Abstract
Bovine herpesvirus 1 (BHV-1) infection leads to upper respiratory tract infections, conjunctivitis, and the infection predisposes cattle to secondary bacterial infections. The infected cell protein 0 (bICP0) encoded by BHV-1 suppresses antiviral innate immune signaling by interfering with expression of interferon beta (IFN-β). In contrast to humans or mice, cattle contain three IFN-β genes that have distinct transcriptional promoters. We previously cloned and characterized all three bovine IFN-β promoters. In this study, we provide evidence that bICP27; a viral early protein that shuttles between the nucleus and cytoplasm inhibits transcriptional activity of two bovine IFN-β gene promoters (IFN-β1 and IFN-β3). Conversely, the BHV-1 infected cell protein 0 (bICP0) early promoter was not inhibited by bICP27. C-terminal mutants lacking the bICP27 zinc RING finger-like motif did not efficiently inhibit IFN-β3 promoter activity but inhibited IFN-β1 promoter activity as efficiently as wild type bICP27. An N-terminal mutant lacking the nuclear localization signal (NLS) and nucleolar localization signal (NoLS) was localized to the cytoplasm and this mutant had no effect on IFN-β promoter activity. In summary, these studies provided evidence that bICP27 inhibited IFN-β1 and IFN-β3 promoter activity in transiently transfected cells.
INTRODUCTION
Bovine herpesvirus 1 (BHV-1) is an alphaherpesvirinae subfamily member that causes significant economical losses to the cattle industry. Infection of cattle with BHV-1 can lead to conjunctivitis, genital disorders, abortions and bovine respiratory disease complex, a life threatening upper respiratory tract infection (Jones, 2009; Jones et al., 2007). The ability of BHV-1 to induce immune suppression in cattle is important for its pathogenic potential, reviewed in (Jones, 2009). Following acute replication in mucosal epithelium, BHV-1 establishes lifelong latency in ganglionic neurons within the peripheral nervous system (Winkler et al., 1999). During productive infection, all viral genes are abundantly expressed and infectious virus is readily detected, reviewed by (Jones, 2009; Jones et al., 2007). In latently infected neurons, the only viral gene that is abundantly expressed is latency related RNA, and infectious virus is not detected.
Stimulation of beta-interferon (IFN-β) transcription is an early response to virus infection (Au et al., 1995; Goodbourn et al., 1985; King and Goodbourn, 1994; Munshi et al., 1998; Sharma et al., 2003; Yoneyama et al., 1998). A complex signaling cascade, which involves activation of existing transcription factors by protein kinases stimulates IFN-β transcription. In contrast to humans or mice, cattle contain three IFN-β genes with distinct promoters (Valarcher et al., 2003; Wilson et al., 1983). BHV-1 infection of low passage bovine cells inhibits expression of all three bovine IFN-β genes (da Silva and Jones, 2011). Blocking viral protein expression by cycloheximide, a de novo protein synthesis inhibitor, prevented BHV-1 from suppressing IFN-β response (da Silva and Jones, 2011). These studies suggest that one or more viral genes suppress the IFN signaling pathway during productive infection. To date, bICP0 is the only BHV-1 gene that has been demonstrated to inhibit the activity of human (Henderson et al., 2005; Saira et al., 2007, 2009) and bovine (da Silva et al., 2011) IFN-β promoters.
Herpes simplex virus type 1 (HSV-1), a human alphaherpesvirinae subfamily member encodes several proteins that inhibit IFN signaling, including ICP0 (Eidson et al., 2002; Lin et al., 2004; Melroe et al., 2007; Paladino et al., 2010), ICP34.5 (Verpooten et al., 2009), Us11 (Sanchez and Mohr, 2007), and ICP27 (Johnson and Knipe, 2010; Johnson et al., 2008). The ICP27 protein inhibits IFN signaling by inhibiting STAT-1 phosphorylation and nuclear accumulation (Johnson and Knipe, 2010; Johnson et al., 2008). The BHV-1 ICP27 homologue (bICP27) and ICP27 share only 32% amino acid sequence homology and have different expression kinetics (Ackermann et al., 1984; Chalifour et al., 1996; Singh et al., 1996) suggesting they have similar but not identical functions. Both proteins contain a C-terminal zinc RING finger, a N-terminal nuclear localization signal (NLS), and a nucleolar localization signal (NoLS) (Guo et al., 2009; Mears et al., 1995). Furthermore, both proteins translocate between the nucleus and cytoplasm during the course of productive infection (Ding et al., 2010; Mears and Rice, 1998; Soliman et al., 1997).
In this study, we found that bICP27 interferes with IFN-β promoter activity in transiently transfected cells. Localization of bICP27 to the nucleus of transfected cells correlated with inhibiting IFN-β promoter activity. bICP27 mutants also had differential effects on IFN-β1 or IFN-β3 promoter activity suggesting bICP27 interferes with more than one step in the IFN-β signaling pathway.
