Abstract
For cells to adapt to different tissues and changes in tissue mechanics, they must be able to respond to mechanical cues by changing their gene expression patterns. Biochemical signaling pathways for these responses have been elucidated, and recent evidence points to the involvement of force-induced deformation of the nucleus. However, it is still unclear how physical cues received at the plasma membrane (PM) spatiotemporally integrate to the functional chromatin organization of the cell nucleus. To investigate this issue, we applied mechanical forces through magnetic particles adhered to the PM of single cells and mapped the accompanying changes in actin polymerization, nuclear morphology, chromatin remodeling, and nuclear transport of soluble signaling intermediates using high-resolution fluorescence anisotropy imaging. Using this approach, we show the timescales associated with force-induced polymerization of actin and changes in the F/G actin ratio resulting in nuclear translocation of the G-actin-associated transcriptional cofactor, megakaryoblastic acute leukemia factor-1 (MKL). Further, this method of measuring nuclear organization at high spatiotemporal resolution with simultaneous force application revealed the physical propagation of forces to the nucleus, resulting in changes to chromatin organization, followed by nuclear deformation. We also describe a quantitative model that incorporates active stresses and chemical kinetics to evaluate the observed timescales. Our work suggests that mechanical activation of cells is accompanied by distinct timescales involved in the reorganization of actin and chromatin assembly, followed by translocation of transcription cofactors from the cytoplasm to the nucleus.
Introduction
Living cells, both in culture and in physiological conditions, are sensitive to mechanical cues from their local microenvironment (1). Cells respond to mechanical cues of fluid shear stress (2), compressive loading (3), and substrate stretching (4) by changing their morphology and modulating their gene expression profiles in response to these mechanical cues (5). Forces applied on the cell surface in the form of geometrical constraints (6), micropipette pulling (7,8), or magnetic beads (9) have been observed to transmit within the cytoplasm and to the nucleus, and induce nuclear deformations much faster than diffusion-limited processes. For this purpose, focal adhesions on the plasma membrane (PM), which act as mechanosensors (10), link the extracellular matrix with actin and microtubule cytoskeleton. Further downstream, SUN and KASH domain proteins form LINC (Linking Nucleus to the Cytoskeleton) complexes that couple the inner and outer nuclear membranes to the cytoskeleton (11–15). A variety of adaptor proteins, including Lamin B receptors (LBR), Emerin, and LAP2β, link the nuclear lamina and nuclear membrane (16–18). These proteins are further involved in linking the lamins with the chromatin structure through HP1 and other proteins (16). In this connected architectural framework, chromatin is nonrandomly organized in the nucleus, with interconnected chromatin structures and spatially organized transcriptionally active euchromatin and inactive heterochromatin structures (19–21). A number of experiments have suggested the possibility of physical transmission of forces from extracellular matrix to the nucleus and have also identified important nucleo-cytoskeletal links mediating such force transmission (22,23). In addition to physical transmission of forces to the nucleus, soluble signaling molecules have also been shown to shuttle between cytoplasm and the nucleus to activate gene expression (24). In this context, transcription cofactors such as megakaryoblastic acute leukemia factor-1 (MKL) have been shown to depend on actin polymerization states for nuclear signaling to invoke specificity in mechanotransduction (25,26). MKL is a transcription cofactor that is involved in the expression of genes mostly related to cytoskeletal architecture and dynamics (27). MKL localization in the nucleus is dependent on F/G-actin ratios that are regulated by cell-mechanics cues (28–30). However, the spatiotemporal integration of mechanical signals that are received at the PM and impinge on the physical architecture of actin and chromatin assembly and MKL signaling to the nucleus has not been explored.
In this work, we developed a single-cell force manipulation method that employs magnetic tweezers combined with fluorescence anisotropy imaging to study the spatial and temporal aspects of nuclear mechanotransduction. With this method, we probed the transduction of mechanical cues from PM to chromatin assembly through the actin cytoskeleton. Using time-lapse, live-cell imaging combined with application of forces on 100 nm paramagnetic beads adhered on the PM, we mapped the force-induced polymerization of actin and subsequent translocation of transcription cofactor MKL to the nucleus. Further fluorescence anisotropy imaging revealed that force-induced reversible remodeling of chromatin assembly preceded deformation of the nucleus, as evidenced by changes in the projected nuclear area and its height. Such remodeling of chromatin required the presence of intact nucleo-cytoplasmic actin links and actomyosin contractility. In addition, we computationally modeled the dynamic changes in nuclear projected area, actin reorganization, and cytoplasmic-to-nuclear translocation of MKL, and were able to delineate the spatiotemporal alterations in nuclear deformation and translocation of transcription cofactor MKL. Taken together, our results suggest that nuclear mechanotransduction is accompanied by distinct timescales of remodeling actin and chromatin assembly, followed by translocation of transcription cofactor MKL.
Materials and Methods
Cell culture, plasmids, and perturbations
HeLa cells and HeLa cells stably transfected with fusion plasmid for core histone H2B tagged with enhanced green fluorescent protein (EGFP) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; both from Life Technologies, Grand Island, NY) at 37°C in 5% CO2. Actin and MKL were visualized in HeLa cells using EGFP-actin (Clonetech, Mountain View CA) and mCherry-MKL (OriGene Technologies, Rockville, MD) fusion plasmids. LINC complex was disrupted by expressing dominant-negative KASH fused with EGFP (a kind gift from Dr. Brian Burke, Institute of Medical Biology, Singapore). Transfections were performed using Jetprime transfection reagent (Polyplus Transfection SA, Illkirch, France). Actin was disrupted by incubation in 1 μM solution of cytochalasin D (CytoD; Sigma Aldrich, St. Louis, MO) in imaging medium (DMEM without Phenol Red and with 3% FBS) for 1 h. The F/G-actin ratios in cells were perturbed by treatment with various concentrations (0 nM–1000 nM) of latrunculin A (Lat A; Sigma Aldrich) for 1 h. Actin filaments were stabilized by treatment with 500 nM Jasplakinolide (Sigma Aldrich) for 0.5 h. Myosin activity was perturbed by treating cells with 50 μM blebbistatin (Calbiochem, Darmstadt, Germany) for 2.5 h. Microtubules were disrupted by incubation in 1 μg/ml solution of Nocodazole (Sigma Aldrich) in DMEM with 10% FBS for 16 h or 10 μg/ml solution for 3 h.
