Abstract
Adenosine and its endogenous precursor ATP are main components of the purinergic system that modulates cellular and tissue functions via specific adenosine and ATP receptors (P1 and P2 receptors), respectively. Although adenosine inhibits excitability and ATP functions as an excitatory transmitter in the central nervous system, little is known about the ability of P1 and P2 receptors to form new functional structures such as a heteromer to control the complex purinergic cascade. Here we have shown that Gi/o protein-coupled A1 adenosine receptor (A1R) and Gq protein-coupled P2Y1 receptor (P2Y1R) coimmunoprecipitate in cotransfected HEK293T cells, suggesting the oligomeric association between distinct G protein-coupled P1 and P2 receptors. A1R and P2Y2 receptor, but not A1R and dopamine D2 receptor, also were found to coimmunoprecipitate in cotransfected cells. A1R agonist and antagonist binding to cell membranes were reduced by coexpression of A1R and P2Y1R, whereas a potent P2Y1R agonist adenosine 5′-O-(2-thiotriphosphate) (ADPβS) revealed a significant potency to A1R binding only in the cotransfected cell membranes. Moreover, the A1R/P2Y1R coexpressed cells showed an ADPβS-dependent reduction of forskolin-evoked cAMP accumulation that was sensitive to pertussis toxin and A1R antagonist, indicating that ADPβS binds A1R and inhibits adenylyl cyclase activity via Gi/o proteins. Also, a high degree of A1R and P2Y1R colocalization was demonstrated in cotransfected cells by double immunofluorescence experiments with confocal laser microscopy. These results suggest that oligomeric association of A1R with P2Y1R generates A1R with P2Y1R-like agonistic pharmacology and provides a molecular mechanism for an increased diversity of purine signaling.
Adenosine and ATP are two major neurotransmitter and neuromodulating systems that share a number of structural and functional characteristics. These purinergic systems modulate many physiological processes, including smooth muscle contraction, immune response, platelet aggregation, pain, cardiac function, cardioprotection, and neurotransmission (1). Pharmacological and molecular cloning studies have identified two purinergic receptor families, named adenosine receptor or P1 receptor and ATP receptor or P2 receptor. P1 receptors have been further subdivided into A1R, A2AR, A2BR, and A3R, all of which are G protein-coupled receptors (GPCRs). A1 and A3 adenosine receptors are coupled to the inhibition of adenylyl cyclase via Gi/o proteins. A2A and A2B adenosine receptors are coupled to stimulation of adenylyl cyclase via Gs proteins. P2 receptors also are subclassified as P2X or P2Y receptors. To date, seven mammalian P2X receptors (P2X1–7R) that are ligand-gated ion channels and five mammalian P2Y receptors (P2Y1R, P2Y2R, P2Y4R, P2Y6R, and P2Y11R) have been cloned. P2Y receptors are GPCRs that are mainly coupled to phospholipase C via Gq proteins. Although the individual pharmacological and biochemical profile of cloned P1 and P2 receptor subtypes have been defined, the assignment of each receptor type to the various purinergic functions in tissues or cells has been limited by the low selectivity and cross-reactivity of available purinergic ligands. The diverse aspects of purinergic functions also may predict a greater number of purinergic receptor subtypes than expected from cloning studies (2–9). In addition, these receptor-mediated events can be modulated either by cross-talk with other receptor systems (10, 11).
Recently, a significant amount of GPCR has been reported to exist in a homomeric (12–17) and heteromeric assembly (18–25). Although most heteromeric assemblies consist of different subtypes of the same receptor family, several combinations such as somatostatin receptor/D2R and β2-adrenergic receptor/opioid receptor (26, 27) have been reported to form heterodimers between truly different GPCRs (with only ≈30% sequence homology). We therefore predicted that, like other GPCRs, P1 receptors can potentially form heteromeric complexes with distinct types of GPCRs through direct association. In fact, previous radioligand binding and biochemical studies (24, 25, 28–30) indicated that A1R could be arranged in dimeric complexes with related proteins or GPCRs. However, to date, no evidence of direct interaction between G protein-coupled P1 and P2 receptors that induces functional changes in cells or tissues has been obtained, although functional interactions between A1R and P2YR has been previously described (31). A recent study showed that P2Y1R localized in neuronal cells of the hippocampus, midbrain, and subthalamic nucleus and associated regions (32) and that A1R localized in the cerebral cortex, hippocampus, and thalamus, especially in the neuronal cells of these regions (33). Therefore, a significant portion of A1R and P2Y1R distributed in the central nervous system is likely to colocalize in the overlap regions and thereby exert new functions. The purpose of this study is to determine whether P1 and P2 receptors, in this case A1R and P2Y1R, can form a heterooligomer that exerts novel pharmacological and functional characteristics with a potential role in the purinergic-signaling cascade.
Materials and Methods
cDNA Construction and Cell Transfection.
The incorporation of sequences encoding the hemagglutinin (HA) epitope tag (YPYDVPDYA) and the Myc epitope tag (EQKLISEEDL) into rat A1R and rat P2Y1R or P2Y2R genes, respectively, was performed by PCR. Each epitope was positioned immediately before the first methionine of the appropriate gene. Purified full-length cDNA of HA-A1R was subcloned into pcDNA3 and purified full-length cDNAs of Myc-P2Y1R and Myc-P2Y2R were subcloned into pcDNA3.1. cDNAs encoding rat P2Y1R and P2Y2R were gifts of G. I. Bell (University of Chicago, Chicago, IL) and W. R. Rice (Children's Hospital Medical Center, Cincinnati, OH), respectively. Myc-dopamine D2 receptor (D2R) cDNA was kindly donated by T. Haga (Tokyo University, Tokyo). The generation of each construct was confirmed by sequencing analysis. DNA (2 μg) was mixed with Effectene transfection reagent (Qiagen, Chatsworth, CA), and the mixture was diluted with DMEM and added to 30–50% confluent HEK293T cells plated on 100-mm dishes. The transfected HEK293T cells were cultured in DMEM with 10% FBS. Cell membranes for immunoprecipitation and Western blotting were prepared from the cells 48 h after the transfection. For adenylyl cyclase and inositol 1,4,5-trisphosphate (IP3) assays, the cells were passaged to 24-well and 12-well plates, respectively, 48 h after the transfection and cultured for another 24 h at 37°C. When indicated, cells were pretreated with pertussis toxin (PTX) for 16–20 h at a concentration of 100 ng/ml.
