Abstract
Genetic defects in breast cancer (BC) susceptibility genes, most importantly BRCA1 and BRCA2, account for ∼40% of hereditary BC and ovarian cancer (OC). Little is known about the contribution of constitutive (soma-wide) epimutations to the remaining cases. We developed bisulfite pyrosequencing assays to screen >600 affected BRCA1/BRCA2 mutation-negative patients from the German Consortium for Hereditary Breast and Ovarian Cancer for constitutive hypermethylation of ATM, BRCA1, BRCA2, RAD51C, PTEN and TP53 in blood cells. In a second step, patients with ≥6% promoter methylation were analyzed by bisulfite plasmid sequencing to demonstrate the presence of hypermethylated alleles (epimutations), indicative of epigenetic gene silencing. Altogether we identified nine (1.4%) patients with constitutive BRCA1 and three (0.5%) with RAD51C hypermethylation. Epimutations were found in both sporadic cases, in particular in 2 (5.5%) of 37 patients with early-onset BC, and familial cases, in particular 4 (10%) of 39 patients with OC. Hypermethylation was always confined to one of the two parental alleles in a subset (12–40%) of the analyzed cells. Because epimutations occurred in cell types from different embryonal layers, they most likely originated in single cells during early somatic development. We propose that analogous to germline genetic mutations constitutive epimutations may serve as the first hit of tumor development. Because the role of constitutive epimutations in cancer development is likely to be largely underestimated, future strategies for effective testing of susceptibility to BC and OC should include an epimutation screen.
INTRODUCTION
Breast cancer (BC) is a major cause of death in women worldwide and the leading cancer-related death among women aged 20–59 years (factsheet no. 297; http://www.who.int). Several percent, in particular, of familial and/or early-onset BC and ovarian cancer (OC) cases are caused by highly (to moderately) penetrant germline mutations in BRCA1, BRCA2 and other BC susceptibility genes (1). These tumor suppressor genes are important for maintaining genome integrity and cell cycle checkpoint control (2). In addition, numerous (>20) susceptibility loci with low penetrance may modify the risk for BC and OC (3). However, rare genetic mutations and common sequence variants alone cannot account for the majority of familial BC and/or OC cases. In this context, it is important to emphasize that dysregulation of DNA repair pathways and genomic instability, which are hallmarks of cancer cells, can also be achieved by epigenetic mechanisms (4,5). Epigenetic information is not encoded by the DNA sequence itself but by reversible modifications of DNA and/or histones, which can be transmitted from cells to daughter cells. In contrast to non-coding regions of the genome where most CpGs (the p refers to the phosphodiester bond between the nucleotides) are methylated, CpG islands in 5′ cis-regulatory regions of genes are usually unmethylated. Methylation of these CpG islands during the development or disease processes is associated with post-translational histone modifications that lead to a locally condensed inactive chromatin structure and gene silencing (6,7).
During tumorigenesis, there is a progressive loss of global DNA methylation and at the same time regional hypermethylation (8). Genome-wide demethylation of repetitive elements can lead to reactivation of retrotransposons and promote genome instability. Tumor-specific hypermethylation of CpG islands in 5′ promoters can inactivate genes for DNA repair, cell cycle control and other mechanisms that prevent neoplastic transformation in a normal cell (9). Epigenetic abnormalities do not only occur as secondary changes at all stages of tumor evolution, but can also act as initiating events (10). In sporadic tumors, epigenetic silencing of tumor suppressor genes can serve as the first and/or the second hit in Knudson's model of tumor development. Approximately 20% of sporadic BCs display hypermethylation of the BRCA1 promoter. These tumors are mainly estrogen receptor (ER)-, progesterone receptor (PR)- and HER2-negative and display similar pathological features as hereditary tumors with BRCA1 germline mutations (11–14). BRCA1 promoter methylation is also seen in a subset of OCs with poor prognosis (15–17). Somatic epimutations are restricted to the tumor tissue and precursor lesions. However, accumulating evidence suggests that constitutive epimutations, involving soma-wide hypermethylation of tumor suppressor genes in normal body cells, may increase cancer susceptibility. Constitutive epimutations in the DNA mismatch repair genes MLH1 and MSH2 have been identified in a small number of mutation-negative cases of hereditary non-polyposis colorectal cancer (HNPCC) (18–21). Constitutive epimutations in the DAPK1 gene predispose to B-cell chronic lymphocytic leukemia (22). We described a constitutive BRCA1 epimutation in the affected twin of a monozygotic pair discordant for childhood leukemia and secondary cancer (23).
Recent studies suggest that constitutive BRCA1 promoter methylation (24–28) and ATM gene body methylation (29,30) in normal tissues (blood) may be involved in the pathogenesis of hereditary BC. However, since BRCA1 hypermethylation was also observed in a considerable percentage (3.5–13.5%) of unaffected women without family history of BC, these data should be interpreted with caution. The average methylation levels (of genomic DNA representing a large number of DNA molecules) of functionally important cis-regulatory regions can show remarkable (both biological and technical) variation (31–33). Moreover, methylation patterns can change in response to a variety of internal and environmental factors (34,35).
In our opinion, it is very crucial to distinguish between single CpG methylation errors and allele methylation errors. A single or a few methylated CpGs within an overall unmethylated promoter region represent either technical (bisulfite conversion) errors or stochastic biological errors without functional consequences. Because individual CpGs cannot stably maintain methylation states that differ from those of the neighboring CpGs, usually the entire promoter is either methylated or not (7,36). Epigenetic silencing, i.e. of the somatostatin gene in gastric cancers, is characterized by an inverse correlation between the promoter methylation level and mRNA expression (37). It is the density of methylated CpGs rather than individual CpGs that turns a promoter off. In vitro methylation and transfection assays showed that that MeCP2-mediated repression of the BRCA1 promoter depends on the number of methylated CpGs (38). The methylation patterns of individual DNA molecules of the BRCA1 promoter in BC and OC are highly heterogeneous (12,16). Because previous studies (11–17) have analyzed different CpG sites and relied on different techniques to determine the methylation status of the BRCA1 promoter, it is difficult to define a universally valid absolute threshold for gene silencing. Here, only alleles with >50% methylated CpGs (located +402 to −80 bp relative the BRCA1 transcription start site) were considered as epimutations.
This study provides a comprehensive epimutation screen of BRCA1, BRCA2 and several other BC susceptibility genes in a large number (>600) of BRCA1/BRCA2 mutation-negative BC and/or OC patients with a high risk for hereditary cancer. Methylated BRCA1 and RAD51C alleles explain a small percentage (1–2%) of these cases.
