SUMMARY
DNA methylation and histone modification exert epigenetic control over gene expression. CHG methylation by CHROMOMETHYLASE3 (CMT3) depends on histone H3K9 dimethylation (H3K9me2), but the mechanism underlying this relationship is poorly understood. Here, we report multiple lines of evidence that CMT3 interacts with H3K9me2-containing nucleosomes. CMT3 genome locations nearly perfectly correlated with H3K9me2 and CMT3 stably associated with H3K9me2-containing nucleosomes. Crystal structures of maize CMT3 homologue, ZMET2, in complex with H3K9me2 peptides, showed that ZMET2 binds H3K9me2 via both BAH- and chromo-domains. The structures reveal an aromatic cage within both BAH- and chromo-domains as interaction interfaces that capture H3K9me2. Mutations that abolish either interaction disrupt CMT3 binding to nucleosomes, and show a complete loss of CMT3 activity in vivo. Our study establishes dual recognition of H3K9me2 marks by BAH- and chromo-domains, and reveals a novel mechanism of interplay between DNA methylation and histone modification.
INTRODUCTION
DNA methylation and histone modification are two major epigenetic marks regulating gene expression and chromatin state. Eukaryotic DNA is wrapped around a histone octamer that consists of two of each of the four core histones H2A, H2B, H3 and H4 and forms the basic structural unit of chromatin: the nucleosome (Luger et al., 1997). The formation of heterochromatin involves the recruitment of silencing complexes containing histone-binding and -modifying proteins and the generation of specific silent epigenetic modification patterns. In animals and Schizosaccharomyces pombe, methylation of histone H3 at lysine 9 (H3K9) is associated with silenced heterochromatic regions and is bound by the silencing protein Heterochromatin-associated protein 1 (Jenuwein and Allis, 2001). In plants, di-methylation of H3K9 (H3K9me2) is required for the silencing of transposable elements (TEs) and other repetitive DNA, which are enriched in heterochromatic regions (Bernatavichute et al., 2008; Jackson et al., 2004; Jackson et al., 2002; Malagnac et al., 2002).
DNA methylation in mammals is mainly found at CG sites and is initially catalyzed by the de novo methyltransferase DNMT3A/B and subsequently maintained by DNMT1. In contrast, DNA methylation in plants exists in three sequence contexts: CG, CHG (where H is either C, T or A), and CHH (or asymmetric) (Cokus et al., 2008). While METHYLTRANSFERASE1 (MET1, an ortholog of mammalian DNMT1) maintains CG methylation and DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2, an ortholog of mammalian DNMT3) primarily maintains CHH methylation, CHROMOMETHYLASE3 (CMT3) is a plant-specific methyltransferase responsible for CHG methylation in A. thaliana (Bartee and Bender, 2001; Law and Jacobsen, 2010; Lindroth et al., 2001). The CMT3 ortholog in maize, the Zea mays methyltransferase2 (ZMET2), was also shown to be required for CHG DNA methylation (Papa et al., 2001).
DNA methylation and histone modifications have been correlated in multiple organisms. In mammals, de novo methyltransferase DNMT3A and its cofactor DNMT3L form a complex that specifically recognizes unmodified H3K4 through the ADD domains (Ooi et al., 2007; Otani et al., 2009). UHRF1 is a key factor connecting DNMT1 and H3K9 methylation by binding methylated DNA through the SRA domain (Bostick et al., 2007), H3K9me3 through a tandem Tudor (Nady et al., 2011; Xie et al., 2012) and unmodified H3R2 through an atypical PHD finger domain (Rajakumara et al., 2011b; Xie et al., 2012). In plants, H3K9me2 is mainly associated with heterochromatic TEs and correlated with CHG methylation (Bernatavichute et al., 2008). The SRA domain of the H3K9 methyltransferase KRYPTONITE (KYP; also called SUVH4) directly binds to methylated CHG containing oligonucleotides (Johnson et al., 2007). Based on these results, a self-reinforcing loop was proposed, in which CMT3 is recruited by H3K9me2 deposited by KYP and its close homolog (SUVH5 and SUVH6), to methylate CHG. In turn, methylated CHG DNA recruits KYP to maintain methylation of H3K9 (Johnson et al., 2007; Law and Jacobsen, 2010).
To investigate the molecular mechanism underlying the relationship between CHG methylation and histone H3K9 methylation, we carried out structural and functional studies on CMT3 and ZMET2. We found that CMT3 is stably associated with heterochromatic nucleosomes. Consistent with its role in DNA methylation maintenance, we found that CMT3 was predominantly expressed in actively replicating cells and was specifically associated with replication dependent histone H3 variants. Genome-wide mapping of CMT3 binding sites demonstrated that CMT3 is nearly perfectly correlated with H3K9me2 in vivo. Chromatin association of CMT3 is dependent on specific binding of H3K9me2 via the bromo adjacent homology (BAH) and chromo domains as shown by our crystal structures of ZMET2 in complex with H3K9me2 peptides. The structures identify an aromatic cage within both the BAH and the chromo domains as interaction interfaces that capture H3K9me2. Mutations that abolish either interaction cause a failure of CMT3 binding to nucleosomes, and a complete loss of CMT3 activity in vivo. Together, these results suggest that CMT3 associates with H3K9me2-containing nucleosomes through binding of its BAH and chromo domains to H3K9me2 in order to target DNA methylation.
RESULTS
CMT3 is Associated with H3K9me2-containing Nucleosomes
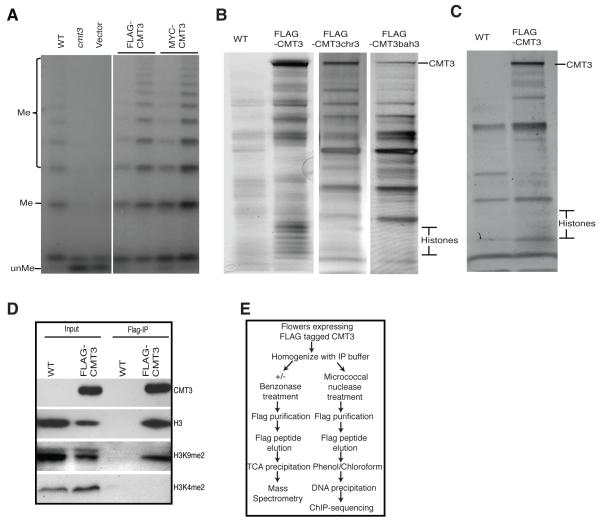
To identify CMT3-interacting proteins, we generated epitope-tagged CMT3 lines. As shown in Figure 1A, both pCMT3:BLRP-3xFLAG-CMT3 and pCMT3:BLRP-9xMYC-CMT3, designated as FLAG-CMT3 and MYC-CMT3, respectively, complemented the cmt3 mutation and restored DNA methylation at the 5S rDNA locus, demonstrating that epitope-tagged CMT3 was functional in vivo. We found a major band at the size of CMT3 and a series of smaller bands that corresponded to the size of individual histone proteins (Figure 1B). As shown in Table 1 from mass spectrometry (MS) analysis, peptides corresponding to CMT3 were the most abundant proteins with 63.4% coverage of the CMT3 protein. We also identified all four core histones: H3, H4, H2A, and H2B at roughly equivalent levels and observed several linker H1 histones, as well as Ku70/80 proteins (Table 1 and S1) that were likely bound to the sheared DNA ends, suggesting that CMT3 interacts with bona fide nucleosomes.