Membrane Preparation, Coimmunoprecipitation, and Western Blotting.
For HEK293T cell membrane preparation, cells expressing single or combinations of receptors (≈2 × 107 cells) were washed twice with PBS and collected with a rubber policeman in hypotonic lysis buffer containing 50 mM Tris-acetate buffer, pH 7.4, with a protease-inhibitor mixture (Roche Diagnostics). Cells were disrupted by sonication and subjected to low-speed centrifugation to remove organelles and nuclei. The resulting supernatant was subjected to centrifugation at 30,000 × g for 20 min, and precipitated cell membranes were collected, washed twice, resuspended in the lysis buffer, and stored at −80°C. The membranes were solubilized by incubation with Tx buffer (50 mM Tris⋅HCl buffer, pH 7.4, containing 1% Triton X-100, 300 mM NaCl, 100 mM iodoacetamide, and a protease-inhibitor mixture) for 60 min at 4°C on a rotator. The mixture was centrifuged at 18,500 × g for 20 min, and the supernatant was collected as the cell membrane lysate. In some instances, the extracted cell membrane lysate was treated with 0.7 units N-glycosidase F for 3 h at 37°C. An aliquot of the cell membrane lysate (500 μg protein) was precleared with 30 μl of Protein G-agarose (50% suspension in PBS) at 4°C for 30 min on a rotator. The Protein G-agarose was then removed by centrifuging the lysate at 18,500 × g for 5 min at 4°C. Subsequently, the precleared cell membrane lysate was incubated with 1 μg of anti-Myc 9E10 mAb (Roche Diagnostics) or anti-HA 3F10 mAb (Roche Diagnostics) for 60 min at 4°C on a rotator, and then 50 μl of Protein G-agarose was added to the mixture. The incubation was continued for an additional 120 min at 4°C. The immune-complex was washed three times with Tx buffer, and subsequently it was eluted from Protein G-agarose by the addition of 50 μl of the sample buffer used for SDS/PAGE. An appropriate amount of immunoprecipitated proteins was subjected to SDS/PAGE, after which the protein on the gel was electrotransferred to a nitrocellulose membrane. After blocking with 5% skim milk dissolved in washing buffer (0.1% Tween 20 in Tris⋅HCl-buffered saline), HA-A1R, Myc-P2Y1R, Myc-P2Y2R, or Myc-D2R on the blot were detected by using anti-HA 3F10 mAb (50 ng/ml) or anti-Myc PL14 mAb (1 μg/ml, Medical and Biological Laboratories), followed by horseradish peroxidase-conjugated goat anti-rat IgG antibody (for anti-HA mAb) or goat anti-mouse IgG antibody (for anti-Myc mAb). The reactive bands were visualized with enhanced chemiluminescent substrates (Pierce).
Receptor Binding and Functional Assays.
For the assay of A1R antagonist binding, 10 μg of cell membranes was incubated with 2 nM [3H]8-cyclopentyl-1, 3-dipropylxanthine (DPCPX) (87.0 Ci/mmol, New England Nuclear) containing 2 units/ml adenosine deaminase (Sigma), 5 mM MgCl2, and 50 mM Tris-acetate buffer (pH 7.4) for 60 min at 25°C in the absence or presence of various concentrations of unlabeled ligands. For agonist binding, 30–50 μg of membrane proteins was incubated with 40 nM [3H]5′-N-ethylcarboxamidoadenosine (NECA) (27.0 Ci/mmol, Amersham Pharmacia) under the same conditions described above. Saturation and competition binding assays were performed as described (3). In some cases, nonspecific binding of [3H]NECA was determined in the presence of cold NECA (10 μM). Values for the estimated concentration for dissociation constant (Ki) were determined from displacement curves by using GRAPHPAD PRISM 2.0 (GraphPad, San Diego).
cAMP production was measured by a cAMP EIA system (Amersham Pharmacia). Briefly, transfected HEK293T cells (1 × 105 cells/well) in serum-free DMEM were preincubated with 50 μM Ro 20–1724 for 10 min and then stimulated with the indicated concentrations of agonists for 10 min in the presence of 10 μM forskolin (FSK). The reactions were terminated by adding HCl (0.1 M final concentration). cAMP extracted from cells was quantified as described in the manufacturer's manual. The production of IP3 was determined by using an IP3 assay kit (Amersham Pharmacia). The transfected cells (3 × 105 cells/well) were preincubated for 30 min at 37°C with serum-free DMEM containing 20 mM LiCl (to inhibit inositol 1-phosphatase) in the presence or absence of antagonists. The cells then were incubated with either N6-cyclopentyladenosine (CPA) or adenosine 5′-O-(2-thiotriphosphate) (ADPβS) at various concentrations in 500 μl of Na-Hepes-buffered saline [140 mM NaCl, 4.7 mM KCl, 1.13 mM MgCl2, 10 mM glucose, 1 mM CaCl2, and 10 mM Hepes (pH 7.4)]. The reaction was terminated at various time points by aspiration of the solution followed by the addition of 500 μl of 4% (vol/vol) HClO4 and incubation for 20 min on ice. After the cell suspension was centrifuged, the supernatant was neutralized with 1.5 M KOH in 10 mM Hepes. The supernatant was assayed for IP3 by a competitive radioreceptor assay according to the manufacturer's instruction.
Immunocytochemistry.