Force protocols
A custom-made electromagnet was mounted on either an Olympus IX 81 or a Nikon eclipse Ti microscope to apply force on single cells. To apply force on the PM, cells plated on 35 mm coverslip dishes (IWAKI brand; Asahi Glass, Japan) were incubated with 10 μl of 100 nm paramagnetic beads (Micromod Partikeltechnologie, Rostock-Warnemuende, Germany) in 1 ml of cell culture medium for 2 h. Before the experiment, the culture medium containing beads was removed and cells were washed twice with 1X phosphate-buffered saline (PBS) to remove unbound beads. Cells were imaged in 3 ml of imaging medium to dissipate heat generated from the electromagnet. The electromagnet was positioned 30 μm away from the PM of cells. Calibration of the electromagnet is described in the Supporting Material. The first 75 s of the experiment were treated as the baseline, when no force was applied. This was followed by 225 s of force for prolonged-force experiments and 75 s for pulse-force experiments. Finally, the cells were allowed to recover for the next 225 s with no force being applied. The magnitude of force was changed by varying the current passed through the electromagnet. Because the magnetic field of a solenoid varies linearly with the current passed through it, the force magnitudes were scaled accordingly. For currents of 1 A, 1.5 A, and 2 A, forces were estimated to be 0.8 nN, 1.25 nN and 1.7 nN, respectively. All experiments were carried out at 25°C.
Fluorescence Anisotropy Imaging Microscopy
To obtain anisotropy images, we acquired fluorescence images on an inverted fluorescence microscope (model IX81, Olympus, Japan; or Eclipse Ti, Nikon Instruments). A linearly polarized excitation obtained by passing the arc lamp beam through a sheet polarizer (New Foucs, Santa Clara, CA) and excitation filter (HQ470/40x; Chroma Tech, Bellows Falls, VT) was used to excite H2BEGFP or actin EGFP-labeled cells, and emissions were collected through an emission filter (HQ 525/50m; Chroma Tech). Rhodamine phalloidin was excited with an excitation filter (HQ 560/40x; Chroma Tech) and emissions were collected with an HQ 630/75m emission filter (Chroma Tech). Each emission was split into parallel and perpendicular polarizations using a U-SIP polarizing module (Olympus, Japan) or Roper Dual View polarization module (Photometrics, Tucson, AZ). The parallel and perpendicular images were simultaneously acquired on two halves of an Andor iXon 897 BV EMCCD camera (Andor Technology, South Windsor, CT). Time-lapse imaging was achieved by using a custom-written program in LabVIEW (National Instruments, Austin, TX) or NIS Elements (Nikon Instruments, Melville, NY) to control the microscope. Data acquisition was done using DAQ (PCI-MIO-16XE-10; National Instruments, Austin, TX), LabVIEW, and NIS Elements. Images were acquired at 7.5 s/frame for slow time-lapse imaging, and 0.1 s/frame for fast time-lapse imaging. A custom-written program in LabVIEW or MATLAB (The MathWorks, Natick, MA) was used to align the parallel and perpendicular images obtained on the two halves of the EMCCD camera and compute anisotropy from the split image on a pixel by pixel basis after performing background subtraction and 3×3 smoothing of the original image. For each pixel, anisotropy was calculated as
where I∥ and I⊥ are background-subtracted intensities of the emitted light with polarization parallel and perpendicular to the excitation, respectively. The g factor arises due to variable sensitivity of the detection system for the parallel and the perpendicular components of the light. The g factor is estimated from the anisotropy of small FITC dye molecules at different concentrations of glycerol (see Fig. S1 in the Supporting Material).
Fluorescence recovery after photobleaching
A fluorescence recovery after photobleaching (FRAP) experiment was performed on MKL-mCherry-labeled cells. A small region (∼3 μm diameter) in either cytoplasm or nucleus was photobleached. Images were acquired at 300 ms/frame to capture the dynamics of the fluorescence recovery. Analysis was performed offline using a custom-written program in MATLAB. Fluorescence intensity in the photobleached region was computed at each time frame, before and after photobleaching. The obtained intensity was normalized as
where Inorm is the normalized intensity, and Ipre-bleach and Ipost-bleach are the fluorescence intensities before and after photobleaching, respectively. The extent of fluorescence recovery in FRAP experiments is generally underestimated due the overall bleaching of the cell. To correct for this, a reference region of interest (ROI) was selected away from the photobleached region, and its normalized intensity was estimated as a function of time. The intensity of the bleached region was normalized by the intensity of the reference region to obtain the actual recovery fraction and recovery rate for the nucleus and cytoplasm.
Theoretical modeling and simulation
The nucleus and stress fibers are modeled as viscoelastic solids (springs and dashpots in parallel) (31–33), whereas the lamellar network is a viscoelastic fluid in which elastic element on the timescale of tens of seconds can be neglected (34). The nucleus is ∼10 times stiffer than the cytoplasm actin network (35), and we assume that stress fibers are an order of magnitude stiffer than the effective spring constants usually measured for the lamellar network. Modeling realistic geometry and viscoelastic deformation of the nucleus and cytoskeleton is complicated, and for comparison with a simple measure such as a normalized nucleus area does not give better results than simplistic modeling of the nucleus and cytoskeleton as viscoelastic springs and dashpots (see Fig. 7 a). The orders of magnitude of the mechanical properties of the actin network and nucleus can be gleaned from previous works (33,36,37). We obtained the spring constants used in the simulations by multiplying Young’s modulus magnitudes reported in the literature by the characteristic nuclear size of ∼10 μm. The respective viscous drag coefficients can be estimated from measured characteristic relaxation times ∼100 s (37) and spring constants (viscous drag is of the order of the time constant multiplied by the spring constant). Definitions and values of the model parameters and variables are summarized in the Table S1 and shown in Fig. 5 a.
Figure 7.
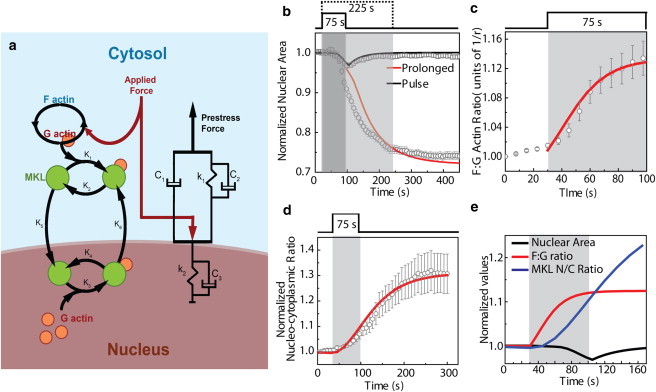
Theoretical model describing the force transduction timescales. (a) Diagram of the mechanical (right), actin turnover (upper left), and MKL transport (lower left) submodels. (b) Predicted time series and data for the normalized nuclear area for prolonged force (open circles: experiment; red line: model fit) and pulse force (open squares: experiment; black line: model fit). (c) Predicted time series and data for the F/G-actin ratio (open circles: experiment; red line: model fit). The F/G-actin ratio is shown in units of 1/r of EGFP-actin. (d) Predicted time series and data for the ratio of the MKL concentrations in the nucleus and cytoplasm (open circles: experiment; red line: model fit). (e) The graph shows the timescales involved in change in area, F/G-actin ratio, and MKL nucleocytoplasmic ratio for a pulse-force application. Error bars indicate the experimental mean ± SE.
Figure 5.