Figure 1. CMT3 is Associated with Histones in vivo.
(A) Complementation analysis by southern blot at 5S rDNA locus. Genomic DNA was digested with the methylation-sensitive MspI restriction enzyme. The (unMe) DNA fragment was the unmethylated DNA and (Me) DNA fragments were generated when MspI digestion was blocked by DNA methylation. The two lanes indicated by a horizontal line are from two independent transgenetic plant lines. Wild type Columbia ecotype (WT) is a control.
(B) Silver stained SDS-PAGE gel showed the presence of histone proteins in CMT3, but not in CMT3chr3 and CMT3bah3.
(C) Histone proteins were absent from protein complexes after benzonase treatment.
(D) CMT3 specifically pulls down H3K9me2.
(E) Outline of CMT3 affinity purification procedure for mass spectrometry and ChIP-seq.
See also Tables S1-S2.
Table 1.
Summary of Proteins Associated with CMT3 Identified by Mass Spectrometric Analyses.
| Protein | AGI Code | Spectra | Unique Peptides |
%Coverage | NSAFe5 | % CMT3 |
|---|---|---|---|---|---|---|
| CMT3 | At1g69770 | 756 | 131 | 63.4 | 7240 | 100 |
|
| ||||||
| H1 | At1g06760 | 19 | 11 | 31.8 | 557 | 7.7 |
|
| ||||||
| H1 | At3g18035 | 29 | 17 | 22.3 | 485 | 6.7 |
|
| ||||||
| H1/H5 | At1g48620 | 28 | 12 | 17.3 | 470 | 6.5 |
|
| ||||||
| H1.2 | At2g30620 | 46 | 16 | 47.3 | 1353 | 18.7 |
|
| ||||||
| HTA13 | At3g20670 | 50 | 7 | 21.2 | 3043 | 42.0 |
|
| ||||||
| HTA6 | At5g59870 | 18 | 4 | 17.3 | 964 | 13.3 |
|
| ||||||
| HTA9 | At1g52740 | 15 | 4 | 30.6 | 899 | 12.4 |
|
| ||||||
| HTA7 | At5g27670 | 15 | 3 | 8.0 | 803 | 11.1 |
|
| ||||||
| HTA12 | At5g02560 | 5 | 2 | 7.8 | 262 | 3.6 |
|
| ||||||
| HTA8 | At2g38810 | 15 | 4 | 32.4 | 251 | 3.5 |
|
| ||||||
| H2B1 | At5g02570 | 59 | 13 | 32.6 | 3675 | 50.8 |
| At3g09480 | ||||||
|
| ||||||
| H2B | At2g37470 | 62 | 15 | 46.4 | 3610 | 49.9 |
|
| ||||||
| H2B | At3g53650 | 32 | 8 | 30.4 | 1863 | 25.7 |
|
| ||||||
| At5g59910 | ||||||
| At1g07790 | ||||||
| H2B1 | At5g22880 | 79.6 | 16.6 | 42.2 | 4327 | 59.8 |
| At3g45980 | ||||||
| At3g46030 | ||||||
|
| ||||||
| At5g10400 | ||||||
| At1g09200 | ||||||
| H31 | At5g10390 | 63 | 21 | 52.9 | 3674 | 50.7 |
| At5g65360 | ||||||
| At3g27360 | ||||||
|
| ||||||
| HTR12 | At1g01370 | 2 | 2 | 25.3 | 90.3 | 1.2 |
|
| ||||||
| At1g07820 | ||||||
| At3g3730 | ||||||
| At2g28740 | ||||||
| H41 | At5g59970 | 26 | 13 | 62.1 | 2028 | 28.0 |
| At3g46320 | ||||||
| At3g45930 | ||||||
| At5g59690 | ||||||
| At1g07660 | ||||||
|
| ||||||
| Ku80 | At1g48050 | 24 | 19 | 33.8 | 284 | 3.9 |
|
| ||||||
| Ku70 | At1g16970 | 10 | 8 | 16.7 | 129 | 1.8 |
The percentage of CMT3 (% CMT3) column indicates the approximate stoichiometry of each co-purifying protein as a function of the Normalized Spectral Abundance Factor (NSAF) (Law et al., 2010).
Unique peptides map to the group of these genes. See also Table S1.
To test whether CMT3 specifically interacts with histone H3, we treated plant extracts with benzonase, a non-specific nuclease, to remove DNA prior to affinity purification. We found that benzonase treatment eliminated the low molecular weight histone bands (Figure 1C). MS analysis further confirmed that H2A, H2B, H3, H4 and Ku70/80 were no longer detectable (Table S2). This suggests the integrity of nucleosomes is critical for CMT3 binding, and that CMT3 does not interact with H3 strongly on its own in vivo.
To investigate whether CMT3 is associated with H3K9me2-containing nucleosomes, we tested for the presence of H3K9me2 marks in histones co-purified with CMT3 and found that H3K9me2, but not H3K4me2, was precipitated with CMT3 (Figure 1D). This indicates that CMT3 preferentially binds to H3K9m2-containing nucleosomes.