MATERIALS AND METHODS
Cells
Mouse neuroblastoma cells (Neuro-2A) and low passage bovine turbinate cells (BT) were cultured in Earle’s modified Eagle’s medium (EMEM) supplemented with 10% fetal bovine serum, penicillin (10 U/ml), and streptomycin (100 μg/ml) in a humidified 5% CO2 atmosphere at 37°C.
bICP27 promoter constructs
The wt bICP27 expression plasmid was generated by PCR-amplification using the BHV-1 genome as a template and a high fidelity and proof reading Taq polymerase (Ambion, catalog no. AM2052). The forward primer (5’-CGGGATCCGCGAGTCGCAGCGGC-3’) was upstream of the first ATG of the bICP27 ORF. The reverse primer (5’- CCCAAGCTTGGGTTCAAATCAATGTTGACAGG-3’) was specific for the 3’ end of the bICP27 gene and included the consensus PolyA addition site. Restriction enzyme sites for BamHI and HindIII were also included in the forward and reverse primer respectively to facilitate cloning. PCR was initiated at 95°C for 7 minutes. This was followed by thirty cycles of denaturing at 95°C for 45 seconds, annealing at 59°C for 60 seconds and extending at 72°C for 60 seconds. Final extension was done at 72°C for 10 minutes.
Amplified bICP27 ORF was then cloned into Flag-tagged pCMV2B and pCMV2C expression vectors (Stratagene, La Jolla, CA USA) at unique BamHI and HindIII restriction sites to generate bICP27B and bICP27C, respectively. The Flag epitope is located at the 5’ terminus of the multiple cloning site. Cloning the bICP27 fragment into pCMV2C generates a frame shift mutant that does not express the protein, but will express the mRNA. The bICP27 deletion mutants were generated by PCR using the same conditions as wt bICP27 and the plasmid bICP27B as a template. To amplify the C-terminal deletion mutants (ΔC1, ΔC2 and ΔC3), the forward primer was the same used for amplifying wt bICP27 (5’-CGGGATCCGCGAGTCGCAGCGGC-3’) and the reverse primers were the following: ΔC1 (5’-GGAATTCCAGGGTGAAATTAGCGACGAG-3’); ΔC2 (5’-GGAATTCCGCCAACATCACGAACATGTACG-3’) and ΔC3 (5’-GGAATTCCTTGCACCACGTTAGGCACTCG-3’). The annealing temperatures were 60°C, 63°C and 59°C, respectively. The N-terminal deletion mutant (ΔN) was amplified by the forward primer (5’-CGGGATCCGGAGAGCAGCGGTCCCGC-3’) and the same reverse primer used to amplify the wt bICP27 (5’-CCCAAGCTTGGGTTCAAATCAATGTTGACAGG-3’). The annealing temperature was 58°C. The bICP27 deletion mutants were then inserted into Flag-tagged pCMV2B expression vector at unique BamHI and EcoRI restriction sites. The bovine beta-interferon (IFN-β) CAT reporter plasmids were previously described (da Silva and Jones, 2011). The IRF7 expression plasmid was obtained from Luwen Zhang (School of Biological Sciences, University of Nebraska, Lincoln, NE).
Chloramphenicol acetyltransferase (CAT) reporter assay
Approximately forty hours after transfection, cells were lysed by three freeze-thaw cycles in 250 mM Tris-HCL (pH7.4). CAT assays were performed with 0.2 μCi (7.4 KBq) {14C}-chloramphenicol (Amersham Biosciences, catalog no. CFA754) and 0.5 mM acetyl coenzyme A (Sigma, catalog no. A2181). Chloramphenicol and its acetylated forms were separated by thin-layer chromatography and CAT activity measured with a PhosphorImager (Molecular Dynamics, CA). Transfection experiments for CAT assays were repeated at least three times to confirm the results.
Western blot analysis
Whole-cell lysate was prepared from Neuro-2A cells. Cells were washed with phosphate-buffered saline (PBS) and suspended in RIPA buffer (1X PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1 % SDS and a proteinase inhibitor). Cell lysate was incubated on ice for 30 min and then clarified by centrifugation at 13,000 rpm at 4°C for 15 min. Protein concentrations were quantified by Bradford assay. Equal amounts of proteins in each sample were mixed with an equal volume of 1x sample loading buffer (62.5 mM Tris-HCL pH6.8, 2% sodium dodecyl sulfate, 50 mM dithiothreitol, 0.1% bromophenol blue, 10% glycerol) and boiled for 5 min. Proteins were separated in an 8% SDS-PAGE gel. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P transfer membrane, Millipore) using semidry gel electrophoresis. Membranes were then blocked for 2 hours in 5% nonfat dry milk with Tris-buffered saline-0.1% Tween 20 (TBS-T) and incubated with a monoclonal Flag antibody (Sigma) or β-actin (Santa Cruz Biotechnology) overnight at 4°C. After 3 washes of 15 min with TBS-T, the membranes were incubated with horseradish peroxidase-conjugated anti-IgG (Amersham Biosciences). Blots were washed 3 times, 15 min each with TBS-T and subsequently exposed to Amersham ECL reagents. Autoradiography was then performed.
