Abstract
L1 elements are mammalian non–long terminal repeat retrotransposons, or long interspersed elements (LINEs), that significantly influence the dynamics and fluidity of the genome. A series of observations suggest that plant L1-clade LINEs, just as mammalian L1s, mobilize both short interspersed elements (SINEs) and certain messenger RNA by recognizing the 3′-poly(A) tail of RNA. However, one L1 lineage in monocots was shown to possess a conserved 3′-end sequence with a solid RNA structure also observed in maize and sorghum SINEs. This strongly suggests that plant LINEs require a particular 3′-end sequence during initiation of reverse transcription. As one L1-clade LINE was also found to share the 3′-end sequence with a SINE in a green algal genome, I propose that the ancestral L1-clade LINE in the common ancestor of green plants may have recognized the specific RNA template, with stringent recognition then becoming relaxed during the course of plant evolution.
Keywords: L1, LINE, parallel evolution, SINE
L1 elements are mammalian non–long terminal repeat (LTR) retrotransposons, or long interspersed elements (LINEs), that drive genome evolution in diverse ways. They constitute a large proportion of the genome, shaping both individual genes and the genome as a whole (Weiner et al. 1986; Brosius 1991). L1s mobilize nonautonomous sequences such as short interspersed element (SINE) RNA and cytosolic messenger RNA (mRNA) by recognizing the 3′-poly(A) tail of the template RNA, resulting in enormous SINE amplification (Dewannieux et al. 2003) and processed pseudogene formation (Esnault et al. 2000; Ohshima et al. 2003; Babushok et al. 2007; Ohshima and Igarashi 2010). In other words, L1s seem to initiate reverse transcription in a “relaxed” manner (Okada et al. 1997). The 3′-end sequences of various SINEs originated from corresponding LINEs other than L1 (Ohshima et al. 1996), however, and to date, ∼20 of these SINE/LINE pairs have been identified (Ohshima and Okada 2005). As the 3′-untranslated regions (UTRs) of several LINEs have been shown to be essential for retroposition, these LINEs presumably require “stringent” recognition of the 3′-end sequence of the RNA template (Okada et al. 1997; Kajikawa and Okada 2002).
A systematic database and literature survey identified 58 SINEs, more than twice the number already identified, each sharing a common 3′-end sequence with the partner LINE (supplementary table S1, supplementary fig. S1, Supplementary Material online). Although more than 800 L1-clade LINEs appeared in the database, only three SINEs with L1 tails were found in this study. This observation suggests that, in general, L1-clade LINEs differ from other LINEs with respect to 3′-end recognition (supplementary fig. S2, Supplementary Material online).
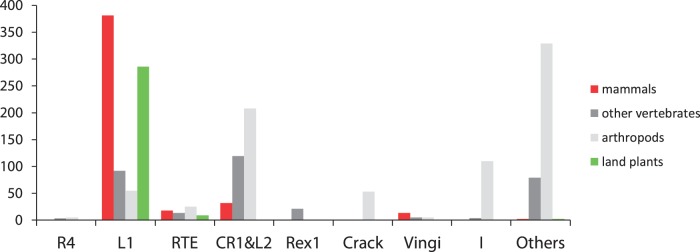
Figure 1 shows the number of LINEs belonging to each LINE clade according to biological taxa (supplementary table S2, Supplementary Material online). The genomes of land plants (mainly flowering plants) exclusively harbor only L1-clade LINEs (RTE-clade LINEs are also found in several species). Moreover, although a significant number of SINEs, more than half of which end in poly(A) repeats, have been identified in the genomes of flowering plants (supplementary table S3, Supplementary Material online), only three SINE/LINE pairs have been discovered: namely, maize ZmSINE2 and ZmSINE3 (LINE1-1_ZM; Baucom et al. 2009) and tobacco TS SINE (RTE-1_STu; this study; supplementary fig. S3, Supplementary Material online). Interestingly, many processed pseudogenes have been reported in flowering plants (Faris et al. 2001; Zhang et al. 2005; Benovoy and Drouin 2006; Nurhayati et al. 2009). As mammalian L1s are thought to recognize the 3′-poly(A) tail of RNA when forming processed pseudogenes (Esnault et al. 2000), it is possible that plant LINE machinery is similar to mammalian L1s (Lenoir et al. 2001). That is, by presumably recognizing the 3′-poly(A) tail of RNA, plant L1-clade LINEs thereby mobilize SINEs with a poly(A) tail and mRNA. In accordance with this hypothesis, almost all L1-clade LINEs in flowering plants were shown to end in poly(A) repeats and all RTE-clade LINEs in (TTG)n or (TTGATG)n (table 1). Poly(T)-ending SINEs: p-SINEs and Au-like SINEs (supplementary table S3, Supplementary Material online) would be mobilized by the LINE machinery that recognize a poly(U) repeat of RNA at the 3′-terminus, although such LINE has never been reported in plants.