RESULTS
Epimutation screening by bisulfite pyrosequencing
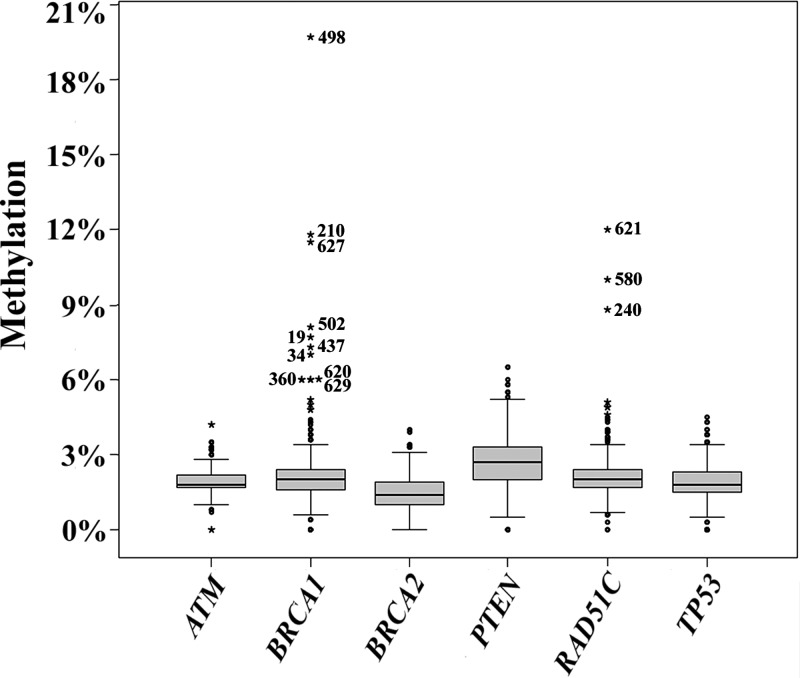
We analyzed the promoter methylation patterns of several tumor suppressor genes (ATM, BRCA1, BRCA2, PTEN, RAD51C and TP53) in normal body cells of BRCA1/BRCA2 mutation-negative patients from independent pedigrees. Bisulfite pyrosequencing is a rapid and highly accurate method for epimutation screening. It can exactly (±1%) quantify the methylation of individual CpG sites located in the 30–50 bp 3′ from the sequencing primer (39). Our pyrosequencing assays are based on a small number (4–7) of adjacent CpG sites in the 5′ promoter regions (Supplementary Material, Table S1). The box plots in Figure 1 present the methylation values of six cancer susceptibility genes in blood samples of 641 affected BC and/or OC patients, including 575 familial and 66 isolated cases (Table 1). The promoter methylation values (determined by bisulfite pyrosequencing) reflect the average methylation level of all analyzed CpGs in a large number of DNA molecules of a given sample. In several hundred studied patients, all genes displayed rather small inter-quartile ranges (IQRs), indicating that both biological variation of promoter methylation and technical variation of our assays are low. The mean BRCA1 methylation (±standard deviation) was 2.1 ± 1.3%, the middle 50 were between 1.6 and 2.4%. The promoter methylation values of the other genes were very similar: 1.4 ± 0.7% (IQR 1.0–1.9%) for BRCA2, 1.9 ± 0.5% (1.7–2.2%) for ATM, 2.2 ± 0.9% (1.7–2.4%) for RAD51C, 2.7 ± 1.1% (2.0–3.3%) for PTEN and 1.9 ± 0.8% (1.5–2.3%) for TP53.
Figure 1.
Box plots show the distribution of methylation values of six tumor suppressor genes (ATM, BRCA1, BRCA2, PTEN, RAD51C and TP53) in 641 affected patients. The average methylation of all analyzed CpG sites in a give gene and sample was used an epigenetic marker for promoter methylation. The bottom and the top of the boxes represent the 25th and 75th percentiles, respectively. The median is represented by vertical lines. Bars extend from the boxes to at most 1.5 times the height of the box. Outliers are indicated by open circles and extreme outliers by stars. The numbers indicate patients with ≥6% promoter methylation.
Table 1.
Studied patients
| Cancer |
No. of cases | No. (%) with epimutations in | ||
|---|---|---|---|---|
| Patient | Pedigree | BRCA1 | RAD51C | |
| BC | FBC | 460 | 3 (0.7%) | 1 |
| BC | EO + FBC | 24 | 0 | 0 |
| BC | FBOC | 52 | 1 | 0 |
| BC | EO + FBOC | 2 | 0 | 0 |
| EO + BC | Isolated case | 37 | 2 (5.5%) | 0 |
| BM + BC | Isolated case | 8 | 0 | 0 |
| M + BC | Isolated case | 15 | 0 | 1 |
| M + BC | FBC | 4 | 0 | 0 |
| OC + BC | Isolated case | 3 | 0 | 0 |
| OC + BC | FBOC | 8 | 0 | 0 |
| OC | FOC | 8 | 0 | 0 |
| EO + OC | Isolated case | 3 | 0 | 0 |
| OC | EO + FBOC | 1 | 1 | 0 |
| OC | FBOC | 16 | 2 (12.5%) | 1 |
| All patients | 641 | 9 (1.4%) | 3 (0.5%) | |
| BC (all cases) | 613 | 6 (1%) | 2 (0.3%) | |
| OC (all cases) | 39 | 3 (8%) | 1 (2.5%) | |
| FBC (all cases) | 488 | 3 (0.6%) | 1 (0.2%) | |
| FBOC (all cases) | 79 | 4 (5%) | 1 (1.3%) | |
| EO (BC or OC) | 40 | 2 (5%) | 0 | |
| M + BC | 19 | 0 | 1 (5%) | |
BC, breast cancer; BM, bilateral/metachronous; EO, early onset; FBC, familial breast cancer; FBOC, familial breast and ovarian cancer; M, male; OC, ovarian cancer.
One important goal of this epimutation screening was to select potentially abnormal samples for further analyses, using a feasible empirical threshold. To this end, extreme outliers which have values more than three times the IQR away from the 75th percentile were considered as hypermethylated. Altogether, only 20 such extreme methylation values were observed, most of them in BRCA1 (13 extreme outliers) and RAD51C (6 extreme outliers). When looking at the distribution of gene-specific methylation values in Figure 1, 6% promoter methylation represents a convincing absolute threshold to select patients with putative epimutations. Ten of the 13 extreme outliers in BRCA1 and 3 of the 6 extreme outliers in RAD51C had methylation values of 6% or higher (Table 2). We therefore concentrated our further analysis on these 13 patients with constitutive BRCA1 and RAD51 promoter hypermethylation (indicated by star symbols and patient numbers in Fig. 1).
Table 2.
Results of bisulfite pyrosequencing and plasmid sequencing in selected patients
| Patient ID | Cancer |
Methylation level (%) | Allele-specific methylation | |||
|---|---|---|---|---|---|---|
| Patient | Pedigree | Pyrosequencinga | Plasmid sequencingb | Methylated allele | Allele ratio | |
| BRCA1 | C/G | |||||
| 433 | BC | FBC | 4.3 | 0 (0/79) | n.i. | n.i. |
| 642 | OC | FOC | 5.0 | 0 (0/81) | n.i. | n.i. |
| 413 | BC | FBC | 5.2 | 0 (0/80) | n.i. | n.i. |
| 360 | BC | EO + FBC | 6.0 | 0 (0/64) | n.i. | n.i |
| 620 | OC | EO + FBOC | 6.0 | 4 (3/76) | G | 1.4 |
| 629 | OC | FBOC | 6.0 | 4.5 (3/66) | n.i. | n.i |
| 34 | EO + BC | Isolated case | 7.0 | 6 (4/70) | n.i. | n.i |
| 437 | BC | FBC | 7.3 | 12 (5/42) | n.i. | n.i. |
| 19 | EO + BC | Isolated case | 7.7 | 18 (12/67) | C | 0.8 |
| 502 | BC | FBC | 8.1 | 6 (4/70) | C | 0.8 |
| 627 | OC | FBOC | 11.5 | 20 (6/30) | n.i. | n.i. |
| 210 | BC | FBOC | 11.8 | 31 (10/32) | C | 1.1 |
| 498 | BC | FBC | 19.7 | 30 (18/60) | G | 0.7 |
| RAD51C | A/G | |||||
| 240 | BC | FBC | 8.8 | 4 (2/50) | G | 1.6 |
| 580 | M + BC | Isolated case | 10.0 | 4 (2/51) | A | 0.6 |
| 621 | OC | FBOC | 12.0 | 8 (5/61) | n.i. | n.i. |
BC, breast cancer; EO, early onset; FBC, familial breast cancer; FBOC, familial breast and ovarian; FOC, familial ovarian cancer; M, male; n.i., not informative; OC, ovarian cancer.
aAverage methylation level of all analyzed CpG sites in a cell population.
bPercentage of single DNA molecules with >50% methylated CpG sites.