For fluorescence immunocytochemistry, 48 h after transfection cells were fixed for 30 min in 4% paraformaldehyde in PBS, permeabilized with 0.25% Triton X-100, and incubated with primary antibody against HA tag or Myc tag for 90 min at room temperature. Rat anti-HA 3F10 mAb was visualized with Cy3-conjugated goat anti-rat IgG antibodies (Jackson ImmunoResearch). Mouse anti-Myc 9E10 mAb was detected by FITC-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch). Fluorescent images were obtained with a Zeiss LSM 410 confocal microscope. The extent of overlap of the two signals was determined by the software for the Carl Zeiss LSM 4 Laser Scan Microscope.
Results
Association of A1R and P2Y1R in Coexpressed HEK293T Cells.
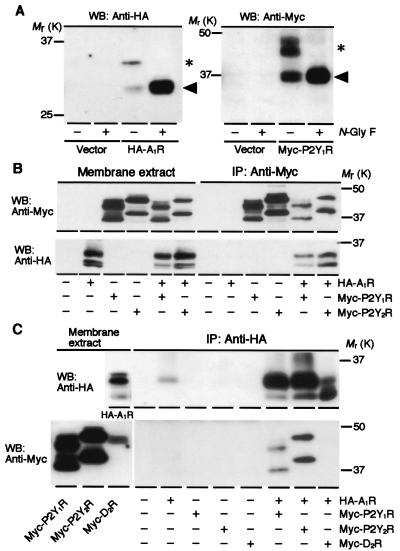
We transiently cotransfected HA-A1R and Myc-P2Y1R cDNAs into HEK293T cells and examined whether A1R and P2Y1R associate with each other as a heteromeric complex by conducting immunoprecipitation experiments using whole-cell membrane lysates (Fig. 1). The addition of an epitope, HA or Myc, to the NH2 terminus of these receptors had no effect on the electrophoretic mobility of the receptors visualized on SDS/PAGE (data not shown). Western blots of cell membranes expressing HA-A1R exhibited anti-HA reactive bands of predicted molecular mass (A1R, 35 kDa; ref. 34) in addition to a band of lower molecular mass of 31 kDa (Fig. 1A). Western blots of cell membranes expressing Myc-P2Y1R showed anti-Myc reactive bands of 45, 42, and 37 kDa (Fig. 1A). The predicted molecular mass of P2Y1R has been reported as 42 kDa (35). The lower molecular mass bands were likely to be the deglycosylated form of the receptors, because most bands shifted to a position of the low molecular mass band after treatment with N-glycosidase F (Fig. 1A). We found that anti-Myc antibody precipitated HA-A1R in addition to Myc-P2Y1R from cells coexpressing HA-A1R/Myc-P2Y1R (Fig. 1B). Conversely, anti-HA antibody precipitated both Myc-P2Y1R and HA-A1R from cells coexpressing HA-A1R and Myc-P2Y1R (Fig. 1C). Such counterimmunoprecipitation was not observed with the admixture of cell membranes expressing each receptor individually (data not shown). For comparison, Myc-P2Y2R or Myc-D2R instead of Myc-P2Y1R was cotransfected with HA-A1R. Anti-HA antibody precipitated Myc-P2Y2R along with HA-A1R from Myc-P2Y2R/HA-A1R coexpressed cells (Fig. 1 B and C). However, Myc-D2R was not immunoprecipitated along with HA-A1R by anti-HA antibody (Fig. 1C). It also was confirmed that neither Myc-P2Y1R nor Myc-P2Y2R was immunoprecipitated by the anti-HA antibody or vice versa for the HA-A1R from cell membrane extracts expressing only Myc-P2Y1R or Myc-P2Y2R. These findings indicate that A1R can form heteromeric complexes with P2Y1R or P2Y2R when transfected simultaneously in HEK293T cells.
Figure 1.
Association of A1R and 2Y1R in A1R/P2Y1R-transfected HEK293T cells. (A) Western blot (WB) analysis of cell membranes expressing HA-A1R (Left) and Myc-P2Y1R (Right). Cell lysates that had been treated by N-glycosidase F (+) for 3 h at 37°C were subjected to Western blotting. HA-A1R and Myc-P2Y1R were detected by anti-HA and anti-Myc antibodies, respectively. The control (−) without treatment by N-glycosidase F also was subjected to Western blotting. The apparent molecular masses of the glycosylated (*) and deglycosylated (arrowheads) HA-A1R are 35 and 31 kDa, respectively. The apparent molecular masses of the glycosylated Myc-P2Y1R are 45 and 42 kDa (*), and the deglycosylated Myc-P2Y1R is 37 kDa (arrowheads). (B) Coimmunoprecipitation of cell lysates by anti-Myc antibody. Anti-Myc antibody precipitated Myc-P2Y1R (Upper, the 9th and 11th lanes from the left) and Myc-P2Y2R (Upper, lanes 10 and 12 from the left), and coimmunoprecipitated HA-A1R with Myc-P2Y1R (Lower, lane 11 from the left) or Myc-P2Y2R (Lower, lane 12 from the left). (C) Coimmunoprecipitation of cell lysates by anti-HA antibody. In addition to HA-A1R, anti-HA antibody coimmunoprecipitated Myc-P2Y1R from the cell membrane lysates coexpressing HA-A1R/Myc-P2Y1R (Lower, lane 9 from the left). Myc-P2Y2R also was coimmunoprecipitated by anti-HA antibody along with HA-A1R from the cell lysates coexpressing HA-A1R/Myc-P2Y2R (Lower, lane 10 from the left). In contrast, Myc-D2R was not immunoprecipitated from the cell lysates coexpressing HA-A1R/Myc-D2R (Lower, lane 11 from the left) by anti-HA antibody. Data are representative of 2–4 independent experiments.
Coexpression with P2Y1R Modulates A1R Binding Pharmacology.