CMT3 is Enriched in Heterochromatic Regions and Highly Correlated with H3K9me2
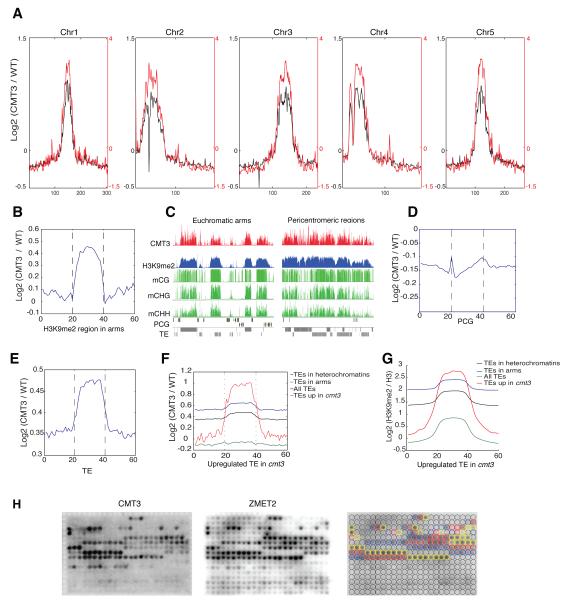
To further explore the in vivo binding pattern of CMT3, we performed chromatin immunoprecipitation coupled with sequencing (ChIP-seq, Figure 1E). We found that CMT3 is highly enriched in Arabidopsis pericentromeric regions and is nearly perfectly co-localized with H3K9me2 (Figure 2A). CMT3 is also enriched in heterochromatic patches in the euchromatic arms, where high levels of H3K9me2 were also observed (Figure 2B). CMT3 tends to bind large uninterrupted blocks in pericentromeric regions, while in chromosome arms CMT3 forms smaller and isolated patches (Figure 2C). These results are consistent with the general trend of H3K9me2 distribution in these regions (Bernatavichute et al., 2008). Furthermore, CMT3 was not enriched over protein coding genes (Figure 2D), consistent with previous data showing that the majority of genes are devoid of CHG methylation and H3K9me2 (Cokus et al., 2008). We also found a strong enrichment of CMT3 over TEs (Figure 2E). To complement the ChIP-seq data, we examined the transcriptome profile of cmt3 mutants by whole genome RNA sequencing (RNA-seq) analysis. As expected, CMT3 was strongly enriched over the TEs that were upregulated in cmt3 mutants (Figure 2F), as were H3K9me2 and CHG methylation (Figure 2G, Figure S1A).
Figure 2. CMT3 Is Enriched in Heterochromatic Regions and Highly Correlated with H3K9me2.
(A) Distribution of CMT3 (black) and H3K9me2 (red) along the five Arabidopsis chromosomes.
(B) CMT3 ChIP-seq signal is enriched in heterochromatic patches in the arms.
(C) Genome browser views of pericentromeric and euchromatic regions. mCG: CG methylation; mCHG: CHG methylation; mCHH: CHH methylation; TE: transposable element; PCG: protein coding gene.
(D,E) CMT3 ChIP-seq signal is not enriched in the protein coding gene regions (D), but is enriched in TEs (E).
(F,G) CMT3 (F) and H3K9me2 (G) are enriched in TEs that are upregulated in cmt3 null mutants.
(H) CMT3 and ZMET2 specifically bind to mono-, di- and tri-methylation of H3K9. Blue, red and yellow circles in right panel represent the locations of mono-, di- and tri-methylation at lysine 9-containing peptides on the array, respectively.
See also Figure S1A.
To determine the specificity of CMT3 binding to histone marks in vitro, we screened full length CMT3 on a peptide array and found that CMT3 can bind to mono-, di- and tri-methylated H3K9 peptides, with preference for di- and tri-methylated H3K9 peptides (Figure 2H). In plants, H3K9me2 is highly enriched in heterochromatin and TEs, and functionally associated with DNA methylation. In contrast, H3K9me3 is found in low abundance in euchromatic regions in plants and its functional role is still unknown (Roudier et al., 2011). In addition, CMT3 binding was blocked by phosphorylation of Ser10 and Thr11 on H3 (Figure 2H).
Collectively, the nearly perfect correlation between CMT3 binding sites with H3K9me2 sites in the genome and the direct interaction of CMT3 and H3K9me2-containing nucleosomes and histone tail peptides support the role of H3K9me2 in recruiting CMT3 to chromatin in vivo.
CMT3 is Primarily a CHG Methyltransferase in vitro and is Predominantly Expressed in Actively Replicating Cells
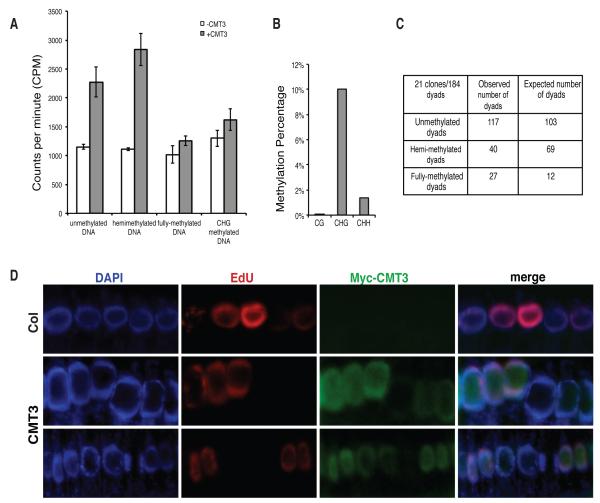
To determine whether CMT3 is active in vitro, recombinant CMT3 protein was assayed on a set of oligonucleotide substrates with cytosines either unmethylated or premethylated at CG, CHG, or CHH sites (Table S3). As shown in Figure 3A, CMT3 methylated oligonucleotides that were either unmethylated or hemimethylated at CHG sites, while it lost the majority of its activity when CHG sites were blocked by premethylation on both strands. We also performed bisulfite sequencing of an in vitro methylated plasmid DNA and observed 10% CHG methylation, 1.4% CHH methylation, and 0% CG methylation, with an error rate of 0.4% (Figure 3B). These results provide direct evidence that CMT3 is indeed a methyltransferase that preferentially methylates CHG sites.
Figure 3. CMT3 Is Primarily a CHG Methyltransferase and Predominantly Expressed in Actively Replicating Cells.
(A) CMT3 has activity on both unmethylated and hemi-methylated DNA oligos, but not fully methylated DNA oligos containing methylations at all cytosines. Error bars represent the standard deviation of three replicates.
(B) Bisulfite sequencing of in vitro methylated plasmid DNA.
(C) Hemimethylated dyads at the Ta3 locus were significantly lower than expected (Chi square value: 32.84). Probability of obtaining the observed distribution by random chance is less than 0.1%.
(D) Accumulation of CMT3 in cells with active replication. DAPI staining shows the localization of nuclei (Blue). Actively replicating cells were labeled by 5-ethynul-2′-deoxyuridine (EdU, Red) and CMT3 was immuno-stained by MYC antibody (Green).
See also Figures S1B and Tables S3 and S4.
During DNA replication, fully methylated DNA becomes hemi-methylated, and can become completely unmethylated in the following round of replication if methylation is not properly maintained. If CMT3 efficiently converts hemi-methylated DNA to fully methylated DNA in vivo, then one should observe a large number of fully methylated CHG sites and a smaller number of hemi-methylated sites in the genome. To test this, we utilized Hairpin Bisulfite Sequencing (Laird et al., 2004) at the Ta3 transposon. We found that the number of observed fully methylated dyads exceeded the number of expected fully methylated dyads occurring at random by a small but significant number (Figure 3C). These results suggest that CMT3 has a significant in vivo preference for conversion of hemi-methylated sites to fully methylated sites and in this way acts as a maintenance methyltransferase.
