Abstract
Purpose
We conducted a case-parent triad study evaluating the role of maternal and offspring genotypes in the folate metabolic pathway on childhood acute lymphoblastic leukemia (ALL) risk.
Methods
Childhood ALL case-parent triads (N = 120) were recruited from Texas Children’s Hospital. DNA samples were genotyped using the Sequenom iPLEX MassARRAY for 68 tagSNPs in six folate metabolic pathway genes (MTHFR, MTRR, MTR, DHFR, BHMT, and TYMS). Log-linear modeling was used to examine the associations between maternal and offspring genotypes and ALL.
Results
After controlling for the false discovery rate (<0.1), there were 20 significant maternal effects in the following genes: BHMT (N = 3), MTR (N = 12), and TYMS (N = 5). For instance, maternal genotypes for BHMT rs558133 (relative risk [RR] = 0.51, 95% confidence interval [CI]: 0.30–0.87, P = 0.008, Q = 0.08) and MTR rs2282369 (RR = 0.46, 95% CI: 0.27–0.80, P = 0.004, Q = 0.08) were associated with ALL. There were no significant offspring effects after controlling for the false discovery rate.
Conclusions
This is one of the few studies conducted to evaluate maternal genetic effects in the context of childhood ALL risk. Furthermore, we employed a family-based design that is less susceptible to population stratification bias in the estimation of maternal genetic effects. Our findings suggest that maternal genetic variation in the folate metabolic pathway is relevant in the etiology of childhood ALL. The observed maternal genetic effects support the need for continued research of how the uterine environment may influence risk of ALL.
Keywords: Acute lymphoblastic leukemia, case-parent triad, folate, genetic epidemiology, pediatric cancer
INTRODUCTION
Leukemia is the most frequent malignancy of childhood, accounting for one out of three cases of childhood cancer. In the United States (US), approximately 4,900 children develop leukemia per year [1]. Acute lymphoblastic leukemia (ALL) is the most common subtype of childhood leukemia, which accounts for 80% of all cases of leukemia [2]. Although ten-year survival is greater than 80%, there is concern over the long term morbidities related to treatment, and little is known about the etiology of ALL [3], which limits prevention efforts. The incidence of ALL is higher among Hispanic children compared to non-Hispanic children and higher in non-Hispanic white children compared to non-Hispanic black children [2]. Generally, ALL peaks in incidence between ages two and four years, suggesting that the etiology lies early in development, possibly in utero [4].
The absence of consistent environmental risk factors and the presence of modest familial associations suggest ALL is a complex trait with an underlying genetic component [5]. Although previous genome-wide association studies (GWAS) and candidate gene approaches have identified susceptibility loci contributing to the genetic basis of ALL, they only explain a small fraction of the heritability [6–9,5]. The folate metabolic pathway is suspected to play an important role in the development of childhood ALL as it is critical for the synthesis, repair, and methylation of DNA [10]. A majority of reports have focused on genetic variants of methylenetetrahydrofolate reductase (MTHFR), a key enzyme in the regulation of folate metabolism [11]. Other genes in the folate metabolic pathway have been assessed in relation to ALL risk [12], but the results have been equivocal. However, only a few studies have taken an approach evaluating multiple variants within candidate genes involved in the folate metabolic pathway [12,10]. Additionally, few have examined the role of maternal genetic effects (a proxy for the intrauterine environment) [13], which are likely to be important in relation to ALL risk [14]. Finally, as the majority of studies have used a case-control approach, population stratification bias may explain mixed findings [5].
The present study was undertaken to evaluate the association between childhood ALL and 68 single nucleotide polymorphisms (SNPs) in six folate metabolic genes: betaine-homocysteine S-methyltransferase (BHMT), dihydrofolate reductase (DHFR), methylenetetrahydrofolate reductase (MTHFR), 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR), 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (MTRR), and thymidylate synthetase (TYMS). Previous literature was used in our candidate gene selection strategy, along with suspected involvement in DNA synthesis, repair, and methylation [10,12]. We used a case-parent triad approach with log-linear modeling in order to eliminate population stratification bias in the evaluation of offspring genetic effects, while also accounting for maternal genetic effects [15].