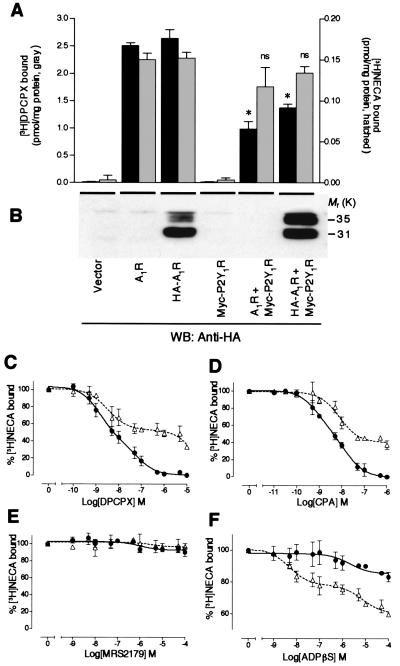
We examined the ligand-binding properties of HEK293T cell membranes expressing A1R and P2Y1R by using A1R selective antagonist [3H]DPCPX or nonselective adenosine receptor agonist [3H]NECA. It should be noted that no significant specific binding of these radioligands was observed with cell membranes expressing Myc-P2Y1R alone or mock plasmid. As shown in Fig. 2A, cell membranes expressing HA-A1R showed [3H]DPCPX and [3H]NECA binding activities similar to those of cell membranes expressing intact A1R, suggesting that N-terminal modification of A1R with HA tag did not alter ligand-binding activities. In contrast, cell membranes coexpressing HA-A1R/Myc-P2Y1R or A1R/Myc-P2Y1R showed significantly lower [3H]DPCPX binding activity than did cell membranes expressing HA-A1R alone (P < 0.05, Student's t test), despite the fact that the expression level of HA-A1R protein in cotransfected cell membranes was equal or even higher than that of cell membranes transfected with HA-A1R alone, as judged by Western blotting (Fig. 2B). The decrease in [3H]DPCPX binding activity observed with the HA-A1R/Myc-P2Y1R-transfected cell membranes was mainly due to the decrease in Bmax values from saturation binding assays (Table 1). In contrast, no significant differences in [3H]NECA binding were observed between HA-A1R-transfected and HA-A1R/Myc-P2Y1R-transfected cell membranes, as shown in Fig. 2A and Table 1. Furthermore, a significant reduction of a selective A1R agonist, [3H]R-N6-phenylisopropyladenosine (R-PIA), binding with HA-A1R/Myc-P2Y1R-transfected cell membranes also was observed. The decrease in [3H]R-PIA binding was mainly due to the increase in KD values, as determined from saturation binding assays (Table 1). Because these changes in ligand-binding pharmacology were not observed with the admixture of cell membranes expressing each receptor individually (data not shown), the expression of this novel binding activity might require in situ direct association of A1R with P2Y1R and it may not result from nonspecific aggregation between these receptors. The difference in the Bmax values between antagonist and agonist interactions is likely due to the uncoupling of A1R and G protein in the transfected cells, which is often reported in other receptor systems. We further examined ligand-binding pharmacology of the cotransfected cell membranes using [3H]NECA by competition experiments with other purinergic ligands (Fig. 2 C–F). The apparent binding potency and efficacy of both A1R-selective antagonist DPCPX (Fig. 2C) and A1R-selective agonist CPA (Fig. 2D) to the [3H]NECA binding site were reduced in the cotransfected cells. Selective P2Y1R antagonist N6-methyl-2′-deoxyadenosine-3′,5′-bisphosphate (MRS2179) failed to displace [3H]NECA bound to HA-A1R-transfected and HA-A1R/Myc-P2Y1R-transfected cell membranes (Fig. 2E). A potent P2Y1R agonist, ADPβS, was found to be quite active in displacing the ligands from the [3H]NECA binding site of cotransfected cell membranes with Ki values of 0.38 ± 0.05 nM (high-affinity site) and 610 ± 85 nM (low-affinity site) (Fig. 2F, ▵). In contrast, ADPβS at the 10−6 M range slightly inhibited [3H]NECA binding of cell membranes expressing HA-A1R alone (Ki = 1,670 ± 98 nM, Fig. 2F, ●). Breakdown products of ADPβS that might have been produced during the incubation may explain the modest inhibitory effect of ADPβS on [3H]NECA binding with cell membranes expressing HA-A1R.
Figure 2.
Coexpression with P2Y1R modulates A1R binding pharmacology. (A) The [3H]DPCPX (gray columns, left y axis) and [3H]NECA (hatched columns, right y axis) binding activities of cell membranes expressing A1R (nontagged and HA-tagged) together with Myc-P2Y1R. The binding affinity of adenosine receptor antagonist [3H]DPCPX was significantly reduced by the coexpression of A1R/P2Y1R, whereas [3H]NECA binding activity was not significantly affected. Data represent the means ± SEM of the [3H]DPCPX or [3H]NECA-specific bound values. Results from three independent experiments performed in duplicate are shown. * indicate statistically significant difference from respective cells expressing A1R or HA-A1R alone (n = 3, P < 0.05, Student's t test). ns, not significant. (B) Western blotting (WB) of transfected cell lysates using anti-HA antibody. The blot showed the slightly higher expression of HA-A1R protein in cotransfected cells (lane 6 from the left) than that in HA-A1R-transfected cells (lane 3 from the left). Displacement of [3H]NECA (40 nM) binding with transfected cell membranes by DPCPX (C), CPA (D), MRS2179 (E), and ADPβS (F). Membranes from HA-A1R-transfected (C–F, ●) or HA-A1R/P2Y1R-transfected (C–F, ▵) cells were incubated with indicated concentrations of each ligand. The [3H]NECA concentrations were selected to ensure maximal saturation binding. The Ki values for A1R ligands, CPA and DPCPX, are shifted about 2-fold toward lower potencies, whereas the Ki value for potent P2Y1R agonist ADPβS is shifted 400-fold toward higher potency in cotransfected cells. The heteromeric complex also reduces the binding efficacy to DPCPX (70% versus 100% at 10 μM) and to CPA (60% versus 100% at 1 μM). Data represent the means ± SEM of the percentage of [3H]NECA-specific bound values. Results from three independent experiments performed in duplicate are shown.
