Abstract
As the first example of hydrogelator derived from aminoglycoside antibiotics, the hydrogel of kanamycin indicates that the hydrogel of aminoglycosides preserve the specific interaction with their macromolecular targets (e.g., 16S rRNA), thus illustrating a simple approach to explore and identify possible biological targets of supramolecular nanofibers/hydrogels.
This communication reports the first hydrogelator derived from an aminoglycoside antibiotic (e.g., kanamycin1) and the resulted hydrogel for sequestering 16S rRNA selectively via divalent interaction. Hydrogelators are small molecules that self-assemble in water to yield supramolecular nanofibers to imbibe water and to afford hydrogels.2 The early development of hydrogelators,3 as a curiosity of physical organic chemistry,4 has demonstrated that carefully balanced hydrophobic interactions and hydrogen bonding induce the aggregations of small molecules in water to form nanofibers and afford hydrogels. After that, the successful integration of biofunctional or bioactive molecules into the hydrogelators5 has led to rapid development of supramolecular hydrogels6, 7 as a new class of biomaterials that have precise control on molecular structures and supramolecular organization. For example, the hydrogelator of vancomycin has exhibited high activities to against vancomycin resistance enterococci (VRE).8, 9 Although a vast pool of bioactive molecules, ranging from peptides10 to clinical used drug molecules,9, 11 are able to serve as build blocks for constructing supramolecular assemblies that could exhibit a superior effectiveness that is unattainable by monomeric epitopes or individual drug molecules, there is little exploration on the self-assembly of functional small molecules in water, partially due to the lack of a general approach to promote molecular self-assembly and to verify the interactions between the supramolecular assemblies and their target(s).
We hypothesized that the conversion of a functional molecule to a hydrogelator, actually, offers a simple way to create the self-assembly of the functional molecule and verify the interactions between the nanofibers and the potential targets. Scheme 1 illustrates the concept. The self-assembly of the hydrogelators leads to the formation of supramolecular nanofibers, which entangle to result in a network, stop the flow of water, and afford a hydrogel. Thus, hydrogelation can serve as a simple visual assay to indicate the self-assembly of small molecules in water. After hydrogel reaches steady state, it is possible to add the solution containing potential target and proper control molecules on the top of the hydrogel. The gel phase is in contact with the solution, but the two phases remain immiscible. While the nanofibers remain in the gel phase, the target and control molecules, however, can enter the gel phase by diffusing through the pores of the network. The specific interaction of the nanofibers with the targets would retain more target molecules than the control molecules in the gel phase. Since it is easy to separate the gel phase from the solution phase by aspiration the supernatant, the analysis of the remaining molecules in the solution phases (i.e., the supernatant) should be able to identify the target molecules sequestered in the hydrogel due to the specific interaction between the target molecules and the supramolecular nanofibers.
Scheme 1.
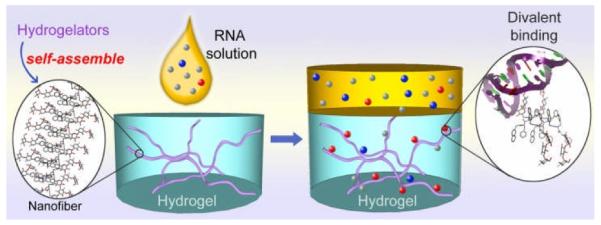
Supramolecular nanofibers sequester the potential targets (e.g., 16S rRNA (in red color)) in the gel phase.
To demonstrate the concept illustrated in Scheme 1, we chose to make the hydrogelator of kanamycin (1) by attaching multiple phenylalanine residues to the 6′-N amino group of kanamycin A, which is a member of aminoglycoside antibiotics that bind 16S rRNA in ribosome to inhibit protein synthesis and act as bactericides.1 We found that 1 not only acts as an effective hydrogelator at neutral pH with critical gelation concentrations, cgc, at 0.3 wt%, but the hydrogel of 1 also selectively sequesters to 16S rRNA—the receptor of kanamycin A. Fluorescent titration confirms two molecules of 1 bind to the same site on the receptor (i.e., A-site of 16S rRNA1, 12). This work not only indicates that the hydrogel of aminoglycosides preserve the specific interaction with their macromolecular targets (e.g., 16S rRNA1), but also illustrates a feasible approach for exploring molecular self-assembly in water and screening the binding of the resulted supramolecular nanofibers/hydrogels to the potential target(s).