In one case, family members were available for investigation. Patient 502 (one of the extreme BRCA1 outliers) was diagnosed with BC at the age of 46 and her sister at the age of 48 years. However, neither the 18-year-old healthy daughter nor the affected sister of Patient 502 displayed ≥6% methylation of the BRCA1 promoter.
Identification of methylated BRCA1 and RAD51C alleles by plasmid sequencing
Because bisulfite pyrosequencing cannot distinguish between single CpG and allele methylation errors, validation of epimutations requires the analysis of individual DNA molecules. Classic bisulfite plasmid sequencing has the added advantage that it allows one to look at a larger number of CpG sites. Here, we analyzed a BRCA1 amplicon with 27 CpG sites including the 5 sites (nos 16–20) targeted by the pyrosequencing assay and a common single-nucleotide polymorphism (SNP) (Fig. 2). BRCA1 single-molecule analysis was performed on 13 affected patients, 10 with promoter methylation values ≥6% and 3 with values below the 6% threshold, as well as on 10 healthy female controls (mean age 54 years, range 49–64 years). By pyrosequencing, all control samples displayed <6% promoter methylation. On average, 63 (range 30–81) independent plasmid clones were analyzed per patient (Table 2) and 65 (45–100) per control. Nine of the 10 patients with ≥6% BRCA1 promoter methylation displayed methylated alleles, whereas the 3 patients with <6% methylation and the 10 controls had only unmethylated alleles with single CpG methylation errors. Overall, 65 of 577 analyzed BRCA1 alleles were classified as epimutations in patients with ≥6% promoter methylation, 0 of 240 in patients with <6% promoter methylation and 0 of 646 in healthy controls. Epimutations were significantly (χ2 test; P < 10−7) more frequent in patients with ≥6% promoter methylation than in the two other groups. Evidently, the selected threshold is useful for the identification of constitutive BRCA1 epimutations. The methylation level (determined by quantitative bisulfite pyrosequencing) of the nine patients with validated epimutations ranged from 6 to 20%, implying that 12–40% of normal body cells are endowed with one epigenetically silenced copy of the BRCA1 tumor suppressor gene.
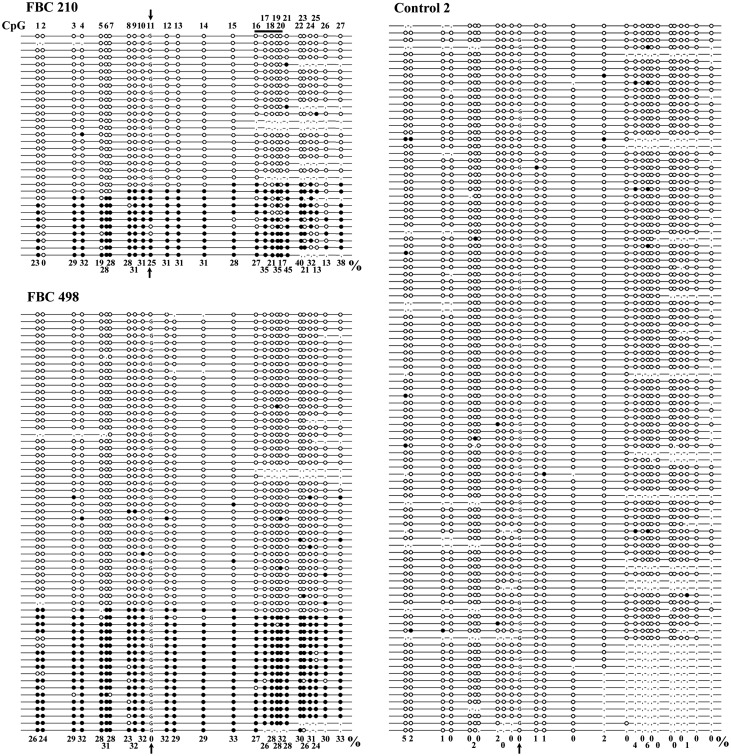
Figure 2.
Single-molecule methylation patterns of the BRCA1 promoter in familial BC (FBC) patients 210 and 498 and control 2. Each line represents an individual allele (DNA molecule) analyzed by bisulfite plasmid sequencing. Filled circles indicate methylated CpG and open circles unmethylated CpG sites. Missing circles (commata) indicate CpG sites that could not be analyzed because of poor sequence quality. The amplicon contains 27 CpG sites (indicated on top of the first allele of patient 210) extending from +402 to −80 bp of the transcription start site. CpG no. 11 (indicated by a vertical arrow) represents a C/G SNP, which allows one to distinguish the two parental alleles. The horizontal bar indicates the five CpG sites analyzed by pyrosequencing. All three individuals display alleles with single CpG (stochastic) methylation errors. Ten (31%) of 32 analyzed alleles in FBC 210 and 18 (30%) of 60 alleles in FBC 498 represent epimutations with the majority of CpGs being aberrantly methylated, whereas all 100 alleles in the healthy control are hypomethylated. Note that only the C allele in Patient 210 and only the G allele in Patient 498 are aberrantly methylated. The single CpG methylation percentages of all analyzed alleles of a given individual are indicated at the bottom. Overall, 28% of all analyzed CpGs were methylated in Patient 210 and 30% in Patient 498, compared with 1% in control 2.
In six informative patients with BRCA1 epimutations, a heterozygous C/G SNP (rs799905), which leads to loss of CpG no. 11 after bisulfite conversion, allowed us to distinguish the two parental alleles. BRCA1 hypermethylation was always confined to a single parental allele. This is consistent with the view that the epimutation arose in a single cell during early development, leading to somatic mosaicism. The C allele in three patients (nos 19, 502 and 210) and the G allele in two patients (nos 498 and 620) were methylated. In three (nos 19, 502 and 620) of these five patients, there was an amplification/cloning bias toward the unmethylated allele (Table 2).
The methylation entropy (ME) is a measure to assess the randomness of DNA methylation patterns in a given sample, considering the total number of analyzed CpG sites, the number of analyzed DNA molecules and the frequency of each observed distinct methylation pattern (33). If all analyzed CpGs in all analyzed DNA molecules of a given sample are demethylated (or the other way round, methylated), the ME is zero (maximum certainty). A totally random distribution of 50% unmethylated and 50% methylated CpGs would result in ME = 1 (maximum uncertainty). The calculated ME = 0.037 in the 100 sequence reads of control sample 2 (Fig. 2) is consistent with demethylation of all but a few CpGs in all individual DNA molecules. The higher ME = 0.074 in Patient 498 reflects semi-randomness in the distribution of methylation patterns associated with more or less complete demethylation in most cells (similar to the control) and allele-specific methylation in a subset of cells. The number of sequence reads of patient 210 was too low for ME calculation.
In addition, single-molecule methylation analysis was performed in the three patients with ≥6% RAD51C methylation. The RAD51C amplicon contained 26 CpGs including the 7 sites (nos 2–8) of the pyroassay and a heterozygous A/G SNP (rs16943176) between CpG sites 2 and 3 (Fig. 3). All three patients exhibited (4–8%) hypermethylated RAD51C alleles, consistent with RAD51C epimutations (Table 2). Interestingly, the methylated CpGs were concentrated in the promoter region (CpGs 1–17), whereas exon 1 (CpGs 18–26) appeared to be unmethylated in some alleles (for example, see Patient 621 in Fig. 3). This possibly suggests the existence of a methylation boundary. When counting alleles with >50% methylated CpGs 1–17 as epimutations, the frequency of abnormal alleles would even be somewhat higher (4–11.5%). The A allele in Patient 580 and the G allele in Patient 240 were methylated (Table 2). In both cases, the unmethylated allele was preferentially amplified/cloned.