Fig. 1.
The number of LINE families belonging to each LINE clade according to biological taxa. LINE clades in which the partner LINE of a SINE was identified are shown. Remaining clades are grouped as “Others” (Repbase 16.10). “other vertebrates”: nonmammalian vertebrates; “land plants”: mostly flowering plants.
Table 1.
3′-Repeats of Plant LINE Families.
| Species | LINE clade | Families | 3′-repeat |
||
|---|---|---|---|---|---|
| (A)n | Other repeats | None | |||
| Flowering plants | L1 | 233 | 224 | 0 | 9 |
| RTE | 7 | 0 | 7a | 0 | |
| Green algae | L1 | 15 | 2b | 8c | 5 |
| RandI | 8 | 0 | 8d | 0 | |
| RTEX | 6 | 0 | 6e | 0 | |
a(TTG)n and (TTGATG)n.
bL1-1_CR (Chlamydomonas) and Zepp (Chlorella).
c(CATA)n, (CA)n, (CAA)n, and (TAA)n.
d(ATT)n and (CTATTT)n.
e(CA)n, (CAA)n, (CCAT)n, (ACAATG)n, and (CTTGTAA)n.
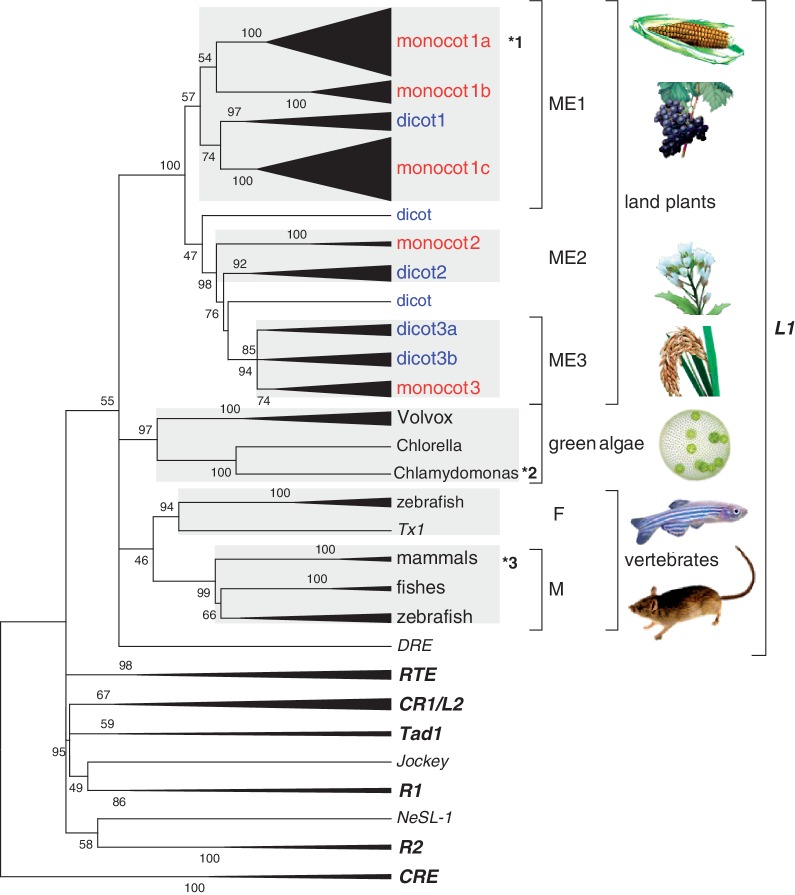
Figure 2 shows the results from comprehensive phylogenetic analysis of L1-clade LINEs (supplementary fig. S4 and supplementary table S4, Supplementary Material online). Three important points were revealed. First, L1-clade LINEs from distinct taxa, namely, land plants, green algae, and vertebrates, formed monophyletic groups. Statistical support for the monophyly of land plants and green algae was high, with bootstrap values of 100 and 97, respectively (82 and 83; maximum likelihood [ML] method; supplementary fig. S5, Supplementary Material online). Monophyly of the vertebrate F and M lineages (Ichiyanagi et al. 2007), however, was not supported by the ML method (supplementary fig. S5, Supplementary Material online). Second, the L1 lineages from these three taxa formed a monophyletic group (55/45; neighbor-joining [NJ]/ML methods) among diverged LINE clades such as RTE and CR1. The Tx1 LINE, with target-specific insertion, was also found in this clade, as observed in previous studies (Kojima and Fujiwara 2004; Ichiyanagi et al. 2007). The Tx1 and vertebrate F lineage formed a monophyletic group with high confidence (94/85). Third, comparison with the species phylogeny revealed that plant L1-clade LINEs consist of at least three deeply branching lineages that have descended from the common ancestor of monocots and eudicots (ME1-3; supplementary fig. S6, Supplementary Material online). These three lineages must have arisen more than 130 million years ago, around the approximate divergence of monocots and eudicots (Moore et al. 2007).
