Background: Mkx is a transcriptional repressor that regulates muscle and tendon differentiation.
Results: MKX binds to nnACA recognition sites as a homodimer. Mkx regulates transcription through recognition sites in the Mkx and Sox6 loci.
Conclusion: MKX has a novel DNA recognition mode and promotes slow muscle fiber type specification through Sox6.
Significance: We provide insight into Mkx regulation of musculoskeletal-specific transcription.
Keywords: DNA-binding Protein, Homeobox, Skeletal Muscle, Transcription Enhancers, Transcription Repressor, Mkx, Mohawk, Sox6
Abstract
The homeobox transcription factor Mohawk (Mkx) is a potent transcriptional repressor expressed in the embryonic precursors of skeletal muscle, cartilage, and bone. MKX has recently been shown to be a critical regulator of musculoskeletal tissue differentiation and gene expression; however, the genetic pathways through which MKX functions and its DNA-binding properties are currently unknown. Using a modified bacterial one-hybrid site selection assay, we determined the core DNA-recognition motif of the mouse monomeric Mkx homeodomain to be A-C-A. Using cell-based assays, we have identified a minimal Mkx-responsive element (MRE) located within the Mkx promoter, which is composed of a highly conserved inverted repeat of the core Mkx recognition motif. Using the minimal MRE sequence, we have further identified conserved MREs within the locus of Sox6, a transcription factor that represses slow fiber gene expression during skeletal muscle differentiation. Real-time PCR and immunostaining of in vitro differentiated muscle satellite cells isolated from Mkx-null mice revealed an increase in the expression of Sox6 and down-regulation of slow fiber structural genes. Together, these data identify the unique DNA-recognition properties of MKX and reveal a novel role for Mkx in promoting slow fiber type specification during skeletal muscle differentiation.
Introduction
Mohawk (Mkx)3 is a transcriptional repressor expressed in the embryonic progenitor cell populations of skeletal muscle, tendon, cartilage, and bone (1–4). Mkx is a conserved homeobox gene that has recently been shown to be a key regulator of skeletal muscle and tendon differentiation (2, 5, 6). Targeted disruption of Mkx in the mouse results in severe tendon hypoplasia and a significant decrease in the expression of extracellular matrix proteoglycans critical for musculoskeletal function, including collagen type I and decorin (5). Parsing the mechanism(s) by which Mkx regulates the development of the musculoskeletal system is dependent on characterizing its DNA-binding properties and ultimately identifying its direct genetic targets.
Mkx is a homeobox gene that belongs to the three-amino acid loop extension superclass of atypical homeobox genes (includes PBC, MEIS, TGIF, IRO, and MKX) (1, 3 4, 7). Homeobox genes contain a conserved ∼180-base pair (bp) sequence that encodes an ∼60-amino acid three α helix DNA-binding domain, called the homeodomain. The homeodomain interacts directly with DNA such that the third α helix, the recognition helix, interacts with nucleotides in the major groove and the N-terminal arm makes contact with the minor groove. DNA recognition occurs through specific residues in the N-terminal arm (amino acids 3, 5–8) and the recognition helix (amino acids 47, 50, 51, 54, and 55), such that an ∼6-bp recognition motif is formed (8–11).
Among the atypical three-amino acid loop extension genes, Mkx is most closely related to the Iroquois family. The Iroquois genes were first identified in Drosophila as a complex of three clustered and highly related genes (Iro-C), including araucan (ara), caupolican (caup), and mirror (mirr). In mouse, six Iroquois genes (Irx1–6) are involved in the development of a diverse set of tissues, including the central nervous system, ventricles of the heart, somites, lungs, gonads, and cartilage (12–16). Mouse Mkx shares 56% homology with Irx2 over the entire homeodomain (35/63 residues) and 82% homology (14/17 residues) specifically within Helix III (1). Among the amino acids important for DNA recognition, mouse Mkx and Irx factors are identical at positions 8, 50, 51, 54, and 55, but differ at positions 2, 3, 5, 6, 7, and 47 (1). Because it has been shown previously that closely related homeobox genes recognize similar DNA recognition motifs, Mkx and Irx homeodomains may have similarity in their DNA-binding specificity (11, 17).
The minimal Iroquois-binding site (IBS) for Drosophila Iro-C family members has been identified using in vitro site selection assays (11, 18). The IBS is composed of an inverted repeat (ACAnnTGT) that consists of two monomeric recognition motifs for each Iro-C homeodomain (ACA). The arrangement of the two monomeric recognition motifs appears to be flexible, as Drosophila ara has been shown to recognize an opposite inverted repeat conformation (TGTnnACA), but not a direct repeat ACAnnACA (17–19). The monomeric recognition motif of the Drosophila Mkx ortholog CG11617 has been characterized and is similar to Irx family members, except for a strong preference for a thymine (T) at position 1 of the binding motif (TnACA) (11).
The genetic networks through which Mkx functions in the normal differentiation and growth of the musculoskeletal system are currently unknown. To identify direct targets of MKX regulation, we characterized the DNA-binding specificity of mouse Mkx; its regulation of its own promoter sequence; and its ability to form a homodimer. Additionally, we have identified two Mkx-responsive elements within the locus of Sox6, a key repressor of slow fiber type specification during skeletal muscle differentiation. These characterized recognition elements provide a basis for identifying additional direct targets of MKX regulation and further our understanding of its role as a regulator of musculoskeletal development.
MATERIALS AND METHODS
Homeodomain-binding Site Selection
A fragment of mouse Mkx containing the homeodomain and adjacent conserved domain CD-A (amino acids 70–161) was amplified from the expression plasmid CS2MT-Mkx (2) (see supplemental Tables S1 and S2 for primers). Additionally, a stop codon, TAA (italicized), was incorporated into the reverse primer. The Mkx fragment was subcloned into the pB1H2ω2-12 vector as a fusion to the Zif12 and the ω subunit of bacterial RNA polymerase. These selections were performed as described previously, where the recognition motif was compiled from the sequences of 10-bp library members recovered from the binding site selection (11, 19). The web-based Motifsampler program was used to develop a position probability matrix for the Mkx-binding site (20, 21).
