Abstract
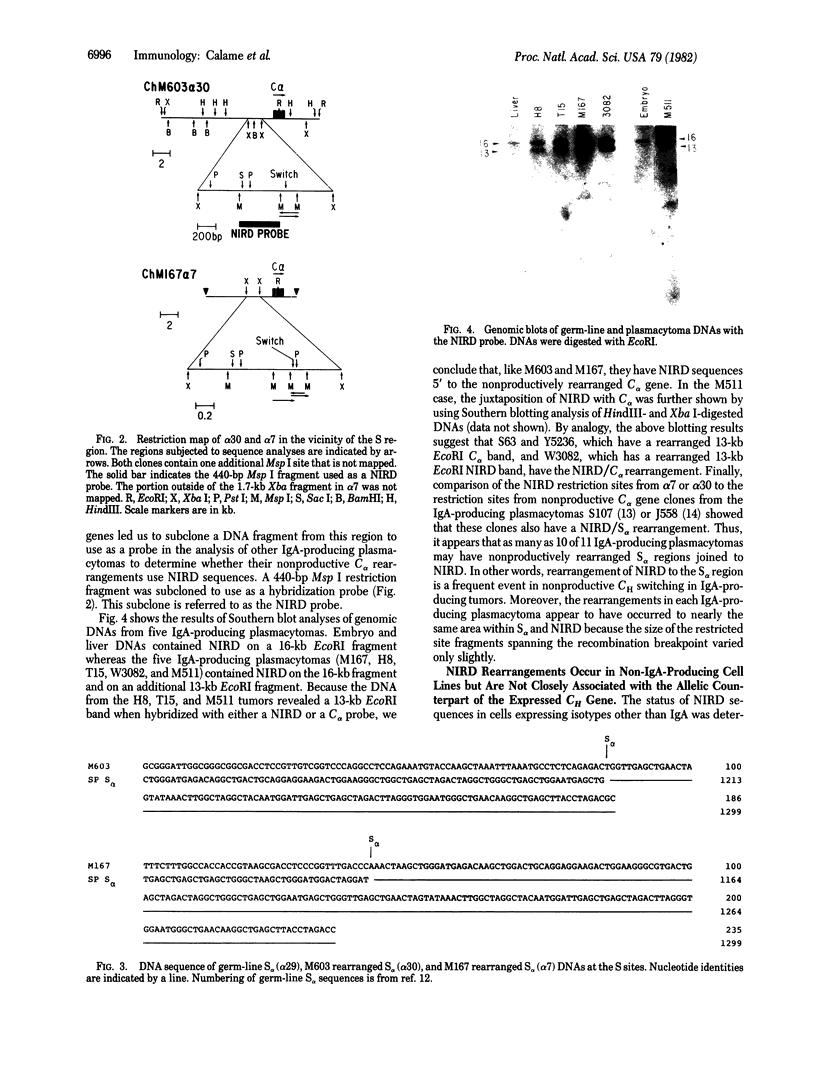
Expression of IgA by plasmacytomas occurs as a result of a DNA rearrangement that brings the variable region gene, VH, a few kilobases 5' to the constant region gene, C alpha. In this study, we show that the allelic nonexpressed C alpha gene also is rearranged in most plasmacytomas. Cloning, restriction mapping, heteroduplex analyses, and sequence analyses of the nonproductively rearrange C alpha genes from two plasmacytomas, M603 and M167, have demonstrated that the nonproductive rearrangement occurs within the alpha switching region, S alpha. In each case, the same DNA sequence has been joined to the 5' side of C alpha and we have termed this DNA "NIRD" (for nonimmunoglobulin rearranged DNA). Southern blotting analyses of genomic DNAs from various IgG-, IgM-, or IgA-producing plasmacytomas suggest that NIRD is rearranged in almost all plasmacytomas. However, NIRD rearranges to the S alpha region only in IgA-producing cells, not in IgM or IgG producers. Cytogenetic evidence has shown that T(12;15) translocations are common in murine plasmacytomas. Immunoglobulin heavy chain genes are located on chromosome 12, and the translocation breakpoint in plasmacytomas occurs near the immunoglobulin genes. NIRD has been mapped to chromosome 15 by Southern blotting analysis of mouse-hamster cell lines, suggesting that the nonproductively rearranged C alpha clones represent the T(12;15) translocations identified cytogenetically. Therefore, we have identified a region of DNA on chromosome 15 that is commonly rearranged in transformed mouse lymphocytes. We speculate on the significance of NIRD in neoplastic transformation of mouse lymphocytes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Davis M. M., Kim S. K., Hood L. E. DNA sequences mediating class switching in alpha-immunoglobulins. Science. 1980 Sep 19;209(4463):1360–1365. doi: 10.1126/science.6774415. [DOI] [PubMed] [Google Scholar]
- Harris L. J., D'Eustachio P., Ruddle F. H., Marcu K. B. DNA sequence associated with chromosome translocations in mouse plasmacytomas. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6622–6626. doi: 10.1073/pnas.79.21.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L. J., Lang R. B., Marcu K. B. Non-immunoglobulin-associated DNA rearrangements in mouse plasmacytomas. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4175–4179. doi: 10.1073/pnas.79.13.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Kim S., Davis M., Sinn E., Patten P., Hood L. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell. 1981 Dec;27(3 Pt 2):573–581. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- Kirsch I. R., Ravetch J. V., Kwan S. P., Max E. E., Ney R. L., Leder P. Multiple immunoglobulin switch region homologies outside the heavy chain constant region locus. Nature. 1981 Oct 15;293(5833):585–587. doi: 10.1038/293585a0. [DOI] [PubMed] [Google Scholar]
- Klein G. The role of gene dosage and genetic transpositions in carcinogenesis. Nature. 1981 Nov 26;294(5839):313–318. doi: 10.1038/294313a0. [DOI] [PubMed] [Google Scholar]
- Lalley P. A., Francke U., Minna J. D. Homologous genes for enolase, phosphogluconate dehydrogenase, phosphoglucomutase, and adenylate kinase are syntenic on mouse chromosome 4 and human chromosome 1p. Proc Natl Acad Sci U S A. 1978 May;75(5):2382–2386. doi: 10.1073/pnas.75.5.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalley P. A., Minna J. D., Francke U. Conservation of autosomal gene synteny groups in mouse and man. Nature. 1978 Jul 13;274(5667):160–163. doi: 10.1038/274160a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Minna J. D., Marshall T. H., Shaffer-Berman P. V. Chinese hamster X mouse hybrid cells segregating mouse chromosomes and isozymes. Somatic Cell Genet. 1975 Oct;1(4):355–369. doi: 10.1007/BF01538667. [DOI] [PubMed] [Google Scholar]
- Ohno S., Babonits M., Wiener F., Spira J., Klein G., Potter M. Nonrandom chromosome changes involving the Ig gene-carrying chromosomes 12 and 6 in pristane-induced mouse plasmacytomas. Cell. 1979 Dec;18(4):1001–1007. doi: 10.1016/0092-8674(79)90212-5. [DOI] [PubMed] [Google Scholar]
- Shepard J. S., Pettengill O. S., Wurster-Hill D. H., Sorenson G. D. A specific chromosome breakpoint associated with mouse plasmacytomas. J Natl Cancer Inst. 1978 Jul;61(1):255–258. doi: 10.1093/jnci/61.1.255. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]