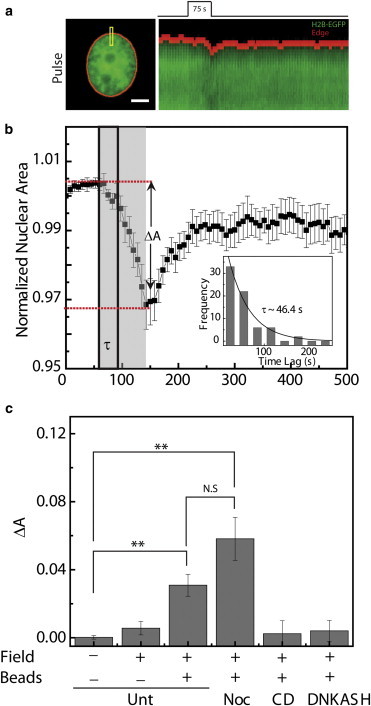
Modulation of nucleo-cytoskeletal coupling by force. (a) Image of an H2B-EGFP HeLa cell before force. The kymograph of the ROI shown in yellow represents the deformation of the nucleus upon application of pulse force. (b) Plot showing the change in normalized nuclear area upon application of pulse force (n = 20). The gray region shows the duration of application of force and black ROI represents the lag time involved in nuclear deformation. ΔA represents the change in nuclear area after application of force. Inset shows the distribution of lag times. (c) Plot showing ΔA under different conditions and perturbations. “Field” represents the on or off state of magnetic field, and “Beads” denotes the presence or absence of beads on the PM. Noc and CD denote cells treated with nocodazole (n = 20) and CytoD (n = 20), respectively, before application of force. DNKASH represents cells expressing dominant-negative KASH fused with EGFP (n = 10). Error bars denote the mean ± SE; scale bar: 5 μm; ∗∗p < 0.01; N.S., not significant.
The displacement of the nuclear membrane, Z, satisfies the equation
which, taking into account the prestressed elongations of the nuclear and cytoskeletal springs, can be simplified to the forms shown in the text. The spring lifetimes are chosen to fit the observations. We solved the equations in the main text using the Virtual Cell software. The projected area of the nucleus is computed as , where R is the unstressed nuclear radius. For all submodels, we ran 20 simulations in which all model parameters were varied randomly within 30% of their baseline value (the average and mean ± SE are shown).
The submodels for the F-G-actin turnover and MKL transport are described in the Supporting Material and are solved and presented in similarity to the mechanical submodel. We chose the characteristic times of F-G-actin turnover based on previous measurements. The order of magnitude of the MKL transport model can be gleaned from a previous work (25); exact numbers were chosen to fit the data.
Statistical analysis
Student’s t-test was performed for all quantitative comparisons in which the SDs were overlapping. The respective p-values are mentioned in the figure legends.
Results
Direct measurement of force-induced changes in F/G-actin ratio
We investigated force-induced actin polymerization dynamics and its role in nuclear mechanotransduction using fluorescence anisotropy imaging. To that end, we expressed EGFP-actin in HeLa cells, which colocalized well with the endogenous actin networks (Fig. S2 a). Fluorescence anisotropy of EGFP-actin was obtained from parallel and perpendicular images, as shown in Fig. 1. Cells expressing EGFP-actin showed lower values of anisotropy at stress fibers with a higher density of actin subunits (Fig. 1 b and Fig. S2 b). Further, the anisotropy of EGFP-actin was inversely dependent on the expression of EGFP-actin in cells (Fig. S2 c). This suggested the possibility of homo-Förster resonance energy transfer (homoFRET) between EGFP-actin molecules (38). The presence of homoFRET between proteins can be confirmed by photobleaching of the molecules, which effectively increases the distance between them, leading to decreased FRET and an increase in anisotropy (39). Hence, we performed photobleaching experiments on cells labeled with EGFP-actin to validate the presence of homoFRET between actin monomers within the stress fiber. When a small region of the cell was photobleached, an increase in the anisotropy of EGFP-actin was observed (Fig. 2, a and c), suggesting homoFRET between EGFP-labeled actin molecules (38). Similar results were obtained by photobleaching rhodamine phalloidin-stained cells (Fig. S3, a and b), confirming homoFRET between actin molecules on the stress fibers.
Figure 1.
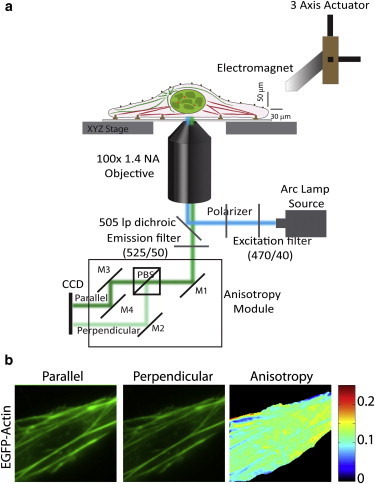
Experimental set up for fluorescence anisotropy imaging and micromanipulation. (a) Schematic of experimental setup for measuring fluorescence anisotropy combined with application of force on single living cells. (b) Representative images showing intensity of EGFP-actin in parallel and perpendicular channels, and anisotropy of EGFP-actin.
Figure 2.
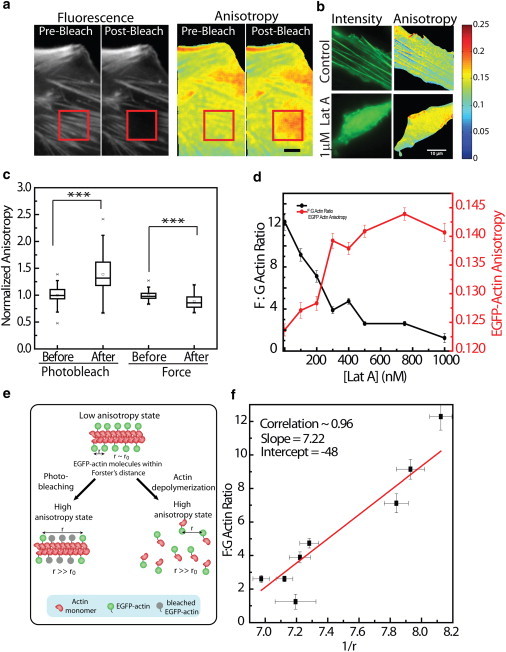
Visualization of actin polymerization by homoFRET anisotropy. (a) Intensity and anisotropy of EGFP-actin before and after photobleaching. Photobleaching was performed in the region shown by the red ROI. (b) Intensity and anisotropy images of EGFP-actin in control and in Lat A-treated cells. (c) Box plot showing normalized anisotropy before and after photobleaching(n = 50) or upon application of force (n = 16). (d) Plot showing a decrease in the F/G-actin ratio (black) and an increase in EGFP-actin anisotropy (red) upon treatment with various concentrations of Lat A (n > 60). (e) Schematic describing the measurement of F/G-actin ratio by fluorescence anisotropy. (f) Scatter plot between 1/r and F/G-actin ratio. The red line indicates linear fit. Error bars indicate the mean ± SE; ∗∗∗p < 10−5; scale bar: 10 μm.