Scheme 2 shows the structure of the hydrogelators of kanamycin A, 1 and 2. The synthesis of 1 is straightforward. Being activated by N-hydroxysuccinimide (NHS), 2-(naphthalen-2-yl)acetamido)-3-phenylpropanamido)-3-phenylpropanoic acid (NapFF)13 reacts with kanamycin in the mixture of acetone and water (pH~8.0) for 24 hours to afford the crude product, which undergoes chromatographic purification to give 1 as white powders. Typically, 2 mg of 1 dissolves in 0.4 mL of water at pH 2.0 to result in a clear solution, which turns into a transparent hydrogel (Gel I) after the pH of the solution being adjusted to 7.0. The transparent hydrogel easily changes back to a clear solution when the pH is lowered to less than 6.5 or the temperature is increased to higher than 46 °C. The resulting transparent hydrogel (Fig. 1A) is stable at room temperature for months without visible changes.
Scheme 2.
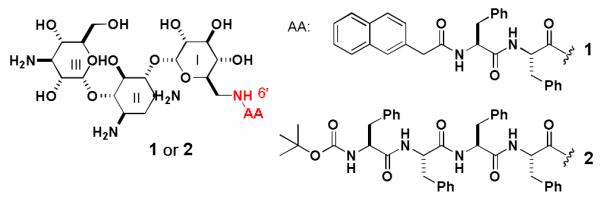
Structures of the hydrogelators based on kanamycin A.
Figure 1.
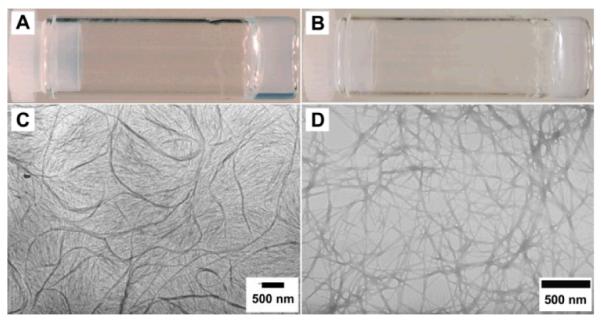
Optical images of (A) Gel I formed by 1 (0.5 wt%, pH7.0); (B) Gel II formed by 2 (1.0 wt%, pH7.0); TEM of (C) Gel I and (D) Gel II.
To study the nanostructures or morphology of the hydrogel, we obtained its transmission electron micrograph (TEM) (Fig. 1C), which shows small fibrils with the diameters from 50 nm to 60 nm. The elongated small fibrils (longer than 5 μm) tend to tangle with each other and form a dense network, which serves as the matrices of Gel I. According to the dynamic frequency sweep of Gel I, the values of the storage moduli (G’) exceed the loss moduli (G”) by a factor of about 5 (Fig. S2), indicating that the sample behaves like a solid.7 The value of G’ of the sample is higher than 600 Pa, indicating that the sample is viscoelastic. Both the values of G’ and G” of the hydrogel exhibit weak dependence with frequency (from 0.1 to 100 rad/s), suggesting that the matrices of the gel have a good tolerance to external force.
We used circular dichroism (CD) and fluorescent spectroscopy to investigate the molecular arrangement of the small molecule in Gel I (Fig. S1†). A positive band near 196 nm (ππ* transition) and a negative peak at about 208 nm (nπ* transition) reveal the β-sheet like conformation of the peptidic backbones. The broad peak centered at 246 nm (ππ* transition of phenyl groups), two small peaks at 286 nm (ππ* transition of naphthyl aromatics) and 296 nm (nπ* transition of naphthyl aromatics) originate from the aromatic parts of the molecules, which adopt helical arrangements induced by the peptidic backbone and kanamycin A. The emission spectrum of 1 (Fig. S1†) shows a peak centered at 336 nm in solution and at 342 nm in gel phase, indicating the monomeric naphthalene moieties. The additional peaks at 381 and 400 nm suggest strong interactions between the aromatic parts (naphthyl and phenyl aromatics) of the molecules, which is consistent with its self-assembled nanofibers observed in TEM image. Based on the above information and the crystal structure of kanamycin A12 and NapFF,13 we propose a possible molecular arrangement of 1 in the gel phase. As shown in Figure 2, NapFF motif plays an important role for the formation of supramolecular structures: the π-π interactions of aromatic rings and the intermolecular hydrogen bonds between peptide bonds help 1 self-assemble into a β-sheet like chain. The part of kanamycin A stick out to the aqueous environment and form hydrogen bonds with water molecules and other kanamycin molecules to help the formation of nanofibers. Although other modes of molecular arrangements may exist in the gel phase, the current evidences indicate that the arrangement in Figure 2A likely dominates.