Confocal microscopy
Cell monolayers were fixed with 4% para-formaldehyde for 10 min, followed by three washes with PBS. Cells were then permeabilized with 100% ethanol at -20°C for 5 min, washed three times and blocked in 3% bovine serum albumin (BSA) in PBS for 1 hour to reduce nonspecific binding. Primary antibody (anti-Flag monoclonal antibody; Sigma) was diluted in PBS with 0.05% Tween 20 and 1% BSA and added to coverslips for 2 hours at room temperature. After three washes, coverslips were incubated with Cy5-conjugated secondary antibody (Alexa Fluor, Invitrogen) for 1 h in the dark. After three washes, nuclear DNA was stained with DAPI (4’,6-diamidino-2-phenylindole). Coverslips were then mounted on slides and images were obtained with a Bio-Rad confocal laser-scanning microscope (MRC-1024ES) with excitation/emission at 488/520 nm. Images are representative of three or more independent experiments.
RESULTS
The bICP27 gene interferes with IFN-β promoter activity
To construct a plasmid that expresses bICP27 in mammalian cells, we PCR-amplified the bICP27 ORF from the BHV-1 genome and inserted it into a Flag-tagged mammalian expression vector pCMV2B (bICP27B) and pCMV2C (bICP27C). A Flag-tagged protein was used because the Flag tag is only 13 amino acids long and thus it is less likely to have dramatic effects on the structure of the protein. Only the pCMV2B construct contains the bICP27 ORF in frame with the Flag epitope. Cloning the same fragment into pCMV2C generates a frame shift mutant and consequently the protein will not be expressed. In transfected mouse neuroblastoma cells (Neuro-2A), the pCMV2B construct expressed a Flag-tagged protein that migrated at approximately 50 KDa, which is near the predicted position of bICP27. In contrast, the bICP27C construct did not express detectable levels of a Flag-tagged protein (Figure 1A). DNA sequencing confirmed that the bICP27B plasmid contains an intact bICP27 ORF in frame with the Flag peptide (data not shown).
Figure 1. bICP27 inhibits IFN-β promoter activity in transiently transfected cells.
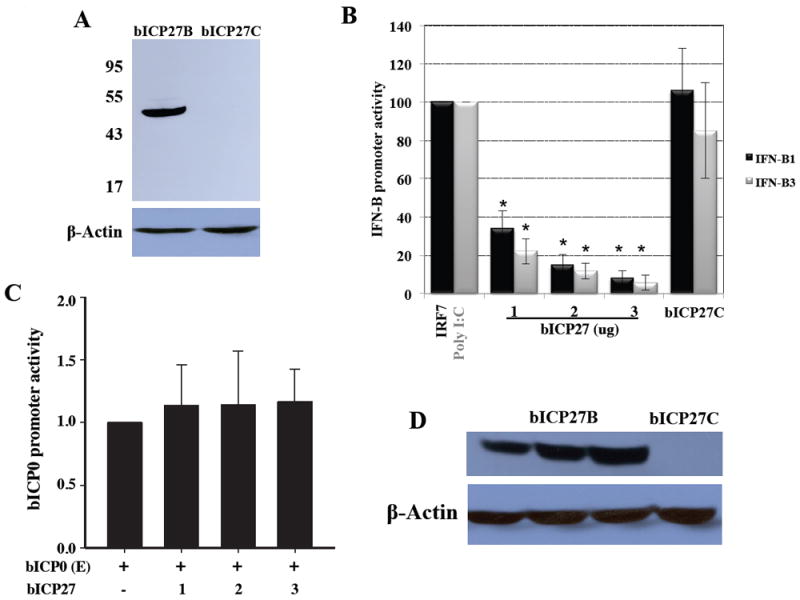
Panel A: Expression of Flag-tagged bICP27 in transfected Neuro-2A cells. The bICP27 coding sequences were PCR-amplified from the BHV-1 genome (Cooper strain) with a forward primer specific for a DNA sequence flanking the first bICP27 ORF ATG and a reverse primer annealing a sequence flanking the TATAA box termination signal, as described in the material and methods. The amplified PCR product was inserted at unique BamHI and HindIII restriction sites of the Flag-tagged expression vectors pCMV2B and pCMV2C to generate bICP27B and bICP27C, respectively. Subsequently, Neuro-2A cells were transfected with 1 μg of the indicated plasmids. Twenty-four hours after transfection whole cell lysate was prepared. A total of 200 μg of protein was separated in an 8% SDS-PAGE gel, and Flag-tagged bICP27 detected by Western blot analysis using an anti-Flag monoclonal antibody. β-Actin protein levels were analyzed in the respective samples as a loading control.
Panel B: Neuro-2A cells were cotransfected with 1 μg of IFN-β1 or IFN-β3 CAT reporter plasmids plus an IRF7 expression plasmid (1 μg) or PolyI:C (0.5 μg) and increasing amounts of a bICP27B (1, 2, or 3 μg) or 1 μg of bICP27C expression constructs. Plasmid DNA was maintained at the same concentration by inclusion of a blank expression vector (pcDNA3.1). Approximately 40 hours after transfection, cells were harvested and analyzed for CAT expression as previously described (da Silva, 2011; da Silva and Jones, 2011). Activation of the designated IFN-β promoters in the presence of IRF7 or Poly I:C, relative to cells transfected only with IFN-β CAT reporter plasmids plus empty vector was arbitrarily set at 100%. The results are the mean of four independent experiments. An asterisk denotes significant differences (P < 0.05) relative to the value of cells transfected with IFN-β CAT reporter plasmids plus the IRF7 expression construct or Poly I:C treated cells, as determined by the Student t test.