Our MS results showed that the replication-dependent H3 (H3.1), known to be expressed in S-phase (Okada et al., 2005), but not the replication-independent H3 (H3.3), co-purified with CMT3 (Table S4). In addition, an analysis of publicly available microarray data shows that CMT3 expression is highly correlated with proteins that are actively expressed during DNA replication (Figure S1B). We also directly examined the localization of CMT3 in replicating root cells. To detect cells undergoing replication, 5-ethynul-2′-deoxyuridine (EdU), a thymidine analogue that is incorporated into dividing cells during S-phase (Chehrehasa et al., 2009), was used to label newly synthesized DNA. We observed high accumulations of CMT3 in actively replicating cells as labeled by EdU (Figure 3D). CMT3 also accumulated in cells not undergoing DNA replication, but at a much lower level than that in replicating cells (Figure 3D).
Together, these results suggest that CMT3 is an active DNA methyltransferase that is predominately expressed in the S phase of actively replicating cells and binds to nucleosomes that contain replication dependent histone variants.
Structure of SAH-bound ZMET2
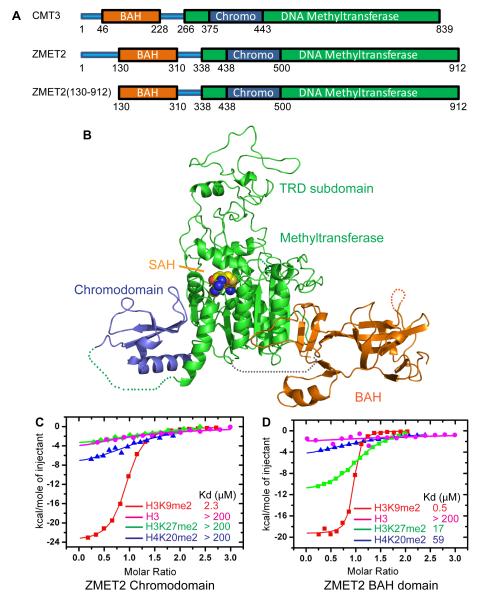
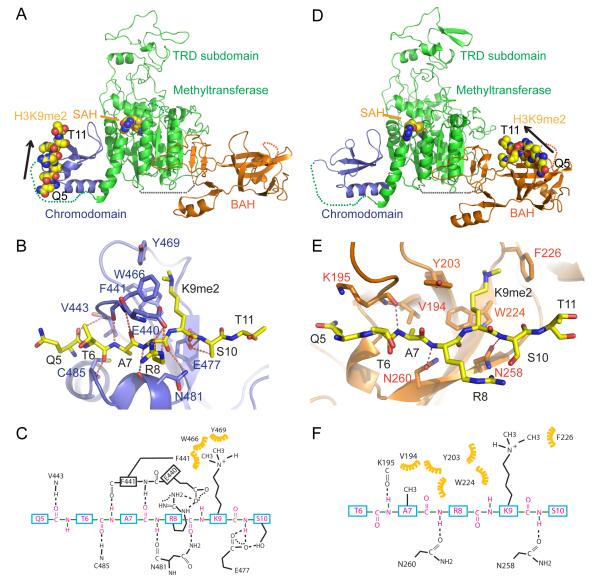
The CMT3 homologue in Zea mays, ZMET2 (Figure 4A and S2A), has a similar binding specificity to methylated H3K9 on the peptide array (Figure 2H) and similar methyltransferase activity on CHG sites (Figure S2B). Although we were unable to crystallize CMT3, we were successful in obtaining diffraction quality crystals for an N-terminal truncated version of ZMET2 (residues 130-912, Figure 4A), containing all the functional domains, along with the cofactor SAH and the structure was refined to 3.2 Å resolution (Figure 4B and Table S5). The BAH and chromo domains project outwards in opposite directions relative to the methyltransferase domain (Figure 4B). The overall topology is that of an equilateral triangle with each edge of about 85-100 Å in length. The BAH domain, the target recognition subdomain (TRD) of the methyltransferase domain, and the chromodomain are positioned at the three vertices, while the catalytic subdomain of the methyltransferase domain is positioned within the interior of the triangular architecture (Figure 4B).
Figure 4. Binding to Methylated Peptides and the Structure of ZMET2 in the Free State.
(A) Color-coded domain architecture of full length CMT3, ZMET2, and the ZMET2 (130-912) construct used to grow crystals.
(B) Ribbon representation of the structure of ZMET2 with bound SAH. The BAH, methyltransferase, and chromo domains are colored in orange, green, and blue, respectively, with the bound SAH molecule shown in a space filling representation. Some disordered regions were not built in the final model and are shown as dashed lines.
(C, D) ITC binding curves for complex formation between ZMET2 chromo (C) and BAH (D) domains and H3K9me2, H3K27me2, H4K20me2, and unmodified H3 peptides. Kd values are listed as an insert. See also Figure S2 and Table S5.
The BAH domain of ZMET2, composed of a twisted β barrel with some extended segments, resembles BAH1 domain of mouse DNMT1. Although they only share a sequence identity of 26%, the superposition of the BAH domains of ZMET2 and mouse DNMT1 yields an rmsd of 1.9 Å for 121 aligned Cα atoms, showing a similar folded topology (Figure S2C). The position of the BAH domain relative to its methyltransferase domain in ZMET2 is also similar to that of BAH1 domain of DNMT1 relative to its methyltransferase domain, indicating a plausible similar function for the BAH domains of these two proteins.
The methyltransferase domain of ZMET2 adopts a typical class-I methyltransferase fold composed of catalytic and TRD subdomains. The catalytic subdomain of ZMET2 resembles the corresponding folds adopted by DNMT1 (Song et al., 2011) and DNMT3A (Jia et al., 2007), with a central seven-stranded β-sheet flanked by two layers of α-helical segments on either side. Similar to its topology in DNMT1, the central seven-stranded β-sheet of the catalytic subdomain of ZMET2 forms a continuous nine-stranded β-sheet by pairing with two additional β-strands from the BAH domain (Figure 4B), thereby probably stabilizing the relative positioning of the BAH and methyltransferase domains of ZMET2. A SAH molecule is positioned in the active site of the catalytic subdomain. The TRD subdomain of ZMET2consisting of 206 residues (612-818), has a well-defined continuous peptide backbone, but poor density for some side chains, which were consequently modeled as poly-alanine.
Although the primary sequence implies that the chromodomain of ZMET2 is embedded within the methyltransferase domain, it buds out from one side of the methyltransferase domain in the structure (Figure 4B), therefore forming an independent structural module that does not affect the topology of the methyltransferase domain. The chromodomain uses several hydrophobic residues to form a continuously hydrophobic surface that interacts with its hydrophobic counterpart on the surface of the methyltransferase domain (Figure S2D). This interaction positions the chromodomain on one side of the methyltransferase domain, indicative of a relatively rigid alignment between chromo and methyltransferase domains. The ZMET2 chromodomain adopts a typical chromodomain topology, which resembles that of methylated H3K9-binding chromodomains, such as the MPP8 chromodomain (rmsd of 1.1 Å for 57 aligned Cα rmsd of 1.2 Å for 52 aligned Cα atoms) (Chang et al., 2011; Jacobs and Khorasanizadeh, 2002).