Figure 2.
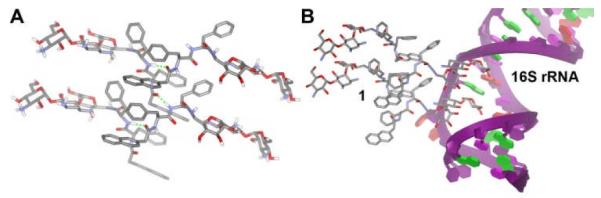
(A) A possible molecular arrangement of the hydrogelator (1) in the gel phase (some hydrogen bonds are shown as the dot lines). (B) A hypothetical binding between two self-assembled molecules of 1 and their receptor (A-site of 16S rRNA) derived from the crystal structure of kanamycin A12.
It is well established that kanamycin A bound to the A-site of 16S rRNA to lead to the misreading of mRNA and the death of the bacteria.1, 12, 14 To prove further that 1 binds to the A-site of 16S rRNA, we add a commercial oligonucleotide, 5μ́-UUGCGUCACACCGGUGAAGUCGC-3μ́ (i.e., A-site of 16S rRNA12), to the solution of 1 and observe the change of fluorescence upon binding. We choose to evaluate the binding in the PBS buffer to minimize the influence of the self-quenching of the naphthalene of 1 in the gel phase. Figure 3A clearly shows that the intensity of the fluorescent peak at around 336 nm decreases after the addition of the oligonucleotide. The fluorescent peak almost disappears after the addition of 7.33 equivalent of the oligonucleotide. Based on this fluorescent titration (Fig. 3A),15 the calculated binding number is 1.97 and the associate constant 4.0 × 109 M between 1 and the A-site of 16S rRNA. This result indicates two molecules of 1 bind to the same A-site of 16S rRNA, which agrees with the recently reported crystal structures of the interaction between ribosomal subunit and aminoglycoside antibiotics12, 14 that reveals two aminoglycosides to bind to the same A site of 16S rRNA.12 The association constant is much higher than the binding constant of kanamycin with 16S rRNA (5.6 × 105 M),16 suggesting cooperative binding.
Figure 3.
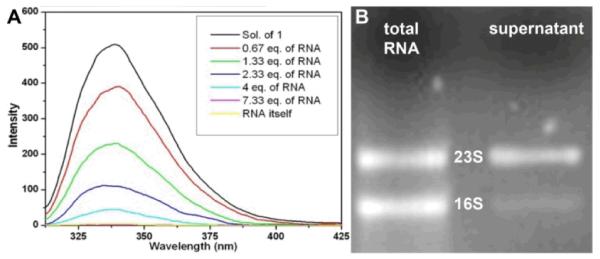
(A) Fluorescence spectra of solution of 1 (1 μg/mL) and mixtures of 1 with different equivalent of oligonucleotide (A site of 16S rRNA) in pbs buffer solution. (B) Gel electrophoresis of 16S rRNA and 23S rRNA in the extract before and after incubation with hydrogel of 1 for 4 hours.
To verify whether the hydrogel of 1 is able to bind selectively to the 16S rRNA, we extracted total RNAs from E. Coli and added the solution containing total RNAs on the top of the hydrogel of 1. Gel electrophoresis shows that 5S, 16S, and 23S rRNA are the major components of the total RNAs (Fig. S2, †), in which the initial ratio of 16S:23S:5S rRNA is about 1:1:8. The large quantity of 5S rRNA in the total RNA, together with 23S rRNA, serves as a useful control. After being incubated with the hydrogel of 1 for 4 hours, the amounts of RNAs in the supernatant decrease due to the absorption of RNA in the hydrogel of 1, as indicated by gel electrophoresis.† Quantitative analysis of the RNA bands in Figure 3B confirms that 20% of 16S rRNA and 70% of 23S rRNA remain in the supernatant. Despite the initial amount of 5S rRNA is about eight times of that of 16S rRNA (or 23S rRNA), 70% of 5S rRNA remains in the supernatant. This result suggests that the hydrogel of 1 selectively sequesters 16S rRNA. Because only 30% of 5S sRNA† being uptake by the hydrogel of 1 (though the size of 5S rRNA is much smaller and the quantity of 5S rRNA in the total RNA much larger than those of 16S rRNA), the observed selectivity unlikely arises from the smaller size of 16S rRNA compared to 23S rRNA. The above results clearly indicate that 1 binds to the A-site of 16S rRNA, thus leading to the hydrogel of 1 binds selectively to 16S rRNA. If the hydrogel of 1 bound extracted RNAs via only electrostatic attractions, the hydrogel of 1 would bind 23S rRNA that carries the most negative charges among the extracted RNAs. Therefore, the hydrogel of 1 selectively bind 16S rRNA, which also indicates that the derivatization at N6μ́ position unlikely completely eliminate the binding of 1 to A-site of 16S rRNA. Thus, we suggest that the self-assembled molecules of 1 to bind with 16S rRNA as shown in Fig. 2B, which is a possible interaction of 1 with A-site of 16S rRNA derived from the binding of kanamycin to the RNA groove without involving the ammonium N6μ́.12 Although the suggested model in Fig. 2B agrees with the 2:1 stoichiometry (between 1 and A-site of 16S rRNA) obtained from Fig. 3A, the atomistic description of the interactions between 1 and A-site of 16S rRNA has yet to be established.