Panel C: Neuro-2A cells were cotransfected with 1 μg of a bICP0 early reporter construct (EP-943) plus increasing amounts of the bICP27B (1, 2, or 3 μg) construct. EP-943 contains 943 base pairs of the bICP0 early promoter and was previously described (Workman et al., 2009). Levels of DNA in the transfection mixture were kept at the same concentration by adding appropriate amounts of empty vector (pCMV2B).
Panel D: Increasing levels of Flag tagged bICP27 expression was detected when Neuro-2A cells were transfected with bICP27B but not with bICP27C. As a loading control, the blots were probed with a β-actin antibody.
To test whether the bICP27 gene interferes with IFN-β promoter activity, Neuro-2A cells were cotransfected with reporter plasmids containing the bovine IFN-β1 or IFN-β3 promoters and increasing amounts of the bICP27B expression vector. The transcription factor interferon regulatory factor 7 (IRF7) is the most efficient stimulator of IFN-β1 promoter activity in Neuro-2A cells (da Silva and Jones, 2011). The synthetic double-stranded RNA (Poly I:C), but not IRF7, strongly activates IFN-β3 promoter activity in Neuro-2A cells (da Silva and Jones, 2011). Therefore these cells were used to test whether bICP27 interferes with IRF7-induced IFN-β1 promoter activity or Poly I:C-induced IFN-β3 promoter activity. We did not examine the IFN-β2 promoter because it has little or no activity in transient transfection assays and it is not stimulated by IRF7 or Poly I:C (da Silva and Jones, 2011). Increasing amounts of bICP27B reduced IFN-β1 and IFN-β3 promoter activity in a dose-dependent fashion (Figure 1B). The bICP27C construct had little to no effect on IFN-β1 and IFN-β3 promoter activity because this construct does not express detectable levels of bICP27 protein in transfected cells (Figure 1A and D).
As a control, we compared the effect of bICP27 on the bICP0 early promoter. The bICP0 early promoter was used because it is cloned in the same vector as the bovine IFN-β1 and IFN-β3 promoter constructs. This may be important because a previous study provided evidence that bICP27 trans-activates the human CMV IE promoter more efficiently when it contains the BHV-1 gB polyA addition site versus the bovine growth hormone polyA addition site (Singh et al., 1996). Although bICP27 did not significantly increase bICP0 early promoter activity (Figure 1C), it was clear that bICP0 early promoter activity was not reduced by bICP27. As expected, increasing concentrations of the bICP27B construct resulted in increasing levels of the Flag-tagged bICP27 protein in transfected cells (Figure 1D). In summary, these studies indicated that bICP27 had an inhibitory effect on IFN-β promoter activity, but not bICP0 promoter activity.
Localization of bICP27 sequences necessary for inhibiting IFN-β promoter activity
Several conserved functional domains within bICP27 and HSV-1 encoded ICP27 have been identified. These include a nuclear localization signal (NLS), a nucleolar localization signal (NoLS), a nuclear export signal (NES) (Ding et al., 2010; Guo et al., 2009), and a zinc RING finger-like domain (Singh et al., 1996) (see Figure 2A for a schematic of the location of these domains). To investigate whether these bICP27 domains are necessary for inhibiting IFN-β promoter activity, bICP27 deletion mutants were constructed. Primers were designed for regions flanking determined bICP27 domains and were used to PCR-amplify truncated bICP27 sequences. Truncations in the C-terminal region generated the mutant ΔC1 that lacked sequences up to, but not including, the zinc RING finger. The mutant ΔC2 lacked the zinc RING finger domain. A mutant lacking the zinc RING finger-like domain plus NES was also constructed (ΔC3). Finally, a mutant lacking the NLS and NoLS was prepared (ΔN mutant). The respective mutants migrated at the expected sizes when compared to wt bICP27 (Figure 2B). The ΔC1 mutant was expressed at higher levels than wt bICP27, whereas the ΔN mutant was expressed at slightly lower levels relative to wt bICP27 (Figure 2B). The ΔC2 and ΔC3 mutants were expressed at similar levels as wt bICP27.
Figure 2. Analysis of bICP27 deletion mutants.