Structural Basis for H3(1-15)K9me2 Peptide Recognition by ZMET2 Chromodomain
To investigate the molecular basis for H3K9me2 recognition by ZMET2, we first performed isothermal calorimetric (ITC) binding studies and established that the chromodomain of ZMET2 bound H3K9me2 and H3K9me3 with a Kd of 2.3 μM (Figure 4C) and 2.0 μM (data not shown), respectively, whereas it bound only weakly to unmodified H3, as well as H3K27me2 and H4K20me2 peptides (Figure 4C). Unexpectedly, ITC binding studies also established that the BAH domain of ZMET2 also exhibits strong binding affinity for H3K9me2 and H3K9me3 with a Kd of 0.5 μM (Figure 4D) and 0.7 μM (data not shown) and weak binding for unmodified H3 and H4K20me2 peptides, and exhibited very reduced affinity to H3K27me2 peptide (Kd of 17 μM) (Figure 4D).
We then crystallized ZMET2 in complex with H3(1-15)K9me2 peptide in the presence of SAH at 3.2 Å resolution (Figure 5A and Table S5). There are two molecules of ZMET2 and one molecule of H3K9me2 peptide bound to one of the two chromodomains in the asymmetric unit (Figures S3A-B). The overall structure of ZMET2 bound to the H3(1-15)K9me2 peptide resembles the structure of ZMET2 in the free state (Figure 4B and Figure 5A). The H3(1-15)K9me2 peptide binds to the chromodomain in a classic chromodomain-binding mode (Figure 5A) and the bound peptide can be traced from Gln5 to Thr11 (Figure S3C). The peptide is clamped within the chromodomain in a β-strand like conformation. The K9me2 side chain inserts into an aromatic cage, which is formed by Phe441, Trp466 and Tyr469, and stabilized by extensive hydrophobic interactions (Figures 5B-C). These three aromatic cage-forming residues are conserved with CMT3 and the classic H3K9me3-recognizing HP1 chromoodmain (Figure S3D-E), suggesting a similar methylated lysine binding properties within this family. The residues adjacent to K9me2 are also involved in sequence specific recognition, with Arg8 of the histone peptide forming two intermolecular hydrogen bonds, as well as electrostatic interactions with the side chain of Glu440 of the protein. The main chain of Arg8 also forms two hydrogen bonds with Asn481 and the backbone and side chain of Ser10 forms hydrogen bonds with the side chain of Glu477 of the protein (Figures 5B-C), consistent with our observation that Ser10 phosphorylation inhibits its binding to CMT3 (Figure 2H). In addition, the backbone of Gln5, Thr6 and Ala7 form intermolecular hydrogen bonds with the protein (Figures 5B-C).
Figure 5. Structure of Chromo and BAH Domains of ZMET2 with Bound H3K9me2 Peptide.
(A, D) Ribbon representation of the structure of ZMET2(130-912) complexes with H3(1-15)K9me2 (A) and H3(1-32)K9me2 peptide (D). The color-coding is the same as in Figure 4B. The bound peptide and SAH are shown in a space-filling representation, with the orientation of the bound peptide shown by an arrow.
(B, E) Intermolecular interactions between the chromodomain of ZMET2 and H3(1-15)K9me2 peptide (B) and between the BAH domain of ZMET2 and H3(1-32)K9me2 peptide (E). The dimethylammonium group of K9 is accommodated within an aromatic cage. Intermolecular hydrogen bonds are designated by dashed red lines.
(C, F) A schematic representation of the intermolecular interactions between the chromodomain of ZMET2 and the bound H3(1-15)K9me2 peptide (C) and between the BAH domain of ZMET2 and the bound H3(1-32)K9me2 peptide (F).
See also Figures S3, S4, and S5 and Table S5.
Structural Basis for H3(1-32)K9me2 Peptide Recognition by ZMET2 BAH Domain
We further determined the structure of another crystal form of ZMET2 in complex with a longer H3(1-32)K9me2 peptide at 2.7 Å resolution that revealed a distinct binding mode of the H3K9me2 peptide to the BAH domain of ZMET2 (Figure 5D and Table S5). Due to the different packing arrangement in this crystal, the peptide-binding site within the chromodomain was blocked by the other molecule in the asymmetric unit (Figures S4A-B) and hence no peptide was found to bind to the chromodomain.
The structure contains two molecules of ZMET2, with H3K9me2 peptides bound to each of the BAH domains, in the asymmetric unit (Figure S4A). The packing results in the chromodomain of one molecule being clamped by the TRD subdomain and the catalytic loop of the second molecule (Figure S4A). These intermolecular interactions stabilize the conformation of the TRD subdomain and catalytic loop, and hence it became possible to trace all the side chains of the TRD subdomain and the catalytic loop, while they could not be fully built in the H3(1-15)K9me2 complex. The electron density was traceable from Gln5 to Thr11 for the H3(1-32)K9me2 peptide bound to the BAH domain in one complex (Figure S4C), while a somewhat shorter segment was traceable in the other complex. The side chain of K9me2 inserts into an aromatic cage formed by Tyr203, Trp224 and Phe226 of the BAH domain (Figure 5E). These aromatic residues are also conserved in CMT3, suggestive of a conserved function (Figure S4D). By contrast, flanking peptide residues form fewer interactions with the protein in the H3(1-32)K9me2 complex (Figures 5E-F) than they do in the H3(1-15)K9me2 complex (Figures 5B-C). The methyl group Ala7 is recognized within a small hydrophobic pocket formed by Val194, Tyr203, and Trp224 in the H3(1-32)K9me2 complex (Figures 5E-F). In addition, the backbone amide protons of Ala7, Arg8 and Ser10 form intermolecular hydrogen bonds with the BAH domain (Figures 5E-F). These studies establish the BAH domain of ZMET2 also as a reader of H3K9me2, with its aromatic cage playing a key role in methyllysine recognition.
ZMET2 has a relatively large TRD subdomain of around 206 residues with a unique fold (Figure S5A). In regions proximal to the catalytic center (middle and bottom segments of Figure S5A), the ZMET2 TRD subdomain adopts α-helixes and loops similar to those observed for DNMT1 (Song et al., 2011), indicating the likelihood of a similar DNA substrate recognition mechanism. However, in regions further from the catalytic center (top segment of Figure S5A), the TRD subdomains of DNMT1 and ZMET2 show different structural features. For DNMT1, regions of the TRD subdomain further from the catalytic center are enriched with loops and a coordinated zinc ion. In addition, a long loop extending outwards from BAH2 domain of DNMT1 (Figure S5A) interacts at its tip with the TRD subdomain, suggestive of a plausible regulatory role. By contrast, ZMET2 has neither a second BAH domain, nor a bound zinc ion. The space occupied by the BAH2 loop-interacting region on the TRD subdomain in DNMT1 is partially occupied by a two-stranded anti-parallel β-sheet in ZMET2 (Figure S5A). These observations imply that though ZMET2 might have a potentially similar DNA recognition mode as DNMT1, its regulatory mechanism could be different.