Web-based Sequence Analysis
A genome-scale DNA pattern search for Mkx-binding sites was performed using the Regulatory Sequence Analysis Tools (22).
Plasmids
The Mkx luciferase reporter, 3.5kbMkx-luc, was constructed by cloning nucleotide −3576 to +127 of the Mkx locus into the XhoI and HindIII cloning sites of pGL3Basic (Promega, Madison, WI). SmaI digest of the 3.5kbMkx-luc removed most of the 5′ flanking sequence, creating the 113Mkx-luc (nucleotide −113 to +127 bp). Domain mapping of the −113 to +127-bp promoter region was done by cloning each fragment in reverse orientation into the KpnI/XhoI sites in front of the SV40 promoter in pGL3Promoter. CS2MT-Mkx(1–204)-VP16 was created by cloning the Mkx fragment into the StuI/XhoI sites in CS2MT and the activation domain of VP16 into the XhoI/XbaI sites (24). Primer sequences are located in supplemental Tables S1 and S2.
Transient Transfections and Luciferase Reporter Assays
Transcriptional activity of the Mkx and Sox6 genomic sequences was measured in NIH3T3-transfected mouse fibroblast cells as described in Anderson et al. (2). Luciferase activity was measured using an FLx800 microplate reader (BioTek Instruments, Inc., Winooski, VT). Experiments were performed in triplicate, and each experiment was repeated at least three times.
Electrophoretic Mobility Shift Assays (EMSA)
Recombinant Myc-epitope-tagged proteins used in EMSA were expressed in mouse NIH3T3 cells using transient transfection with Lipofectamine and Plus reagent (Invitrogen), as described in Ref. 2. Double-stranded EMSA probes were created by annealing complementary oligonucleotides in sodium chloride-Tris-EDTA buffer (50 mm NaCl, 10 mm Tris, pH 8.0, and 1 mm EDTA). Probe sequences are listed in supplemental Tables S1 and S2. Binding reactions were carried out at room temperature in 4× binding buffer and separated using polyacrylamide gel electrophoresis as described by Wilson-Rawls et al. (25).
Mkx Knock-out Mouse Genotyping
Targeted disruption of the Mkx locus has been reported previously (6). Mkx knock-out mice were genotyped using a duplex PCR with the primers (see supplemental Tables S1 and S2). Mice were maintained as heterozygotes for the Mkx-null allele and wild type, and null littermates from heterozygous crosses were used for experimental analysis.
Primary Satellite Cell Isolation and Culture
Satellite cells were isolated from hindlimb muscle from six 4-month-old mice. Satellite cells were maintained in 20% fetal bovine serum (FBS) in Ham's F-10 (supplemented with 10 ng/ml basic fibroblast growth factor (BD Biosciences) and Primocin (InvivoGen, San Diego, CA). To induce differentiation, satellite cells were plated in 60-mm dishes and changed to media containing DMEM containing 2% horse serum and Primocin. Differentiation medium was changed every day, and satellite cells were allowed to differentiate for 3 days. Expression of fast twitch myosin heavy chain isoforms (Myh1, 2, 4, and 8) and slow twitch myosin heavy chain (Myh7) was detected by immunohistochemistry using the MY32 and NOQ7.5.4D antibodies (Sigma-Aldrich), respectively. Immunohistochemistry was performed as described in Ref. 26.
Analysis of Satellite Cell Gene Expression Using Real-time PCR
Total RNA was isolated and purified from satellite cells cultured in 60-mm dishes using 1 ml of ice-cold TRIzol (Invitrogen) according to the manufacturer's protocols. Reverse transcription of 2 μg of total RNA was performed using Superscript III (Invitrogen). Real-time quantitative PCR analysis of the cDNA was performed using qPCR MasterMix Plus w/o UNG (Eurogentec, Fremont, CA) on an ABI 7900HT quantitative Real-Time PCR Machine (Applied Biosystems, Foster City, CA).
RESULTS
The Mouse Mkx Homeodomain Binds a Recognition Motif Common to the Irx Family
To characterize the DNA-binding specificity of the mouse Mkx homeodomain, we utilized a modified bacterial one-hybrid assay to select recognition sequences from a library of random decamer sequences. A fragment of mouse Mkx including the homeodomain and adjacent conserved domain CD-A (amino acids 70–161) was cloned into the pB1H2ω2-12 vector, as a fusion with the ω subunit of RNA polymerase and the first and second zinc fingers of Zif268. This construct was co-transformed with a reporter plasmid that contains a region of random decamers adjacent to the binding site of the Zif268, and transformants were selected for their ability to grow under nutrient-deficient conditions (11, 19).
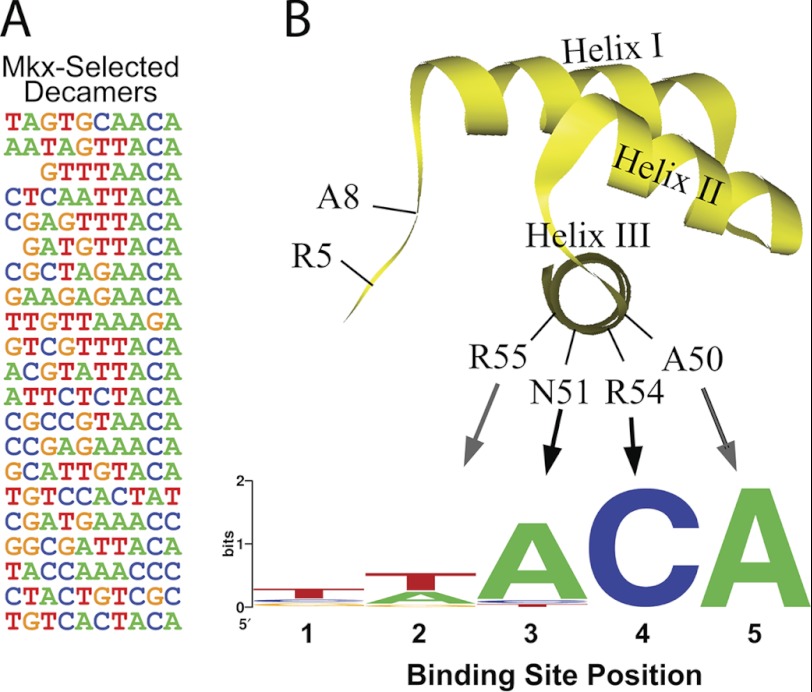
A total of 21 sequences were recovered and analyzed using the web-based program Motifsampler (Fig. 1A). The motif-detecting algorithm of this program uses Gibbs sampling to find the position probability matrix of the motif (20, 21). The preferred recognition sequence of mouse MKX was determined to be A-C-A (Fig. 1B). Thus, mouse MKX shares the same recognition motif as previously characterized Irx family members but diverges from the recently reported binding site (T-n-A-C-A) of the Drosophila MKX ortholog, which has a strong affinity for a T at position 1 (11).