Table 1.
Comparison of the ligand-binding properties of A1R and its heteromers
| Radioligand
|
||||||
|---|---|---|---|---|---|---|
| [3H]DPCPX
|
[3H]NECA
|
[3H]R-PIA
|
||||
| KD, nM | Bmax, pmol/mg | KD, nM | Bmax, pmol/mg | KD, nM | Bmax, pmol/mg | |
| A1R | 1.3 ± 0.14 | 3.5 ± 0.12 | — | — | — | — |
| HA-A1R | 1.2 ± 0.12 | 3.6 ± 0.11 | 3.4 ± 1.2 | 0.11 ± 0.07 | 0.85 ± 0.3 | 0.09 ± 0.02 |
| HA-A1R + Myc-P2Y1R | 1.2 ± 0.1 | 1.9 ± 0.05* | 3.5 ± 1.8 | 0.09 ± 0.03 | 3.1 ± 1.7* | 0.08 ± 0.03 |
For an A1R antagonist [3H]DPCPX saturation experiment, 10 μg of membrane protein was incubated with 0.2–10 nM [3H]DPCPX containing 2 units/ml adenosine deaminase, 5 mM MgCl2, and 50 mM Tris-acetate buffer, pH 7.4 for 60 min at 25°C. Nonspecific binding was measured in the presence of 1 μM XAC. For agonist [3H]NECA and [3H]R-PIA saturation experiment, 30–50 μg of membrane proteins was incubated with 2–50 nM radioligands in the same condition described above. The binding of these ligands to Myc-P2Y1R was not detected.
, P < 0.05 (Student's t test, n = 3).
Adenylyl Cyclase Coupling in Cotransfected Cells.
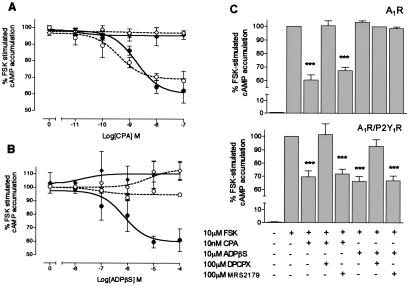
Next we attempted to determine whether heteromerization leads to alterations in cellular functional coupling. To that end, we studied A1R agonist-induced adenylyl cyclase inhibition, a main index of A1R function, in cotransfected cells (Fig. 3). The cells expressing A1R alone revealed an inhibition of FSK-stimulated cAMP accumulation by CPA in a dose-dependent manner, with the estimated concentration for half-maximal response (IC50) of 0.42 ± 0.1 nM to a maximum inhibition of 70 ± 6%. This activity was completely abolished by pretreatment of the cells with PTX (Fig. 3A). CPA-induced inhibition of FSK-stimulated adenylyl cyclase activity also was detected with the estimated IC50 value of 1.0 ± 0.12 nM in the cells coexpressing A1R/P2Y1R. This activity also was abolished by PTX treatment (Fig. 3A). The potency of adenylyl cyclase attenuation by CPA was reduced significantly in the coexpressing cells compared with cells expressing A1R alone (P < 0.05, Student's t test). The treatment of cells expressing A1R alone with ADPβS revealed no changes in FSK-stimulated cAMP production (Fig. 3B). Activation of P2Y1R-transfected cells with ADPβS did not lead to a significant change in FSK-evoked cAMP levels (data not shown). In cells coexpressed with A1R and P2Y1R, ADPβS markedly reduced FSK-evoked adenylyl cyclase activity in a concentration-dependent manner, with the estimated IC50 value of 730 ± 35 nM, to a maximum inhibition of 62 ± 9%. PTX treatment resulted in complete loss of the dose-dependent activity of ADPβS, suggesting the involvement of a PTX-sensitive Gi/o protein (Fig. 3B). We next examined whether the ADPβS-induced adenylyl cyclase inhibition in coexpressed cells was mediated through the ligand-binding site of A1R (Fig. 3C). In both A1R-expressing cells and A1R/P2Y1R-coexpressing cells, CPA (10 nM) maximally inhibited the FSK-evoked adenylyl cyclase activity to virtually identical extents. This inhibitory effect was blocked in the presence of A1R antagonist DPCPX. When cells coexpressing A1R/P2Y1R were pretreated with DPCPX, however, the ADPβS-evoked adenylyl cyclase inhibition was decreased by ≈95%, whereas MRS2179 had no effect on the ADPβS-evoked adenylyl cyclase inhibition. Taken together, these results suggest that ADPβS exerts the adenylyl cyclase inhibitory activity through xanthine-sensitive ligand-binding sites of A1R via Gi/o protein-linked effector system.
Figure 3.
Generation of P2Y1R agonist-sensitive adenylyl cyclase inhibition of A1R. (A and B) Concentration-dependent reduction of maximal FSK (10 μM)-stimulated intracellular cAMP accumulation by CPA (A) or ADPβS (B) in A1R/P2Y1R-transfected cells. The attenuation was blocked by the PTX pretreatment (100 ng/ml, 16 h). Dotted line, cells expressing HA-A1R alone; solid line, cells coexpressing HA-A1R and Myc-P2Y1R; circles, nontreated cells; diamonds, PTX-pretreated cells. The 100% values of cAMP for the cells transfected with HA-A1R and HA-A1R plus Myc-P2Y1R were 72 ± 14 and 67 ± 19 pmol/105 cells, respectively (mean ± SEM, n = 5). Estimated IC50 values are shown in the text. (C) Pretreatment of cells with A1R antagonist DPCPX, but not P2Y1R antagonist MRS2179, significantly inhibited maximal ADPβS-induced adenylyl cyclase attenuation in the A1R/P2Y1R-transfected cells. (Upper) HA-A1R transfected cells. (Lower) HA-A1R/Myc-P2Y1R cotransfected cells. The 100% values of cAMP for the cells transfected with HA-A1R and HA-A1R/Myc-P2Y1R were 70 ± 12 and 71 ± 17 pmol/105 cells, respectively (mean ± SEM, n = 5). Data represent the means ± SEM of the percentage of FSK-induced cAMP accumulation values. Results from 3–5 independent experiments performed in duplicate are shown. ***, P < 0.01, Student's t test.
