Abstract
In this paper we propose and provide evidence for a mechanism, oxidative nitrogen scavenging (ONS), whereby seedlings of some grass species may extract nitrogen from symbiotic diazotrophic bacteria through oxidation by plant-secreted reactive oxygen species (ROS). Experiments on this proposed mechanism employ tall fescue (Festuca arundinaceae) seedlings to elucidate features of the oxidative mechanism. We employed 15N2 gas assimilation experiments to demonstrate nitrogen fixation, direct microscopic visualization of bacteria on seedling surfaces to visualize the bacterial oxidation process, reactive oxygen probes to test for the presence of H2O2 and cultural experiments to assess conditions under which H2O2 is secreted by seedlings. We also made surveys of the seedlings of several grass species to assess the distribution of the phenomenon of microbial oxidation in the Poaceae. Key elements of the proposed mechanism for nitrogen acquisition in seedlings include: 1) diazotrophic bacteria are vectored on or within seeds; 2) at seed germination bacteria colonize seedling roots and shoots; 3) seedling tissues secrete ROS onto bacteria; 4) bacterial cell walls, membranes, nucleic acids, proteins and other biological molecules are oxidized; 5) nitrates and/or smaller fragments of organic nitrogen-containing molecules resulting from oxidation may be absorbed by seedling tissues and larger peptide fragments may be further processed by secreted or cell wall plant proteases until they are small enough for transport into cells. Hydrogen peroxide secretion from seedling roots and bacterial oxidation was observed in several species in subfamily Pooideae where seeds possessed adherent paleas and lemmas, but was not seen in grasses that lacked this feature or long-cultivated crop species.
Keywords: Grasses, Nitrogen fixation, Nitrogen scavenging, Reactive oxygen species, Symbiosis
Introduction
Non-pathogenic microbes are abundant on plants as epiphytes and endophytes (Klein et al. 1990; Stone et al. 2000; Tsavkelova et al. 2004). The benefits to plants that host these microbes are generally subtle, but they are gradually being discerned and clarified (Redman et al. 2002; Rodriguez et al. 2009; Puente et al. 2009; Torres et al. 2012). Often benefits are found to be defensive with microbes protecting plants from biotic stresses (White et al. 2001; Clay et al. 2005; Clarke et al. 2006; Selosse and Schardl 2007; Álvarez-Loayza et al. 2011; Bacon and Hinton 2011) or abiotic stresses (Redman et al. 2002; Malinowski et al. 2005; Waller et al. 2005; Kuldau and Bacon 2008; White and Torres 2010; Torres et al. 2012; Hamilton et al. 2012). Some bacterial epiphytes and endophytes are capable of nitrogen fixation (Reinhold-Hurek et al. 1993; Döbereiner 1992; Döbereiner et al. 1994; James et al. 1994; Hurek et al. 1994; Kloepper 1994; Puente and Bashan 1994; Feng et al. 2006; Magnani et al. 2010). Many of these microbes stimulate development of the plant host (e.g. Plant-Growth Promoting Rhizobacteria (PGPR)) (Kloepper 1993, 1994).