To further validate force-induced changes in anisotropy values as a measure of the F/G-actin ratio, we treated cells with Lat A, which depolymerizes actin filaments and alters the F/G-actin ratio. F-actin was visualized by Alexa-568 phalloidin, and G-actin was visualized by DNaseI conjugated to Alexa-488 (Fig. S4 a). Titration of Lat A concentration (0–1000 nM) showed a significant decrease in the F/G-actin ratios (Fig. 2 d and Fig. S4 b). Concomitantly, a significant increase in anisotropy of EGFP-actin was observed for the same concentration of Lat A, suggesting that the increase in anisotropy values represents a shift from polymeric to monomeric actin (Fig. 2, b, d, and e, and Fig. S4 c). Further, the increase in anisotropy was similar to that observed in the photobleaching experiments, suggesting a decrease in homoFRET between the monomers (Fig. S4 d). This was also confirmed by observing the changes in EGFP-actin anisotropy upon release from Lat A treatment. EGFP-actin expressing cells were treated with 1 μM Lat A for 30 min. After the Lat A was washed off, a decrease in anisotropy was observed corresponding to polymerization of actin (Fig. S4 e). A scatter plot between 1/r and F/G-actin ratios showed a linear correlation, with a Pearson correlation of 0.96 (Fig. 2 f). This established that the F/G-actin ratio is inversely proportional to the anisotropy of EGFP-actin, as
where Factin and Gactin represent the concentrations of F-actin and G-actin, respectively, and A = 7.22 and B = −48 are the constants obtained from the slope and intercept of the linear fit of the calibration curve, respectively.
Next, we used anisotropy of EGFP-actin to observe the changes in actin polymerization upon application of force. The schematic of the experimental geometry is shown in Fig. 1 a. Controlled step forces were applied on 100 nm paramagnetic carboxyl beads adhered onto the PM (∼1800 beads per cell) of HeLa cells (Fig. S5) using an electromagnet placed 30 μm away from the cell surface at an angle of 30° with the horizontal (shown in the schematic of Fig. 1). This resulted in a significant component of force in the horizontal direction, although a smaller component was observed in the vertical direction. The distribution of beads on the cell surface and the variation of the force with distance from the electromagnet, obtained from the calibration curve (Fig. S6 and Supporting Material), resulted in a force similar to the application of shear force on cells. When such a calibrated pulse force (1.25 nN averaged over the cell, for 75 s) was applied, a decrease in anisotropy of EGFP-actin was observed, which was evident from the time course images and the line profiles of EGFP-actin stress fibers (Fig. 3, a–c). Fig. 3 d shows the observed increase in 1/anisotropy of EGFP-actin upon application of the pulse force. When the observed values of 1/anisotropy of EGFP-actin were mapped to the calibration curve, changes in F/G-actin ratios were obtained that showed an increase upon application of force, suggesting enhanced actin polymerization (Fig. 3 e). To further validate that the observed increase in the F/G-actin ratio was due to increased actin polymerization, we rendered G-actin nonpolymerizable by treating the cells with Lat A or CytoD. This resulted in insignificant changes in F/G-actin ratios, suggesting that these changes were indeed due to enhanced actin polymerization (Fig. 3 e). Because MKL, a transcription cofactor of SRF, has F/G-actin ratio-dependent nuclear localization (26), we directly probed the effect of a force-induced elevated F/G-actin ratio on MKL nuclear translocation.
Figure 3.
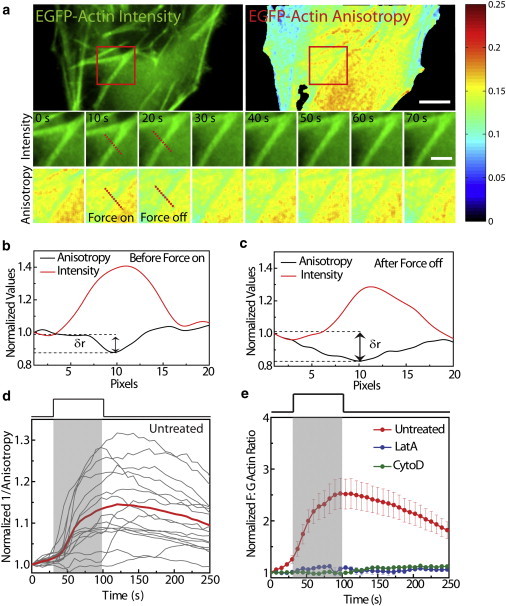
Force-induced actin polymerization. (a) Images showing EGFP-actin intensity and anisotropy. The collage of images shows the changes in intensity and anisotropy of EGFP-actin upon application of force, in the region shown by the red ROI. (b and c) Line profile of intensity and anisotropy of EGFP-actin across the dotted lines shown in panel a, before (b) and after (c) application of force. δr shows the drop in anisotropy over the actin stress fiber. (d) Increase in 1/anisotropy of EGFP-actin upon application of pulse force. Gray traces represent individual cell response and the red line indicates the mean response (n = 16). (e) Changes in F/G-actin ratio upon application of pulse force in untreated cells (n = 16) and cells treated with Lat A (n = 10) or CytoD (n = 10). Scale bar: 5 μm. Scale bar in inset: 2 μm. Error bars indicate the mean ± SE.
Mechanical force-induced nuclear translocation of MKL
We probed force-induced nuclear translocation of MKL by applying a pulse force on cells expressing MKL-mCherry. To estimate the amount of nuclear translocation, we computed the nucleo-cytoplasmic (N/C) ratio of MKL at each time frame and normalized it with the N/C ratio before application of force. A very small change in the N/C ratio was observed during the first 40 s after the onset of force, which was not significant from the N/C ratio before the application of force (Fig. 4, a and c). However, a significant increase in the N/C ratio was observed beyond 40 s, and the translocation of MKL continued even after the cessation of the force (Fig. 4, a and c, inset i). In contrast, control cells with no force applied showed insignificant translocation of MKL during the period of observation of 500 s (Fig. 4 c, inset i).
Figure 4.