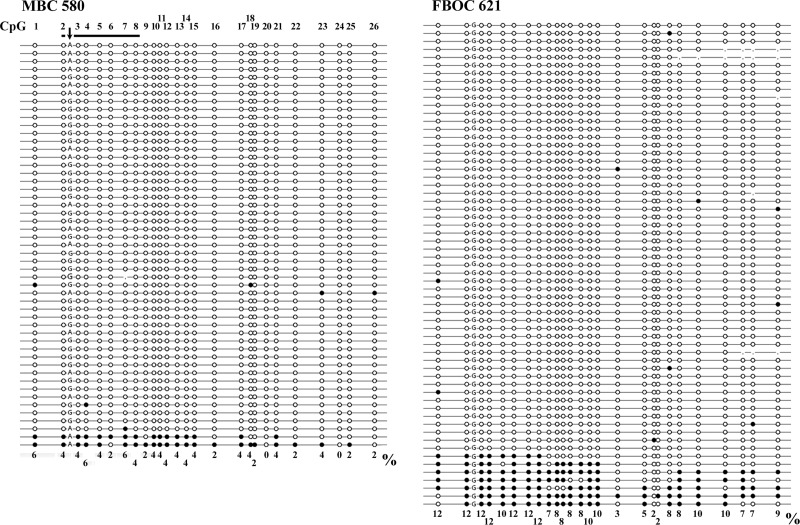
Figure 3.
Single-molecule methylation patterns of the RAD51C promoter in male BC (MBC) Patient 580 and familial BC and OC (FBOC) Patient 621. Each line represents an individual allele (DNA molecule) analyzed by bisulfite plasmid sequencing. Filled circles indicate methylated CpG and open circles unmethylated CpG sites. Missing circles (commata) indicate CpG sites that could not be analyzed because of poor sequence quality. The amplicon contains 26 CpG sites (indicated on top of the first allele of MBC 580) extending from −189 to +89 bp of the transcription start site. The vertical arrow indicates a G/A SNP, which allows one to distinguish the two parental alleles. The horizontal bar indicates the seven CpG sites analyzed by the pyrosequencing assay. Two (4%) of 51 analyzed alleles in MBC 580 and 5 (8%) of 61 alleles in FBOC 621 represent epimutations. In addition to the five fully methylated alleles, FBOC 621 displays three alleles with methylation of CpGs 1–17 (promoter region). Note that in MBC 580, only the A allele is aberrantly methylated (FBOC 621 is not informative for the SNP). The single CpG methylation percentages of all analyzed alleles of a given individual are indicated at the bottom. Overall, 3% of all analyzed CpGs were methylated in MBC 580 and 8% in FBOC 621.
Collectively, our two-step protocol with bisulfite pyrosequencing and plasmid sequencing identified 9 (1.4%) patients with BRCA1 epimutations and 3 (0.5%) with RAD51C epimutations in 641 BRCA1/BRCA2 mutation-negative patients from the German Consortium for Hereditary Breast and Ovarian Cancer (Table 1). Six of the nine index patients with BRCA1 epimutations suffered from BC and three from OC. Immunohistochemistry had been done on five of the six BCs: two sporadic cases (nos 19 and 34) with early-onset BC were ER-, PR- and HER2-negative, one patient (no. 437) with familial BC was ER-positive and PR-negative, one patient (no. 502) ER- and PR-positive and another one patient (no. 210) with a family history of BC and OC was ER- and PR-positive but HER2-negative. In particular, early-onset BCs in patients with BRCA1 epimutation showed the typical features of tumors with BRCA1 germline mutations.
Epimutation frequencies in different patient cohorts
BRCA1 epimutations were significantly (Fisher's test; P = 0.016) more frequent in index patients with OC (3/39; 8%) than with BC (6/613; 1%). The same appeared to be true for RAD51C epimutations (1/39; 2.5% versus 2/613; 0.3%, respectively); however, this difference was not significant. In families with a history of BC and OC, both BRCA1 (4/79; 5%) and RAD51C epimutations (1/79; 1.3%) were significantly (P = 0.01 and P = 0.02) more frequent than in families with BC alone (3/488; 0.6% and 1/488; 0.2%, respectively). Thus, constitutive epimutations are predominantly associated with an increased risk for OC. Interestingly, BRCA1 epimutations were also significantly (P = 0.047) enriched in women with early-onset BC (2/37; 5.5%) without a family history of cancer, compared with women with familial BC after the age of 36 years (3/460; 0.7%).
Of the three patients with RAD51C epimutations, one suffered from OC (with a family history of BC and OC), one from familial BC and another one was an isolated case of male BC. Although one epimutation (5%) in 19 males compared with one (0.2%) in 594 females with BC is conspicuous, this is not sufficient to claim a significant association with male BC. Similar to RAD51C germline mutations (40,41), the breast tumors of both the male (no. 580) and the female patient (no. 240) with constitutive RAD51C hypermethylation were ER- and PR-positive.
Sequence analysis of the BRCA1 and RAD51C promoter regions
Because constitutive hypermethylation in different tumor suppressor genes has been linked to sequence variants in the 5′ cis-regulatory regions (19,21,22), we sequenced a 3.2 kb upstream fragment including the BRCA1 promoter and exon 1 (Supplementary Material, Table S2). Only one of eight patients (nos 19, 34, 210, 360, 437, 498, 502 and 627) with 7–20% BRCA1 promoter methylation showed a novel sequence variant: a heterozygous G>A substitution was present in Patient 34 at −1479 bp relative to the transcription start site (corresponding to position 161 bp in amplicon 6 and 459 bp in amplicon 7, respectively). Most likely, this represents a rare SNP without functional implications. In addition, the studied patients displayed SNPs (i.e. rs799905, rs111292942, rs3092986, rs799906, rs11655505, rs144412026, rs799908, rs4793204, rs799910, rs12947782, rs35981166, rs34410138, rs35931760 and rs33945274), which are known to form discrete haplotypes in the German population (Supplementary Material, Table S3). In four patients (nos 19, 210, 502 and 629), the hypermethylated allele belonged to the frequent haplotype 1 and in two (437 and 498) to haplotype 3. Patient 34 with haplotypes 1 and 2 and Patient 360 with haplotypes 3 and 4 were not informative.
Similarly, we sequenced 1.7 kb of the 5′ UTR of the RAD51C gene in the three patients (nos 240, 580 and 621) with RAD51C epimutations and found only frequent SNPs (rs16943176, rs12946397 and rs302873). The three SNPs appear to form two different haplotypes (Supplementary Material, Table S3). Haplotype 1 was methylated in two (nos 240 and 580) and haplotype 2 in one patient (no. 621).
BRCA1 methylation analysis in different cell types
Magnetic-activated cell sorting (MACS) was used to separate different leukocyte types from whole blood of Patient 502 who showed 8.1% BRCA1 methylation in our screening assay. CD3-positive T-cells were enriched from 23% in the whole-blood sample to 80% in the respective MACS fraction. CD19-positive B-cells were enriched from 7 to 93%, CD14-positive monocytes from 11 to 73% and CD15-positive granulocytes from 52 to 97%. Similar to whole blood, the B-cell, monocyte and granulocyte fractions showed 7.1–7.6% methylation by bisulfite pyrosequencing. In contrast, the T-cell fraction displayed only 4%. Considering that the T-cell fraction contains 20% contaminating granulocytes and few monocytes, the real T-cell methylation may be even lower. The fact that all analyzed myeloid cell types and B-lymphocytes were hypermethylated suggests that the epimutation is present in blood stem (myeloid and lymphoid) cells, which are derived from the mesoderm. Due to somatic mosaicism, the epimutation may have a lower frequency or be absent in the T-cell progenitors.