It has been proposed and often assumed that nitrogen fixed from the atmosphere by endophytic or epiphytic microbes may be utilized by host plants for growth (Döbereiner et al. 1994; Dakora et al. 2008). However, these microbes are frequently capable of producing plant growth regulators such as auxins and cytokinins (e.g., Glick 1995; Feng et al. 2006). Because of the production of plant growth regulators by many symbiotic microbes of plants, growth stimulation effects due to diazotrophic microbes may be attributed to effects of growth regulators rather than to nitrogen fixation by microbes (e.g., Bashan et al. 1989; Mantelin and Touraine 2004; Feng et al. 2006; Ortíz-Castro et al. 2008). The absence of any clear mechanism by which nitrogen fixed by bacteria may be transferred to the plant host further adds to the uncertainty as to whether plants obtain nitrogen from endophytic or epiphytic diazotrophic bacteria (Lethbridge and Davidson 1983; James and Olivares 1998; James 2000; Steenhoudt and Vanderleyden 2000). Without a nitrogen transfer mechanism, nitrogen assimilated into organic molecules by the bacterial populations on plants may simply remain in the microbial nitrogen pool rather than move to plants to support plant growth (Bremer et al. 1995; James and Olivares 1998; Zogg et al. 2000; James 2000). This is due in part to the nitrogen scavenging capacity of bacteria and the tenacity by which bacterial populations within or on the surfaces of plants hold onto fixed nitrogen (Hill and Postgate 1969; Hurek et al. 1988; Steenhoudt and Vanderleyden 2000). In this paper we report work we have conducted on grass seedlings with associated seed-transmitted bacteria and provide evidence for a mechanism for the transfer of fixed nitrogen from bacteria to seedlings via oxidation of the microbes and/or their protein products. We term this mechanism, oxidative nitrogen scavenging (ONS).
Materials and methods
Source of tall fescue seeds
For experiments described in 2.2–2.6 below, we used one-year old seeds of tall fescue grass (Festuca arundinaceae) that were originally collected in Morocco and subsequently maintained at the Rutgers University Turfgrass Research Station in Adelphia, New Jersey. We replicated experiments using various commercially available tall fescue seeds to confirm the presence of diazotrophic bacteria on seeds and H2O2 secretion from seedlings.
Isolation and identification of bacteria from tall fescue seeds
To isolate and identify bacteria, we placed 4 non-disinfected seeds (caryopses) onto each of five petri plates (15 cm. diam.) containing potato dextrose agar (PDA; Difco, Becton, Dickenson and Company, Sparks, MD). Using 980 base pairs of the 16S rDNA region (Baker et al. 2003) we identified four of five morphologically unique isolates to Pantoea agglomerans; and a fifth isolate to Pseudomonas sp. We submitted representative sequences to NCBI (GenBank JX089400 and JX089401).
Effects of light and bacteria on H2O2 secretion of tall fescue seedlings
To assess the effect of light and microbial presence on seedling H2O2 secretion, we conducted peroxide secretion experiments using the following treatments: 1.) 12 h alternating light/dark with surface disinfected seeds; 2.) 12 h alternating light/dark with non-disinfected seeds; 3.) dark (24 h) with surface disinfected seeds; 4.) dark (24 h) with non-disinfected seeds. In each of these treatments we used 5 plates (approximately 20 seeds/plate) containing 1 % agarose media. For the disinfected seed treatments, seeds were disinfected for 30 min using 50 % Clorox® followed by a 30 min rinse in sterile distilled water. We incubated all plates at ambient temperature for 7 days to facilitate seed germination. On the eighth day after plating, we flooded plates with 5 ml solution of 100 mM potassium phosphate buffer, pH 6.9, 2.5 mM diaminobenzidine tetrachloride and 5 purpurogallin units/ml of horseradish peroxidase (Type VI, Sigma Chemical Company, St. Louis, MO) (Munkres 1990; Pick and Keisari 1980). Plates were incubated at 30 °C in dark for 1 h before the solution was discarded. After 10 h, we rinsed the plates in sterile, distilled water, and examined them for red diffusible zones due to H2O2 around the seedlings. We then rated the intensity of the H2O2 zones as follows: 0 = no zones; + = low intensity or lighter zones; ++ = moderate intensity zones; +++ = high intensity or larger zones).