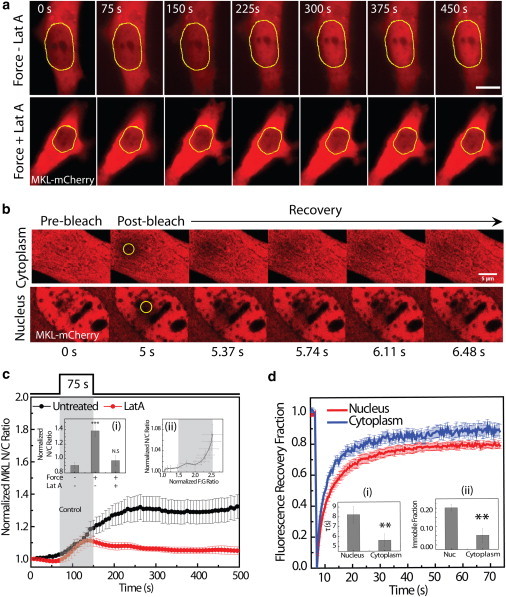
Force-induced nuclear translocation of transcription cofactor MKL. (a) Images showing cytoplasmic and nuclear localization of MKL-mCherry in untreated cells and cells treated with 500 nM Lat A, upon application of force. Force was applied from 75 s - 150 s. (b) Collage of images showing cytoplasmic and nuclear fluorescence intensity of MKL-mCherry upon photobleaching and recovery. The yellow ROI represents the photobleached region. (c) Normalized N/C ratio of MKL mCherry in untreated cells (n = 8) and cells treated with Lat A (n = 4), and upon application of force. The gray region represents the duration of force. Inset i shows the normalized N/C ratio at t = 300 s, without application of force and with application of force on untreated cells and cells treated with Lat A. Inset ii shows the plot between normalized F/G-actin ratio and normalized MKL N/C ratio. Gray box indicates application of force. (d) Fluorescence recovery curve for cytoplasmic and nuclear MKL. Inset i shows the recovery timescales, and inset ii shows the immobile fraction for cytoplasm and the nucleus. Error bars denote the mean ± SE. Scale bars: 5 μm; ∗∗p < 0.01; ∗∗∗p < 0.001.
Although the nuclear translocation of MKL was in agreement with the force-induced changes in the F/G-actin ratio, we addressed the possibility that this could also have been a direct effect of force. To that end, we measured force-induced MKL translocation in cells treated with Lat-A, and MKL translocation upon treatment with the actin-stabilizing agent jasplakinolide without application of force. Whereas application of force in the presence of Lat-A showed an insignificant increase in the N/C ratio of MKL (Fig. 4 c and inset i), treatment of cells with jasplakinolide without force application showed a significant nuclear translocation of MKL (Fig. S7, a and c). These results validated that the observed increase in the MKL N/C ratio was due to the force-induced changes in F/G-actin ratio. The sigmoidal behavior of MKL nuclear translocation with an initial delay in response suggests that this process may be triggered by an increase in the F/G-actin ratio (elaborated in the theoretical modeling). To investigate this, we measured the sensitivity of the MKL translocation to the F/G-actin ratio in cells. When the N/C ratio of MKL was plotted against the F/G-actin ratio, no significant increase in the MKL N/C ratio was observed until the F/G-actin ratio increased by ∼2-fold (Fig. 4 c, inset ii). A significant increase in the N/C ratio was observed only when the F/G-actin ratio increased beyond this threshold value, and the translocation was sustained even when the F/G-actin ratio ceased to increase, suggesting that a threshold value of F/G-actin ratio may be required to trigger MKL nuclear translocation.
To probe the mobility of nuclear fraction of MKL, we carried out fluorescence recovery after photobleaching (FRAP) of MKL-mCherry transfected cells in both the cytoplasm and the nucleus. We observed a significantly slower recovery of MKL in the nucleus as compared with the cytoplasm (τ ∼ 8.2 s for the nucleus and τ ∼ 5.5 s for the cytoplasm; Fig. 4 d and 4 d, inset i). Further, the immobile fraction of MKL, estimated from the extent of fluorescence recovery, was significantly larger in the nucleus than in the cytoplasm (Fig. 4 d, inset ii). These results suggested that the nuclear fraction of MKL was less mobile due to diffusion barriers resulting from macromolecular crowding, or to transient interactions with the chromatin. Although these soluble intermediates provide specificity to the process of mechanotransduction, physical forces could also modulate the nuclear morphology and chromatin organization to bring about changes in transcription. Because the dynamics of actin cytoskeleton and its link to the nucleus play a critical role in nuclear organization, we next investigated the effect of force on nucleo-cytoplasmic links.
Force-induced modulation of cytoplasmic to nuclear coupling
We probed the nature of the coupling between the nucleus and the cytoskeleton by dynamically modulating both the duration and magnitude of forces applied on the PM. Upon application of a pulse force, we observed a small decrease in the projected nuclear area (ΔA, ∼5%; Fig. 5, a and b) that was completely reversible upon cessation of force, suggesting that the active remodeling of the nucleo-cytoplasmic links maintain the nuclear architecture (Fig. S8 e). In contrast, an insignificant decrease in the nuclear cross-sectional area was observed under zero force conditions (Fig. 5 c). When the duration of force was increased to 225 s, keeping the magnitude of force invariant (prolonged force), the nuclei showed an irreversible (30% ± 6%) decrease in the cross-sectional area (Fig. S8, a, b, and d) with an overall deformation of the nucleus, indicated by an increase in the nuclear height (Fig. S8 c).
Upon modulation of the force magnitude, keeping the duration of the force constant (75 s), similar responses were observed. Application of 0.8 nN force resulted in an insignificant change in nuclear area (Fig. S8 f), 1.25 nN force resulted in a reversible change in the nuclear deformation (Fig. S8 f), and 1.7 nN force induced irreversible deformation of the nucleus (Fig. S8 f). These results suggested that changes in the duration or magnitude of force resulted in dynamic modulation of the nucleo-cytoplasmic links. As a control, we estimated the contribution from the pleiotropic effect of applying forces using an electromagnet by observing the change in size of the nucleus upon application of magnetic field on cells lacking the paramagnetic beads. In this case, we observed a very small change in the nuclear area unlike that seen when force was applied on cells with magnetic beads (Fig. 5 c).
This dynamic force-induced modulation of the nuclear area was abrogated upon chemical perturbation of nucleo-cytoplasmic links by inhibiting actin polymerization using 1 μM CytoD (Fig. S9). The change in projected nuclear area (ΔA) with microtubule depolymerization using nocodazole was similar to that observed for untreated cells (Fig. 5 c), whereas actin perturbation using CytoD resulted in a smaller ΔA compared with untreated cells (Fig. 5 c and Fig. S8 g), which suggests the role of actin in linking the nucleus to the cytoskeleton, and is consistent with previous reports (6–8,40). To further probe the role of the LINC complex in this dynamic modulation, we perturbed the localization of Nesprin 2, a component of the LINC complex, by expressing dominant-negative KASH fused with EGFP (DNKASH) (7). In accord with previous reports (7), we observed that expression of DNKASH-EGFP displaced endogenous Nesprins from the nuclear envelope to the endoplasmic reticulum (Fig. S10 a) (7).When prolonged force was applied on cells expressing DNKASH, we observed a very small change in the projected nuclear area (Fig. 5 c and Fig. S10 b).
Perturbation of the LINC complex has been shown to affect perinuclear actin organization without changing the overall F/G-actin ratio (7), suggesting that DNKASH expression should not affect MKL nuclear translocation. To further address the possibility that DNKASH expression modulates MKL nuclear translocation, we performed experiments by applying force on cells expressing both MKL-mCherry and DNKASH-EGFP. A significant translocation of MKL was observed upon force application, similar to what was observed for the untreated cells (Fig. S7, b and d). These observations further suggest that the application of force results in modulation of the nucleocytoplasmic links, which does not impinge on the overall F/G-actin ratios and MKL nuclear translocation.