IP3 Production in Cotransfected Cells.
To analyze the effect of A1R/P2Y1R heteromeric formation on the P2Y1R-effector systems, we examined the production of intracellular IP3 stimulated by P2Y1R agonist ADPβS in cotransfected cells. ADPβS at its maximum effective dose of 10 μM induced a 3.1 ± 0.5-fold (n = 3, duplicates in each experiment) increase in IP3 production over basal levels in cells expressing A1R/P2Y1R. The time course of the ADPβS-induced IP3 production was similar to that of the cells expressing P2Y1R alone. Both responses peaked at 15 s after the addition of ADPβS and declined rapidly toward basal levels in 2 min. The dose-dependent potency of ADPβS (EC50 = 3.5 ± 0.6 μM), however, slightly decreased in cotransfected cells (P < 0.05, Student's t test, n = 3) compared with the potency of ADPβS in P2Y1R-transfected cells (EC50 = 1.4 ± 0.4 μM), whereas there were no significant differences in the maximum responses induced by ADPβS between cells expressing P2Y1R and cells expressing A1R/P2Y1R. We confirmed that the amount of P2Y1R as determined by Western blotting did not change upon coexpression with A1R. ADPβS (10 μM) did not stimulate IP3 production in A1R-transfected cells, and CPA (10 nM) did not stimulate P2Y1R- and A1R/P2Y1R-transfected cells. Also, the simultaneous addition of CPA (10 nM) and ADPβS (10 μM) in the cotransfected cells did not stimulate IP3 production any more than did the stimulation by ADPβS alone.
Double-Immunostaining of A1R/P2Y1R in Cotransfected Cells.
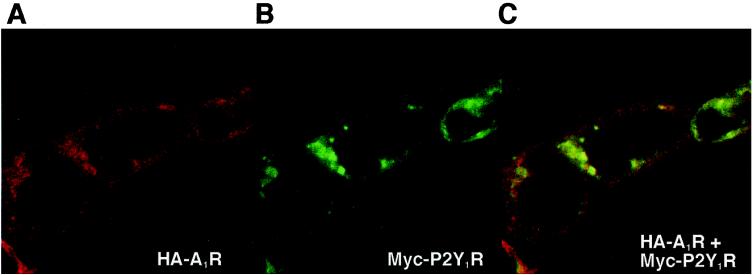
We studied the subcellular distribution of HA-A1R and Myc-P2Y1R in cotransfected cells by confocal laser microscopy (Fig. 4). When expressed in HEK293T cells individually, HA-A1R and Myc-P2Y1R were localized in the vicinity of the plasma membranes (data not shown). Images taken at the microscopic level with a ×63 objective of cotransfected cells that were double labeled for HA-A1R (red) and Myc-P2Y1R (green) are shown (Fig. 4 A and B). Both receptors were expressed prominently near the plasma membranes. When the images are merged by using the confocal assistant software, there is a striking overlap (intense yellow spots) in the distribution of the two receptors (Fig. 4C). The extent of overlap pixels of the two signals was 35.4 ± 9.6% (n = 3). The immunostaining of unpermealized cells also was performed with similar results (data not shown). The fact that this colocalization occurred over plasma membranes supports the heteromeric association of A1R and P2Y1R.
Figure 4.
Confocal imaging of HEK293T cells expressing HA-A1R/Myc-P2Y1R. HA-A1R (A, Cy3, red) and Myc-P2Y1R (B, FITC, green) were detected by using double fluorescent immunohistochemistry. (C) The product of merging A and B, showing the colocalization of HA-A1R and Myc-P2Y1R in cotransfected HEK293T cells (yellow).
Discussion
The present study provides biochemical, pharmacological, and functional evidence for the existence of a heteromeric complex between P1 and P2 receptors. We also report on heteromer formation between distinct G protein-coupled purinergic receptors with very low amino acid sequence homology (less than 5% amino acid sequence homology between A1R and P2Y1R), although there is increasing biochemical and functional evidence for oligomerization of GPCRs (36, 37).
Immunoprecipitation (Fig. 1) and double immunostaining (Fig. 4) experiments showed the existence of A1R/P2Y1R heteromer in HEK293T cells when cotransfected with A1R and P2Y1R. Ligand binding (Fig. 2, Table 1) and functional experiments (Fig. 3) indicate that the A1R/P2Y1R heteromeric complex altered the pharmacology of these receptors, i.e., the A1R was altered to have P2Y1R-like agonistic pharmacology. The structural requirements for the A1R/P2Y1R heteromeric association are not known, although several other studies reported that the C tails of γ-aminobutyric acid type B receptor (21), the extracellular amino-terminal domain for the bradykinin B2 receptors (17), and the intracellular third loop for the β-adrenergic receptors (12) may represent monomeric or oligomeric interfaces. In the case of A1R/P2Y1R heteromer, the C tail deletion mutant of A1R was still able to associate with P2Y1R and also with P2Y2R in HEK293T cells (unpublished results), which suggests that the C tail of A1R is not required for the heteromeric association with P2Y1R or P2Y2R. The precise structural requirements for the association of A1R with P2Y1R remain to be elucidated. It also remains to be further investigated whether direct receptor association between P1 and P2 receptor is restricted to the A1R and P2Y1R subtypes. We found that the complex between A1R and P2Y2R can be formed, although the pharmacology or cellular effector systems of A1R/P2Y2R heteromeric complex have not been studied. This finding suggests that hetero-oligomerization between subclasses of purinergic receptors may be a widespread phenomenon.