METHODS
Study Population
The study population included 120 ALL case-parent triads recruited from the Childhood Cancer Epidemiology and Prevention Center at Texas Children’s Hospital (Houston, TX) between 2003 and 2010. Both males and females, and individuals of all racial/ethnic groups were eligible to participate. After written informed consent was obtained from the parent, we obtained a blood sample from each participant. Additionally, saliva samples were collected from parents. Participation of both parents was not required for our analysis [16]. These samples were used to obtain DNA for genotyping. Demographic and clinical data were abstracted from medical records. The study protocol was approved by the Baylor College of Medicine Institutional Review Board.
SNP Selection and Genotyping Methods
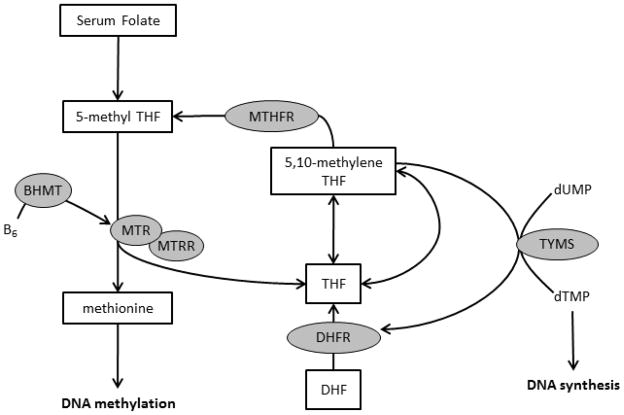
Six genes in the folate metabolic pathway (BHMT [N = 8], DHFR [N = 1], MTHFR [N = 12], MTR [N = 28], MTRR [N = 11], and TYMS [N = 8]) were selected because of their role in DNA synthesis, repair, and methylation (Figure 1). Previous literature was also used in our selection strategy [10,12]. TagSNPs for the six genes were selected using an r2 threshold of 0.80 and the MultiPop-TagSelect Algorithm in the Genome Variation Server [17,18]. SNPs with minor allele frequencies of <10% were not included in the analysis due to the available sample size. Based on these criteria, 68 SNPs were available for analysis.
Figure 1.
Folate metabolic pathway. Genes included in the analysis are represented in grey circles. Abbreviations: betaine-homocysteine S-methyltransferase (BHMT), dihydrofolate reductase (DHFR), methylenetetrahydrofolate reductase (MTHFR), 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR), 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (MTRR), and thymidylate synthetase (TYMS).
DNA was extracted from peripheral blood lymphocytes and saliva using the QIAmp DNA Blood Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Genotyping was done using the Sequenom MassARRAY iPLEX platform (Sequenom, San Diego, CA) in the Human Genetics Center at The University of Texas School of Public Health according to the manufacturer’s instructions.
Statistical Analysis
The characteristics of case subjects and their parents were summarized using counts and proportions. For each analyzed variant, samples for which a genotype could not be assigned and triads that had genotype combinations that were inconsistent with Mendelian inheritance were determined. For each subject, the number of genotyping failures (i.e., genotypes that could not be assigned) was determined. These analyses were performed using Intercooled Stata, version 12.1 (StataCorp LP, College Station, TX).
Log-linear models were used to assess the association between childhood ALL and both the offspring and maternal genotypes for each variant [15]. Specifically, genotype relative risks (RR) and 95% confidence intervals (CI) were estimated using a log-additive model of inheritance. Therefore, the RR represents the increase or decrease in risk with each additional copy of the minor allele. The log-additive model of inheritance was selected as it provides greater power compared to the genotypic model when determining associations and is commonly used in an agnostic approach [19]. A P-value for offspring and maternal genetic effects was determined using a likelihood ratio test (LRT) that compared the model that included terms for both offspring and maternal genotypes (i.e., full model), to models that included terms for only the offspring or only the maternal genotype (i.e., reduced models). According to Wilcox et al., the LRT allows a test of whether the offspring’s genotype carries any predictive information once the maternal allele count has been accounted for, and vice versa [20]. These analyses were run using the MI-GWAS platform for LEM [21,22], which allows for the inclusion of incompletely genotyped triads [22,16]. Due to concerns about population stratification bias when assessing maternal genetic effects, analyses were repeated among non-Hispanic Whites, however the potential for bias in the case-parent triad design in the estimation of maternal genetic effects is not as great as for the case-control design [15,23]. Finally, due to the number of comparisons, we used the Benjamini and Hochberg method to calculate a corrected P-value (Q-value) and control for the false discovery rate (FDR) at 0.1 [24].