Interestingly, deformation of the nucleus upon application of force was not immediate. Instead, it was preceded by a lag time, measured as the time between the application of force and onset of the collapse when the nuclear area decreases by >2 standard deviations (SDs) from its mean value before application of force (Fig. 5 b). This lag time was exponentially distributed with a mean of ∼46 s (Fig. 5 b, inset), probably due to the viscoelastic behavior of the cytoskeleton. Upon deletion of the LINC complex by expression of DNKASH, large lag times were observed, further suggesting that these lag timescales are due to the stochastic rupture of the nucleo-cytoplasmic links. The observed lag time may provide a temporal window for propagation of physical forces to the chromatin organization, before the nucleocytoplasmic links are perturbed. Hence, we next probed the spatiotemporal integration of these forces to chromatin assembly and assessed the timescales associated with its remodeling.
Visualization of rapid force-induced chromatin remodeling
To probe changes in chromatin assembly upon application of forces, we used fluorescence anisotropy imaging, a method that we have established to visualize chromatin compaction states (41). The changes in H2B-EGFP anisotropy arise from rotational diffusion, in contrast to homoFRET in EGFP-actin. This was validated by photobleaching experiments on H2B-EGFP-expressing cells. Upon stepwise photobleaching, an insignificant change in anisotropy of EGFP-actin was observed (Fig. S11), negating the presence of homoFRET. To further ensure that the fluorescence anisotropy imaging method was sensitive to changes in chromatin compaction states in the HeLa cell nucleus, we visualized regions showing higher anisotropy of H2B-EGFP (one of the core histone proteins that compact DNA into chromatin) and correlated them with regions of compact heterochromatin regions identified by localization of the heterochromatin-binding protein HP1α in the nucleus (Fig. 6 a), a known marker for heterochromatin (42,43). The scatter plot between H2B-EGFP anisotropy and HP1α intensity shows a strong correlation, with a Pearson correlation coefficient of 0.82 (Fig. 6 b). In addition, our earlier experiments showed that trichostatin A, a histone deacetylase inhibitor that decompacts chromatin (44), results in a decrease in anisotropy of H2B-EGFP compared with control cells (45). These results suggest that fluorescence anisotropy of H2B-EGFP is a measure of chromatin compaction states.
Figure 6.
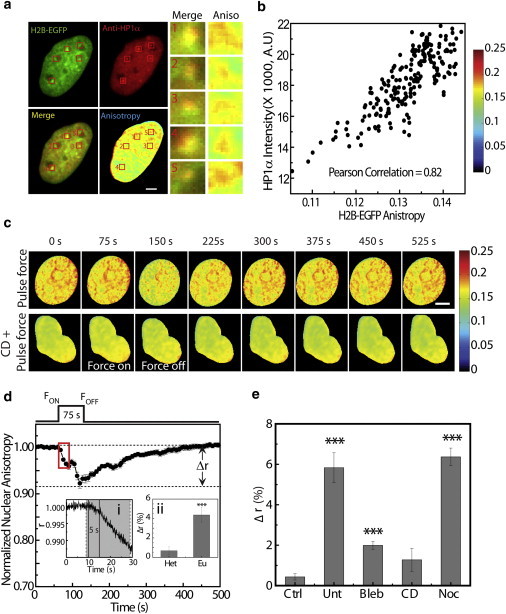
Force-induced rapid chromatin remodeling visualized by fluorescence anisotropy. (a) Image of HeLa cell expressing H2B-EGFP stained for anti-HP1α antibody. H2B-EGFP is shown in green, anti-HP1α antibody is shown in red, and the merged regions are shown in yellow. Enlarged images of the ROIs marked in red show colocalization between regions with high intensity of HP1α and high anisotropy of H2B-EGFP. (b) Scatter plot between H2B-EGFP anisotropy and HP1α intensity. (c) Time-course images showing the change in anisotropy of H2B-EGFP upon application of force in untreated cells and cells treated with CytoD (CD + pulse force). (d) Plot showing normalized nuclear anisotropy upon application of pulse force (n = 20). The decrease in anisotropy upon application of force is represented by Δr. Inset i shows anisotropy at faster temporal resolution of the duration shown in red ROI. Inset ii shows Δr in heterochromatin and euchromatin regions. (e) Bar plot showing Δr upon zero force (Ctrl) and upon application of pulse force in untreated cells (Unt, n = 20) and cells treated with blebbistatin (Bleb, n = 10), CytoD (CD, n = 10) or nocodazole (Noc, n = 10). Error bars denote the mean ± SE; ∗∗∗p < 10−5; scale bar: 5 μm.
To probe the remodeling of chromatin through the nucleo-cytoskeletal links, we subjected cells to pulse force and measured the changes in chromatin compaction. Pulse force resulted in a rapid decompaction of chromatin that was completely reversible upon cessation of force (Fig. 6, c and d). The timescale involved in initiation of the chromatin remodeling process was captured by fast time-lapse imaging at 10 frames per second. A significant change in chromatin decompaction was observed within 5 s of application of force (p < 0.05), suggesting the role of physical transduction of forces in remodeling the chromatin structure (Fig. 6 d, inset i). Further, when multiple pulses of force were applied, a similar response was observed, with rapid decompaction of chromatin and recovery upon cessation of the force (Fig. S12), reflecting the direct transmission of forces to the chromatin organization. Interestingly, the chromatin decompaction was observed within the lag time of 46 s that was required for nuclear deformation due to modulation of nucleo-cytoplasmic links. Moreover, Δr (change in anisotropy value of H2B-EGFP upon application of force) was observed to be dependent on the state of chromatin compaction before application of force. Highly condensed heterochromatin regions showed smaller changes in anisotropy, i.e., smaller Δr, whereas larger Δr was observed in less-condensed euchromatin regions (Fig. 6 d, inset ii, and Fig. S13). This suggests differential transmission of forces through the chromatin structure, making euchromatin more sensitive to force-dependent changes.
When adherent cells are spread on a substrate, they are maintained in a prestressed state by nucleo-cytoskeletal links. Decompaction of chromatin through these links suggests that the chromatin should be partially decondensed in adherent cells that are already under prestress. To probe this, we treated cells with CytoD, which relieves the nuclear prestress by disrupting the actin cytoskeleton. We found that when the prestress was relieved, the anisotropy increased, further validating chromatin remodeling through the nucleo-cytoskeletal links (Fig. S14 a). The change in anisotropy value after application of force (Δr) is sensitive to perturbations to the actin cytoskeleton. Force applied on untreated cells resulted in a Δr of ∼6%, which decreased to ∼2% upon treatment with CytoD and to 3% upon treatment with blebbistatin (Bleb; Fig. 6 e and Fig. S14). Depolymerization of microtubules by nocodazole resulted in Δr similar to that of untreated cells, suggesting that microtubules may be less important for transmitting forces from the PM to the chromatin organization (Fig. 6 e and Fig. S14). We next developed a simple phenomenological model for nuclear mechanotransduction to evaluate the distinct timescales involved in force-induced remodeling of actin and chromatin assembly, and the resulting changes in nuclear deformation and MKL transport.
