Abstract
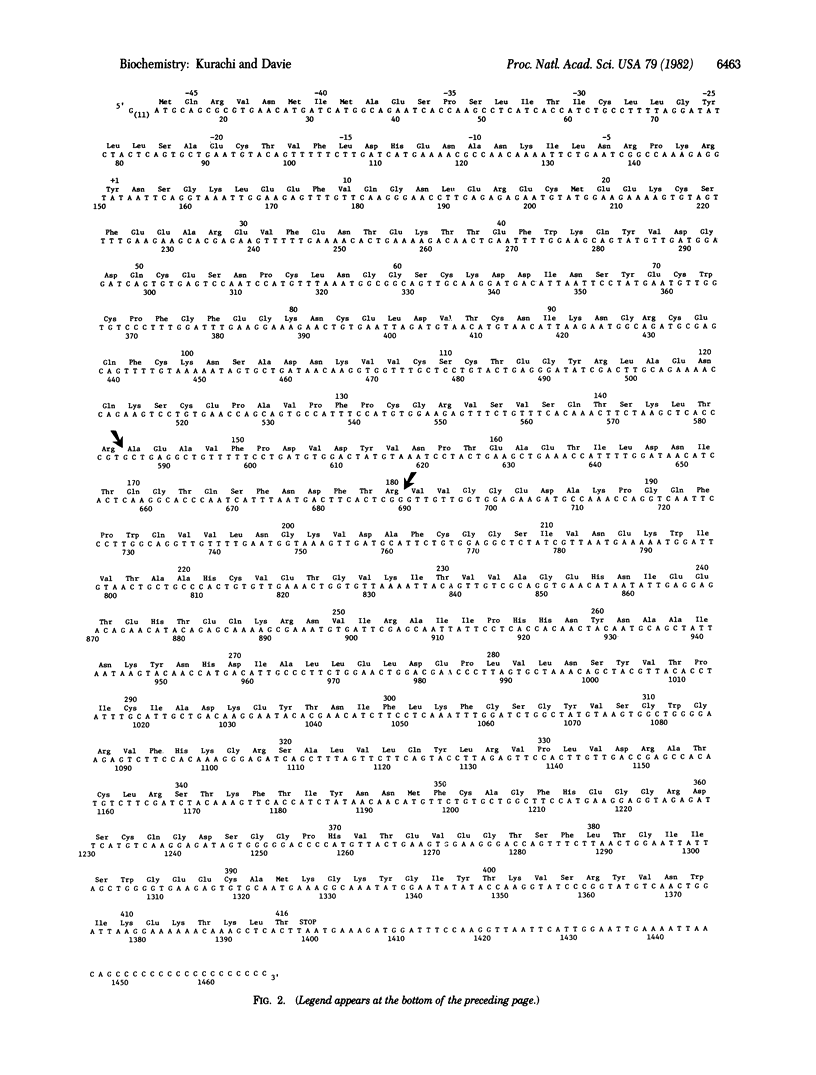
A cDNA library prepared from human liver has been screened for factor IX (Christmas factor), a clotting factor that participates in the middle phase of blood coagulation. The library was screened with a single-stranded DNA prepared from enriched mRNA for baboon factor IX and a synthetic oligonucleotide mixture. A plasmid was identified that contained a cDNA insert of 1,466 base pairs coding for human factor IX. The insert is flanked by G-C tails of 11 and 18 base pairs at the 5' and 3' ends, respectively. It also included 138 base pairs that code for an amino-terminal leader sequence, 1,248 base pairs that code for the mature protein, a stop codon, and 48 base pairs of noncoding sequence at the 3' end. The leader sequence contains 46 amino acid residues, and it is proposed that this sequence includes both a signal sequence and a pro sequence for the mature protein that circulates in plasma. The 1,248 base pairs code for a polypeptide chain composed of 416 amino acids. The amino-terminal region for this protein contains 12 glutamic acid residues that are converted to gamma-carboxyglutamic acid in the mature protein. These glutamic acid residues are coded for by both GAA and GAG. The arginyl peptide bonds that are cleaved in the conversion of human factor IX to factor IXa by factor XIa were identified as Arg145-Ala146 and Arg180-Val181. The cleavage of these two internal peptide bonds results in the formation of an activation peptide (35 amino acids) and factor IXa, a serine protease composed of a light chain (145 amino acids) and a heavy chain (236 amino acids), and these two chains are held together by a disulfide bond(s). The active site residues including histidine, aspartate, and serine are located in the heavy chain at positions 221, 270, and 366, respectively. These amino acids are homologous with His57, Asp102, and Ser195 in the active site of chymotrypsin. Two potential carbohydrate binding sites (Asn-X-Thr) were identified in the activation peptide, and these were located at Asn157 and Asn167. The homology in the amino acid sequence between human and bovine factor IX was found to be 83%.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blow D. M., Janin J., Sweet R. M. Mode of action of soybean trypsin inhibitor (Kunitz) as a model for specific protein-protein interactions. Nature. 1974 May 3;249(452):54–57. doi: 10.1038/249054a0. [DOI] [PubMed] [Google Scholar]
- Davie E. W., Fujikawa K., Kurachi K., Kisiel W. The role of serine proteases in the blood coagulation cascade. Adv Enzymol Relat Areas Mol Biol. 1979;48:277–318. doi: 10.1002/9780470122938.ch6. [DOI] [PubMed] [Google Scholar]
- Di Scipio R. G., Hermodson M. A., Yates S. G., Davie E. W. A comparison of human prothrombin, factor IX (Christmas factor), factor X (Stuart factor), and protein S. Biochemistry. 1977 Feb 22;16(4):698–706. doi: 10.1021/bi00623a022. [DOI] [PubMed] [Google Scholar]
- Di Scipio R. G., Kurachi K., Davie E. W. Activation of human factor IX (Christmas factor). J Clin Invest. 1978 Jun;61(6):1528–1538. doi: 10.1172/JCI109073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa K., Legaz M. E., Kato H., Davie E. W. The mechanism of activation of bovine factor IX (Christmas factor) by bovine factor XIa (activated plasma thromboplastin antecedent). Biochemistry. 1974 Oct 22;13(22):4508–4516. doi: 10.1021/bi00719a006. [DOI] [PubMed] [Google Scholar]