Effects of nutrient environment on H2O2 secretion of tall fescue seedlings
We also sought to determine the effect(s) of the medium nutrient environment on seedling H2O2 secretion. We amended 1 % (w/v) agarose (Type 1 Low EEO; Sigma-Aldrich, St. Louis, MO) media with 0.01 % (w/v) and 0.1 % (w/v), respectively, of L-alanine (Sigma, St. Louis, MO), glycine (Sigma, St. Louis, MO), yeast extract (Bacto™; Difco, Becton, Dickenson and Company, Sparks, MD), ammonium nitrate (Sigma, St. Louis, MO), sodium nitrate (Sigma, St. Louis, MO), and ammonium phosphate (Dibasic; Fisher Scientific Company, Fair Lawn, NJ). Media amendments were added after autoclaving to avoid thermal decomposition of amendments. In this experiment we replicated each treatment using 5 plates (20 seeds that had not been surface disinfected on each plate). In a separate experiment we prepared two sets of 5 plates: 1) 1 % (w/v) agarose media; and 2) 1 % agarose amended with 0.1 % (w/v) cellulase enzyme (from Aspergillus niger; Sigma, St. Louis, MO). To inactivate the cellulase enzyme we boiled the medium for 30 min prior to cooling and pouring into plates. In this experiment we used seeds that had been surface disinfected (see Section 2.3) to reduce bacterial populations on seedlings. After 7 days of incubation at laboratory ambient temperature with a 12 h alternating light/dark cycle, we stained the plates with DAB/horseradish peroxidase and evaluated them as described in Section 2.3 above. These results are summarized in Table 1.
Table 1.
Summary of substrate effects on seedling secretion of hydrogen peroxide into agarose media
| Agarose medium amendment | Seed treatment prior to germination | Percentage of seedlings showing H2O2 secretion zonesa | Average intensity of H2O2 zonesb |
|---|---|---|---|
| Nonec | Disinfection | 23.08 | + |
| None | None | 100 | +++ |
| Yeast Extract (0.01 %) | None | 100 | +++ |
| Yeast Extract (0.1 %) | None | 54 | ++ |
| Alanine (0.01 %) | None | 96.3 | +++ |
| Alanine (0.1 %) | None | 21.05 | + |
| Glycine (0.01 %) | None | 88 | +++ |
| Glycine (0.1 %) | None | 40 | + |
| Inactivated Cellulase (0.1 %) | Disinfection | 100 | +++ |
| Ammonium Nitrate (0.01 %) | None | 86.2 | +++ |
| Ammonium Nitrate (0.1 %) | None | 78.13 | +++ |
| Potassium Nitrate (0.01 %) | None | 76.67 | +++ |
| Potassium Nitrate (0.1 %) | None | 86.67 | +++ |
aAverage of 100 seedlings assessed
b+ = low intensity or lighter zones, +++ is higher intensity or darker zones, and ++ are intermediate intensity zones
cControl from cellulase enzyme amendment experiment
Visualization of bacterial oxidation on tall fescue seedlings
To visualize bacterial oxidation on seedlings, we stained plates bearing seedlings with DAB/horseradish peroxidase for a 10 h period. We then excised seedling roots and shoots, placed them on a slide containing 1 % aniline blue/lactic acid stain, and examined the slide using bright field microscopy (Bacon and White 1994). On a second set of slides, we stained seedling roots (to visualize nucleic acids during oxidation) with SYTO9® (Life Technologies, Carlsbad, CA) and observed them using fluorescence microscopy on a Zeiss Axioskop (using the FITC filter system (range 430–520 nm for Fig. 2a and b; and blue violet excitation (range 395–440 nm) for Fig. 2c and d).
Fig. 2.