Theoretical modeling of force-transduction dynamics
We modeled the cytoskeleton as very viscous lamellar actin fluid in parallel with viscoelastic stress fibers connected to the nucleus, which was also modeled as a viscoelastic body (Fig. 7 a and Table S1). We calibrated the model with values of mechanical moduli found in the literature (Supporting Material), and solved equations of the mechanics of such system deformed by the external force, assuming that links between the actin cytoskeleton and nucleus (or mechanical elements within the nucleus) break if the strong external force persists for too long. We found that the simulations reproduced the nuclear deformation (Fig. 7 b and Fig. S15, a and b), indicating that the viscoelastic elements in the nucleus and cytoskeleton are responsible for instantaneous force sensing, and the viscous elements introduce characteristic timescales of tens of seconds, allowing the cell to gauge not only the magnitude but also the duration of the mechanical perturbations.
Furthermore, making the simple assumption that G- to F-actin assembly is linearly accelerated by external stress, we demonstrated mathematically than an immediate, linearly growing shift from monomeric to F-actin in response to external force takes place (Fig. 7 c and Fig. S15 c). Finally, we modeled MKL transport as simple first-order reactions, assuming that the MKL-G-actin association reactions obey mass action kinetics (Fig. 7 a; Supporting Material). Specifically, the reactions took place among four types of molecules: both cytoplasmic and nuclear MKL transited between the states bound to and disassociated from actin monomers. Cytoplasmic MKL disassociated from G-actin was transported into the nucleus with a constant rate, and nuclear MKL associated with G-actin was transported to the cytoplasm with a constant rate (the reaction rates can be gleaned from Fig. 7 a). The molar MKL concentration is much smaller than that of G-actin, so we did not include G-actin transport into and out of the nucleus. The G-actin concentration in the cytoplasm was obtained from the F-G-actin dynamics model. The numerical solution of the model showed that the observed sigmoidal chemical response to the force with the characteristic time lag (Fig. 7 d and Fig. S7 d) is the result of the multistep biochemical relay (Fig. 7 e), and thus the mechanochemical signaling is delayed relative to the mechanical sensing, perhaps indicating the second stage of the cellular response to the force.
Interestingly, the model parameters at which the predictions fit the data are such that reaction between MKL and G-actin in cytoplasm is much faster than all other reactions. Thus, if the cell is treated with Lat A and the equilibrium G-actin concentration in the cytoplasm is lower, the model predicts that when force is applied, the resulting MKL nucleo-cytoskeletal ratio will be lower than in the control case, but the time lag until the steady ratio upon the force application will be the same as that in the control case. Both of these predictions agree with the experimental results (Fig. 4 c). Similarly, the model predicts that in the case of jasplakinolide treatment, when the F-actin is stabilized and G-actin is lowered, the MKL nucleo-cytoskeletal ratio has to be higher than in control, which we found to be the case (Fig. S7 c).
Discussion
In this work, we have demonstrated that nuclear mechanotransduction of force applied on the PM can be modeled as a two-step process. The first step involves the remodeling of actin cytoskeleton and chromatin assembly. The actin cytoskeleton reorganized with the application of force, as visualized by fluorescence anisotropy imaging of EGFP-actin molecules. HomoFRET between EGFP-actin molecules provided a direct measure of changes in F/G-actin ratios upon force application. These changes in F/G-actin ratio could be a consequence of a force-dependent increase in Rho GTPase activity (46). FRET has been shown between actin monomers labeled with different fluorophores (47); however, the competition between these fluorophores for incorporation into actin structure results in a decreased FRET efficiency. Such effects are not observed in homoFRET because the same fluorophore acts as both donor and acceptor.
A recent study showed the rapid dissociation of Cajal bodies interacting with chromatin upon application of force on PM (48). Here, we extend that observation to understand the transmission of force to chromatin organization. Concomitantly with actin remodeling, transmission of force to the nucleus resulted in decondensation of chromatin within 5 s. Actin cytoskeleton played a greater role in force transmission as compared with microtubules, in agreement with recent evidence (48). Interestingly, the fluorescence anisotropy values of H2B-EGFP were altered in relatively more decompact regions (euchromatin) upon force application, suggesting that these regions were more sensitive to changes in physical cues. These heterogeneous responses in chromatin compaction states could provide a mechanism for unfolding local chromatin structure (possibly at regulatory regions) upon application of force. Although our experiments strongly suggest the physical transmission of forces to the chromatin organization, chemical signaling intermediates could play a role in further modulating these responses.
The force-induced decondensation of chromatin was abrogated upon disruption of actin cytoskeleton or actomyosin contractility, suggesting the transmission of force through an elaborate network of actomyosin-dependent physical connections bridging the cytoskeleton and the nucleus, consistent with recent findings (6,7,49). In addition to chromatin remodeling, application of prolonged force on the PM resulted in an irreversible change in the nuclear area. Such an irreversible response could be attributed to the physical plasticity of the nucleus of differentiated cells, as previously observed in experiments involving micropipette aspiration of cells (35). The observed change in nuclear area was due to rupture of nucleocytoplasmic links relaxing the nuclear prestress, which was a stochastic phenomenon. Hence, this resulted in a much slower response (τ ∼ 46 s) in comparison with changes in chromatin compaction, which required an intact cytoskeleton.
The second step of the mechanotransduction process involves translocation of transcription cofactor MKL into the nucleus, perhaps to regions of decondensed chromatin. Translocation of MKL into the nucleus is critically dependent on the concentrations of free MKL and MKL bound to G-actin, as shown in the theoretical modeling. The theoretical model predicted that the amount of nuclear translocation of MKL would be affected by latrunculin treatment, whereas the initial time lag in the sigmoidal response would be unaffected. This was validated by experiments in which force was applied on cells treated with Lat A. The rates of different steps involved in this mechanotransduction process, obtained from the theoretical modeling, suggested a faster timescale in force transmission to chromatin assembly, which may provide accessibility to the transcription machinery. This step was followed by a much slower process of translocation of MKL into the nucleus, perhaps providing specificity for initiation of transcription at MKL target sites (26).
Recent evidence suggests that force integrates to proteinaceous scaffolds, thereby opening up buried amino acid residues for post-translational modifications and further eliciting signaling cascades (50,51). Force has also been shown to modulate chromatin structure in vitro (34,52). In addition, work from our laboratory and others has shown that core histones, the prime modulators of gene expression, are less mobile and exhibit spatiotemporal compaction in their organization (53,54). In this context, integration of forces from the PM to chromatin structure then provides a mechanism to expose local chromatin structures of distinct stiffness due to the heterogeneity in their packaging. Our findings hint at the possibility that force integration to the prestressed cell nucleus, along with translocation of transcription factors and cofactors, can differentially tune the local fluidity (55) and accessibility of chromatin assembly, possibly eliciting differential functional outputs. Taken together, our results offer a model for mechanotransduction that can be tested across different cell types and transcription factor systems. Because cells in tissue constantly adapt to their local microenvironment by modulating their gene expression patterns, the dynamic nucleo-cytoplasmic connections and the distinct timescales in nuclear mechanotransduction that we reveal may be integral components of cell behavior.