The catalytic loop region is disordered for structures of ZMET2 in the free state and in the complex with H3(1-15)K9me2 peptide, but is well defined in the complex with H3(1-32)K9me2 peptide. We observe that the catalytic loop (in red, stereo view in Figure S5B) and catalytic cysteine (labeled C in red, Figure S5B) of ZMET2 in its peptide-bound form is directed outwards from the catalytic center, similar to that of DNMT1 bound to DNA in its autoinhibitory conformation (labeled C in blue, Figure S5B) (Song et al., 2011). By contrast, the catalytic loop (in green) and catalytic cysteine (labeled C in green, Figure S5B) are directed inwards towards the catalytic center in the active form of M.HhaI bound to DNA (Klimasauskas et al., (1994) and the active form of DNMT1 bound to hemi-methylated DNA (Song et al., 2012). These results are consistent with ZMET2 being in an inactive conformation in the peptide bound state in the absence of bound DNA.
Both BAH and Chromo Domains Are Important for CMT3 Function in vivo
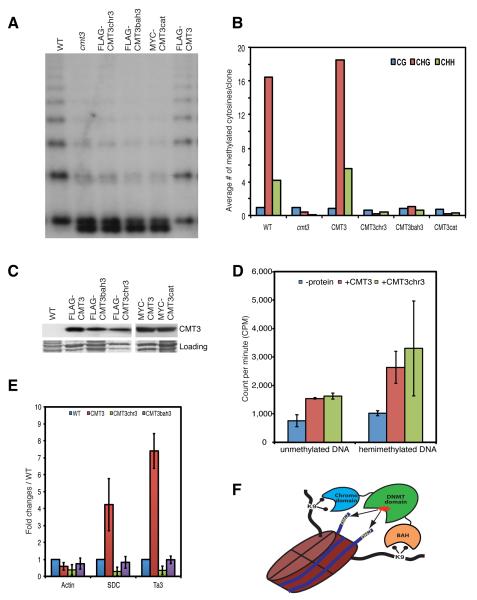
To study the biological significance of CMT3 and H3K9me2 interaction, we mutated the three aromatic residues lining the methyl-lysine-binding aromatic cages in the chromo (Phe382, Trp409 and Tyr412) and the BAH (Phe127, Trp148 and Tyr150) domains to generate triple point mutants named CMT3chr3 and CMT3bah3, respectively. We also generated a catalytically inactive C460A mutant (CMT3cat). Notably, CMT3cat, CMT3chr3 and CMT3bah3 failed to restore DNA methylation at the 5S rDNA (Figure 6A) and at the Ta3 (Figure 6B), even though the mutated proteins were expressed at a similar level (Figure 6C) and had similar catalytic activity as wild-type CMT3 (Figure 6D). In summary, mutations in either BAH or chromo domains of CMT3 abolish its methylation activity in vivo.
Figure 6. Both BAH and Chromo Domains Are Important for CMT3 Function in vivo.
(A) Southern blot of the 5S rDNA locus.
(B) Graphical representation of bisulfite sequencing analysis of Ta3 locus.
(C) Western blots showing the expression of the wild-type and mutant versions of the CMT3 protein. The bottom panel is the Coomassie blue staining and serves as a loading control.
(D) In vitro activity assay. Error bars represent the standard deviation of three biological replicates.
(E) ChIP data showing the failure of CMT3chr3 and CMT3bah3 to bind to chromatin at Ta3 and SDC. Error bars represent the standard deviation of three biological replicates.
(F) A working model of dual recognition mechanism of CMT3 BAH and chromo domains simultaneously reading the H3K9me2 on a single nucleosome to position the methyltransferase domain over the nucleosomal DNA.
See also Figures S6 and Table S2.
We also analyzed the effect of the chromo and BAH mutations on the binding of CMT3 to nucleosomes. Silver staining of proteins co-purifying with CMT3 showed that, unlike the situation in wild-type CMT3, proteins co-purifying with the CMT3chr3 and CMT3bah3 mutants lacked the low molecular weight bands corresponding to histone proteins (Figure 1B), which was further confirmed by MS analysis on affinity-purified CMT3chr3 samples (Table S2) and by ChIP experiments showing that mutations of either BAH or chromodomain abolish CMT3 binding to chromatin (Figure 6E). These results indicate that both the chromo and BAH domains are required for stable association of CMT3 with nucleosomes.
While the CMT3chr3 mutation abolished the interaction of CMT3 with nucleosomes, we found that CMT3chr3 retained similar in vitro catalytic activity as that of wild-type CMT3 (Figure 6D). This suggests that H3K9me2 acts as a recruitment factor for CMT3 in vivo.
DISCUSSION
Nucleosome occupancy is an important determinant of global DNA methylation patterns in vivo (Chodavarapu et al., 2010). The association of CMT3 with nucleosomes suggests that CMT3 may methylate nucleosome-bound DNA, which may explain our previous observations from whole genome shotgun bisulfite sequencing that CHG methylation was enhanced on nucleosomal DNA relative to linker DNA and that CHG sites facing on the outside of nucleosomes were methylated at a higher level than those facing on the inside (Chodavarapu et al., 2010). This model also explains the 167-nt periodicity pattern of CHG methylation previously observed in bulk analysis of whole genome methylation data (Cokus et al., 2008).
The observation that CMT3 is predominately expressed in cells undergoing active replication implies that methylation by CMT3 takes place during DNA replication, when H3.1 is incorporated onto newly synthesized chromatin and modified by the histone H3K9 methyltransferase, KYP. One plausible model is that CMT3 is displaced from nucleosomes during replication. Once replication is completed, it is recruited back to the newly assembled nucleosomes that have been methylated by KYP (or other SUVHs) on H3K9. Alternatively, CMT3 may be recruited to chromatin during DNA replication by pre-modified H3K9me2 marks from parental histones. In turn, CHG methylation would recruit KYP to deposit methylation marks on newly synthesized histones.
We attempted to test whether the H3K9-specific histone methyltransferase KYP (SUVH4) or possibly SUVH5 and SUVH6 (two other H3K9 methyltransferases) would form a complex with CMT3. However, the CMT3 IP-MS analysis did not detect any KYP, SUVH5, or SUVH6 peptides. Reciprocal IP experiments with protein extracts from Nicotiana benthamiana leaves co-expressing MYC-tagged CMT3 and FLAG-tagged KYP also failed to detect interaction between CMT3 and KYP (data not shown). These findings suggest either that KYP is not stably associated with nucleosomes, or that KYP and CMT3 cannot simultaneously remain bound to the same nucleosomes.