In addition, we analyzed saliva DNA, which is mainly derived from buccal mucosa and salivary epithelium cells, as well as urine DNA from exfoliated urinary tract epithelium and renal tubular cells. An increased BRCA1 promoter methylation level was only found in urine DNA (9.4%) but not in saliva DNA (3.6%), although both samples are of endodermal origin.
DISCUSSION
Formation of epimutations and its role for tumor development
During gametogenesis and early embryogenesis, the parental genomes undergo two waves of demethylation and remethylation (42). These are vulnerable time windows where stochastic and/or environmentally induced methylation reprogramming defects may occur. In the first round of genome reprogramming, sex-specific methylation patterns are established at certain loci in the male and female germline, respectively. In the second round after fertilization, the somatic methylation patterns for normal development are created, underlying activation and silencing of specific genes. Tumor suppressor genes must remain unmethylated in all somatic cell types. Somatic BRCA1 epimutations, which arise in tumor cell precursors and are restricted to the tumor tissue, occur in ∼20% of sporadic BC and/or OC (11–17). Our study shows that constitutive epimutations in normal body cells also contribute to both sporadic and familial cases, which often may appear as ‘phenocopies’ of tumors with germline mutations. Constitutive epimutations are usually present in varying percentages of cells in normal tissues and most likely represent stochastic and/or environmentally induced methylation errors during early development (23,32,43). In contrast, germline epimutations, which are transmitted by the sperm or egg into the zygote and then escape genome reprogramming after fertilization, should be present in all cells of the body. So far, there is little, if any hard evidence for germline transmission of epimutations in humans. Interestingly, we found constitutive epimutations in BRCA1 and RAD51C, but not in ATM, BRCA2, PTEN and TP53. A previous study (27) reported that constitutive promoter methylation occurs in BRCA1 but not in ATM, ATR, BRCA2 and TP53.
In our cohort of 641 affected BC and/or OC patients, 1.4 and 0.5%, respectively, displayed constitutive BRCA1 and RAD51C epimutations (allele methylation errors). Although most (575) patients had a positive family history for BC and/or OC, we also included 66 sporadic cases with early-onset, bilateral or male cancer. Constitutive epimutations were enriched in specific subgroups, i.e. in patients with early-onset BC (5.5%), OC (10%) and a family history for both BC and OC (6%). Previous studies (11–17) revealed that BRCA1 promoter hypermethylation in tumor tissue of sporadic BC or OC cases is often combined with a loss of the second BRCA1 allele, but not with classical BRCA1 gene mutations. Experimental demethylation of BRCA1-methylated tumor cell lines re-activated gene expression (14,38). On the other hand, promoter hypermethylation is a rare second hit in hereditary tumors with germline mutations (44). Taken together, these observations suggest that analogous to germline genetic mutations constitutive BRCA1 epimutations may serve as the first hit of tumor development. Inactivation of the second allele most likely occurs by loss of heterozygosity and maybe rarely by genetic mutation of the second gene copy. The genomic caretaker BRCA1 is necessary for faithful rejoining of broken DNA ends (2). The reduced BRCA1 transcripts/proteins in body cells with a heterozygous epimutation may compromise genetic stability and, thus, trigger the genetic changes (second hit) that are necessary for neoplastic transformation. Since our retrospective study design did not allow us to analyze tumor DNA and tissue of our index patients, we do not have information on BRCA1 and RAD51C mRNA or protein expression in the tumors of the identified epimutation carriers. However, earlier studies (24,27,28) already showed an association between blood and tumor BRCA1 methylation, supporting the view that constitutive epimutations contribute to tumorigenesis.
Characteristics of BRCA1 and RAD51C epimutations
The promoter methylation level (measured by bisulfite pyrosequencing) was always smaller than the 50% that is expected for a heterozygous epimutation in all cells. All epimutations occurred in a mosaic state with 12–40% of the analyzed blood cells carrying one epigenetically inactive allele. This relatively high percentage cannot be explained by rare circulating tumor cells or cell-free tumor DNA. Moreover, in most affected patients, genetic counseling and analyses were performed several months to years after tumor therapy. Similar to constitutive MLH1 and MSH2 epimutations in HNPCC families (19,20), BRCA1 epimutations exhibited considerable variation in the promoter methylation levels (proportion of cells carrying the epimutation) between different cell types and tissues. In all informative cases, promoter hypermethylation was restricted to one of the two alleles. Because we did not have parental DNAs of our index patients, the parental origin of the methylated allele could not be determined.
Several previous studies (24–28) reported constitutive BRCA1 promoter methylation in peripheral blood of different groups of BC patients, in particular in patients with early-onset BC and/or a BRCA1 mutation-like pathology. In general, the prevalence of BRCA1 hypermethylation, detected by methylation-specific polymerase chain reaction (PCR) and/or methylation-sensitive high-resolution melting, was much higher than in our study, namely 1 (14%) of 7 (24), 9 (7%) of 132 (25), 2 (28.5%) of 7 (26), 43 (21.5%) of 200 (27) and 28 (11%) of 255 patients (28), respectively. Because promoter hypermethylation was also found in 5 (6%) of 84 (25), 8 (11%) of 73 (26), 27 (13.5%) of 200 (27) and 6 (3.5%) of 169 controls (28), it was concluded that promoter methylation constitutes a risk factor for developing BC. Although we agree with this statement, in our opinion, single-molecule analysis is necessary to distinguish between biological/technical methylation variation and true epimutations. So far, there is no evidence for fully methylated BRCA1 alleles in controls (4,23). Consistent with a preliminary study (45) which did not detect any methylated BRCA1 alleles in 41 women with a family history of BC, the prevalence of constitutive epimutations in our familial BC patients was relatively low (0.9%). BRCA1 epimutations mainly occurred in early-onset BC (5.5%) and familial OC patients (8%) and, therefore, may be associated with specific predisposition for these types of cancer. In this study, we identified for the first time constitutive epimutations in the RAD51C gene. Similar to RAD51C germline mutations (40,41), the RAD51C epimutation rate was significantly higher in families with both BC and OC (1/79; 1.3%) than in families with BC alone (1/488; 0.2%).
In rare HNPCC families, dominant transmission of mosaic MLH1 hypermethylation was linked to a single-nucleotide variant in the 5′ UTR of the gene (19). Similarly, MSH2 epimutations were linked to 3′ end deletions of EPCAM, a gene directly upstream of MSH2 (21). Constitutive hypermethylation and down-regulation of the DAPK1 gene in patients with B-cell chronic lymphocytic leukemia are caused by a point mutation upstream of the DAPK1 promoter (22). However, Sanger sequencing revealed only frequent SNPs in the BRCA1 and RAD51C cis-regulatory regions. Neither BRCA1 nor RAD51C epimutations were confined to a specific haplotype, which also argues against a 5′ promoter sequence variant predisposing to hypermethylation. Of course, we cannot exclude a causative rare genetic variant(s) elsewhere in the genome. With one notable exception, family members were not available of our 12 index patients with epimutations. The affected sister and unaffected daughter of BC patient 502 did not display BRCA1 promoter methylation, which argues against germline transmission in this family. The overall risk of developing BC for a 40- to 50-year-old woman is ∼1%. Therefore, BC in the 46- and 48-year-old sisters may well be a chance coincidence of different etiologies. Since a recent study (28) also failed to detect BRCA1 promoter methylation in family members of nine patients with detectable blood methylation, the heritability of BRCA1 epimutations appears to be low.