FIGURE 1.
Characterizing the monomeric Mkx recognition motif. A fragment encoding the mouse Mkx homeodomain was used to screen a random decamer library using a modified bacterial one-hybrid site selection assay. A, 21 clones from the selection were analyzed for overrepresented sequence motifs. B and C, diagram of key determinants within the homeodomain that are important for specifying the recognition motif for mouse Mkx is shown. Mkx showed little specificity for binding positions 1 and 2 but a strong preference for A-C-A at positions 3–5.
Mkx Is Capable of Negative Autoregulation
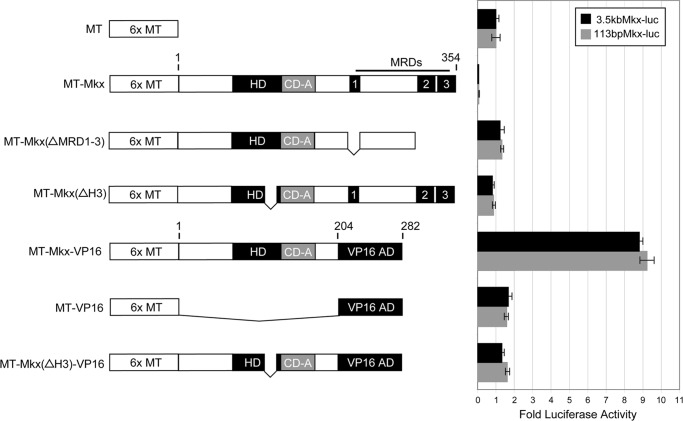
DNA-binding transcription factors often participate in the regulation of their own transcription through autoregulatory feedback loops (27). Negative autoregulation is a well established mechanism for both stabilizing transcription factor levels as well as modulating spatial and temporal expression patterns (23, 28, 29). To determine whether MKX is capable of directly regulating its own transcription, we cloned a fragment of the mouse Mkx promoter (−3,576 to +127 bp) upstream of the luciferase reporter gene in pGL3Basic (3.5kbMkx-luc) (Fig. 2A). Full-length Mkx (MT-MKX) strongly repressed transcription (13.4-fold reduction) when compared with background (Fig. 2B). Deletion of the three MKX repression domains (2) (MT-Mkx(ΔMRD1-3)) or Helix III of the homeodomain (MT-Mkx(ΔH3)) abrogated repression of 3.5kbMkx-luc (Fig. 2B). To confirm the responsiveness of 3.5kbMkx-luc to Mkx, the C-terminal region containing the three Mkx repressor domains (amino acids 205–352) was replaced by the potent transcriptional activation domain of VP16 (MT-Mkx-VP16) (30). The MT-Mkx-VP16 fusion resulted in 8.9-fold activation of 3.5kbMkx-luc. Additionally, neither a deletion mutant of MT-Mkx-VP16 that does not contain Helix III of the homeodomain (MT-Mkx(ΔH3)-VP16), nor VP16 alone, could activate the 3.5kbMkx-luc reporter to the same extent (Fig. 2B). This demonstrates that MKX is capable of regulating its own promoter and that this regulation is dependent upon amino acids within the homeodomain.
FIGURE 2.
Mkx is capable of negative autoregulation. Fragments of the mouse Mkx promoter cloned upstream of a luciferase reporter gene were assayed for their responsiveness to Mkx regulation in NIH3T3 cells. An Mkx promoter-luciferase construct encompassing −3,576 bp to +127 bp (3.5kbMkx-luc) was strongly repressed by co-transfection with a plasmid-encoding Myc tag full-length Mkx (MT-Mkx). Deletion of the three Mkx repressor domains (MT-Mkx(ΔMRD1-3)) or Helix III of the homeodomain (MT-Mkx(ΔH3)) abrogated this effect. An activated form of Mkx (MT-Mkx-VP16) resulted in specific activation of the Mkx promoter reporter. A smaller Mkx promoter construct encompassing −113 bp to +127 bp (113bpMkx-luc) responded in an identical manner.
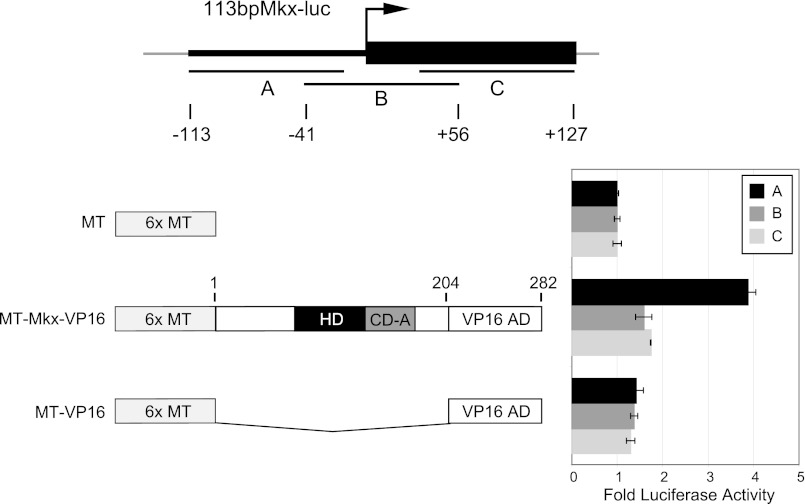
Similar repression levels were observed with a smaller fragment of the Mkx promoter spanning −113 bp to +127 bp (113bpMkx-luc) (Fig. 2). This fragment was therefore further subdivided into three overlapping fragments (A, −113 to −17; B, −41 to +56; and C, +32 to +127) (Fig. 3A). Fragment A alone was responsive to MT-Mkx-VP16, which we will herein refer to as the MRE (Fig. 3B). A multiple sequence alignment revealed that the MRE is highly conserved in the promoters of vertebrate Mkx orthologs and is composed of a large inverted repeat (see supplemental Fig. S1).