RESULTS
Genotyping was performed on DNA samples derived from 120 families (276 individuals). Call rates for the 68 SNPs ranged from 95% to 99%. Based on quality control checks, 2 families (2% of families) were excluded for being inconsistent with Mendelian inheritance on more than five genotypes (7% of genotypes). Additionally, 7 subjects (3%) were excluded for failure on more than 31 genotypes (>40%), leaving a total of 118 case-parent triads (98% of the original sample). Of those, 42 were complete triads, 72 were dyads, and 4 were monads. After these quality control measures were applied, at least 97% of the samples for each variant were available for analyses; therefore the genotypes were considered of sufficiently high quality.
The distributions of key characteristics of childhood ALL cases are presented in Table 1. The majority of cases (55%) were male. Fifty percent of the population was non-Hispanic white, with the second largest group being Hispanic (39%). Age at diagnosis ranged from 0 to 14 years, with a plurality (47%) being diagnosed at less than four years of age.
Table 1.
Population characteristics of childhood acute lymphoblastic leukemia cases, Childhood Cancer Epidemiology and Prevention Center, 2003–2010
| Characteristic | No. (%) |
|---|---|
| Triads included | 118 |
| Case sex | |
| Male | 65 (55.0) |
| Female | 53 (45.0) |
| Race/ethnicity | |
| Non-Hispanic White | 59 (50.0) |
| Non-Hispanic Black | 6 (5.1) |
| Hispanic | 46 (39.0) |
| Other | 7 (5.9) |
| Age (range 0–14 years) | |
| <4 years | 56 (47.4) |
| 4–7 years | 46 (39.0) |
| >7 years | 16 (13.6) |
Table 2 includes estimates of the relative risks (RR estimates represent the increase or decrease in risk with each additional copy of the minor allele) and 95% confidence intervals (CI) for the association between the top 20 offspring genotypes and childhood ALL, as well as the likelihood ratio test (LRT) P-values and FDR Q-values for the model comparisons for these variants. Although there were modest associations between offspring genotypes in BHMT, MTR, MTRR, and TYMS and childhood ALL, only MTR rs2275565 was significantly associated with disease risk (RR = 1.68, 95% CI: 1.01, 2.86; LRT P = 0.049). However, none of the genotypes, including MTR rs2275565, were significantly associated with risk after applying the FDR.
Table 2.
Log-linear results for the association between selected offspring folate metabolic genotypes and childhood acute lymphoblastic leukemia
| Gene | RefSNP | RR1 | 95% CI | P-value2 | Q-value3 |
|---|---|---|---|---|---|
| BHMT | rs7700970 | 1.65 | 0.96, 2.81 | 0.062 | 0.752 |
| BHMT | rs3733890 | 1.31 | 0.83, 2.08 | 0.235 | 0.752 |
| MTR | rs2275565 | 1.68 | 1.01, 2.86 | 0.049 | 0.752 |
| MTR | rs3768150 | 0.68 | 0.43, 1.09 | 0.106 | 0.752 |
| MTR | rs10754584 | 0.69 | 0.43, 1.10 | 0.116 | 0.752 |
| MTR | rs2385511 | 0.70 | 0.44, 1.11 | 0.128 | 0.752 |
| MTR | rs1050996 | 0.71 | 0.45, 1.12 | 0.141 | 0.752 |
| MTR | rs2282369 | 0.72 | 0.45, 1.14 | 0.156 | 0.752 |
| MTR | rs10802569 | 0.74 | 0.48, 1.16 | 0.187 | 0.752 |
| MTR | rs3768142 | 0.74 | 0.48, 1.17 | 0.195 | 0.752 |
| MTR | rs12138911 | 1.44 | 0.82, 2.52 | 0.200 | 0.752 |