Fig. 2.
Phylogenetic relationships among the L1-clade LINEs. LINE-clades are shown in bold italics. Several lineages in which a stringent or relaxed L1 was found are indicated by asterisks: (*1) LINE1-1_ZM (stringent), (*2) L1-1_CR (stringent), and (*3) L1HS (relaxed). The phylogenetic relationships among 146 LINEs were inferred using the amino acid sequences of ORF2 proteins from plant L1 entries in the database (Repbase 15.08; Viridiplantae) and from other LINEs (Ohshima and Okada 2005). A total of 404 positions made up the final data set. The linearized NJ consensus tree obtained from bootstrap analysis with 1,000 replications is shown (an ML consensus tree formed with the same data set is available as supplementary fig. S5, Supplementary Material online). The evolutionary distances were computed using the Jones-Taylor-Thornton (JTT) matrix-based method. For clarity, some clades were collapsed with filled triangles, the widths of which were in proportion to the number of LINEs. The full expanded tree is shown in supplementary figure S4, Supplementary Material online. Bootstrap values are only shown for nodes with scores > 45.
One group of LINEs in a monocot L1 lineage (monocot 1a in fig. 2) retained a conserved 3′-end sequence (supplementary fig. S7, Supplementary Material online). Average pairwise divergence of this region (the last 45 nucleotides) among the LINEs was only 0.144 (standard error [SE], 0.043), whereas that for the entire sequence was 0.570 (SE, 0.012). Interestingly, maize SINEs (ZmSINE2 and ZmSINE3) with 3′-end sequences very similar to that of the above LINE, LINE1-1_ZM, were recently reported (Baucom et al. 2009). This study also revealed possession of similar 3′-end sequences by several sorghum SINEs (supplementary fig. S8, Supplementary Material online). Comparison of the 3′-end sequences from these SINEs and LINEs revealed that part of the sequence (ca., 50 nucleotides) is apparently related, presumably having been derived from a common ancestral L1 sequence (supplementary fig. S9, Supplementary Material online).
The putative transcript from this region was also shown to form a possible hairpin structure (supplementary fig. S10, Supplementary Material online). Compensatory mutations were observed in the stem-forming sequences, confirming a secondary structure (supplementary figs. S7 and S10, Supplementary Material online). Several nucleotides were strongly conserved in the 3′-flanking region of the stem (5′-CGAG-3′) and in the loop (5′-UCU-3′), though the stem-forming nucleotides were variable. This stem-loop structure is commonly observed in the 3′-end sequences of stringent-type LINEs and SINEs (Osanai et al. 2004; Nomura et al. 2006). These results strongly suggest that, at least in this lineage, plant LINEs require a particular 3′-end sequence of stringent type.