In summary, we have designed and synthesized novel molecular hydrogelators based on kanamycin A and demonstrated a feasible approach to explore the potential targets of supramolecular nanofibers/hydrogels. One potential practical application of this hydrogel of kanamycin would be the enrichment of 16S rRNA from cell lysates for typing bacteria. However, the selectivity of the gel towards 16S remains to be improved, possibly by designing and screening new kanaymicin containing hydrogelators. The principle illustrated in this work should help identify new targets of supramolecular nanostructures,17 which may lead to new insights or phenomena that are inaccessible by an individual molecule.18 Our future works will focus on the modifications of other amino-functions of kanamycin A and other aminoglycosides.
Supplementary Material
Footnotes
Electronic Supplementary Information (ESI) available: The details of materials, synthesis, characterization, and measurements are available. See DOI: 10.1039/b000000x/
The authors acknowledge the partial financial support from HFSP and a start-up fund from Brandeis University and thank the assistance of Brandeis EM facility.
references
- 1.Brunton L, Lazo J, Parker K, Buxton I, Blumenthal D. The Pharmacological Basis of Therapeutics. McGraw-Hill; New York: 2005. [Google Scholar]
- 2.Estroff LA, Hamilton AD. Chem. Rev. 2004;104:1201. doi: 10.1021/cr0302049. [DOI] [PubMed] [Google Scholar]
- 3.Estroff LA, Hamilton AD. Angew. Chem. Int. Ed. 2000;39:3447. doi: 10.1002/1521-3773(20001002)39:19<3447::aid-anie3447>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]; Jung JH, John G, Masuda M, Yoshida K, Shinkai S, Shimizu T. Langmuir. 2001;17:7229. [Google Scholar]; Suzuki M, Yumoto M, Kimura M, Shirai H, Hanabusa K. Chem. Commun. 2002;884 doi: 10.1039/b200242f. [DOI] [PubMed] [Google Scholar]
- 4.Terech P, Weiss RG. Chem. Rev. 1997;97:3133. doi: 10.1021/cr9700282. [DOI] [PubMed] [Google Scholar]
- 5.Zhao F, Ma ML, Xu B. Chem. Soc. Rev. 2009;38:883. doi: 10.1039/b806410p. [DOI] [PubMed] [Google Scholar]
- 6.Williams RJ, Mart RJ, Ulijn RV. Biopolymers. 2010;94:107. doi: 10.1002/bip.21346. [DOI] [PubMed] [Google Scholar]; Ulijn RV, Baragana B, Halling PJ, Flitsch SL. J Am Chem Soc. 2002;124:10988. doi: 10.1021/ja026912d. [DOI] [PubMed] [Google Scholar]; Williams RJ, Smith AM, Collins R, Hodson N, Das AK, Ulijn RV. Nat. Nanotech. 2009;4:19. doi: 10.1038/nnano.2008.378. [DOI] [PubMed] [Google Scholar]; Kiyonaka S, Sada K, Yoshimura I, Shinkai S, Kato N, Hamachi I. Nat. Mater. 2004;3:58. doi: 10.1038/nmat1034. [DOI] [PubMed] [Google Scholar]; Adams DJ, Topham PD. Soft Matter. 2010;6:3707. [Google Scholar]; Sangeetha NM, Maitra U. Chemical Society Reviews. 2005;34:821. doi: 10.1039/b417081b. [DOI] [PubMed] [Google Scholar]
- 7.Yan CQ, Pochan DJ. Chem. Soc. Rev. 2010;39:3528. doi: 10.1039/b919449p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing BG, Ho PL, Yu CW, Chow KH, Gu HW, Xu B. Chem. Commun. 2003:2224. doi: 10.1039/b305886g. [DOI] [PubMed] [Google Scholar]