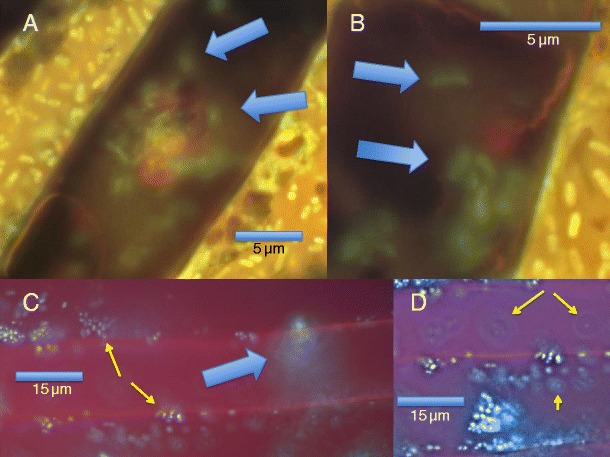
a–d. Root tissues and bacteria stained with SYTO9 florescent nucleic acid stain. a and b Root hairs showing epiphytic swollen oxidizing bacteria (arrows) with intact bacteria in background; c Root parenchyma showing intensely staining intact bacteria (yellow arrows) and masses of oxidizing bacteria (blue arrow); d Root parenchyma showing nucleic acid rings (bulls-eye rings) around oxidizing bacteria (arrows)
Nitrogen assimilation of tall fescue seedlings
To assess nitrogen assimilation by seedlings, we placed surface-disinfected seeds or non-disinfected seeds (approximately 20/plate) onto 5 petri plates for each treatment containing 1 % (w/v) agarose media, and then placed the plates into a 1-liter nitrogen assimilation chamber (constructed using a sealable glass jar with gas input ports) into which we added 15N2 gas and incubated at laboratory ambient temperature. We repeated this experiment three times adding 20 mls of 15N2 gas in run 1, 125 mls of 15N2 gas in run 2, and 250 mls 15N2 gas in run 3. After a 3-week incubation period under 12 h alternating light/dark cycle we removed the plates from the chamber, washed the seedlings with sterile, distilled water and excised shoots from the seedling roots. Experimental controls included a shoot sample and three separate samples of seeds and agarose media that had not been exposed to 15N2 gas. For mass-spectroscopic 14N/15N ratio analysis, we oven dried all samples for 24 h at 80 °C. After drying, we sent 0.9-1.0 g of dried material to the Stable Isotope/Soil Biology Laboratory at the Odum School of Ecology at the University of Georgia for analysis (Radajweski et al. 2000). Results of the isotopic nitrogen analysis are reported in Table 2.
Table 2.
15N isotopic analysis results for 15N enriched and non-enriched plants after 3 weeks growth
| Treatment | Sample wt (mg) | Total % nitrogen | δ15N vs (‰) | Atoms % 15N |
|---|---|---|---|---|
| Agarose media (no15N controls) | 1.268 | 0 | N/Aa | N/Aa |
| 1.750 | 0 | |||
| 1.231 | 0 | |||
| Seeds (no 15N controls) | 1.303 | 2.10 | 0.70 | 0.366729 |
| 1.082 | 2.00 | 0.64 | 0.366705 | |
| 1.325 | 2.11 | 0.79 | 0.366761 | |
| Shoots (no 15N Control) | 1.102 | 3.82 | 2.02 | 0.367209 |
| Run 1 sterilized (20 mls 15N gas) | 1.412 | 3.78 | 17.05 | 0.372697 |
| Run 1 non-sterilized (20 mls 15N gas) | 1.733 | 3.50 | 23.49 | 0.375049 |
| Run 2 sterilized (125 mls 15N gas) | 1.091 | 2.95 | 194.18 | 0.437322 |
| Run 2 non-sterilized (125 mls 15N gas) | 1.049 | 2.91 | 214.09 | 0.444582 |
| Run 3 sterilized (250 mls 15N gas) | 1.962 | 4.21 | 152.92 | 0.422274 |
| Run 3 non-sterilized (250 mls 15N gas) | 1.250 | 3.2 | 385.67 | 0.507094 |
aBelow detection limit
Survey of grass species for seedling root secretion of H2O2 and bacterial oxidation
To evaluate how widespread the phenomenon of microbial oxidation on seedling roots is we screened seedlings of several species in agarose media (Table 3). Species screened included species in genera Bromus, Festuca, Lolium, Phalaris, Poa, Secale, Sorghum, Triticum and Zea. In these experiments 3–6 non-disinfected seeds of each species were placed onto each of 3 plates of agarose media. After 7–14 days of incubation at laboratory ambient temperature with a 12 h alternating light/dark cycle, we stained the plates by flooding with DAB/horseradish peroxidase. After 10 h we rinsed the plates with water, evaluated presence of H2O2 zones, and rated intensity of H2O2 zones as described in Section 2.3, above. We also noted any enhanced H2O2 staining in root or shoot tissues. We then excised seedling roots and placed them on a slide containing 1 % aniline blue/lactic acid stain. We examined root and shoot surfaces using bright field microscopy for any evidence of bacterial oxidation. Changes in bacterial shape, loss of integrity of bacterial cell walls, and loss of capacity to stain using the protein stain aniline blue was taken as evidence of bacterial oxidation. These results are summarized in Table 3.