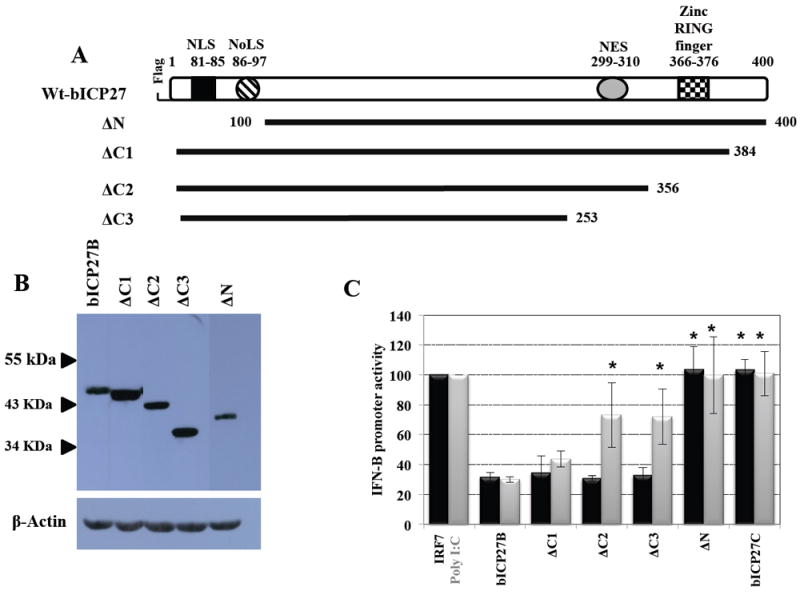
Panel A: Schematic of the bICP27 ORF and bICP27 deletion mutants. The N-terminal nuclear localization signal (NLS) and nucleolar localization signal (NoLS) span amino acids 81 to 85 and 86 to 97, respectively. Near the C-terminus, the zinc RING finger-like motif encompasses amino acids 366 to 376 and the nuclear export signal (NES) is located between amino acids 299 to 310. The bICP27 deletion mutants were generated by PCR amplification using primers specific for sequences flanking the bICP27 domains of interest as described in material and methods. Amplified PCR products were inserted at the unique restriction enzyme sites BamHI and EcoRI within pCMV2B expression vector. The numbers correspond to the amino acid positions within the bICP27 ORF. The position of the Flag epitope is shown for the wt-bICP27 construct. All deletion constructs contain a N-terminus Flag epitope.
Panel B: Western blot analysis comparing the expression of the bICP27 C-terminal mutants (ΔC1, ΔC2 and ΔC3), the N-terminal mutant (ΔN), and wt bICP27. Neuro-2A cells were transfected with 1 μg of the indicated bICP27 constructs. After 24 hours, whole cell lysate was prepared. A total of 200 μg of protein was separated in an 8% SDS-PAGE gel, and expression of Flag-tagged bICP27 detected by Western blot analysis using an anti-Flag monoclonal antibody. β-Actin protein levels were analyzed in the respective samples as a loading control. These results are representative of five independent experiments.
Panel C: Identification of the bICP27 domains necessary for inhibiting IFN-β1 and IFN-β3 promoter activity. Neuro-2A cells were cotransfected with 1 μg of IFN-β1 (black column) or IFN-β3 (grey column) CAT reporter plasmids plus IRF7 (1 μg) or Poly I:C (0.5 μg) respectively and 1ug of the designated bICP27 expression plasmids. Plasmid DNA in the transfection mix was maintained at the same concentration by inclusion of a blank expression vector (pcDNA3.1). Approximately 40 hours after transfection, cells were harvested and analyzed for CAT expression as previously described (da Silva, 2011; da Silva and Jones, 2011). Activation of the designated IFN-β promoters in the presence of IRF7 or Poly I:C, relative to cells transfected only with IFN-β CAT reporter plasmids plus empty vector was arbitrarily set at 100%. The results are the mean of five independent experiments. An asterisk denotes significant differences (P < 0.05) relative to the value of cells transfected with IFN-β CAT reporter plasmids plus the bICP27B construct, as determined by the Student t test.
To examine whether the respective bICP27 sequences had an effect on inhibiting IFN-β promoter activity, Neuro-2A cells were cotransfected with the IFN-β1 or IFN-β3 constructs, and either IFN-β activators IRF7 or Poly I:C, plus 1 ug of the designated bICP27 construct. We used 1 ug of the designated bICP27 constructs for these studies because this concentration of plasmid clearly reduced IFN-β1 and IFN-β3 promoter activity. In contrast to the IFN-β promoters, 1-3 ug of the bICP27 expression plasmid had little or no effect on bICP0 promoter activity (Figure 1C). As expected, wt bICP27 (bICP27B) inhibited the activity of IFN-β1 and IFN-β3 promoters approximately 3 fold, whereas bICP27C did not (Figure 2C). Disruption of the C-terminal region did not have a dramatic effect on bICP27-mediated inhibition of IFN-β1 promoter because all three C-terminal mutants (ΔC1, ΔC2 and ΔC3) inhibited IFN-β1 activity with similar efficiency as wt bICP27. In contrast, deletion of the zinc RING finger-like motif (ΔC2 mutant) or deletion of both zinc RING finger-like motifs and the NES domain (ΔC3 mutant) was significantly different (P < 0.05) compared to bICP27-mediated inhibition of IFN-β3 promoter activity. Furthermore, deletion of the N-terminal NLS and NoLS (ΔN) also interfered with the ability of bICP27 to reduce IFN-β1 and IFN-β3 promoter activity (Figure 2C).
Subcellular localization of bICP27 in transfected cells
The intracellular distribution of bICP27 was initially investigated by biotin-avidin-enhanced immunoadsorbent (black plaque) assay (Singh et al., 1996). This study concluded that bICP27 accumulated in the nucleus of BHV-1 infected cells. The subcellular localization of bICP27 was recently examined using immunofluorescence. These studies demonstrated that bICP27 exhibits predominantly nucleolar localization with faint staining at the nucleus of cells infected with BHV-1 or transfected with plasmids expressing bICP27 (Ding et al., 2010; Guo et al., 2009). bICP27 also translocates between the nucleus and cytoplasm during productive infection (Ding et al., 2010). To compare the localization of the bICP27 deletion mutants relative to wt bICP27, confocal microscopy was performed in Neuro-2A cells (Figure 3) transfected with the respective bICP27 constructs. Disruption of the C-terminal region did not appear to drastically alter the localization of the bICP27 mutants. The wt bICP27 and C-terminal deletion mutants (ΔC1, ΔC2 and ΔC3) localized to the nucleus of transfected cells and accumulated in certain nuclear structures (Figure 3). In general, the ΔC2 mutant was detected throughout the nucleus when compared to wt bICP27. In agreement with previous studies (Ding et al., 2010; Guo et al., 2009), the construct that contained a deletion of the N-terminal NLS and NoLS (ΔN) expressed a protein that was primarily localized to the cytoplasm. Similar results were obtained with low passage bovine turbinate cells (data not shown).