| MTR | rs10925261 | 1.43 | 0.81, 2.51 | 0.208 | 0.752 |
| MTR | rs883396 | 0.75 | 0.47, 1.20 | 0.222 | 0.752 |
| MTRR | rs6555501 | 0.64 | 0.39, 1.05 | 0.069 | 0.752 |
| MTRR | rs161871 | 1.58 | 0.93, 2.69 | 0.085 | 0.752 |
| MTRR | rs162036 | 1.62 | 0.86, 3.03 | 0.123 | 0.752 |
| MTRR | rs162039 | 1.55 | 0.84, 2.84 | 0.145 | 0.752 |
| MTRR | rs3776467 | 0.61 | 0.29, 1.30 | 0.190 | 0.752 |
| TYMS | rs2612100 | 0.69 | 0.41, 1.14 | 0.151 | 0.752 |
| TYMS | rs699517 | 0.71 | 0.43, 1.16 | 0.171 | 0.752 |
RR estimates represent the increase or decrease in risk with each additional copy of the minor allele
One-degree of freedom likelihood ratio test
False discovery rate adjusted P-value
A total of 20 maternal SNPs in BHMT, MTR, and TYMS were significantly associated with disease risk (Table 3). Specifically, 12 of the 28 maternal SNPs (43%) in MTR had a Q <0.1. The less common alleles in the 12 MTR SNPs showed a 42 to 54% reduction of risk. This trend was similar for the three maternal SNPs in BHMT with Q <0.1, whereby the less common alleles were associated with a 44 to 51% reduction of risk. When analyses were repeated among non-Hispanic Whites (to evaluate the potential for population stratification bias when assessing maternal genetic effects) the direction and magnitude of the RRs for the MTR and BHMT SNPs remained the same (data not shown).
Table 3.
Log-linear results for the association between selected maternal folate metabolic genotypes and childhood acute lymphoblastic leukemia
| Gene | RefSNP | RR1 | 95% CI | P-value2 | Q-value3 |
|---|---|---|---|---|---|
| BHMT | rs585800 | 0.49 | 0.27, 0.88 | 0.010 | 0.076 |
| BHMT | rs558133 | 0.52 | 0.30, 0.90 | 0.011 | 0.076 |
| BHMT | rs506500 | 0.56 | 0.33, 0.93 | 0.017 | 0.076 |
| MTR | rs2282369 | 0.46 | 0.27, 0.80 | 0.004 | 0.076 |
| MTR | rs4659745 | 0.51 | 0.30, 0.87 | 0.010 | 0.076 |
| MTR | rs10802564 | 0.54 | 0.33, 0.90 | 0.012 | 0.076 |
| MTR | rs10754584 | 0.51 | 0.29, 0.90 | 0.013 | 0.076 |
| MTR | rs10733117 | 0.54 | 0.33, 0.90 | 0.013 | 0.076 |
| MTR | rs2297965 | 0.55 | 0.33, 0.90 | 0.013 | 0.076 |
| MTR | rs1050996 | 0.51 | 0.29, 0.91 | 0.014 | 0.076 |
| MTR | rs12759827 | 0.46 | 0.23, 0.90 | 0.014 | 0.076 |
| MTR | rs2385511 | 0.52 | 0.30, 0.91 | 0.015 | 0.076 |
| MTR | rs3768150 | 0.52 | 0.30, 0.92 | 0.018 | 0.076 |
| MTR | rs10802569 | 0.57 | 0.33, 0.96 | 0.025 | 0.094 |
| MTR | rs3768142 | 0.58 | 0.34, 0.97 | 0.028 | 0.096 |
| TYMS | rs699517 | 1.96 | 1.19, 3.21 | 0.005 | 0.076 |
| TYMS | rs2612100 | 1.82 | 1.12, 2.95 | 0.011 | 0.076 |
| TYMS | rs3786362 | 7.12 | 0.90, 56.00 | 0.017 | 0.076 |
| TYMS | rs1004474 | 1.64 | 1.04, 2.58 | 0.023 | 0.091 |
| TYMS | rs2847153 | 1.67 | 1.04, 2.69 | 0.026 | 0.094 |
RR estimates represent the increase or decrease in risk with each additional copy of the minor allele
One-degree of freedom likelihood ratio test
False discovery rate adjusted P-value
Five of the eight maternal SNPs (63%) in TYMS had a Q <0.1. For each of these SNPs, the less common allele was associated with increased risk with the RR estimates ranging from 1.64 to 7.12 (Table 3). However, when analyses were restricted to non-Hispanic Whites, the effect estimates approached the null (data not shown).