Acknowledgments
We thank Dr. Brian Burke for providing us with plasmids and antibodies, and Michael P. Sheetz for valuable discussions.
This study used the core facilities at the Mechanobiology Institute, Singapore, and the National Centre for Biological Sciences, Bangalore. K.V.I. received a graduate research fellowship from the Council of Scientific and Industrial Research. This work was funded in part by the Department of Science and Technology Nanoscience Initiative, India and Mechanobiology Institute, Singapore.
Supporting Material
References
- 1.Vogel V., Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 2.Tzima E., Irani-Tehrani M., Schwartz M.A. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 3.Leipzig N.D., Athanasiou K.A. Static compression of single chondrocytes catabolically modifies single-cell gene expression. Biophys. J. 2008;94:2412–2422. doi: 10.1529/biophysj.107.114207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamada M., Sheetz M.P., Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev. Cell. 2004;7:709–718. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Wang J.H., Thampatty B.P., Im H.J. Mechanoregulation of gene expression in fibroblasts. Gene. 2007;391:1–15. doi: 10.1016/j.gene.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khatau S.B., Hale C.M., Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombardi M.L., Jaalouk D.E., Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maniotis A.J., Chen C.S., Ingber D.E. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Na S., Collin O., Wang N. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc. Natl. Acad. Sci. USA. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiger B., Spatz J.P., Bershadsky A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 11.Crisp M., Liu Q., Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostlund C., Folker E.S., Worman H.J. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J. Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haque F., Lloyd D.J., Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzur Y.B., Wilson K.L., Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 15.Nery F.C., Zeng J., Breakefield X.O. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J. Cell Sci. 2008;121:3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruenbaum Y., Margalit A., Wilson K.L. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 17.Lammerding J., Hsiao J., Lee R.T. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lammerding J., Schulze P.C., Lee R.T. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert N., Boyle S., Bickmore W.A. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Lanctôt C., Cheutin T., Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 22.Shivashankar G.V. Mechanosignaling to the cell nucleus and gene regulation. Annu Rev Biophys. 2011;40:361–378. doi: 10.1146/annurev-biophys-042910-155319. [DOI] [PubMed] [Google Scholar]
- 23.Wang N., Tytell J.D., Ingber D.E. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 24.Dupont S., Morsut L., Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 25.Miralles F., Posern G., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 26.Vartiainen M.K., Guettler S., Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 27.Olson E.N., Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connelly J.T., Gautrot J.E., Watt F.M. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat. Cell Biol. 2010;12:711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 29.McGee K.M., Vartiainen M.K., Bailly M. Nuclear transport of the serum response factor coactivator MRTF-A is downregulated at tensional homeostasis. EMBO Rep. 2011;12:963–970. doi: 10.1038/embor.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somogyi K., Rørth P. Evidence for tension-based regulation of Drosophila MAL and SRF during invasive cell migration. Dev. Cell. 2004;7:85–93. doi: 10.1016/j.devcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S., Maxwell I.Z., Ingber D.E. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys. J. 2006;90:3762–3773. doi: 10.1529/biophysj.105.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens A.D., Haase J., Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J. Cell Biol. 2011;193:1167–1180. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaziri A., Mofrad M.R. Mechanics and deformation of the nucleus in micropipette aspiration experiment. J. Biomech. 2007;40:2053–2062. doi: 10.1016/j.jbiomech.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 34.Cui Y., Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pajerowski J.D., Dahl K.N., Discher D.E. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deguchi S., Sato M. Biomechanical properties of actin stress fibers of non-motile cells. Biorheology. 2009;46:93–105. doi: 10.3233/BIR-2009-0528. [DOI] [PubMed] [Google Scholar]
- 37.Lee J.S., Hale C.M., Wirtz D. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys. J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vishwasrao H.D., Trifilieff P., Kandel E.R. In vivo imaging of the actin polymerization state with two-photon fluorescence anisotropy. Biophys. J. 2012;102:1204–1214. doi: 10.1016/j.bpj.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varma R., Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 40.Luxton G.W., Gomes E.R., Gundersen G.G. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banerjee B., Bhattacharya D., Shivashankar G.V. Chromatin structure exhibits spatio-temporal heterogeneity within the cell nucleus. Biophys. J. 2006;91:2297–2303. doi: 10.1529/biophysj.105.079525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischle W., Tseng B.S., Allis C.D. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 43.Schmiedeberg L., Weisshart K., Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol. Biol. Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tóth K.F., Knoch T.A., Rippe K. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 2004;117:4277–4287. doi: 10.1242/jcs.01293. [DOI] [PubMed] [Google Scholar]
- 45.Rao J., Bhattacharya D., Shivashankar G.V. Trichostatin-A induces differential changes in histone protein dynamics and expression in HeLa cells. Biochem. Biophys. Res. Commun. 2007;363:263–268. doi: 10.1016/j.bbrc.2007.08.120. [DOI] [PubMed] [Google Scholar]
- 46.Zhao X.H., Laschinger C., McCulloch C.A. Force activates smooth muscle α-actin promoter activity through the Rho signaling pathway. J. Cell Sci. 2007;120:1801–1809. doi: 10.1242/jcs.001586. [DOI] [PubMed] [Google Scholar]
- 47.Okamoto K., Hayashi Y. Visualization of F-actin and G-actin equilibrium using fluorescence resonance energy transfer (FRET) in cultured cells and neurons in slices. Nat. Protoc. 2006;1:911–919. doi: 10.1038/nprot.2006.122. [DOI] [PubMed] [Google Scholar]
- 48.Poh Y.C., Shevtsov S.P., Wang N. Dynamic force-induced direct dissociation of protein complexes in a nuclear body in living cells. Nat Commun. 2012;3:866. doi: 10.1038/ncomms1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Celedon A., Hale C.M., Wirtz D. Magnetic manipulation of nanorods in the nucleus of living cells. Biophys. J. 2011;101:1880–1886. doi: 10.1016/j.bpj.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson C.P., Tang H.Y., Discher D.E. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawada Y., Tamada M., Sheetz M.P. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kruithof M., Chien F.T., van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharya D., Mazumder A., Shivashankar G.V. EGFP-tagged core and linker histones diffuse via distinct mechanisms within living cells. Biophys. J. 2006;91:2326–2336. doi: 10.1529/biophysj.105.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meshorer E., Yellajoshula D., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roopa T., Shivashankar G.V. Direct measurement of local chromatin fluidity using optical trap modulation force spectroscopy. Biophys. J. 2006;91:4632–4637. doi: 10.1529/biophysj.106.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