Figure 3. Localization of bICP27 in transfected cells.
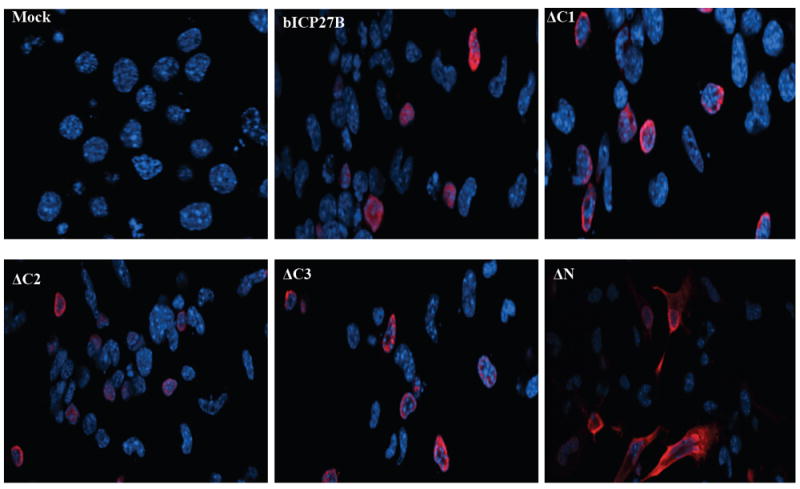
Confocal microscopy was performed to compare the localization of the bICP27 mutants relative to wt bICP27. Low passage bovine turbinate (BT) (Panel A) or Neuro-2A cells (Panel B) were transfected with 1 μg of the indicated bICP27 constructs. After 24 hours, cells were processed for confocal microscopy and stained for bICP27 using an anti-Flag monoclonal antibody (red) or nuclear stained with DAPI (blue). Magnification is 400.
DISCUSSION
In this study, we presented evidence demonstrating that bICP27 reduced IFN-β promoter activity. As previously found for HSV-1 encoded ICP27, the bICP27 protein interfered with bovine IFN-β promoter activity. Nuclear localization of bICP27 was necessary for inhibiting IFN-β promoter activity in transfected cells. The ability of BHV-1 to regulate innate immune responses is crucial for pathogenesis, reviewed in (Jones, 2009), and even host range (Abril et al., 2004). Furthermore, the ability of BHV-1 to interfere with host immune responses increases the incidence of secondary bacterial infections, reviewed in (Jones, 2007), which is important for development of the poly-microbial upper respiratory disease known as bovine respiratory disease complex. Consequently, understanding the mechanism by which BHV-1 interferes with innate immune responses is important and may lead to a modified live vaccine that is not immune-suppressive.
Our previous studies demonstrated that cytoplasmic localized bICP0 inhibits IFN-β signaling (da Silva et al., 2011; Saira et al., 2007, 2009). In contrast, nuclear localization of bICP27 was necessary for inhibiting IFN-β1 and IFN-β3 promoter activity suggesting that bICP27 interfered with cellular factors in the nucleus that stimulate IFN-β promoter activity. A previous study demonstrated that bICP27 regulates 3’ mRNA processing (Singh et al., 1996) suggesting bICP27 may interfere with 3’ processing of cellular factors that are necessary for IFN-β dependent transcription. The Epstein-Barr virus (EBV) SM protein, a homolog of ICP27, mediates alternative splicing of the cellular STAT-1 pre-mRNA (Verma et al., 2010; Verma and Swaminathan, 2008) suggesting bICP27 has similar functions. HSV-1 ICP27 also promotes alternative splicing of the promyelocytic leukemia (PML) mRNA (Nojima et al., 2009), which encodes a protein involved in the innate response to viral infections (Chee et al., 2003; Everett and Chelbi-Alix, 2007). Instead of binding to promyelocytic leukemia mRNAs through its RGG box motif, the binding was mediated by one of its predicted alternative RNA binding domains, the KH domain (Nojima et al., 2009). HSV-1 ICP27 also interferes with IFN signaling by inhibiting STAT-1 phosphorylation and nuclear accumulation (Johnson and Knipe, 2010; Johnson et al., 2008). Since the bICP27 protein and ICP27 protein only share limited amino acid similarity, bICP27 may have novel properties that are necessary for inhibiting IFN-β promoter activity.
