 | 11 | 85 |
| H-4-IIE | 1028 | 115 | 27 | 52 | 82 | 6 | 98 |
| mesenchymal | 1150 | 81 | 49 | 30 | 44 | 6 | 48 |
| BJ | 1157 | 83 | 34 | 16 | 60 | 4 | 54 |
| Jurkat | 1023 | 104 | 30 | 51 | 72 | 1 | 127 |
| MRC-5 | 1204 | 62 | 16 | 22 | 39 | 10 | 55 |
| SK-N-SH | 1144 | 67 | 16 | 23 | 83 | 4 | 71 |
| HEK 293 | 1093 | 94 | 32 | 40 | 86 | 0 | 63 |
| SH-SY5Y | 1110 | 40 | 2 | 52 | 85 | 4 | 115 |
| Primary renal proximal tubule | 1176 | 72 | 14 | 39 | 45 | 7 | 55 |
| HepG2 #1 | 1135 | 83 | 40 | 22 | 72 | 1 | 55 |
| HepG2 #2 | 1118 | 81 | 42 | 41 | 65 | 1 | 60 |
| HepG2 #3 | 1145 | 65 | 31 | 52 | 64 | 2 | 49 |
| Total | 16898 | 1161 | 400 | 576 | 944 | 76 | 1065 |
Discussion
The algorithm used in this paper was able to fit the data well. The normalization was incorporated directly into the estimation of the model parameters, in contrast to previous work in the area of normalization of high-throughput screening data 6,10, and was able to remove unwanted plate location effects from the data. Because of the design of the experiments, the measured effect magnitude is determined by an interaction between concentration-response effects and plate effects. The optimization method, which includes steps that optimize parameters controlling both types of effects, is able to deal with that interaction. To determine whether there is activity for any given compound, we used several different statistical approaches linked with some common sense subjective evaluations. After looking at these approaches both in the real data and for simulated data, we have decided to use a stringent test that greatly reduces the number of false positives. A consequence of that choice is an increase in the number of false negatives. The choice of test in practice should be determined by the intended use of the results. The tests of significance of response, though not exact, showed that, as might be expected, concentration-response curves with stronger responses tended to be more significant than those with weaker responses (Figure 5).
When analyzing experimental data, it is important to know how reproducible the data are. The data used here allow that question to be addressed in two ways: by use of duplicated substances within an assay, and by use of triplicated assays for the HepG2 cells. After normalization, the data at high concentration levels were highly correlated between duplicates or between triplicates. The actual concentration-response parameter values were less highly correlated. Simulation studies suggest that this effect is due to the properties of the Hill function model applied to noisy data. The concentration-response was modeled with a Hill function, which uses three parameters to describe the concentration-response curve. The parameter v (strength of response) showed the greatest consistency across replicates which is not surprising given the strong correlation in the response at the highest concentrations. Tests of parameter equivalence between replicates showed that, while most sets of replicate data could be described by identical parameters, there were some that could not.
Another question raised by the experimental design is how to approach data whose effect significance is dependent on response at the highest concentration. As Table 2 shows, this situation occurs quite often for substances with a response of medium strength (maximum suppression of cell viability between 30% and 80%) but much less often for substances with a strong response. There is a possibility that a single strong response can be an outlier; however, since the number of outliers identified was small (less than one in 2000 data points), that possibility is low. When using screening as a way to identify substances for further testing, attention should be given to selecting concentration levels in such a way as to give information on parts of the concentration-response curve not well specified in the screening.
Simulation studies and comparison of this method’s results under varying classification criteria showed that there can be a high degree of confidence in the method’s determination that substances with a modeled strong response are true positives. There is more uncertainty as to the true status of substances with a weaker response. In those cases, the classification criteria determine whether the substance is considered to have no effect or a weak but still real effect. The simulation found that using replicated data can improve the classification. How strict the criteria need to be in practice and how much replication needs to be done depend on how the classification will be used.
Overall, these analyses support the concept that the current design envisioned by the NTP will be useful in developing a semi-quantitative metric to compare targeted response of a large number of compounds in a HTS framework.
Supplementary Material
Acknowledgments
We thank Menghang Xia for providing the datasets on which the analysis was based. This research was supported [in part] by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
References
- 1.Bard Y. Nonlinear Parameter Estimation. New York City: Academic Press; 1974. [Google Scholar]
- 2.Buxser S, Vroegop S. Calculating the probability of detection for inhibitors in enzymatic or binding reactions in high-throughput screening. Anal Biochem. 2005;340:1–13. doi: 10.1016/j.ab.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 3.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Nat Acad Sci USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohn MC, Portier CJ. Effects Of The Mechanism Of Receptor-Mediated Gene-Expression On The Shape Of The Dose-Response Curve. Risk Anal. 1993;13:565–572. doi: 10.1111/j.1539-6924.1993.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 5.Lundholt BK, Scudder KM, Pagliaro L. A simple technique for reducing edge effect in cell-based assays. J Biomol Screen. 2003;8:566–570. doi: 10.1177/1087057103256465. [DOI] [PubMed] [Google Scholar]
- 6.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 7.Mevissen M, Denac H, Schaad A, Portier CJ, Scholtysik G. Identification of a cardiac sodium channel insensitive to synthetic modulators. J Cardiovasc Pharmacol Ther. 2001;6:201–212. doi: 10.1177/107424840100600212. [DOI] [PubMed] [Google Scholar]
- 8.NTP. A roadmap to achieve the NTP vision. Research Triangle Park, NC: National Toxicology Program / National Institute of Environmental Health Sciences; 2004. A National Toxicology Program for the 21st Century. [Google Scholar]
- 9.Toyoshiba H, Walker NJ, Bailer AJ, Portier CJ. Evaluation of toxic equivalency factors for induction of cytochromes P450 CYP1A1 and CYP1A2 enzyme activity by dioxin-like compounds. Toxicol Appl Pharmacol. 2004;194:156–168. doi: 10.1016/j.taap.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Wu ZJ, Liu DM, Sui YX. Quantitative assessment of hit detection and confirmation in single and duplicate high-throughput screenings. J Biomol Screen. 2008;13:159–167. doi: 10.1177/1087057107312628. [DOI] [PubMed] [Google Scholar]
- 11.Xia M, Huang R, Witt KL, Southall N, Fostel J, Cho MH, Jadhav A, Smith CS, Inglese J, Portier CJ, Tice RR, Austin CP. Compound cytotoxicity profiling using quantitative high-throughput screening. Environmental Health Perspectives. 2008;116:284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.