Table 3.
Survey of grass species for hydrogen peroxide secretion from seedlings and bacterial oxidation
| Plant species/‘Common Name’ | Plant family/subfamily | Collection origin/location | Typical habitat | H2O2 intensity on/or around seedling roots | Evidence of bacterial oxidation on roots |
|---|---|---|---|---|---|
| Bromus tectorum/‘cheatgrass’ | Poaceae/Pooideae | South River, New Jersey | Meadow | +++1 | Yes |
| Festuca arundinaceae/‘Tall fescue grass’ | Poaceae/Pooideae | Adelphia, New Jersey, USA | Meadow | +++ | Yes |
| Festuca ovina/‘sheep’s fescue’ | Poaceae/Pooideae | Helsinki, Finland | Meadow | ++ | Yes |
| Lolium perenne/‘Perennial ryegrass’ | Poaceae/Pooideae | Adelphia, New Jersey, USA | Meadow | +++ | Yes |
| Poa annua/‘Annual bluegrass’ | Poaceae/Pooideae | Adelphia, New Jersey, USA | Meadow | +++ | Yes |
| Poa pratensis ‘Kentucky bluegrass’ | Poaceae/Pooideae | Adelphia, New Jersey, USA | Meadow | +++ | Yes |
| Phalaris arundinacea ‘Reed canary grass’ | Poaceae/Pooideae | Chatsworth, New Jersey, USA | Wetland | 0 | No |
| Secale cereale ‘Rye’ | Poaceae/Pooideae | Chatsworth, New Jersey, USA | Cultivated | 0 | No |
| Tritucum aestivum ‘Wheat’ | Poaceae/Pooideae | Chatsworth, New Jersey, USA | Cultivated | 0 | No |
| Sorghum bicolor/‘Sorghum’ | Poaceae/Panicoideae | New Brunswick, New Jersey, USA | Cultivated | 0 | No |
| Zea diploperennis/‘Teosinte’ | Poaceae/Panicoideae | Tucson, Arizona, USA | Cultivated | 0 | No |
| Zea mays/‘Indian corn’ | Poaceae/Panicoideae | New Brunswick, New Jersey, USA | Cultivated | 02 | No |
| Zea mays/‘Sweet corn’ | Poaceae/Panicoideae | New Brunswick, New Jersey, USA | Cultivated | 0 | No |
10 = no evidence of H2O2 secreted from roots, + = low intensity or lighter zones around roots, +++ is higher intensity or darker zones around roots, and ++ are intermediate intensity zones
2H2O2 staining in the plant tissues but not showing substantial secretion into agarose medium
Results
Effects of light and bacterial presence on H2O2 secretion by tall fescue seedlings
We found the highest production of H2O2 in the non-disinfected seedlings maintained under light conditions (intensity = +++). Non-disinfected seedlings maintained in dark conditions showed a reduction in H2O2 production (intensity = ++). Disinfected seedlings maintained in light conditions showed low levels of H2O2 production (intensity = +). Disinfected seedlings maintained in dark conditions showed no or negligible H2O2 production (intensity = 0). Seeds of tall fescue obtained from commercial sources yielded the same results in this experiment.