FIGURE 3.
Identifying the Mkx response element within the Mkx promoter. A, Mkx promoter sequence (−113 bp to +127) was cloned as three individual fragments (fragment A, −113 to −17; B, −41 to +56; and C, +32 to +127) upstream of SV40 in the luciferase pGL3Promoter. B, of the three smaller Mkx promoter fragments, fragment A was the most responsive to MT-Mkx-VP16 activation. This sequence was renamed the MRE.
MKX Interacts Directly with a Highly Conserved Sequence within Its Promoter
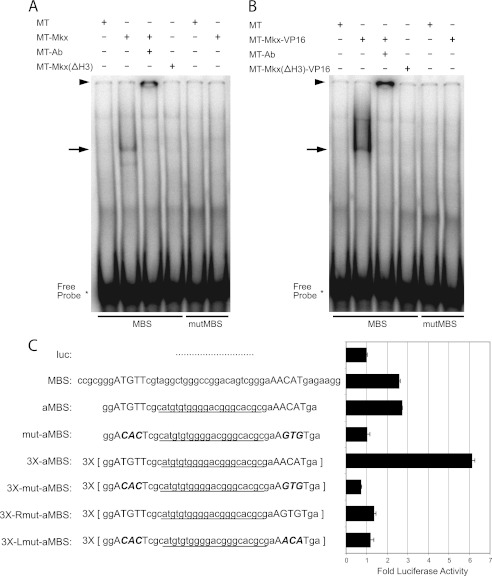
The MRE includes a highly conserved inverted repeat, which contains the recognition motif we characterized, separated by 25 bases (ATGTT-N25-AACAT). To test if MKX can bind this sequence, we performed an EMSA using whole cell lysates from NIH3T3 cells expressing MT-MKX and a double-stranded DNA probe specific for the conserved inverted repeat sequence (Fig. 4). A band was consistently observed in the presence of MT-MKX and could be further supershifted by an anti-Myc antibody (Fig. 4A). A mutant fusion protein, MT-MKX with the Helix III of the homeodomain deleted (MT-Mkx(ΔH3)), was not able to shift the MRE probe (Fig. 4A). Further, MT-MKX-VP16 also formed a complex with the probe and was supershifted by the anti-Myc antibody (Fig. 4B). This demonstrated that MKX can form a complex with this inverted repeat, which we will refer to as the Mkx-binding sequence (MBS), and the homeodomain is necessary for this interaction.
FIGURE 4.
Identification of the MBS within the MRE. A and B, MT-Mkx (A) and MT-Mkx-VP16 (B) were able to bind the MBS sequence but not a mutated MBS sequence in an electrophoresis mobility shift assay. The Mkx-MBS complex could be supershifted with an anti-Myc antibody (MT-Ab). Deletion mutants of Mkx lacking Helix III of the homeodomain (MT-Mkx(ΔH3) and MT-Mkx(ΔH3)-VP16) were not sufficient to shift MBS. EMSA primers are provided in supplemental Table S1. C, cell-based luciferase transcription assays demonstrate that the MBS sequence is responsive to Mkx-VP16 activation. An artificial MBS (aMBS) that removes 5′-flanking sequence and randomizes the central 21 bp was equally responsive. Increasing the copy number of the MBS resulted in increased activation by Mkx-VP16. Mutation of either half-site demonstrated that both are required for Mkx-mediated regulation.
The ability of MKX to regulate the MBS in cells was tested by cloning this sequence upstream in pGL3Promoter (MBS-luc). MT-Mkx-VP16 activated the MBS-luc vector 2.5-fold, when compared with pGL3Promoter alone (luc) (Fig. 4C). Reducing the number of flanking residues and randomizing the intervening 21 bp did not alter the level of activation (artificial MBS-aMBS-luc) (Fig. 4C). Multimerizing three copies of the aMBS in a head-to-tail fashion resulted in additive increases in luciferase activation (3X-aMBS-luc). Point mutation of nucleotides within both inverted repeats (mut-aMBS; ACACT N25 AGTGT) abolished activation by MT-Mkx-VP16 (Fig. 4C). Multimerized versions of the MBS in which one of the half-sites was mutated (Lmut-aMBS or Rmut-aMBS) abrogated activation by MT-Mkx-VP16 (Fig. 4C). This indicated that both half-sites of the MBS were required and given the inverted repeat nature of the MBS, suggested that MKX might bind the MBS as a homodimer.