DISCUSSION
In one of the few assessments evaluating the role of both offspring and maternal genetic effects and childhood ALL risk, we examined 68 maternal and offspring SNPs in six folate metabolic genes (BHMT, DHFR, MTHFR, MTR, MTRR, and TYMS) using a case-parent triad design. Although offspring genotypes in BHMT, MTR, MTRR, and TYMS were associated with childhood ALL, only one SNP, MTR rs2275565, was significantly associated with risk. However, after adjusting for multiple comparisons, the association was not significant. Previous studies evaluating offspring genotypes in MTHFR have suggested an association with ALL risk; however, MTHFR variants were not associated with risk in this population. These differences may be due to sample size or population structure.
In our study population, maternal genotypes in BHMT, MTR, and TYMS were associated with childhood ALL risk after adjusting for the FDR at Q <0.1. In one of the few studies evaluating the association between maternal folate metabolic genotypes and childhood ALL, Lightfoot et al. examined maternal genotypes for the following three SNPs: MTHFR rs1801133 (677 C>T), MTHFR rs1801131 (1298 A>C), and MTR rs1805087 (2756 A>G). The authors determined there was no evidence of an association between these three maternal folate metabolic genotypes and childhood ALL [13]. Additionally, a study by Krajinovic et al. evaluated maternal genotypes for MTHFR rs1801133 (677 C>T) and MTHFR rs1801131 (1298 A>C), comparing case mothers to control mothers, and found no association [25]. These SNPs were included in our analysis, and as with the previous studies, we found no association.
Betaine-homocysteine methyltransferase (encoded by BHMT) catalyzes the transfer of a methyl group from betaine to homocysteine producing dimethylglycine and methionine [26]. Defects in this gene may be associated with increased homocysteine levels, which in turn may lead to oxidative stress-induced carcinogenesis [27]. Although maternal genotypes in this gene have not been evaluated in the context on childhood ALL, maternal minor alleles in BHMT SNPs (rs7356530 and rs600473), which were not assessed in this study, appear to be inversely associated with oral clefts [26], suggesting BHMT variants play a role in fetal development. As childhood ALL is believed to originate in utero, this provides additional support for the role of BHMT in disease risk [13]. In our analysis, the maternal minor alleles of three BHMT SNPs (rs585800, rs558133, and rs506500) were inversely associated with childhood ALL. Of these SNPs, rs558133 and rs506500 are within intronic regions of BHMT [18], however, rs585800 is in the 3′ UTR exonic splicing element of BHMT [28,18].
Five-methyltetrahydrofolate-homocysteine methyltransferase (encoded by MTR) catalyzes regeneration of methionine from homocysteine [13]. This step in the folate metabolic pathway maintains levels of methionine used in DNA methylation. Also, methionine forms S-adenosyl methionine, the universal methyl group donor. A known variant of MTR, rs1805087 (2756A>G), is suggested to induce DNA hypomethylation, important in carcinogenesis [13,12]. While some studies have found associations with offspring MTR SNPs and childhood ALL [13], others have not [12]. To our knowledge, only the maternal MTR rs1805087 (2756A>G) has previously been assessed in relation to childhood ALL risk and there was no association [13], which is consistent with our findings. We assessed additional SNPs (N = 28) in the MTR gene and found maternal minor alleles in 12 SNPs were inversely associated with childhood ALL risk. Eleven of the SNPs are within intronic regions of MTR [18]. Although MTR rs1050996 is in the 3′ UTR region, to our knowledge, this SNP has not been previously assessed (maternal or offspring effects) in relation to childhood ALL risk.
Thymidylate synthase (encoded by TYMS) catalyzes methylation of dUMP (deoxyuridine monophosphate) to dTMP (thymidine monophosphate) using 5,10-methylenetetrahydrofolate as a co-factor. It is the sole pathway for production of dTMP, which incorporates thymine during DNA synthesis. Lowered TYMS activity may increase uracil misincorporation in DNA synthesis and may lead to chromosomal translocations. Specifically, excision of uracil by uracil-DNA glycosylase leads to transient single strand breaks, and if two or more of these occur near each other, a double-strand break may occur. In a state of folate deficiency, the probability of double-strand breaks is increased, thereby increasing the likelihood of translocations [29][30]. One study reported an inverse association between the TYMS 28-basepair triple repeat in offspring and ALL [31], which was in the same magnitude and direction of the offspring TYMS variants assessed in the current study (the 28-basepair triple repeat was not assessed), although not significant. Although maternal minor alleles in TYMS were positively associated with childhood ALL, these associations were null when we restricted our analysis to non-Hispanic Whites, suggesting our findings may be: 1) limited to other race/ethnicity groups or 2) biased by population structure [15].