Our in vitro activity assays show that CMT3 has some activity on unmethylated DNA. We speculate that this “de novo” activity is in fact part of the “maintenance” loop of KYP and CMT3. Unlike the mammalian maintenance DNA methyltransferase DNMT1, which has much greater activity on hemi-methylated DNA than on unmethylated DNA (Bestor and Ingram, 1983), CMT3 only has slightly higher activity on hemimethylated substrates than unmethylated substrates. In addition, our data suggested that CMT3 showed only a small tendency to fully methylate CHG sites in vivo rather than leaving them hemimethylated, which is in contrast to the strong tendency of DNMT1 to fully methylate CG sites in vivo. This suggests that CMT3 is not as good as DNMT1 at restoring hemimethylated sites to fully methylated sites and consequently, methylation at many sites will be completely lost during multiple cycles of replication. The “de novo” activity of CMT3, however, could persistently target methylation to regions marked with H3K9 methylation. CMT3’s inefficiency at maintaining methylation is also reflected by the fact that the overall global levels of CHG methylation (6.7%) are much lower than those of CG methylation (24%) (Cokus et al., 2008). In addition to the preference for hemimethylated DNA as substrate, DNMT1 and MET1 work with the UHRF1/VIM cofactor that specifically recognizes hemimethylated DNA through its SRA domain (Bostick et al., 2007). However, the SRA domain of KYP and SUVH5 bind equally well to hemimethylated and fully methylated DNAs (Johnson et al., 2007; Rajakumara et al., 2011a). Therefore, CMT3’s “maintenance” activity is likely to be driven mainly by the feedback loop between KYP and CMT3, rather than by the inherent preference of CMT3 for hemimethylated DNA.
H3K27me1 is enriched in heterochromatin and was proposed to be involved in CMT3 function based on in vitro data showing that the chromodomain of CMT3 bound to histone H3 peptides only when both H3K9 and H3K27 were methylated (Lindroth et al., 2004). However, these doubly methylated peptides were longer than the singly methylated control peptides, which might be an alternative explanation for this result. More critically, a recent study showed that depletion of H3K27me1 in vivo did not reduce CHG methylation (Jacob et al., 2009). In addition, our peptide array and structural analyses showed that H3K9me2 is necessary and sufficient to bind CMT3. Therefore, H3K27me is unlikely to play a role in CMT3 targeting.
The BAH domain functions as a mediator of protein-protein interactions. Yeast Sir3 BAH domain in complex with mononucleosomes revealed that the BAH domain makes contacts through unmodified H4K16 and H3K79 (Armache et al., 2011), which are important for BAH-nucleosome binding (Onishi et al., 2007). Recent studies of the mouse ORC1 BAH domain bound to H4K20me2 peptide have established that methylated-lysine is recognized through positioning within an aromatic cage on the surface of the BAH domain, and the methyllysine recognition mode is similar as the ZMET2 BAH domain recognizing H3K9me2 peptide reported here (Figure S4E) (Kuo et al., 2012). In the current study, we also observed that the methylated-lysine of H3K9me2 peptide was positioned within an aromatic cage of the BAH domain of ZMET2. The aromatic cage residues in the BAH domain of ZMET2, that are conserved amongst the BAH domains of other plant chromomethylases, are also observed within the BAH1 domain of human DNMT1. These results imply that the BAH1 domain of DNMT1 may also be a reader of methylated-lysine marks using aromatic cage capture. By contrast, no aromatic cage was identified following alignment of the BAH2 domain of DNMT1.
How is CHG methylation precisely controlled and faithfully maintained? We propose a dual recognition mechanism, in which the CMT3 BAH and chromo domains simultaneously read the H3K9me2 on the two tails emanating from a single nucleosome to ensure a higher fidelity of CHG DNA methylation (Figure 6F). In the crystal, we are restricted to one or the other complex most likely due to packing interactions, but there should be no such constraints for simultaneous recognition of H3K9me2 marks by both BAH and chromo domains in solution. Indeed, ITC binding studies yield a stoichiometry of 1.8 for binding of H3(1-15)K9me2 peptide to ZMET2 (Figure S6A). Modeling efforts indicate that it is possible that two H3K9m2 modified tails could extend from the single nucleosome and bind to the chromo and BAH domains simultaneously. The nucleosomal DNA between the two H3 tails could be clamped by the TRD subdomain and the catalytic loop and subsequently methylated (Figures S6B-C). The H3 tail has sufficient length so as to likely allow the enzyme enough flexibility to catalyze on a range of nucleosomal DNA. The two-domain binding mode could potentially increase CMT3’s binding affinity and specificity towards H3K9me2 marks and thus help to recruit CMT3 to H3K9me2-containing nucleosomes. It is also conceivable that the dual binding modules could help CMT3 walk from nucleosomes to adjacent nucleosomes, characteristic of a spreading mechanism. Interestingly, the mammalian de novo DNA methyltransferase DNMT3A/3L complex also binds stably to nucleosomes (Jeong et al., 2009), and DNMT3A and DNMT3L each have an unmodified H3K4-recognizing ADD domain, suggesting that the DNMT3A/3L complex may also have the potential for dual recognition of two unmodified H3K4-containing tails of a nucleosome. By contrast, DNMT1 only contains one BAH domain that has a potential methyl-lysine binding cage and is not tightly associated with nucleosomes (Jeong et al., 2009). Thus a dual histone tail recognition mode may be a common feature of DNA methyltransferase that are stably bound to chromatin.
EXPERIMENTAL PROCEDURES
Immunoprecipitation and Mass Spectrometry
Epitope-tagged CMT3 transgenic plants were generated and used for IP-MS analysis as described previously (Law et al., 2010) and detailed information can be found in the Extended Experimental Procedures.
Western Blot
The FLAG and MYC epitope tags were detected using the anti-Flag M2 monoclonal antibody (Sigma, A8592) and anti-Myc 9E10 monoclonal antibody (Covance, MMS-150B), respectively. Primary antibodies used for histones included: anti-histone H3 antibody (Abcam, ab10799), anti-histone H3K9me2 (Abcam, ab1220-100), and anti-histone H3K4me2 (Abcam, ab32356).
Immunofluorescence
EdU labeling of BLRP-9xMYC-CMT3 transgenic root cells was based on the Click-iT kit (Invitrogen) and immunofluorescence analysis was described in detail in the Extended Experimental Procedures.
Protein Purification and Histone Peptide Array
Recombinant CMT3 and ZMET2 protein expression and purification were based on (Song et al., 2011) and described in detail in the Extended Experimental Procedures. MODified™ histone peptide array slide (Active Motif, 13001) was blocked by incubation in TBS buffer (10 mM Tris-HCl pH 7.5 and 150 mM NaCl) containing 5% milk at RT for 1h and washed 3 times with TTBS buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.05% Tween-20). The array slide was then incubated with 30 μg purified proteins in 3ml binding buffer (50 mM HEPES pH 7.5, 50 mM NaCl, 5% glycerol, 0.4% BSA and 2 mM DTT) overnight at 4°C, washed 3 times with TTBS buffer and incubated with anti-His antibody in TBS buffer for 1h at RT. The array was washed 3 times with TTBS and developed by ECL. The images were analyzed according to the instruction of array analysis software.