- Fujikawa K., Thompson A. R., Legaz M. E., Meyer R. G., Davie E. W. Isolation and characterization of bovine factor IX (Christmas factor). Biochemistry. 1973 Nov 20;12(24):4938–4945. doi: 10.1021/bi00748a019. [DOI] [PubMed] [Google Scholar]
- Gough N. M., Adams J. M. Immunoprecipitation of specific polysomes using Staphylococcus aureus: purification of the immunoglobulin k chain messenger RNA from the mouse myeloma MPC11. Biochemistry. 1978 Dec 12;17(25):5560–5566. doi: 10.1021/bi00618a036. [DOI] [PubMed] [Google Scholar]
- Katayama K., Ericsson L. H., Enfield D. L., Walsh K. A., Neurath H., Davie E. W., Titani K. Comparison of amino acid sequence of bovine coagulation Factor IX (Christmas Factor) with that of other vitamin K-dependent plasma proteins. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4990–4994. doi: 10.1073/pnas.76.10.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGillivray R. T., Degen S. J., Chandra T., Woo S. L., Davie E. W. Cloning and analysis of a cDNA coding for bovine prothrombin. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5153–5157. doi: 10.1073/pnas.77.9.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGillivray R. T., Chung D. W., Davie E. W. Biosynthesis of bovine plasma proteins in a cell-free system. Amino-terminal sequence of preproalbumin. Eur J Biochem. 1979 Aug 1;98(2):477–485. doi: 10.1111/j.1432-1033.1979.tb13209.x. [DOI] [PubMed] [Google Scholar]
- NOMENCLATURE of blood clotting factors; four factors, their characterization and international number. J Am Med Assoc. 1959 May 16;170(3):325–328. [PubMed] [Google Scholar]
- Patterson J. E., Geller D. M. Bovine microsomal albumin: amino terminal sequence of bovine proalbumin. Biochem Biophys Res Commun. 1977 Feb 7;74(3):1220–1226. doi: 10.1016/0006-291x(77)91648-5. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rühlmann A., Kukla D., Schwager P., Bartels K., Huber R. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. Crystal structure determination and stereochemistry of the contact region. J Mol Biol. 1973 Jul 5;77(3):417–436. doi: 10.1016/0022-2836(73)90448-8. [DOI] [PubMed] [Google Scholar]
- Sigler P. B., Blow D. M., Matthews B. W., Henderson R. Structure of crystalline -chymotrypsin. II. A preliminary report including a hypothesis for the activation mechanism. J Mol Biol. 1968 Jul 14;35(1):143–164. doi: 10.1016/s0022-2836(68)80043-9. [DOI] [PubMed] [Google Scholar]
- Staden R. Further procedures for sequence analysis by computer. Nucleic Acids Res. 1978 Mar;5(3):1013–1016. doi: 10.1093/nar/5.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J. P., Catterall J. F., Woo S. L., Means A. R., O'Malley B. W. Molecular cloning of ovomucoid gene sequences from partially purified ovomucoid messenger RNA. Biochemistry. 1978 Dec 26;17(26):5763–5772. doi: 10.1021/bi00619a025. [DOI] [PubMed] [Google Scholar]
- Strauss A. W., Bennett C. A., Donohue A. M., Rodkey J. A., Boime I., Alberts A. W. Conversion of rat pre-proalbumin to proalbumin in vitro by ascites membranes. Demonstration by NH2-TERMINAL SEQUENCE ANALYSIS. J Biol Chem. 1978 Sep 10;253(17):6270–6274. [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Hirose T., Miyake T., Kawashima E. H., Itakura K. The use of synthetic oligonucleotides as hybridization probes. II. Hybridization of oligonucleotides of mixed sequence to rabbit beta-globin DNA. Nucleic Acids Res. 1981 Feb 25;9(4):879–894. doi: 10.1093/nar/9.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]