MKX Can Homodimerize and Requires an Adjacent Conserved Domain to Recognize Its Binding Site
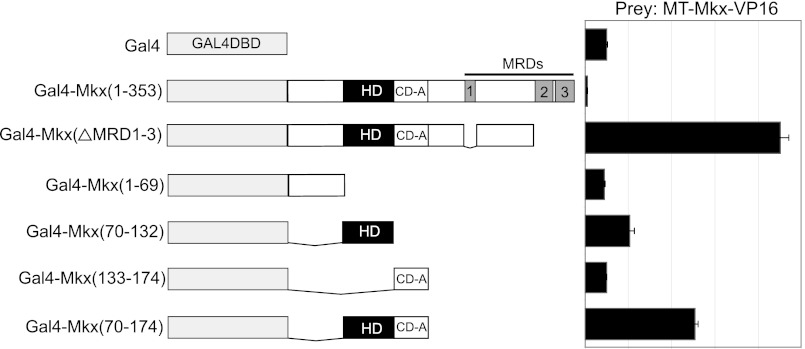
Homeodomain-containing proteins can function as monomers, homodimers, or heterodimers, which can lead to differential DNA-binding specificity (31–33). To test whether MKX is capable of homodimerization, we used a modified mammalian two-hybrid approach using transient transfection in NIH3T3 cells. Gal4 DNA-binding domain (Gal4DBD)-fusion proteins with MKX-specific bait fragments were assayed for their ability to interact with the MT-Mkx-VP16. Dimerization of Mkx bait fragments was scored by the level of activation of the Gal4DBD-responsive luciferase reporter, 5XUASpGL3Promoter, relative to Gal4DBD alone (2). Initially, full-length Mkx resulted in a reduction of reporter activity (Fig. 5A). Because the three Mkx repression domains (MRD1-3) may suppress luciferase transcription, a fusion construct that lacks these domains (Gal4DBD-Mkx(ΔMRD1-3)) was examined. This resulted in a 9-fold activation of the reporter, demonstrating that Mkx is capable of homodimerization. A fusion construct containing the Mkx homeodomain led to modest activation of the reporter (2.1-fold), whereas the inclusion of the adjacent conserved domain (CD-A) resulted in a 5.1-fold activation of the reporter (Fig. 5A). Neither the N-terminal amino acid sequence to the homeodomain nor CD-A domain alone was sufficient to activate the reporter (Fig. 5A). This demonstrates that MKX is capable of dimerization through the homeodomain and that this interaction is enhanced by the presence of the CD-A.
FIGURE 5.
Mkx is able to form a homodimer. The ability of Mkx to form a homodimer was assayed using a cell-based mammalian two-hybrid. Mkx is capable of homodimerization dependent upon amino acids within the homeodomain and CD-A. Data are presented for each Gal4-Mkx bait as -fold activation relative to the level of luciferase activity obtained by co-transfection of the Gal4DBD alone with appropriate prey.
The Sox6 Locus Contains Multiple Mkx-binding Sites
To identify additional targets of MKX, a genome-scale DNA pattern search of the conserved MBS sequence was performed using the Regulatory Sequence Analysis Tools (22). The number of intervening nucleotides was allowed to vary from 0 to 25, and mismatches of 1 bp were allowed in either half-site (ATGTT N0–25 AACAT). A list of putative MRE sites identified by this approach is provided in supplemental Table S2. Within the intronic sequence in the Sox6 locus, we identified a well conserved MBS with an intervening space of 11 nucleotides between the inverted repeat sequences (ATGTT N11 AACAT) (Fig. 6). Because of the overlap in the MKX and IRX DNA-recognition sequences, a similar genome-scale DNA pattern search was performed using the Irx-binding sequence (ACAnnTGT). This revealed an additional well conserved element within the Sox6 promoter (AACATGTGTT). Because Sox6 plays an important role in musculoskeletal development, it represents a strong potential target for MKX (34–37).
FIGURE 6.
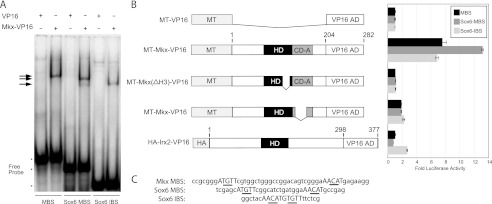
Mkx-binding sites within the Sox6 locus. Mkx (ATGTT-N11-AACAT)- and Irx (ACAnnTGT)-binding sites were identified in the locus of Sox6. A, EMSAs demonstrate that MT-Mkx-VP16 can physically interact with the Sox6 MBS and IBS sequences in vitro. B, cell-based luciferase assays demonstrate that these sequences are responsive to MT-Mkx-VP16 activation and are dependent upon sequences within Helix III of the homeodomain (MT-Mkx(ΔH3)-VP16) and CD-A (MT-Mkx(ΔCDA)-VP16). An activated form of mouse Irx2 (HA-Irx2-VP16) could only regulate the Sox6 IBS.
To validate these sequences as Mkx-binding sites, electrophoretic mobility shift assays were performed. MKX was able to bind to both the Sox6 MBS and IBS sequences similar to the MBS in the Mkx promoter (Fig. 6A). Multimerized (3×) Sox6 target sequences were cloned upstream of the SV40 promoter in the pGL3Promoter vector. Similar to the MBS identified in the Mkx promoter, the Sox6 MBS and the Sox6 IBS were responsive to MT-MKX-VP16 activation (Fig. 6B). Deletion of Helix III (MT-Mkx(ΔH3)-VP16) abrogated the activation at all three sites (Fig. 6B). Transcriptional activation was lost with the deletion of CD-A from the MKX-VP16 fusion (MT-Mkx(ΔCD-A)-VP16), underscoring its importance in the function of Mkx regulation (Fig. 6B). This demonstrates that Mkx is capable of regulating both MBS and IBS motifs within the Sox6 locus.
Because Mkx can recognize a binding site conformation similar to the IBS, it is possible that MKX and IRX proteins may co-regulate a subset of genes. To determine whether IRX proteins can similarly recognize both binding site conformations, we utilized an activated form of mouse IRX2 (HA-IRX2-VP16). The HA-IRX2-VP16 fusion activated modestly the Sox6 IBS (2.6-fold) but did not activate either MBS-containing reporter plasmid (Fig. 6B).