The major limitation of this study is the sample size, which did not allow us to detect modest associations and precluded our assessment of haplotypes. For instance, with our current sample size (N = 118), using a log-additive model and assuming a minor allele frequency of 10%, we had 80% power to detect a RR of 2.12 (inverse 0.47). Additionally, we were not able to stratify our results by ALL subtypes (e.g., B-lineage or T-lineage) or age at diagnosis. However, in spite of these limitations, we were able to identify significant associations, even after adjusting for the FDR. An important strength of our study was the use of the case-parent triad design, which provides increased power over the case-control approach in the assessment of genetic effects [20]. Additionally, the case-parent triad design is immune to confounding by race/ethnicity (i.e., population stratification) in the assessment of offspring genotypes [15]. Additionally, we conducted sub-analyses among non-Hispanic Whites to evaluate the potential for population stratification in the assessment of maternal genetic effects. In these sub-analyses the results for BHMT and MTR remained the same; however, the effects for the maternal TYMS genotypes were attenuated. However, it should be noted that even though we restricted our analyses to non-Hispanic Whites, hidden population structure may explain the observed maternal associations. A final strength is the log-linear modeling approach to analyses also allowed us to include data from incomplete triads (i.e., genotype data is missing for one or two individuals) [16].
In conclusion, our findings suggest that maternal genetic variation in the folate metabolic pathway is relevant in the etiology of childhood ALL. Previous studies of other outcomes have found maternal genetic effects to be important independent of offspring genetic effects [32,33]. One potential explanation may be the importance of the fetal environment independent of the fetal genotype. For example, a maternal allele may damage a fetus through effects on the intrauterine milieu, regardless of the fetal genotype [20]. The observed maternal genetic effects support the need for continued research of how the in utero environment may influence risk of childhood ALL. Replication of these findings in other populations and investigation of additional genes is warranted. Furthermore, the exploration of gene-environment interactions, including how maternal nutritional factors during pregnancy (e.g., folate supplementation) are modified by maternal genotypes may be important in future assessments.
Supplementary Material
Acknowledgments
The genotyping for this work was supported by an Inter-Institutional Pilot Project (to M.E.S. and P.J.L.) from the Dan L. Duncan Cancer Center at Baylor College of Medicine, P30CA125123 (PI: Osborne). M.E.S. was also supported in part by an NCI Career Development Award, K07CA131505. The authors would also like to thank Ms. Megan Grove-Gaona for her technical assistance and the families who participated in this study.
Footnotes
CONFLICTS OF INTEREST STATEMENT
The authors declare they have no conflict of interest.
References
- 1.Margolin JF, Rabin KR, Steuber CP, Poplack DG. Acute Lymphoblastic Leukemia. In: Pizzo PA, Poplack DG, editors. Principles and practice of pediatric oncology. Lippincott Williams & Wilkins; Philadelphia: 2011. pp. 518–565. [Google Scholar]
- 2.Surveillance Epidemiology and End Results. [Accessed January 22 2012];Childhood Cancer. 2011 http://seer.cancer.gov/csr/1975_2008/results_merged/sect_28_childhood_cancer.pdf.