Technical aspects of epimutation detection
An increased methylation level can be due to an increased rate of single aberrantly methylated CpG sites in overall hypomethylated alleles (DNA molecules) or to the presence of hypermethylated alleles in addition to hypomethylated alleles. In our experience with single-molecule methylation analyses (23,43), the rate of single CpG errors which can be due to stochastic methylation errors (biological variation) or bisulfite conversion errors (technical variation) is in the order of several (0–5) percent, depending on the analyzed gene and cell type. It is plausible to assume that single CpG errors without functional implications account for most promoter methylation values <6%. However, only allele methylation errors (epimutations) with a high density of methylated CpGs in the cis-regulatory region can be expected to act as inactivating mechanism for BRCA1 expression.
To distinguish between single CpG methylation errors and inactivating allele methylation errors, we developed a two-step protocol. Bisulfite pyrosequencing is a screening technique which allows one to determine the average methylation level of a large number of DNA molecules. Subsequently, the more labor-intensive bisulfite plasmid sequencing was used for single-molecule methylation analysis of patients with putative promoter hypermethylation. Because of the enormous costs and expenditure of time, it is not possible to sequence a large number of plasmids in all patients. Box plot analyses revealed a clustering of the promoter methylation values determined by bisulfite pyrosequencing around a central value. Extreme outliers which do not seem to belong to the rest of the data set are often the most interesting data points that can provide novel insights. For practical reasons, we empirically choose an absolute threshold of 6% methylation to select patients who are worthy of further investigation. Most (13 of 20; 65%) extreme outliers in different tumor suppressor genes displayed ≥6% promoter methylation. The vast majority (12 of 13; 92%) of these patients exhibited hypermethylated alleles indicative of a heterozygous epimutation. This clearly demonstrates that the extreme outliers in our data set are not just bad data points due to technical problems, but can be associated with an underlying biological phenomenon. It is likely that patients with ≥6% promoter methylation will continue to appear in future pyrosequencing analyses (i.e. in a diagnostic setting) and also exhibit true epimutations. It is not unexpected that in some cases, the methylation percentages determined by bisulfite pyrosequencing and plasmid sequencing varied. First, plasmid sequencing of individual DNA molecules is not a very accurate quantitative method, unless very high numbers of clones are analyzed. Secondly, it is well known that preferential amplification/cloning of methylated or unmethylated DNA molecules from the starting sample can lead to a bias toward higher or lower methylation levels (46).
Although we cannot exclude the presence of epimutations in individuals with <6% promoter methylation, the rate of such false-negative cases may be low. In three patients with <6% promoter methylation and 10 healthy controls, we did not find a single (0 of 866) hypermethylated allele. We did not perform an extensive epimutation screen in controls, because a considerable percentage of apparently healthy women may develop BC or OC during lifetime, and even in our affected patient cohort, the number of suspect (≥6%) promoter methylation values was relatively low (13/641; 2%). In addition, not all samples with ≥6% promoter methylation may be endowed with a detectable number of hypermethylated alleles, which is required to validate epimutations. Bisulfite plasmid sequencing of several hundred or thousand controls is not feasible.
Outlook
Our study demonstrates that constitutive epimutations in BRCA1 and RAD51C are relevant to OC and BC pathogenesis. On an adverse genetic background, indicated by early-onset cancer or familial aggregation, such epimutations may be more likely to become penetrant. Genetic variants or environmental factors may predispose to the de novo arisal of epimutations throughout the soma. We recommend a two-step constitutive epimutation screening (using blood DNA) in all BRCA1/BRCA2 mutation-negative patients with a high risk of developing cancer, in particular women with familial OC and early-onset BC. BRCA1 promoter methylation appears to be a frequent first hit in sporadic BC (11–14) and OC (15–17). So far, nothing is known about the frequency of RAD51C epimutations in sporadic tumors. It is certainly worthwhile to systematically look for RAD51C promoter hypermethylation in primary BC and OC. Considering that bisulfite pyrosequencing is a relatively inexpensive high-throughput technique, screening of larger patient populations is feasible. Prospective studies are needed to determine the life-long cancer risk of unaffected women with promoter hypermethylation.
MATERIALS AND METHODS
Study participants
Two centers (at the Universities of Würzburg and Münster) from the German Consortium for Hereditary Breast and Ovarian Cancer recruited 641 affected patients (Table 1) from independent families through a genetic counseling program. All patients had given informed consent to use their diagnostic DNA samples (excess materials) for research purposes. All patients participating in this study belonged to one or more of the following risk groups: three or more affected females with BC, at least two of them diagnosed before the age of 51 years; three or more affected females with BC, independent of age; two females with BC diagnosed before the age of 51 years; two females with BC, one of them diagnosed before the age of 51 years; at least one case of BC and one case of OC, independent of age; one case of early-onset BC, diagnosed before the age of 36 years; one case of bilateral/multifocal BC, the first diagnosed before the age of 51 years; one case of early-onset OC, diagnosed before the age of 30 years; at least one case of male BC. All individuals did not carry pathogenetic germline mutations in BRCA1 and BRCA2.
Bisulfite pyrosequencing
Bisulfite conversion of genomic DNA from whole blood was performed with the EpiTect 96 Bisulfite Kit (Qiagen, Hilden, Germany). Genomic DNA from saliva and urine was isolated with the High Pure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany) and converted with the EZ DNA Methylation Direct Kit (Zymo Research, Irvine, CA, USA), which is particularly suitable for working with small amounts of DNA. Bisulfite pyrosequencing (39) was used for quantitative methylation analysis of the tumor suppressor genes ATM, BRCA1, BRCA2, PTEN, RAD51C and TP53. DNA sequences of the promoter regions were retrieved from the Ensembl genome browser (http://www.ensembl.org/Homo_sapiens/). CpG islands were predicted using the default settings of the MethPrimer program (47).
PCR and sequencing primers (Supplementary Material, Table S1) were designed within the identified CpG islands using the PyroMark Assay Design 2.0 software (Qiagen). PCR amplification of bisulfite-treated DNA was performed in 25 µl reactions containing 2 μl (1 U) FastStart Taq DNA Polymerase (Roche Diagnostics), 2.5 μl 10× PCR buffer, 20 mm MgCl2, 0.5 μl 10 mm dNTP mix, 1.0 μl (10 pmol) of each forward and reverse primer, 1 μl (∼100 ng) bisulfite treated template DNA and 18 μl PCR-grade water. PCR was carried out with an initial denaturation step at 95°C for 5 min, 35 cycles (for BRCA1 and BRCA2) or 40 cycles (for ATM, PTEN, RAD51C and TP53) of 95°C for 30 s, primer-specific annealing temperature (55°C for BRCA1, BRCA2 and TP53; 58°C for PTEN; 60°C for ATM and RAD51C) for 30 s, elongation at 72°C for 45 s and a final extension step at 72°C for 5 min. Bisulfite pyrosequencing was performed on a PyroMark Q96 MD Pyrosequencing System with the PyroMark Gold Q96 CDT Reagents Kit (Qiagen). Pyro Q-CpG software (Biotage, Uppsala, Sweden) was used for data analysis.
To demonstrate the reliability of our quantitative methylation assays, we performed technical replicates (including bisulfite conversion, PCR and pyrosequencing) on DNA samples of >60 patients. The average methylation difference between duplicate measurements was 0.6% for ATM, 1.1% for BRCA1, 1.0% for BRCA2, 1.3% for PTEN, 1.2% for RAD51C and 1.6% for TP53. In addition, we analyzed independent blood DNA samples of the same patients which were collected at intervals of several years and found the methylation levels to be stable over time.