Sox6 Is Up-regulated in Satellite Cells Isolated from Mkx−/− Mice
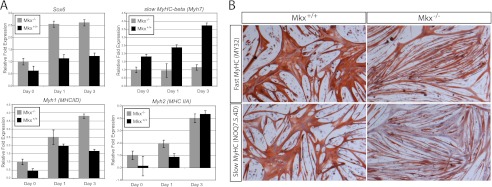
To validate that Mkx was a repressor of Sox6 transcription in vivo, the levels of endogenous Sox6 mRNA were examined in primary muscle cells deficient for Mkx. Skeletal muscle satellite cells were isolated from the hindlimb muscles of 5-month-old Mkx−/− and wild type littermate mice. Quantitative RT-PCR using Sox6-specific primers revealed that Sox6 is expressed at a similar level in freshly isolated satellite cells from Mkx−/− and control muscle (Fig. 7A). However, satellite cells differentiating into myotubes demonstrated that Sox6 expression was 2.2-fold higher in the Mkx−/− mice (Fig. 7A). Sox6 has been shown to participate in fiber type specification in skeletal muscle by repressing the transcription of the slow twitch myosin heavy chain (MHC) isoform (Myh7) in fast twitch muscle fibers (34–36). Myh7 transcription was decreased in differentiating Mkx−/− myotubes but not the fast twitch myosin heavy chain isoforms (Myh1 and Myh2) (Fig. 7A). The differential reduction of the slow isoform was confirmed by immunostaining with antibodies specific to slow and fast MHC isoforms (Fig. 7B). This is consistent with Mkx regulating the transcription of Myh7 through repression of Sox6 and predicts a role in fiber type specificity.
FIGURE 7.
Sox6 and MHC expression in Mkx−/− satellite cells. Primary cultures of adult muscle satellite cells were isolated from Mkx knock-out (Mkx−/−) and wild type (Mkx+/+) mice and cultured in vitro. Total RNA was isolated from proliferating cultures (day 0) or cultures that were differentiated under low mitogenic conditions for 1 or 3 days (day 1 and day 3). A, quantitative real-time PCR revealed that Sox6 transcription is up-regulated in Mkx−/− satellite cells. Transcription of Myh7 was reduced in differentiated Mkx−/− myotubes, whereas fast MHC isoforms were up-regulated (Myh1) or unaffected (Myh2). B, expression of fast MHC (My32) and slow MHC (NOQ7.5.4D) were detected in differentiated myoblasts isolated from Mkx−/− and Mkx+/+ mice.
DISCUSSION
The genetic pathways controlling the patterning and differentiation of the vertebrate musculoskeletal system require the combined actions of multiple regulatory factors. The Mkx homeobox gene has been shown to be an important regulator of skeletal muscle and tendon differentiation (5, 6). We have characterized the DNA recognition motif of the mouse Mkx homeodomain and identified an Mkx-responsive element within the proximal promoter of Mkx. Using the identified Mkx regulatory element as a model, functional MREs within the Sox6 locus were identified, suggesting that MKX can directly repress the expression of Sox6. Combined with the observation of up-regulation of Sox6 in Mkx−/− satellite cells, this defines a potential pathway by which Mkx promotes the expression of slow MHC during muscle development.
The DNA recognition motif for members of the homeodomain-containing transcription factor family is largely dependent on a core set of amino acids in Helix III and residues at the N terminus of the homeodomain that make direct contact with the DNA. A comprehensive analysis performed in Drosophila organizes this superfamily of genes into 11 distinct specificity groups (11). Consistent with sequence conservation in the homeodomain, the Drosophila Mkx ortholog (CG11617) falls into the Iroquois group that is characterized by a monomeric nnACA motif. Interestingly, CG11617 diverges from Iroquois genes such as mirr through the preference for T at position 1 (TnACA) (11). Our data demonstrate that mouse Mkx has lost specificity for a T and therefore can bind half-sites such as that found in the vertebrate MBS (GAACAT and AAACAT). Mouse and Drosophila Mkx orthologs may recognize different recognition motifs as the result of specific amino acid changes at critical residues of the homeodomain. At position 8 of the N-terminal arm of the homeodomain, mouse Mkx contains an alanine (Ala) and the Drosophila ortholog contains a phenyalanine (Phe) (7). Large hydrophobic residues in position 8 have been shown to influence binding specificity at position 1 of the recognition motif (11). The Iroquois factors, which share an affinity for the same monomeric site as mouse Mkx, similarly contain an Ala at position 8 of the N-terminal arm. Replacement of the Ala with the large hydrophobic Phe residue in Drosophila caup partially added a preference for T at binding position 1 (11). Whereas the amino acids critical for DNA recognition are identical among vertebrate Mkx orthologs, position 8 in the N-terminal arm is different among cephalochordates, hemichordates, and echinoderms. This predicts that the alteration in position 8, coupled with the adaptation of dimerization in vertebrates, expands the functionality of Mkx by increasing the complexity of binding sites and thereby target genes. Future studies will be required to determine whether changes in the amino acid identity at position 8 have altered Mkx DNA recognition among distantly related metazoans.
Whereas the mouse MKX homeodomain shares a common recognition motif with Irx family members, we have demonstrated that they differ in their ability to recognize half-site conformations. Our data show that Mkx can recognize and bind both MBS- and IBS-like conformations, whereas an IRX2-VP16 fusion protein is only capable of regulating the IBS conformation. MBS and IBS conformations are different in both the number of nucleotides separating the half-sites and the orientation of the half-sites. Inter-site spacing has been shown to be a critical parameter for homeobox gene regulation. The Drosophila bicoid homeobox transcription factor can bind strongly to sites that are spaced 25 bp apart but only weakly to sites separated by 11 bp (38). This reveals an additional layer of Mkx regulation by which the level repression could be mediated by specific MBS conformations that differ in spacer length between each half-site.
The inverted repeat arrangement of the MBS and IBS sequences is consistent with Mkx-binding DNA as dimers. The Iroquois family has been shown previously to bind DNA in both homo- and heterodimer arrangements, and these interactions are dependent upon sequences N-terminal to the homeodomain (18). Similarly, MKX was capable of homodimerization, although this interaction was dependent upon a unique sequence immediately C-terminal to the MKX homeodomain. Deletion of this conserved domain also diminished the ability of MKX to regulate gene expression through the MBS, suggesting that MKX binds DNA as a requisite homodimer. It is interesting to speculate on whether members of the Irx family and MKX form functional heterodimers because their expression patterns overlap in the somites, limb buds, and muscle during development. Future studies will be required to determine the extent to which this interaction occurs in vivo and whether these closely related family members share functional redundancy during development.