DNA methyltransferase assay
Baculovirus-mediated CMT3 expression and purification were performed as previously described (Patnaik et al., 2004). DNA methyltransferase activity assay was described in detail in the Extended Experimental Procedures.
Hairpin Bisulfite Sequencing
Genomic DNA was digested with Bgl II restriction enzyme and ligated with oligo (/5Phos/GATCTGCGATCGDDDDDDDGCATCGCA) by T4 DNA ligase (NEB). JP3200 and JP1615 primers were used to amplify Ta3 hairpin bisulfite treated DNA. Methylation on both strands of the DNA was assayed in 21 clones from wild-type plants. Chi-square value was calculated by using standard chi-square formula (the sum of squared observed minus expected over expected). The expected number of dyads was calculated based on the observed cytosine methylation percentage and four possible combinations of methylated and unmethylated cytosines in a dyad. Calculated chi-square value, together with degree of freedom of two, was used to obtain probability based on the chi-square distribution table.
Chromatin immunoprecipitation
ChIP was based on IP with the following modifications. Micrococcal nuclease was included in crude extracts to shear chromatin before incubation with beads. The eluted protein-DNA complexes were treated by proteinase K followed by phenol:chlorophorm purification. The enriched DNA was ethanol precipitated and subjected to library generation following Illumina’s manufacturer instructions (see Extended Experimental Procedures).
ChIP-seq and RNA-seq data analyses
Illumina base-called reads were mapped with Bowtie (Langmead et al., 2009) allowing up to 2 mismatches. For ChIP-seq, identical reads were collapsed into one read. Both gene and transposon expressions were measured by calculating reads per kilobase per million mapped reads (RPKM). P-values were calculated using Fisher’s exact test and Benjamini corrected for multiple testing. Differentially expressed elements in wild-type and mutants were defined by applying log2(mutant >wild-type)>2 and P<0.05 cutoffs.
Crystallization, structure determination and refinement
Crystals were grown using the hanging drop vapor diffusion method and the diffraction data were collected at the NE-CAT beamline 24ID-E, Advanced Photon Source (APS) at the Argonne National Laboratory, Chicago, and processed with the program HKL2000 (Otwinowski Z, 1997). The structure of Se-substituted ZMET2 in the presence of SAH and the structures of ZMET2 in complex with H3K9me2 peptides in the presence of SAH were solved using SAD method and molecular replacement method, respectively, as implemented in the program Phenix (Adams et al., 2010). The model building was carried out using the program Coot (Emsley et al., 2010). The statistics of the diffraction data and the refinement are summarized in Table S5. Additional details are provided in the Extended Experimental Procedures.
Supplementary Material
Highlight (4 bullet points with each no more than 85 characters)
CMT3 stably associates with H3K9me2-containing nucleosomes (62 characters)
CMT3 has CHG MTase activity and is expressed in actively replicating cells (77 characters)
Structural and ITC studies reveal both BAH and chromo domains recognize H3K9me2 (82 characters)
Dual recognition model establishes mechanism targeting DNA methylation in plants (84 characters)
ACKNOWLEDGMENTS
We thank Anders Lindroth and Debasis Patnak for initiating CMT3 in vitro activity assays, staff members at APS for support in diffraction data collection and at UCLA BSCRC BioSequencing core for high-throughput sequencing, Nathan Springer and Shawn Kaeppler for providing ZMET2 gene. We also grateful for Lianna Johnson for help with histone peptide array experiments, Greg Horwitz for help with baculoviral protein expression and their critical reading of this manuscript. X. Z. is supported by Ruth L. Kirschstein National Research Service Award (F32GM096483-01). S.F. is supported by Leukemia & Lymphoma Society. This work was supported by the Abby Rockefeller Mauze Trust and the Maloris and STARR foundations to D.J.P., NIH grant GM089778 and the UCLA Jonsson Cancer Center to J.A.W. and NIH grant GM60398 to S.E.J. S.E.J. is an Investigator of the Howard Hughes Medical Institute. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Coordinates and structure factors for Se-ZMET2 in the free state (PDB code: 4FSX), and in complex with H3(1-15)K9me2 (PDB code: 4FT2) and H3(1-32)K9me2 (PDB code: 4FT4) peptides, all in the presence of SAH, have been deposited in the RCSB Protein Data Bank. Sequencing data have been deposited at GEO (GSE39097 and GSE38286).
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L, Bender J. Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family. Nucleic Acids Res. 2001;29:2127–2134. doi: 10.1093/nar/29.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome- wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Chang Y, Horton JR, Bedford MT, Zhang X, Cheng X. Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding. J Mol Biol. 2011;408:807–814. doi: 10.1016/j.jmb.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehrehasa F, Meedeniya AC, Dwyer P, Abrahamsen G, Mackay-Sim A. EdU, a new thymidine analogue for labelling proliferating cells in the nervous system. J Neurosci Methods. 2009;177:122–130. doi: 10.1016/j.jneumeth.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, Jacobsen SE. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol. 2009;16:763–768. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson IR, Callis J, Jacobsen S. The SRA Methyl-Cytosine-Binding Domain Links DNA and Histone Methylation. Curr Biol. 2007;17:379. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird CD, Pleasant ND, Clark AD, Sneeden JL, Hassan KM, Manley NC, Vary JC, Jr., Morgan T, Hansen RS, Stoger R. Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc Natl Acad Sci U S A. 2004;101:204–209. doi: 10.1073/pnas.2536758100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21:6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, et al. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem. 2011;286:24300–24311. doi: 10.1074/jbc.M111.234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Endo M, Singh MB, Bhalla PL. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 2005;44:557–568. doi: 10.1111/j.1365-313X.2005.02554.x. [DOI] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX- DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z MW. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Papa CM, Springer NM, Muszynski MG, Meeley R, Kaeppler SM. Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell. 2001;13:1919–1928. doi: 10.1105/TPC.010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik D, Chin HG, Esteve PO, Benner J, Jacobsen SE, Pradhan S. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J Biol Chem. 2004;279:53248–53258. doi: 10.1074/jbc.M409604200. [DOI] [PubMed] [Google Scholar]
- Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011a;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011b;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Rechkoblit O, Bestor TH, Patel DJ. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331:1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335:709–712. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Jakoncic J, Qian C. UHRF1 Double Tudor Domain and the Adjacent PHD Finger Act Together to Recognize K9me3-Containing Histone H3 Tail. J Mol Biol. 2012;415:318–328. doi: 10.1016/j.jmb.2011.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.