Bisulfite plasmid sequencing
Classical bisulfite plasmid sequencing was performed to determine the methylation patterns of individual BRCA1 and RAD51C DNA molecules. The BRCA1 amplicon was extended from the promoter into intron 1, using forward primer 5′-AGGTTGGTTTGGAATTTTTGATTTTATGA-3′ and the pyrosequencing reverse primer (Supplementary Material, Table S1). The 552 bp fragment (chromosome 17: 41 276 972–41 277 523 bp; Ensembl release 65) contains 27 CpGs sites for methylation analysis and a frequent intronic SNP (rs799905), which allows one to distinguish the parental alleles in informative cases. Forty cycles with an annealing temperature of 61°C were performed. The RAD51C fragment amplified by the pyrosequencing primers (Supplementary Material, Table S1) was also used for plasmid sequencing. It contains an informative SNP (rs16943176) and 26 CpG sites. BRCA1 and RAD51C PCR products, respectively, were cloned into pCR2.1-TOPO vector using T4-DNA ligase, the TA cloning kit and One Shot TOP10 chemically competent Escherichia coli (Invitrogen, Karlsruhe, Germany). Plasmid DNA of individual clones was isolated with the ZR Plasmid Miniprep Classic Kit (Zymo Research). Clones containing inserts of the right size were sequenced using dye terminator cycle sequencing with M13 primers on an ABI 3730 automated sequencer. Sequences were analyzed with the BiQ Analyzer software tool (48).
The ME was calculated according to Xie et al. (33). Briefly, sliding windows of four CpGs each were analyzed and the average value of all windows in a given region was used as an estimate of the ME of a given sample.
BRCA1 and RAD51C promoter sequencing
For Sanger sequencing of the 5′ UTR and exon 1 of the BRCA1 gene, a 3.2 kb fragment on chromosome 17 (41 277 112–41 280 328 bp, Ensembl release 64) was divided into 10 PCR amplicons (Supplementary Material, Table S2). The 5′ UTR of the RAD51C gene was divided into five amplicons, representing 1.7 kb on chromosome 17 (56 768 708–56 770 376 bp). For BRCA1 amplicons 1–5, 8 and RAD51C amplicons 1–5, we used FastStart Taq DNA Polymerase and the same reaction mixture as for bisulfite pyrosequencing (see above). For BRCA1 amplicons 6, 7, 8 and 10, the PCR mixture (25 μl) contained 25 mm ammonium sulfate, 750 mm Tris–HCl, 0.1% Tween 20, 240 µM dNTPs, 2.4 mm magnesium sulfate, 2.4× PCRX Enhancer Solution (Invitrogen), 0.4 µM of each primer, 1 U of Platinum Taq (Invitrogen) and 1 μl (∼100 ng) template DNA. All PCR amplifications were carried out with an initial denaturation step at 95°C for 5 min, 40 cycles of 95°C for 30 s, primer-specific annealing temperature (Supplementary Material, Table S2) for 45 s, 72°C for 45 s and a final extension step at 72°C for 7 min. Sequencing of the resulting PCR products was done with M13 sequencing primers on an ABI 3730 automated sequencer.
Magnetic-activated cell sorting
MACS was performed to separate different leukocyte types from peripheral blood of Patient 502, using the Whole Blood Column Kit and a QuadroMACS Separator (Miltenyi Biotec, Bergisch Gladbach, Germany). Ten milliliters of fresh ethylene-diamine-tetraacetic-acid blood were incubated on a rotator for 15 min at 4°C with 500 µl Whole Blood Microbeads carrying anti-CD3 and anti-CD19 antibodies. Following a washing and centrifugation step, CD13-positive T-cells and CD19-positive B-cells were isolated by magnetic separation with Whole Blood Columns. The flow-through containing unlabelled cells was collected and incubated with antibodies against CD14 (most monocytes and macrophages) and CD15 (neutrophil and eosinophil granulocytes), respectively, followed by a second round of magnetic separation. Finally, the four isolated cell fractions were purified using MS Columns and the OctoMACS Separator (Miltenyi). About one million cells per fraction were analyzed by FACS to evaluate the purity (epitope-specific enrichment) after MACS. Genomic DNA of ∼100 000 cells was isolated and bisulfite converted using the EZ DNA Methylation Direct Kit (Zymo Research).
SUPPLEMENTARY MATERIAL
Supplementary material is available at HMG online.
ACKNOWLEDGEMENTS
We thank Dr David Hehuang Xie for calculation of the methylation entropy and Richard Friedl for help with the FACS analyses.
Conflict of Interest statement. None declared.
FUNDING
Funding to pay the Open Access publication charges for this article was provided by the operating revenues of the Würzburg University Institute of Human Genetics.
REFERENCES
- 1.Antoniou A.C., Chenevix-Trench G. Common genetic variants and cancer risk in Mendelian cancer syndromes. Curr. Opin. Genet. Dev. 2010;20:299–307. doi: 10.1016/j.gde.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 2.O'Donovan P.J., Livingston D.M. BRCA1 and BRCA2: breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis. 2010;31:961–967. doi: 10.1093/carcin/bgq069. [DOI] [PubMed] [Google Scholar]
- 3.Ghoussaini M., Fletcher O., Michailidou K., Turnbull C., Schmidt M.K., Dicks E., Dennis J., Wang Q., Humphreys M.K., Luccarini C., et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat. Genet. 2012;44:312–318. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baylin S.B., Esteller M., Rountree M.R., Bachman K.E., Schuebel K., Herman J.G. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 6.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 7.Weber M., Hellmann I., Stadler M.B., Ramos L., Pääbo S., Rebhan M., Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr. Top. Microbiol. Immunol. 2006;10:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 9.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 10.Dobrovic A., Kristensen L.S. DNA methylation, epimutations and cancer predisposition. Int. J. Biochem. Cell Biol. 2009;41:34–39. doi: 10.1016/j.biocel.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Bianco T., Chenevix-Trench G., Walsh D.C., Cooper J.E., Dobrovic A. Tumour-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis. 2000;21:147–151. doi: 10.1093/carcin/21.2.147. [DOI] [PubMed] [Google Scholar]
- 12.Rice J.C., Ozcelik H., Maxeiner P., Andrulis I., Futscher B.W. Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis. 2000;21:1761–1765. doi: 10.1093/carcin/21.9.1761. [DOI] [PubMed] [Google Scholar]
- 13.Esteller M., Silva J.M., Dominguez G., Bonilla F., Matias-Guiu X., Lerma E., Bussaglia E., Prat J., Harkes I.C., Repasky E.A., et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl. Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 14.Wei M., Grushko T.A., Dignam J., Hagos F., Nanda R., Sveen L., Xu J., Fackenthal J., Tretiakova M., Das S., et al. BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res. 2005;65:10692–10699. doi: 10.1158/0008-5472.CAN-05-1277. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin R.L., Nemeth E., Tran H., Shvartsman H., Cass I., Narod S., Karlan B.Y. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 2000;60:5329–5333. [PubMed] [Google Scholar]
- 16.Wilcox C.B., Baysal B.E., Gallion H.H., Strange M.A., DeLoia J.A. High-resolution methylation analysis of the BRCA1 promoter in ovarian tumors. Cancer Genet. Cytogenet. 2005;59:114–122. doi: 10.1016/j.cancergencyto.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Seeber L.M., van Diest P.J. Epigenetics in ovarian cancer. Methods Mol. Biol. 2012;863:253–269. doi: 10.1007/978-1-61779-612-8_15. [DOI] [PubMed] [Google Scholar]