In this study, we observed significant changes in the ligand-binding properties in the heteromers. Ligand-binding studies revealed a significant reduction of A1R-agonist and A1R-antagonist binding in the cotransfected cell membranes. In contrast, we observed a significant 400-fold increase in the binding affinity of ADPβS, a potent P2Y1R agonist, for the A1R/P2Y1R-transfected cell membranes. It is likely that a physical association of A1R with P2Y1R induced ligand-binding sites with A1R-P2Y1R hybrid selectivity. In other words, the modified ligand-binding pocket of A1R in the heteromer now appears to fit well to a P2Y1R agonist but slightly less well to A1R ligands.
It should be interesting to examine functional changes in the cotransfected cells, because A1R is a GPCR coupled to adenylyl cyclase via Gi/o proteins, whereas P2Y1R is a GPCR coupled to phospholipase C via Gq proteins. The heteromerization resulted in a significant modification of cellular functions (cAMP, IP3), as shown in Fig. 3. We showed that a potent P2Y1R agonist ADPβS was able to couple with a PTX-sensitive adenylyl cyclase system only when A1R and P2Y1R were coexpressed in HEK293T cells. Because an ADPβS-evoked response in adenylyl cyclase activity was blocked by either A1R antagonist or PTX but was not blocked by P2Y1R antagonist, ADPβS is likely to exert its activity via the A1R ligand-binding site. In contrast, no major alterations in ADPβS-evoked phospholipase C activity (IP3 production) was induced by the heteromer formation, although we did observe a slight decrease in the affinity of P2Y1R to the cotransfected cells. These results indicate again that the heteromeric formation between A1R and P2Y1R produces functional changes that are preferential to adenylyl cyclase coupling.
Although several studies indicate the presence of atypical subtypes of P2YRs (4–9) that are sensitive to P1 receptor antagonist theophylline or PTX, a molecular basis for these observations has not been well described. A P2Y-like receptor coupled to the Gi/o family of G proteins has been observed in rat glioma C6 cells, which can be selectively activated by AMP derivatives, although its identity is unclear (9). Mendoza-Fernandez et al. (4) demonstrated that ATP inhibited the synaptic release of glutamate by direct activation of P2Y receptors that are PTX- and 8-cyclopentyltheophylline (P1 receptor antagonist)-sensitive, and suramin-, pyridoxalphosphate-6-azophenyl-2′,4′-disulfonic acid-, and reactive blue 2 (P2 receptor antagonists)-insensitive. They proposed to classify them as theophylline-sensitive P2Y receptors (4). Thus, heteromerization between purinergic receptors that produce a hybrid pharmacology as shown in this study may help to explain undefined physiological functions of purines in various tissues and cells.
In conclusion, this work shows that signal modification is triggered by receptor heteromerization of P1 and P2 purinergic receptors. Heteromerization of G protein-coupled purinergic receptors may be a mechanism for the control of purinergic functions, although it remains to be established whether such heteromerization occurs in a living organism, because artifacts may arise from the aggregation of GPCRs in overexpression experiments.
Acknowledgments
We are grateful to Dr. G. I. Bell for providing rat P2Y1R cDNAs and Dr. W. R. Rice for rat P2Y2R cDNA. We thank Dr. T. Haga and R. Takahashi for donating Myc-tagged dopamine D2 receptor cDNA and HEK293T cells, respectively. We also thank Dr. K. Nishi for his help with the confocal microscopy experiments. This work was supported in part by grants for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (to O.S. and H.N.), by the Kato Memorial Bioscience Foundation (to O.S.) and also by a Research Grant from the Yamanouchi Foundation for Research on Metabolic Disorders (to H.N.).
Abbreviations
- GPCR
G protein-coupled receptor
- A1R
A1 adenosine receptor
- P2Y1R
P2Y1 receptor
- P2Y2R
P2Y2 receptor
- D2R
dopamine D2 receptor
- HA
hemagglutinin
- PTX
pertussis toxin
- NECA
5′-N-ethylcarboxamidoadenosine
- DPCPX
8-cyclopentyl-1,3-dipropylxanthine
- CPA
N6-cyclopentyladenosine
- ADPβS
adenosine 5′-O-(2-thiotriphosphate)
- MRS2179
N6-methyl-2′-deoxyadenosine-3′,5′-bisphosphate
- FSK
forskolin
- IP3
inositol 1,4,5-trisphosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ralevic V, Burnstock G. Pharmacol Rev. 1998;50:413–492. [PubMed] [Google Scholar]
- 2.Shinozuka K, Bjur R A, Westfall D P. Naunyn-Schmiedeberg's Arch Pharmacol. 1988;338:221–227. doi: 10.1007/BF00173391. [DOI] [PubMed] [Google Scholar]
- 3.Saitoh Y, Nakata H. Biochem Biophys Res Commun. 1996;219:469–474. doi: 10.1006/bbrc.1996.0257. [DOI] [PubMed] [Google Scholar]
- 4.Mendoza-Fernandez V, Andrew R D, Barajas-López C. J Pharmcol Exp Ther. 2000;293:172–179. [PubMed] [Google Scholar]
- 5.Song S L, Chueh S H. Brain Res. 1996;734:243–251. [PubMed] [Google Scholar]