Effects of nutrient environment on H2O2 secretion by tall fescue seedlings
The results of these experiments are summarized in Table 1. We found that the amino acids alanine, glycine and yeast extract, reduced H2O2 production at the 0.1 % concentrations as evidenced by reductions in the percentage of seedlings showing H2O2 secretion and intensity of H2O2 secretion zones when compared to agarose only controls or the 0.01 % concentrations of each nitrogen source (Table 1). None of the inorganic nitrogenous compounds that we evaluated showed notable differences in overall seedling H2O2 secretion intensity between the 0.01 and 0.1 % concentrations, although there was a slight decrease (ranging from 13 to 23 % lower than agarose alone controls) in the percentage of seedlings showing H2O2 secretion in either 0.01 % or 0.1 % levels depending on the compound (Table 1). When compared to agarose alone controls, we found that 0.1 % cellulase enzyme stimulated a marked increase in the secretion of H2O2 with dense zones forming around seedling roots (Table 1). This stimulation of H2O2 secretion was enhanced in seedlings generated from surface disinfected seeds. Media containing agarose alone gave only diffuse H2O2 zones around roots.
Visualization of bacterial oxidation on tall fescue seedlings
In microscopic examinations of seedlings using DAB/peroxidase we detected areas of high oxidation activity around bacteria. By counterstaining tissues with aniline blue, it was possible to visualize intact bacteria on plant tissues. During oxidation, bacteria lost capacity to stain with aniline blue/lactic acid. Instead, the bacteria swelled and became spherical or amorphous with diffuse walls. Eventually the bacteria vanished on the surface of plant tissues (Fig. 1). Experiments using seeds of tall fescue obtained from commercial sources gave the same results in this study. Through application of the florescent nucleic acid stain SYTO9®, we were able to observe swollen, oxidizing bacteria become amorphous, with nucleic acids dimming and diffusing away from the original site of the oxidized bacteria. We frequently visualized ‘bulls-eye rings’ around degraded bacteria (Fig. 2d). We characterize these rings as being composed of partially degraded fragments of nucleic acids that adhere to plant cell walls. More intense secretion of H2O2 in the vicinity of the bacterial cells may result in the clearing of nucleic acids nearest to the original bacterial sites, thus producing the ‘bulls-eye ring’ effect.
Fig. 1.
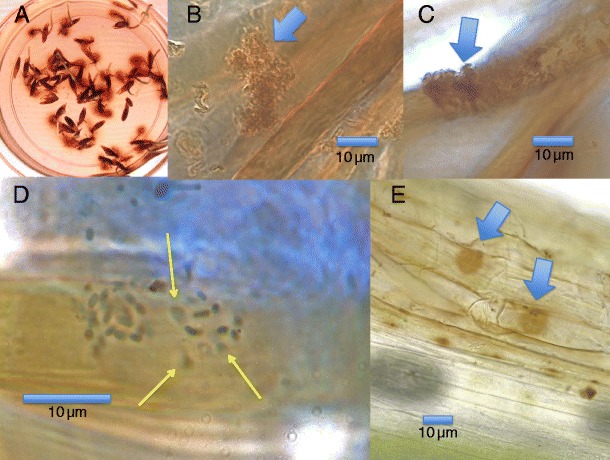
a–e Tall fescue seedlings stained with DAB. a Fescue seedlings on agarose showing H2O2 secretion (red coloration) from roots; b and c Fescue root hair showing bacterial cells staining brown due to H2O2 concentration (arrows); d Fescue root hair showing bacterial cells losing rod structure and capacity to stain with aniline blue during oxidation (arrows; stained with DAB/peroxidase, then counterstained with 0.1 % aniline blue; e Seedling mesocotyl showing brown areas (arrows) that mark sites of bacterial oxidation
Nitrogen assimilation by tall fescue seedlings
In 15N2 gas assimilation experiments (Table 2), mass spectroscopic analysis showed that tall fescue seedlings incorporated 15N into tissues. Greatest incorporation was observed in seedlings that had not been surface sterilized in all three runs of the experiment. Incorporation of 15N into seedlings seemed to correlate with the amount of 15N2 gas placed into the experimental chambers. Seeds or seedling material that had not been exposed to 15N2 gas showed minimal 15N content, ranging from 0.64 to 2.02 delta over air. Agarose media analyzed was not found to contain detectible nitrogen and 15N/14N ratios could not be calculated.