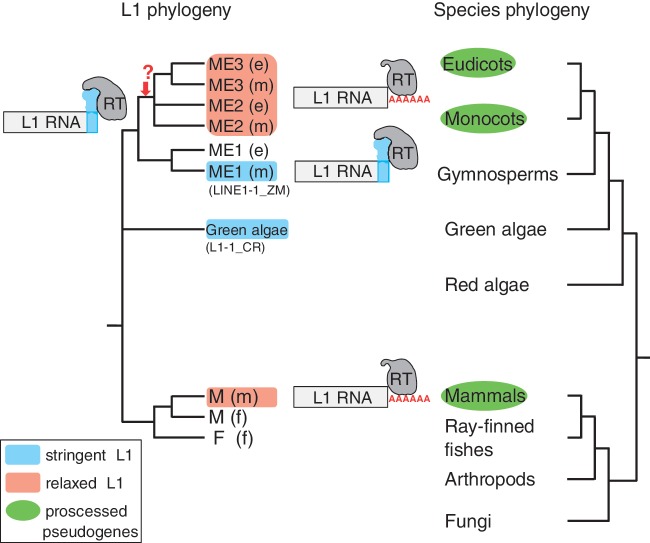
The last example of a SINE/LINE pair in the L1-clade was found in a green alga. The 3′-end sequence (ca., 80 nucleotides) of Chlamydomonas SINEX-3_CR (Cognat et al. 2008) was very similar to that of L1-1_CR, with both ending in poly(A) repeats (supplementary fig. S11, Supplementary Material online). As land plants emerged from green algae (Karol et al. 2001), the following is proposed for 3′-end recognition of plant L1-clade LINEs (fig. 3). It is possible that the ancestral L1-clade LINE in the genome of the common ancestor of green plants possessed stringent, nonmammalian-type RNA recognition properties. During the course of plant evolution, a L1 lineage(s) then lost the ability to specifically recognize the RNA template for reverse transcription, thereby introducing relaxed 3′-end recognition in land (flowering) plants as in mammals. As horizontal transfer of LINEs between eukaryotes is rare (Kordiš and Gubenšek 1998; Malik et al. 1999), the discontinuous distribution of L1-clade LINEs with low specificity (i.e., mammalian L1s and plant ME2/ME3) suggests a type of parallel evolution.
Fig. 3.
Proposed model for the 3′-end recognition of L1-clade LINEs. The ancestral L1-clade LINE in the ancestral green plant possessed a stringent, nonmammalian-type RNA recognition property. During the course of plant evolution, a L1 lineage(s) lost the ability to specifically recognize the RNA template for reverse transcription, introducing relaxed 3′-end recognition in land plants. Processed pseudogenes have been reported in eudicots, monocots, and mammals. ME1-3: plant L1 lineages; (e): eudicots; (m): monocots; M, F: vertebrate L1 lineages; (m): mammals; (f): fish.
The ancestral L1-clade LINE might have required both the 3′-end sequence and the terminal poly(A) repeats. A few L1 lineages might then have lost specific interaction with the 3′-UTR of the template RNA, retaining some role for the 3′-repeats. As listed in table 1, most plant L1-clade LINEs have poly(A) repeats at their 3′-termini as in mammalian L1s. However, 3′-poly(A) repeats are not necessarily a hallmark of relaxed 3′-end recognition. For example, although silkworm SART1, an R1-clade LINE, uses stringent-type recognition (its 3′-UTR is essential for retroposition), it ends in poly(A) repeats (Takahashi and Fujiwara 2002; Osanai et al. 2004), which are necessary for efficient and accurate retroposition (Osanai et al. 2004).
L1 LINEs have contributed significantly to the architecture and evolution of mammalian genomes, whereas LTR retrotransposons are overwhelmingly found in certain flowering plants. Understanding the independent origins of flexible 3′-end recognition may help us determine what distinguishes the fate of retroposons in the eukaryotic genome and why it has succeeded so well in certain genomes (Zhang and Wessler 2004; Heitkam and Schmidt 2009; Hollister et al. 2011).