- 6.Ikeuchi Y, Nishizaki T, Mori M, Okada Y. Eur J Pharmacol. 1996;304:191–199. doi: 10.1016/0014-2999(96)00113-6. [DOI] [PubMed] [Google Scholar]
- 7.Barajas-López C, Muller M J, Prieto-Gómez B, Espinosa-Luna R. J Pharmcol Exp Ther. 1995;274:1238–1245. [PubMed] [Google Scholar]
- 8.Koizumi S, Inoue K. Br J Pharmacol. 1997;122:51–58. doi: 10.1038/sj.bjp.0701344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer J L, Lazarowski E R, Chen X H, Harden T K. J Pharmcol Exp Ther. 1993;267:1140–1146. [PubMed] [Google Scholar]
- 10.Quitterer U, Lohse M J. Proc Natl Acad Sci USA. 1999;96:10626–10631. doi: 10.1073/pnas.96.19.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Z, Chen T, Weber M J, Linden J. J Biol Chem. 1999;274:5972–5980. doi: 10.1074/jbc.274.9.5972. [DOI] [PubMed] [Google Scholar]
- 12.Hebért T E, Moffett S, Morello J P, Loisel T P, Bichet D G, Barret C, Bouvier M. J Biol Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 13.Nimchinsky E A, Hof P R, Janssen W G M, Morrison J H, Schmauss C. J Biol Chem. 1997;272:29229–29237. doi: 10.1074/jbc.272.46.29229. [DOI] [PubMed] [Google Scholar]
- 14.Bai M, Trivedi S, Brown E M. J Biol Chem. 1998;273:23605–23610. doi: 10.1074/jbc.273.36.23605. [DOI] [PubMed] [Google Scholar]
- 15.George S R, Lee S P, Varghese G, Zeman P R, Seeman P, Ng G Y K, O'Dowd B F. J Biol Chem. 1998;273:30244–30248. doi: 10.1074/jbc.273.46.30244. [DOI] [PubMed] [Google Scholar]
- 16.Zeng F Y, Wess J. J Biol Chem. 1999;274:19487–19497. doi: 10.1074/jbc.274.27.19487. [DOI] [PubMed] [Google Scholar]
- 17.AbdAlla S, Zaki E, Lother H, Quitterer U. J Biol Chem. 1999;274:26079–26084. doi: 10.1074/jbc.274.37.26079. [DOI] [PubMed] [Google Scholar]
- 18.Jones K A, Borowsky B, Tamm J A, Craig D A, Durkin M M, Dai M, Yao W J, Johnson M, Gunwaldsen C, Huang L Y, et al. Nature (London) 1998;396:674–679. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- 19.White J H, Wise A, Main M J, Green A, Fraser N J, Disney G H, Barnes A A, Emson P, Foord S M, Marshall F H. Nature (London) 1998;396:679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- 20.Jordan B A, Devi L A. Nature (London) 1999;399:697–700. doi: 10.1038/21441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuner R, Köhr G, Grünewald S, Eisenhardt G, Bach A, Kornau H-C. Science. 1999;283:74–77. doi: 10.1126/science.283.5398.74. [DOI] [PubMed] [Google Scholar]
- 22.Rocheville M, Lange D C, Kumar U, Sasi R, Patel R C, Patel Y C. J Biol Chem. 2000;275:7862–7869. doi: 10.1074/jbc.275.11.7862. [DOI] [PubMed] [Google Scholar]
- 23.AbdAlla S, Lother H, Quitterer U. Nature (London) 2000;407:94–98. doi: 10.1038/35024095. [DOI] [PubMed] [Google Scholar]
- 24.Ferré S, Torvinen M, Antoniou K, Irenius E, Civelli O, Arenas E, Fredholm B B, Fuxe K. J Biol Chem. 1998;273:4718–4724. doi: 10.1074/jbc.273.8.4718. [DOI] [PubMed] [Google Scholar]
- 25.Ginés S, Hillion J, Torvinen M, Crom S L, Casadó V, Canela E I, Rondin S, Lew J Y, Watson S, Zoli M, et al. Proc Natl Acad Sci USA. 2000;97:8606–8611. doi: 10.1073/pnas.150241097. . (First Published July 11, 2000, 10.1073/pnas.150241097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocheville M, Lange D C, Kumar U, Patel S C, Patel R C, Patel Y C. Science. 2000;288:154–157. doi: 10.1126/science.288.5463.154. [DOI] [PubMed] [Google Scholar]
- 27.Jordan B A, Trapaidze N, Gomes I, Nivarthi R, Devi L A. Proc Natl Acad Sci USA. 2001;98:343–348. doi: 10.1073/pnas.011384898. . (First Published December 26, 2000, 10.1073/pnas.011384898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frame L T, Yeung S-M H, Venter J C, Cooper D M F. Biochem J. 1986;235:621–624. doi: 10.1042/bj2350621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciruela F, Casadó V, Mallol J, Canela E I, Lluis C, Franco R. J Neurosci Res. 1995;42:818–828. doi: 10.1002/jnr.490420610. [DOI] [PubMed] [Google Scholar]
- 30.Ciruela F, Saura C, Canela E I, Mallol J, Lluis C, Franco R. FEBS Lett. 1996;380:219–223. doi: 10.1016/0014-5793(96)00023-3. [DOI] [PubMed] [Google Scholar]
- 31.Gerwins P, Fredholm B B. J Biol Chem. 1992;267:16081–16087. [PubMed] [Google Scholar]
- 32.Moore D, Chambers J, Waldvogel H, Faull R, Emson P. J Comp Neurol. 2000;421:374–384. doi: 10.1002/(sici)1096-9861(20000605)421:3<374::aid-cne6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 33.Ochiishi T, Chen L, Yukawa A, Saitoh Y, Sekino Y, Arai T, Nakata H, Miyamoto H. J Comp Neurol. 1999;411:301–316. [PubMed] [Google Scholar]
- 34.Nakata H. Biochim Biophys Acta. 1993;1177:93–98. doi: 10.1016/0167-4889(93)90163-j. [DOI] [PubMed] [Google Scholar]
- 35.Hoffmann C, More S, Nicholas R A, Harden T K, Jacobson K A. J Biol Chem. 1999;274:14639–14647. doi: 10.1074/jbc.274.21.14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hebért T E, Bouvier M. Biochem Cell Biol. 1998;76:1–11. doi: 10.1139/bcb-76-1-1. [DOI] [PubMed] [Google Scholar]
- 37.Salahpour A, Angers S, Bouvier M. Trends Endocrinol Metab. 2000;11:163–168. doi: 10.1016/s1043-2760(00)00260-5. [DOI] [PubMed] [Google Scholar]