Our studies predict that Mkx is able repress its own transcription through the MBS found in its promoter. Such negative autoregulatory loops have been found at the core of several developmental processes, including the anterior-posterior co-linear expression of the Hox genes, the oscillatory genetic networks associated with circadian rhythms, and the segmental clock of somitogenesis, stem cell potency, and cell differentiation (39–41). Further, it has been proposed that negative autoregulation can suppress the transcriptional noise intrinsic in genetic networks that reduces both the switching on and switching off time of gene expression (42). The Mkx-responsive element is highly conserved among vertebrate Mkx orthologs and clustered with other conserved inverted repeat sequences. This suggests that negative autoregulation is a conserved feature among Mkx orthologs and may be modulated in a larger regulatory complex. The significance of the negative autoregulation will become clearer as the Mkx genetic network is better understood.
Our initial characterization of the MKX-binding properties led to the identification of two well conserved MBS within the Sox6 locus. Sox6 has been shown to be an important regulator during musculoskeletal development and fiber type specification (35–37, 43). Sox6 possesses a redundant function with Sox5 during the differentiation of chondrocytes (44) and is required to block cells of the sclerotome from entering the tendon lineage (45). In skeletal muscle, Sox6 suppresses slow twitch fiber development through the direct repression of Myh7 transcription (34, 35). In primary satellite cells isolated from Mkx−/− mice, we observed that endogenous Sox6 transcription was elevated upon differentiation, suggesting it is a direct target of Mkx-mediated repression. Consistent with this result, Myh7 gene expression was reduced in these same cells, whereas fast MHC isoform expression levels were unchanged. This provides the first evidence for a role for Mkx in the regulation of fiber type specificity during skeletal muscle differentiation. Further, it provides a possible mechanism by which Mkx is able to participate in the integration of the differentiation of multiple cell types in the musculoskeletal system.
Supplementary Material
Acknowledgments
We thank Dr. Ron Allen for advice on culturing primary satellite cells, Dr. Anthony Firulli for providing the CS2MT plasmid, Dr. Scott Bingham for assistance in DNA sequencing, and Dr. James Elser for the generous use of the FLx800 microplate reader.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01HG004744 (to S. A. W.) and National Science Foundation (to J. A. R. and J. W. R.).

This article contains supplemental Tables S1 and S2 and Fig. S1.
- Mkx
- Mohawk
- MRE
- Mkx-responsive element
- IBS
- Iroquois-binding site
- MBS
- Mkx-binding sequence
- Gal4DBD
- Gal4 DNA-binding domain
- CD-A
- adjacent conserved domain
- MHC
- myosin heavy chain.
REFERENCES
- 1. Anderson D. M., Arredondo J., Hahn K., Valente G., Martin J. F., Wilson-Rawls J., Rawls A. (2006) Mohawk is a novel homeobox gene expressed in the developing mouse embryo. Dev. Dyn. 235, 792–801 [DOI] [PubMed] [Google Scholar]
- 2. Anderson D. M., Beres B. J., Wilson-Rawls J., Rawls A. (2009) The homeobox gene Mohawk represses transcription by recruiting the sin3A/HDAC co-repressor complex. Dev. Dyn. 238, 572–580 [DOI] [PubMed] [Google Scholar]
- 3. Liu H., Liu W., Maltby K. M., Lan Y., Jiang R. (2006) Identification and developmental expression analysis of a novel homeobox gene closely linked to the mouse twirler mutation. Gene Expr. Patterns 6, 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takeuchi J. K., Bruneau B. G. (2007) Irxl1, a divergent Iroquois homeobox family transcription factor gene. Gene Expr. Patterns 7, 51–56 [DOI] [PubMed] [Google Scholar]
- 5. Ito Y., Toriuchi N., Yoshitaka T., Ueno-Kudoh H., Sato T., Yokoyama S., Nishida K., Akimoto T., Takahashi M., Miyaki S., Asahara H. (2010) The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 10538–10542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu W., Watson S. S., Lan Y., Keene D. R., Ovitt C. E., Liu H., Schweitzer R., Jiang R. (2010) The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol. Cell. Biol. 30, 4797–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mukherjee K., Bürglin T. R. (2007) Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J. Mol. Evol. 65, 137–153 [DOI] [PubMed] [Google Scholar]
- 8. Gehring W. J., Affolter M., Bürglin T. (1994) Homeodomain proteins. Annu. Rev. Biochem. 63, 487–526 [DOI] [PubMed] [Google Scholar]
- 9. Ades S. E., Sauer R. T. (1995) Specificity of minor groove and major groove interactions in a homeodomain-DNA complex. Biochemistry 34, 14601–14608 [DOI] [PubMed] [Google Scholar]
- 10. Wolberger C. (1996) Homeodomain interactions. Curr. Opin. Struct. Biol. 6, 62–68 [DOI] [PubMed] [Google Scholar]
- 11. Noyes M. B., Christensen R. G., Wakabayashi A., Stormo G. D., Brodsky M. H., Wolfe S. A. (2008) Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133, 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bosse A., Zülch A., Becker M. B., Torres M., Gómez-Skarmeta J. L., Modolell J., Gruss P. (1997) Identification of the vertebrate Iroquois homeobox gene family with overlapping expression during early development of the nervous system. Mech. Dev. 69, 169–181 [DOI] [PubMed] [Google Scholar]
- 13. Bellefroid E. J., Kobbe A., Gruss P., Pieler T., Gurdon J. B., Papalopulu N. (1998) Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J. 17, 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bao Z. Z., Bruneau B. G., Seidman J. G., Seidman C. E., Cepko C. L. (1999) Regulation of chamber-specific gene expression in the developing heart by Irx4. Science 283, 1161–1164 [DOI] [PubMed] [Google Scholar]
- 15. Cohen D. R., Cheng C. W., Cheng S. H., Hui C. C. (2000) Expression of two novel mouse Iroquois homeobox genes during neurogenesis. Mech. Dev. 91, 317–321 [DOI] [PubMed] [Google Scholar]