- 18.Hitchins M.P., Ward R.L. Constitutional (germline) MLH1 epimutation as an aetiological mechanism for hereditary non-polyposis colorectal cancer. J. Med. Genet. 2009;46:793–802. doi: 10.1136/jmg.2009.068122. [DOI] [PubMed] [Google Scholar]
- 19.Hitchins M.P., Rapkins R.W., Kwok C.T., Srivastava S., Wong J.J., Khachigian L.M., Polly P., Goldblatt J., Ward R.L. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5′UTR. Cancer Cell. 2011;20:200–213. doi: 10.1016/j.ccr.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Chan T.L., Yuen S.T., Kong C.K., Chan Y.W., Chan A.S., Ng W.F., Tsui W.Y., Lo M.W., Tam W.Y., Li V.S., et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat. Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 21.Ligtenberg M.J., Kuiper R.P., Chan T.L., Goossens M., Hebeda K.M., Voorendt M., Lee T.Y., Bodmer D., Hoenselaar E., Hendriks-Cornelissen S.J., et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 22.Raval A., Tanner S.M., Byrd J.C., Angerman E.B., Perko J.D., Chen S.S., Hackanson B., Grever M.R., Lucas D.M., Matkovic J.J., et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galetzka D., Hansmann T., El Hajj N., Weis E., Irmscher B., Ludwig M., Schneider-Raetzke B., Kohlschmidt N., Beyer V., Bartsch O., et al. Monozygotic twins discordant for constitutive BRCA1 promoter methylation, childhood cancer and secondary cancer. Epigenetics. 2012;7:47–54. doi: 10.4161/epi.7.1.18814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snell C., Krypuy M., Wong E.M. Loughrey M.B., Dobrovic A., editors. kConFab investigators. BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Res. 2008;10:R12. doi: 10.1186/bcr1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kontorovich T., Cohen Y., Nir U., Friedman E. Promoter methylation patterns of ATM, ATR, BRCA1, BRCA2 and p53 as putative cancer risk modifiers in Jewish BRCA1/BRCA2 mutation carriers. Breast Cancer Res. Treat. 2009;116:195–200. doi: 10.1007/s10549-008-0121-3. [DOI] [PubMed] [Google Scholar]
- 26.Al-Moghrabi N., Al-Qasem A.J., Aboussekhra A. Methylation-related mutations in the BRCA1 promoter in peripheral blood cells from cancer-free women. Int. J. Oncol. 2011;39:129–135. doi: 10.3892/ijo.2011.1021. [DOI] [PubMed] [Google Scholar]
- 27.Iwamoto T., Yamamoto N., Taguchi T., Tamaki Y., Noguchi S. BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast Cancer Res. Treat. 2011;129:69–77. doi: 10.1007/s10549-010-1188-1. [DOI] [PubMed] [Google Scholar]
- 28.Wong E.M., Southey M.C., Fox S.B., Brown M.A., Dowty J.G., Jenkins M.A., Giles G.G., Hopper J.L., Dobrovic A. Constitutional methylation of the BRCA1 promoter is specifically associated with BRCA1 mutation-associated pathology in early-onset breast cancer. Cancer Prev. Res. 2011;4:23–33. doi: 10.1158/1940-6207.CAPR-10-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flanagan J.M., Munoz-Alegre M., Henderson S., Tang T., Sun P., Johnson N., Fletcher O., Dos Santos Silva I., Peto J., Boshoff C., et al. Gene-body hypermethylation of ATM in peripheral blood DNA of bilateral breast cancer patients. Hum. Mol. Genet. 2009;18:1332–1342. doi: 10.1093/hmg/ddp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennan K., Garcia-Closas M., Orr N., Fletcher O., Jones M., Ashworth A., Swerdlow A., Thorne H. Riboli E., et al., editors. KConFab Investigators. Intragenic ATM methylation in peripheral blood DNA as a biomarker of breast cancer risk. Cancer Res. 2012;72:2304–2313. doi: 10.1158/0008-5472.CAN-11-3157. [DOI] [PubMed] [Google Scholar]
- 31.Bock C., Walter J., Paulsen M., Lengauer T. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping. Nucleic Acids Res. 2008;36:e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider E., Pliushch G., El Hajj N., Galetzka D., Puhl A., Schorsch M., Frauenknecht K., Riepert T., Tresch A., Müller A.M., et al. Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res. 2010;38:3880–3890. doi: 10.1093/nar/gkq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie H., Wang M., de Andrade A., Bonaldo M.F., Galat V., Arndt K., Rajaram V., Goldman S., Tomita T., Soares M.B. Genome-wide quantitative assessment of variation in DNA methylation patterns. Nucleic Acids Res. 2011;39:4099–4108. doi: 10.1093/nar/gkr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feinberg A.P. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 35.Alegría-Torres J.A., Baccarelli A., Bollati V. Epigenetics and lifestyle. Epigenomics. 2011;3:267–377. doi: 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sontag L.B., Lorincz M.C., Georg Luebeck E. Dynamics, stability and inheritance of somatic DNA methylation imprints. J. Theor. Biol. 2006;242:890–899. doi: 10.1016/j.jtbi.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Jackson K., Soutto M., Peng D., Hu T., Marshal D., El-Rifai W. Epigenetic silencing of somatostatin in gastric cancer. Dig. Dis. Sci. 2011;56:125–130. doi: 10.1007/s10620-010-1422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magdinier F., Billard L.M., Wittmann G., Frappart L., Benchaïb M., Lenoir G.M., Guérin J.F., Dante R. Regional methylation of the 5′ end CpG island of BRCA1 is associated with reduced gene expression in human somatic cells. FASEB J. 2000;14:1585–1594. doi: 10.1096/fj.14.11.1585. [DOI] [PubMed] [Google Scholar]
- 39.Tost J., Dunker J., Gut I.G. Analysis and quantification of multiple methylation variable positions in CpG islands by pyrosequencing. Biotechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 40.Meindl A., Hellebrand H., Wiek C., Erven V., Wappenschmidt B., Niederacher D., Freund M., Lichtner P., Hartmann L., Schaal H., et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 2010;42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 41.Thompson E.R., Boyle S.E., Johnson J., Ryland G.L., Sawyer S., Choong D.Y. Chenevix-Trench G., Trainer A.H., Lindeman G.J., et al., editors. kConFab. Analysis of RAD51C germline mutations in high-risk breast and ovarian cancer families and ovarian cancer patients. Hum. Mutat. 2012;33:95–99. doi: 10.1002/humu.21625. [DOI] [PubMed] [Google Scholar]
- 42.Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 43.El Hajj N., Trapphoff T., Linke M., May A., Hansmann T., Kuhtz J., Reifenberg K., Heinzmann J., Niemann H., Daser A., et al. Limiting dilution bisulfite (pyro)sequencing reveals parent-specific methylation patterns in single oocytes and early embryos. Epigenetics. 2011;6:1176–1188. doi: 10.4161/epi.6.10.17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dworkin A.M., Spearman A.D., Tseng S.Y., Sweet K., Toland A.E. Methylation not a frequent ‘second hit’ in tumors with germline BRCA mutations. Fam. Cancer. 2009;8:339–346. doi: 10.1007/s10689-009-9240-1. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y., Toland A.E., McLennan J., Fridlyand J., Crawford B., Costello J.F., Ziegler J.L. Lack of germ-line promoter methylation in BRCA1-negative families with familial breast cancer. Genet. Test. 2006;10:281–284. doi: 10.1089/gte.2006.10.281. [DOI] [PubMed] [Google Scholar]
- 46.Warnecke P.M., Mann J.R., Frommer M., Clark S.J. Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics. 1988;51:182–190. doi: 10.1006/geno.1998.5371. [DOI] [PubMed] [Google Scholar]
- 47.Li L.C., Dahiya R. Methprimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 48.Bock C., Reither S., Mikeska T., Paulsen M., Walter J., Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.