- 3.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. The New England journal of medicine. 2006;354(2):166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 4.Bas Suarez MP, Lopez Brito J, Santana Reyes C, Gresa Munoz M, Diaz Pulido R, Lodos Rojas JC. Congenital acute lymphoblastic leukemia: a two-case report and a review of the literature. Eur J Pediatr. 2011;170(4):531–534. doi: 10.1007/s00431-010-1339-8. [DOI] [PubMed] [Google Scholar]
- 5.Sherborne AL, Hemminki K, Kumar R, Bartram CR, Stanulla M, Schrappe M, Petridou E, Semsei AF, Szalai C, Sinnett D, Krajinovic M, Healy J, Lanciotti M, Dufour C, Indaco S, El-Ghouroury EA, Sawangpanich R, Hongeng S, Pakakasama S, Gonzalez-Neira A, Ugarte EL, Leal VP, Espinoza JP, Kamel AM, Ebid GT, Radwan ER, Yalin S, Yalin E, Berkoz M, Simpson J, Roman E, Lightfoot T, Hosking FJ, Vijayakrishnan J, Greaves M, Houlston RS. Rationale for an international consortium to study inherited genetic susceptibility to childhood acute lymphoblastic leukemia. Haematologica. 2011;96(7):1049–1054. doi: 10.3324/haematol.2011.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijayakrishnan J, Houlston RS. Candidate gene association studies and risk of childhood acute lymphoblastic leukemia: a systematic review and meta-analysis. Haematologica. 2010;95(8):1405–1414. doi: 10.3324/haematol.2010.022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, Kinsey SE, Lightfoot T, Roman E, Irving JA, Allan JM, Tomlinson IP, Taylor M, Greaves M, Houlston RS. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nature genetics. 2009;41(9):1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J, Papaemmanuil E, Bartram CR, Stanulla M, Schrappe M, Gast A, Dobbins SE, Ma Y, Sheridan E, Taylor M, Kinsey SE, Lightfoot T, Roman E, Irving JA, Allan JM, Moorman AV, Harrison CJ, Tomlinson IP, Richards S, Zimmermann M, Szalai C, Semsei AF, Erdelyi DJ, Krajinovic M, Sinnett D, Healy J, Gonzalez Neira A, Kawamata N, Ogawa S, Koeffler HP, Hemminki K, Greaves M, Houlston RS. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nature genetics. 2010;42(6):492–494. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, Willman C, Neale G, Downing J, Raimondi SC, Pui CH, Evans WE, Relling MV. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nature genetics. 2009;41(9):1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koppen IJ, Hermans FJ, Kaspers GJ. Folate related gene polymorphisms and susceptibility to develop childhood acute lymphoblastic leukaemia. British journal of haematology. 2010;148(1):3–14. doi: 10.1111/j.1365-2141.2009.07898.x. [DOI] [PubMed] [Google Scholar]
- 11.Yan J, Yin M, Dreyer ZE, Scheurer ME, Kamdar K, Wei Q, Okcu MF. A meta-analysis of MTHFR C677T and A1298C polymorphisms and risk of acute lymphoblastic leukemia in children. Pediatric blood & cancer. 2012;58(4):513–518. doi: 10.1002/pbc.23137. [DOI] [PubMed] [Google Scholar]
- 12.Metayer C, Scelo G, Chokkalingam AP, Barcellos LF, Aldrich MC, Chang JS, Guha N, Urayama KY, Hansen HM, Block G, Kiley V, Wiencke JK, Wiemels JL, Buffler PA. Genetic variants in the folate pathway and risk of childhood acute lymphoblastic leukemia. Cancer Causes Control. 2011;22(9):1243–1258. doi: 10.1007/s10552-011-9795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lightfoot TJ, Johnston WT, Painter D, Simpson J, Roman E, Skibola CF, Smith MT, Allan JM, Taylor GM. Genetic variation in the folate metabolic pathway and risk of childhood leukemia. Blood. 2010;115(19):3923–3929. doi: 10.1182/blood-2009-10-249722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Infante-Rivard C, Vermunt JK, Weinberg CR. Excess transmission of the NAD(P)H:quinone oxidoreductase 1 (NQO1) C609T polymorphism in families of children with acute lymphoblastic leukemia. Am J Epidemiol. 2007;165(11):1248–1254. doi: 10.1093/aje/kwm022. kwm022 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62(4):969–978. doi: 10.1086/301802. S0002-9297(07)60990-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinberg CR. Allowing for missing parents in genetic studies of case-parent triads. Am J Hum Genet. 1999;64(4):1186–1193. doi: 10.1086/302337. AJHG980963 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–120. doi: 10.1086/381000. S0002-9297(07)61949-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Heart L, and Blood Institute. [Accessed January 24 2012];GVS: Genome Variation Server version 5.11. 2010 http://gvs.gs.washington.edu/GVS/