The bICP27 zinc RING finger-like domain was necessary for inhibiting IFN-β3 promoter activity but not IFN-β1, which was surprising because one would expect that IFN-β signaling pathways would be similar. However, previous studies found that BHV-1 infection led to increased IFN-β3 RNA levels in established bovine kidney cells but completely inhibited IFN-β RNA expression in low passage bovine cells (da Silva and Jones, 2011; Perez et al., 2008). When protein synthesis was inhibited in low passage bovine cells and then these cells were infected with BHV-1, IFN-β1, but not IFN-β3, RNA expression was induced (da Silva and Jones, 2011) indicating that cell type dependent factors differentially regulate activation of the bovine IFN-β promoters. Further evidence for differential activation of the IFN-β1 versus IFN-β3 promoter activity comes from the finding that IRF3 trans-activates the IFN-β3 promoter but not the IFN-β1 promoter (da Silva and Jones, 2011). Conversely, IRF7 strongly activated both promoters. The bICP27 zinc RING finger-like motif is similar to ICP27 (Singh et al., 1996). The ICP27 zinc RING finger motif is important for interacting with several host proteins, including RNA polymerase II and proteins involved in mRNA processing (Chen et al., 2005; Dai-Ju et al., 2006). It is currently not known how the zinc RING finger-like motif regulates the known functions of bICP27. In summary, we predict that the ability of bICP27 to interfere with IFN-β promoter activity plays a role in promoting BHV-1 productive infection and viral pathogenesis.
Research Highlights.
Bovine herpesvirus 1 (BHV-1) infection interferes with innate immune responses.
BHV-1 infected cell protein 27 (bICP27) reduces beta-interferon promoter activity.
Nuclear entry of bICP27 correlates with reducing the beta-interferon promoter.
Acknowledgments
This research was supported by grants from the USDA (09-01653). Devis Sinani was partially supported by a fellowship from a Ruth L. Kirschstein National Research Service Award 1 T32 AIO60547 (National Institute of Allergy and Infectious Diseases). A grant to the Nebraska Center for Virology (1P20RR15635), in particular the microscopy core facility, also supported certain aspects of these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abril C, Engels M, Limman A, Hilbe M, Albini S, Franchini M, Suter M, Ackerman M. Both viral and host factors contribute to neurovirulence of bovine herpesvirus 1 and 5 in interferon receptor-deficient mice. J Virol. 2004;78:3644–3653. doi: 10.1128/JVI.78.7.3644-3653.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman M, Braun DK, Pereira L, Roizman B. Characterization of herpes simplex virus 1 alpha proteins 0, 4, and 27 with monoclonal antibodies. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Moore PA, Lowther W, Juang YT, Pitha PM. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalifour A, Basso J, Gagnon N, Trudel M, Simard C. Transcriptional and translational expression kinetics of the early gene encoding the BICP27 protein of bovine herpesvirus type 1. Virology. 1996;224:326–329. doi: 10.1006/viro.1996.0536. [DOI] [PubMed] [Google Scholar]
- Chee AV, Lopez P, Pandolfi PP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77:7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Li L, Silva L, Sandri-Goldin RM. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J Virol. 2005;79:3949–3961. doi: 10.1128/JVI.79.7.3949-3961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva LF, Gaudreault N, Jones C. Cytoplasmic localized infected cell protein 0 (bICP0) encoded by bovine herpesvirus 1 inhibits beta interferon promoter activity and reduces IRF3 (interferon response factor 3) protein levels. Virus Res. 2011;160:143–149. doi: 10.1016/j.virusres.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva LF, Jones C. Infection of cultured bovine cells with bovine herpesvirus 1 (BHV-1) or Sendai virus induces different beta interferon subtypes. Virus Res. 2011;157:54–60. doi: 10.1016/j.virusres.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai-Ju JQ, Li L, Johnson LA, Sandri-Goldin RM. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J Virol. 2006;80:3567–3581. doi: 10.1128/JVI.80.7.3567-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Guo H, Lin F, Pan W, Ye B, Zheng AC. Characterization of the nuclear import and export mechanisms of bovine herpesvirus-1 infected cell protein 27. Virus Res. 2010;149:95–103. doi: 10.1016/j.virusres.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Eidson KM, Hobbs WE, Manning BJ, Carlson P, DeLuca NA. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J Virol. 2002;76:2180–2191. doi: 10.1128/jvi.76.5.2180-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Goodbourn S, Zinn K, Maniatis T. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell. 1985;41:509–520. doi: 10.1016/s0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- Guo H, Ding Q, Lin F, Pan W, Lin J, Zheng AC. Characterization of the nuclear and nucleolar localization signals of bovine herpesvirus-1 infected cell protein 27. Virus Res. 2009;145:312–320. doi: 10.1016/j.virusres.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson G, Zhang Y, Jones C. The Bovine herpesvirus 1 gene encoding infected cell protein 0 (bICP0) can inhibit interferon-dependent transcription in the absence of other viral genes. J Gen Virol. 2005;86:2697–2702. doi: 10.1099/vir.0.81109-0. [DOI] [PubMed] [Google Scholar]