- 16. Houweling A. C., Dildrop R., Peters T., Mummenhoff J., Moorman A. F., Rüther U., Christoffels V. M. (2001) Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech. Dev. 107, 169–174 [DOI] [PubMed] [Google Scholar]
- 17. Berger M. F., Badis G., Gehrke A. R., Talukder S., Philippakis A. A., Peña-Castillo L., Alleyne T. M., Mnaimneh S., Botvinnik O. B., Chan E. T., Khalid F., Zhang W., Newburger D., Jaeger S. A., Morris Q. D., Bulyk M. L., Hughes T. R. (2008) Variation in homeodomain DNA binding revealed by high resolution analysis of sequence preferences. Cell 133, 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bilioni A., Craig G., Hill C., McNeill H. (2005) Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 14671–14676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Noyes M. B., Meng X., Wakabayashi A., Sinha S., Brodsky M. H., Wolfe S. A. (2008) A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 36, 2547–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thijs G., Lescot M., Marchal K., Rombauts S., De Moor B., Rouzé P., Moreau Y. (2001) A higher order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics 17, 1113–1122 [DOI] [PubMed] [Google Scholar]
- 21. Thijs G., Marchal K., Lescot M., Rombauts S., De Moor B., Rouzé P., Moreau Y. (2002) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of co-expressed genes. J. Comput. Biol. 9, 447–464 [DOI] [PubMed] [Google Scholar]
- 22. van Helden J. (2003) Regulatory sequence analysis tools. Nucleic Acids Res. 31, 3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bateman E. (1998) Autoregulation of eukaryotic transcription factors. Prog. Nucleic Acids Res. Mol. Biol. 60, 133–168 [DOI] [PubMed] [Google Scholar]
- 24. Rupp R. A., Snider L., Weintraub H. (1994) Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8, 1311–1323 [DOI] [PubMed] [Google Scholar]
- 25. Wilson-Rawls J., Rhee J. M., Rawls A. (2004) Paraxis is a basic helix-loop-helix protein that positively regulates transcription through binding to specific E-box elements. J. Biol. Chem. 279, 37685–37692 [DOI] [PubMed] [Google Scholar]
- 26. Wilson-Rawls J., Hurt C. R., Parsons S. M., Rawls A. (1999) Differential regulation of epaxial and hypaxial muscle development by paraxis. Development 126, 5217–5229 [DOI] [PubMed] [Google Scholar]
- 27. Crews S. T., Pearson J. C. (2009) Transcriptional autoregulation in development. Curr. Biol. 19, R241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Irvine K. D., Botas J., Jha S., Mann R. S., Hogness D. S. (1993) Negative autoregulation by Ultrabithorax controls the level and pattern of its expression. Development 117, 387–399 [DOI] [PubMed] [Google Scholar]
- 29. Semsey S., Krishna S., Erdossy J., Horváth P., Orosz L., Sneppen K., Adhya S. (2009) Dominant negative autoregulation limits steady-state repression levels in gene networks. J. Bacteriol. 191, 4487–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sadowski I., Ma J., Triezenberg S., Ptashne M. (1988) GAL4-VP16 is an unusually potent transcriptional activator. Nature 335, 563–564 [DOI] [PubMed] [Google Scholar]
- 31. Chang C. P., Brocchieri L., Shen W. F., Largman C., Cleary M. L. (1996) Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell. Biol. 16, 1734–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neuteboom S. T., Peltenburg L. T., van Dijk M. A., Murre C. (1995) The hexapeptide LFPWMR in Hoxb-8 is required for cooperative DNA binding with Pbx1 and Pbx2 proteins. Proc. Natl. Acad. Sci. U.S.A. 92, 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Furukawa K., Iioka T., Morishita M., Yamaguchi A., Shindo H., Namba H., Yamashita S., Tsukazaki T. (2002) Functional domains of paired-like homeoprotein Cart1 and the relationship between dimerization and transcription activity. Genes Cells 7, 1135–1147 [DOI] [PubMed] [Google Scholar]
- 34. Hagiwara N., Ma B., Ly A. (2005) Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p100H. Dev. Dyn. 234, 301–311 [DOI] [PubMed] [Google Scholar]
- 35. Hagiwara N., Yeh M., Liu A. (2007) Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev. Dyn. 236, 2062–2076 [DOI] [PubMed] [Google Scholar]
- 36. von Hofsten J., Elworthy S., Gilchrist M. J., Smith J. C., Wardle F. C., Ingham P. W. (2008) Prdm1- and Sox6-mediated transcriptional repression specifies muscle fiber type in the zebrafish embryo. EMBO Rep. 9, 683–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Quiat D., Voelker K. A., Pei J., Grishin N. V., Grange R. W., Bassel-Duby R., Olson E. N. (2011) Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc. Natl. Acad. Sci. U.S.A. 108, 10196–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fu D., Zhao C., Ma J. (2003) Enhancer sequences influence the role of the amino-terminal domain of bicoid in transcription. Mol. Cell. Biol. 23, 4439–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bessho Y., Kageyama R. (2003) Oscillations, clocks, and segmentation. Curr. Opin. Genet. Dev. 13, 379–384 [DOI] [PubMed] [Google Scholar]
- 40. Pan G., Li J., Zhou Y., Zheng H., Pei D. (2006) A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 20, 1730–1732 [DOI] [PubMed] [Google Scholar]
- 41. Wong W. F., Kurokawa M., Satake M., Kohu K. (2011) Down-regulation of Runx1 expression by TCR signal involves an autoregulatory mechanism and contributes to IL-2 production. J. Biol. Chem. 286, 11110–11118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zabet N. R. (2011) Negative feedback and physical limits of genes. J. Theor. Biol. 284, 82–91 [DOI] [PubMed] [Google Scholar]
- 43. Dumitriu B., Dy P., Smits P., Lefebvre V. (2006) Generation of mice harboring a Sox6 conditional null allele. Genesis 44, 219–224 [DOI] [PubMed] [Google Scholar]
- 44. Lefebvre V., Behringer R. R., de Crombrugghe B. (2001) L-Sox5, Sox6, and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage 9, Suppl. A, S69–75 [DOI] [PubMed] [Google Scholar]
- 45. Brent A. E., Braun T., Tabin C. J. (2005) Genetic analysis of interactions between the somitic muscle, cartilage, and tendon cell lineages during mouse development. Development 132, 515–528 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.