- 19.Salanti G, Southam L, Altshuler D, Ardlie K, Barroso I, Boehnke M, Cornelis MC, Frayling TM, Grallert H, Grarup N, Groop L, Hansen T, Hattersley AT, Hu FB, Hveem K, Illig T, Kuusisto J, Laakso M, Langenberg C, Lyssenko V, McCarthy MI, Morris A, Morris AD, Palmer CN, Payne F, Platou CG, Scott LJ, Voight BF, Wareham NJ, Zeggini E, Ioannidis JP. Underlying genetic models of inheritance in established type 2 diabetes associations. Am J Epidemiol. 2009;170(5):537–545. doi: 10.1093/aje/kwp145. kwp145 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilcox AJ, Weinberg CR, Lie RT. Distinguishing the effects of maternal and offspring genes through studies of “case-parent triads”. Am J Epidemiol. 1998;148 (9):893–901. doi: 10.1093/oxfordjournals.aje.a009715. [DOI] [PubMed] [Google Scholar]
- 21.Agopian AJ, Mitchell LE. MI-GWAS: a SAS platform for the analysis of inherited and maternal genetic effects in genome-wide association studies using log-linear models. BMC bioinformatics. 2011;12:117. doi: 10.1186/1471-2105-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermunt JK. LEM: A general program for the analysis of categorical data. Tilberg University; 1997. [Accessed August 20 2012]. http://spitswww.uvt.nl/~vermunt/ [Google Scholar]
- 23.Mitchell LE, Weinberg CR. Evaluation of offspring and maternal genetic effects on disease risk using a family-based approach: the “pent” design. Am J Epidemiol. 2005;162(7):676–685. doi: 10.1093/aje/kwi249. kwi249 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 1995;57 (1):289–300. [Google Scholar]
- 25.Krajinovic M, Lamothe S, Labuda D, Lemieux-Blanchard E, Theoret Y, Moghrabi A, Sinnett D. Role of MTHFR genetic polymorphisms in the susceptibility to childhood acute lymphoblastic leukemia. Blood. 2004;103(1):252–257. doi: 10.1182/blood-2003-06-1794. 2003-06-1794 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Mostowska A, Hozyasz KK, Biedziak B, Misiak J, Jagodzinski PP. Polymorphisms located in the region containing BHMT and BHMT2 genes as maternal protective factors for orofacial clefts. Eur J Oral Sci. 2010;118(4):325–332. doi: 10.1111/j.1600-0722.2010.00757.x. EOS757 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Oikawa S, Murakami K, Kawanishi S. Oxidative damage to cellular and isolated DNA by homocysteine: implications for carcinogenesis. Oncogene. 2003;22(23):3530–3538. doi: 10.1038/sj.onc.1206440. [DOI] [PubMed] [Google Scholar]
- 28.Martinez CA, Northrup H, Lin JI, Morrison AC, Fletcher JM, Tyerman GH, Au KS. Genetic association study of putative functional single nucleotide polymorphisms of genes in folate metabolism and spina bifida. Am J Obstet Gynecol. 2009;201(4):394 e391–311. doi: 10.1016/j.ajog.2009.06.042. S0002-9378(09)00686-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A. 1997;94 (7):3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skibola CF, Smith MT, Hubbard A, Shane B, Roberts AC, Law GR, Rollinson S, Roman E, Cartwright RA, Morgan GJ. Polymorphisms in the thymidylate synthase and serine hydroxymethyltransferase genes and risk of adult acute lymphocytic leukemia. Blood. 2002;99 (10):3786–3791. doi: 10.1182/blood.v99.10.3786. [DOI] [PubMed] [Google Scholar]
- 31.Canalle R, Silveira VS, Scrideli CA, Queiroz RG, Lopes LF, Tone LG. Impact of thymidylate synthase promoter and DNA repair gene polymorphisms on susceptibility to childhood acute lymphoblastic leukemia. Leukemia & lymphoma. 2011;52(6):1118–1126. doi: 10.3109/10428194.2011.559672. [DOI] [PubMed] [Google Scholar]
- 32.Goldmuntz E, Woyciechowski S, Renstrom D, Lupo PJ, Mitchell LE. Variants of folate metabolism genes and the risk of conotruncal cardiac defects. Circ Cardiovasc Genet. 2008;1(2):126–132. doi: 10.1161/CIRCGENETICS.108.796342. CIRCGENETICS.108.796342 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lupo PJ, Mitchell LE, Goldmuntz E. NAT1, NOS3, and TYMS genotypes and the risk of conotruncal cardiac defects. Birth Defects Res A Clin Mol Teratol. 2011;91(1):61–65. doi: 10.1002/bdra.20745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.