- Johnson KE, Knipe DM. Herpes simplex virus-1 infection causes the secretion of a type I interferon-antagonizing protein and inhibits signaling at or before Jak-1 activation. Virology. 2010;396:21–29. doi: 10.1016/j.virol.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Song B, Knipe DM. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology. 2008;374:487–494. doi: 10.1016/j.virol.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. Regulation of innate immune responses by bovine herpesvirus 1 and infected cell protein 0. Viruses. 2009;1:255–275. doi: 10.3390/v1020255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Chowdhury S. A review of the biology of bovine herpesvirus type 1 (BHV-1), its role as a cofactor in the bovine respiratory disease complex, and development of improved vaccines. Adv in Anim Health. 2007;8:187–205. doi: 10.1017/S146625230700134X. [DOI] [PubMed] [Google Scholar]
- King P, Goodbourn S. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J Biol Chem. 1994;269:30609–30615. [PubMed] [Google Scholar]
- Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J Virol. 2004;78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears WE, Lam V, Rice SA. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J Virol. 1995;69:935–947. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears WE, Rice SA. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology. 1998;242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- Melroe GT, Silva L, Schaffer PA, Knipe DM. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: Potential role in blocking IFN-beta induction. Virology. 2007;360:305–321. doi: 10.1016/j.virol.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- Nojima T, Oshiro-Ideue T, Nakanoya H, Kawamura H, Morimoto T, Kawaguchi Y, Kataoka N, Hagiwara M. Herpesvirus protein ICP27 switches PML isoform by altering mRNA splicing. Nucleic Acids Res. 2009;37:6515–6527. doi: 10.1093/nar/gkp633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladino P, Collins SE, Mossman KL. Cellular localization of the herpes simplex virus ICP0 protein dictates its ability to block IRF3-mediated innate immune responses. PLoS One. 2010;5:e10428. doi: 10.1371/journal.pone.0010428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez S, Meyer F, Saira K, Doster A, Jones C. Premature expression of the latency-related RNA encoded by bovine herpesvirus type 1 correlates with higher levels of beta interferon RNA expression in productively infected cells. J Gen Virol. 2008;89:1338–1345. doi: 10.1099/vir.0.83481-0. [DOI] [PubMed] [Google Scholar]
- Saira K, Zhou Y, Jones C. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J Virol. 2007;81:3077–3086. doi: 10.1128/JVI.02064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saira K, Zhou Y, Jones C. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) associates with interferon regulatory factor 7 and consequently inhibits beta interferon promoter activity. J Virol. 2009;83:3977–3981. doi: 10.1128/JVI.02400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Mohr I. Inhibition of cellular 2’-5’ oligoadenylate synthetase by the herpes simplex virus type 1 Us11 protein. J Virol. 2007;81:3455–3464. doi: 10.1128/JVI.02520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou G-P, Lin R, Hiscott J. Trigerring the interferon antiviral response through and IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Singh M, Fraefel C, Bello LJ, Lawrence WC, Schwyzer M. Identification and characterization of BICP27, an early protein of bovine herpesvirus 1 which may stimulate mRNA3’ processing. J Gen Virol. 1996;77:615–625. doi: 10.1099/0022-1317-77-4-615. [DOI] [PubMed] [Google Scholar]
- Singh M, Fraefel C, Bello LJ, Lawrence WC, Schwyzer M. Identification and characterization of BICP27, an early protein of bovine herpesvirus 1 which may stimulate mRNA 3’ processing. J Gen Virol. 1996;77(Pt 4):615–625. doi: 10.1099/0022-1317-77-4-615. [DOI] [PubMed] [Google Scholar]
- Soliman TM, Sandri-Goldin RM, Silverstein SJ. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valarcher J-F, Furze J, Wyld S, Cook R, Conzelman K-K, Taylor G. Role of alpha/beta interferons in the attenuation and immunogenecity of recombinant bovine respiratory syncitial viruses lacking NS proteins. J Virol. 2003;77:8426–8439. doi: 10.1128/JVI.77.15.8426-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Bais S, Gaillard M, Swaminathan S. Epstein-Barr Virus SM protein utilizes cellular splicing factor SRp20 to mediate alternative splicing. J Virol. 2010;84:11781–11789. doi: 10.1128/JVI.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Swaminathan S. Epstein-Barr virus SM protein functions as an alternative splicing factor. J Virol. 2008;82:7180–7188. doi: 10.1128/JVI.00344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpooten D, Ma Y, Hou S, Yan Z, He B. Control of TANK-binding kinase 1-mediated signaling by the gamma(1)34.5 protein of herpes simplex virus 1. J Biol Chem. 2009;284:1097–1105. doi: 10.1074/jbc.M805905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V, Jeffreys AJ, Barrie PA. A comparison of vertebrate interferon gene families deteced by hybridization with human interferon DNA. J Mol Biol. 1983;166:457–475. doi: 10.1016/s0022-2836(83)80281-2. [DOI] [PubMed] [Google Scholar]
- Winkler MT, Doster A, Jones C. Bovine herpesvirus 1 can infect CD4(+) T lymphocytes and induce programmed cell death during acute infection of cattle. J Virol. 1999;73:8657–8668. doi: 10.1128/jvi.73.10.8657-8668.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman A, Perez S, Doster A, Jones C. Dexamethasone treatment of calves latently infected with bovine herpesvirus 1 (BHV-1) leads to activation of the bICP0 early promoter, in part by the cellular transcription factor C/EBP-alpha. J Virol. 2009;83:8800–8809. doi: 10.1128/JVI.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fukuhara M, Fukuda M, Nishida E, Fujita T. Direct triggering of the type 1 interferon system by virus infection: activation of a transcription factors containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]