- 9.Xing BG, Yu CW, Chow KH, Ho PL, Fu DG, Xu B. J. Am. Chem. Soc. 2002;124:14846. doi: 10.1021/ja028539f. [DOI] [PubMed] [Google Scholar]
- 10.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington D, Kessler JA, Stupp SI. Science. 2004;303:1352. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]; Toledano S, Williams RJ, Jayawarna V, Ulijn RV. J. Am. Chem. Soc. 2006;128:1070. doi: 10.1021/ja056549l. [DOI] [PubMed] [Google Scholar]; Gao J, Wang HM, Wang L, Wang JY, Kong DL, Yang ZM. J. Am. Chem. Soc. 2009;131:11286. doi: 10.1021/ja9042142. [DOI] [PubMed] [Google Scholar]; Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. J. Am. Chem. Soc. 2002;124:15030. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]; Micklitsch CM, Knerr PJ, Branco MC, Nagarkar R, Pochan DJ, Schneider JP. Angew. Chem.-Int. Ed. 2011;50:1577. doi: 10.1002/anie.201006652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Y, Kuang Y, Guo ZF, Guo ZH, Krauss IJ, Xu B. J. Am. Chem. Soc. 2009;131:13576. doi: 10.1021/ja904411z. [DOI] [PubMed] [Google Scholar]; Wang HM, Yang CH, Wang L, Kong DL, Zhang YJ, Yang ZM. Chem. Commun. 2011;47:4439. doi: 10.1039/c1cc10506j. [DOI] [PubMed] [Google Scholar]
- 12.Francois B, Russell RJM, Murray JB, Aboul-ela F, Masquida B, Vicens Q, Westhof E. Nucleic Acids Research. 2005;33:5677. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Kuang Y, Gao YA, Xu B. Langmuir. 2011;27:529. doi: 10.1021/la1020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Nature. 2000;407:340. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 15.Chipman DM, Grisaro V, Sharon N. J. Biol. Chem. 1967;242:4388. [PubMed] [Google Scholar]
- 16.Misumi M, Nishimura T, Komai T, Tanaka N. Biochem. Biophys. Res. Commun. 1978;84:358. doi: 10.1016/0006-291x(78)90178-x. [DOI] [PubMed] [Google Scholar]
- 17.Gao Y, Long MJC, Shi JF, Hedstrom L, Xu B. Chem. Commun. 2012;48 doi: 10.1039/c2cc33631f. DOI: 10.1039/c2cc33631f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu ZL, Kurokawa T, Liang S, Furukawa H, Gong JP. J. Am. Chem. Soc. 2010;132:10064. doi: 10.1021/ja101969k. [DOI] [PubMed] [Google Scholar]; Casuso P, Carrasco P, Loinaz I, Grande HJ, Odriozola I. Org. Biomol. Chem. 2010;8:5455. doi: 10.1039/c0ob00311e. [DOI] [PubMed] [Google Scholar]; Kameta N, Yoshida K, Masuda M, Shimizu T. Chem. Mater. 2009;21:5892. [Google Scholar]; Price GM, Chu KK, Truslow JG, Tang-Schomer MD, Golden AP, Mertz J, Tien J. J. Am. Chem. Society. 2008;130:6664. doi: 10.1021/ja711340d. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tang MD, Golden AP, Tien J. Adv. Mater. 2004;16:1345. [Google Scholar]; Lovell JF, Roxin A, Ng KK, Qi QC, McMullen JD, DaCosta RS, Zheng G. Biomacromolecules. 2011;12:3115. doi: 10.1021/bm200784s. [DOI] [PubMed] [Google Scholar]; Liang GL, Ren HJ, Rao JH. Nature Chem. 2010;2:54. doi: 10.1038/nchem.480. [DOI] [PMC free article] [PubMed] [Google Scholar]; Choi SW, Zhang Y, Xia YN. Angew. Chem. Intl Ed. 2010;49:7904. doi: 10.1002/anie.201004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