Supplementary Material
Supplementary figures S1–S11 and tables S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
References
- Babushok DV, Ohshima K, Ostertag EM, Chen X, Wang Y, Mandal PK, Okada N, Abrams CS, Kazazian HH., Jr A novel testis ubiquitin-binding protein gene arose by exon shuffling in hominoids. Genome Res. 2007;17:1129–1138. doi: 10.1101/gr.6252107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, SanMiguel PJ, Bennetzen JL. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 2009;5:e1000732. doi: 10.1371/journal.pgen.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovoy D, Drouin G. Processed pseudogenes, processed genes, and spontaneous mutations in the Arabidopsis genome. J Mol Evol. 2006;62:511–522. doi: 10.1007/s00239-005-0045-z. [DOI] [PubMed] [Google Scholar]
- Brosius J. Retroposons—seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Cognat V, Deragon JM, Vinogradova E, Salinas T, Remacle C, Maréchal-Drouard L. On the evolution and expression of Chlamydomonas reinhardtii nucleus-encoded transfer RNA genes. Genetics. 2008;179:113–123. doi: 10.1534/genetics.107.085688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Faris J, Sirikhachornkit A, Haselkorn R, Gill B, Gornicki P. Chromosome mapping and phylogenetic analysis of the cytosolic acetyl-CoA carboxylase loci in wheat. Mol Biol Evol. 2001;18:1720–1733. doi: 10.1093/oxfordjournals.molbev.a003960. [DOI] [PubMed] [Google Scholar]
- Heitkam T, Schmidt T. BNR—a LINE family from Beta vulgaris—contains a RRM domain in open reading frame 1 and defines a L1 sub-clade present in diverse plant genomes. Plant J. 2009;59:872–882. doi: 10.1111/j.1365-313X.2009.03923.x. [DOI] [PubMed] [Google Scholar]
- Hollister JD, Smith LM, Guo YL, Ott F, Weigel D, Gaut BS. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci U S A. 2011;108:2322–2327. doi: 10.1073/pnas.1018222108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyanagi K, Nishihara H, Duvernell DD, Okada N. Acquisition of endonuclease specificity during evolution of L1 retrotransposon. Mol Biol Evol. 2007;24:2009–2015. doi: 10.1093/molbev/msm130. [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Okada N. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- Karol KG, McCourt RM, Cimino MT, Delwiche CF. The closest living relatives of land plants. Science. 2001;294:2351–2353. doi: 10.1126/science.1065156. [DOI] [PubMed] [Google Scholar]
- Kojima KK, Fujiwara H. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol Biol Evol. 2004;21:207–217. doi: 10.1093/molbev/msg235. [DOI] [PubMed] [Google Scholar]
- Kordiš D, Gubenšek F. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc Natl Acad Sci U S A. 1998;95:10704–10709. doi: 10.1073/pnas.95.18.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir A, Lavie L, Prieto JL, Goubely C, Côté JC, Pélissier T, Deragon JM. The evolutionary origin and genomic organization of SINEs in Arabidopsis thaliana. Mol Biol Evol. 2001;18:2315–2322. doi: 10.1093/oxfordjournals.molbev.a003778. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci U S A. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y, Kajikawa M, Baba S, Nakazato S, Imai T, Sakamoto T, Okada N, Kawai G. Solution structure and functional importance of a conserved RNA hairpin of eel LINE UnaL2. Nucleic Acids Res. 2006;34:5184–5193. doi: 10.1093/nar/gkl664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurhayati N, Gondé D, Ober D. Evolution of pyrrolizidine alkaloids in Phalaenopsis orchids and other monocotyledons: identification of deoxyhypusine synthase, homospermidine synthase and related pseudogenes. Phytochemistry. 2009;70:508–516. doi: 10.1016/j.phytochem.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Ohshima K, Hamada M, Terai Y, Okada N. The 3′ ends of tRNA-derived short interspersed repetitive elements are derived from the 3′ ends of long interspersed repetitive elements. Mol Cell Biol. 1996;16:3756–3764. doi: 10.1128/mcb.16.7.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Hattori M, Yada T, Gojobori T, Sakaki Y, Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Igarashi K. Inference for the initial stage of domain shuffling: tracing the evolutionary fate of the PIPSL retrogene in hominoids. Mol Biol Evol. 2010;27:2522–2533. doi: 10.1093/molbev/msq138. [DOI] [PubMed] [Google Scholar]
- Ohshima K, Okada N. SINEs and LINEs: symbionts of eukaryotic genomes with a common tail. Cytogenet Genome Res. 2005;110:475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- Okada N, Hamada M, Ogiwara I, Ohshima K. SINEs and LINEs share common 3′ sequences: a review. Gene. 1997;205:229–243. doi: 10.1016/s0378-1119(97)00409-5. [DOI] [PubMed] [Google Scholar]
- Osanai M, Takahashi H, Kojima KK, Hamada M, Fujiwara H. Essential motifs in the 3′ untranslated region required for retrotransposition and the precise start of reverse transcription in non-long-terminal-repeat retrotransposon SART1. Mol Cell Biol. 2004;24:7902–7913. doi: 10.1128/MCB.24.18.7902-7913.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Fujiwara H. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J. 2002;21:408–417. doi: 10.1093/emboj/21.3.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM, Deininger PL, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wessler SR. Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc Natl Acad Sci U S A. 2004;101:5589–5594. doi: 10.1073/pnas.0401243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu Y, Liu Y, Han B. Computational identification of 69 retroposons in Arabidopsis. Plant Physiol. 2005;138:935–948. doi: 10.1104/pp.105.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.