Survey of grass species for seedling root secretion of H2O2 and bacterial oxidation
Hydrogen peroxide secretion from seedling roots was seen in pooid grasses Bromus tectorum, Festuca arundinaceae, Festuca ovina, Lolium perenne, Poa annua and Poa pratensis. Bacteria on seedling root hairs were often observed to oxidize as described in 3.3, above. Other grasses screened, including Phalaris arundinacea, Secale cereale, Triticum aestivum, Sorghum bicolor, Zea diploperennis and Zea mays did not show H2O2 secretion in agarose media. Data are summarized in Table 3.
Discussion
Bacterial induction of H2O2 secretion by seedlings
We repeatedly isolated Pantoea agglomerans from non-disinfected tall fescue seeds. However, a Pseudomonas sp. was also occasionally isolated. It is evident that seed coats of the line of tall fescue that we employed in these experiments harbor a mixed population of Proteobacteria. Because many bacteria are capable of colonization of the exterior surfaces of grass plants, many different bacteria could be present on grass seed coats and their adherent lemmas and paleas. Pantoea agglomerans fixes nitrogen and has previously been implicated as a plant growth promoting bacterium (Feng et al. 2006; Yashiro et al. 2011; Johnston-Monje and Raizada 2011). Several diazotrophic pseudomonads that closely match sequences of our fescue seed isolate have also been shown to be associated with the rhizospheres of grasses (Behrendt et al. 2003). Both P. agglomerans and Pseudomonas sp. were capable of growth on Norris Nitrogen-Free Agar (data not shown), suggesting that both are diazotrophic. During germination of the seed, bacteria were seen to colonize seedling roots and shoots. Our experiments show that plant seedling root tissues secrete H2O2 and perhaps other ROS that oxidize bacterial cells, their nucleic acids and likely proteins. After reduction of bacterial populations on surfaces of seeds using sodium hypochlorite disinfection, seedlings grown on agarose medium free of any nutrients appeared to show reduced secretion of H2O2. However, tall fescue seedlings grown in agarose medium amended with heat inactivated cellulase enzyme demonstrated enhanced secretion of H2O2 into the medium around seedling roots (Table 1). We conducted similar experiments using 0.1 % (w/v) Albumen Fraction V (from bovine serum; Merck Chemicals, Darmstadt, Germany) and this protein similarly enhanced H2O2 secretion from seedlings (data not shown). This suggests that enhancement of H2O2 secretion by proteins may occur regardless of the specific protein used. Bacteria on the surface of plant seedlings may secrete enzymes to degrade cellulosic cell walls, pectins, or other components of the plant (Jayani et al. 2005). These enzymes may then be deactivated and degraded by ROS secreted by the grass seedling.
Nitrogen fixation by microbes on grass seedlings
In the three 15N2 gas assimilation experiments involving tall fescue seedlings, we found that bacteria on or within seedlings assimilated 15N2 gas (Table 2). Mass spectroscopic analysis showed that tall fescue seedlings (delta 15N vs air = 17.05 to 385.67) assimilated 15N2 gas when compared to seeds and seedling controls that had not been exposed to 15N2 gas (delta 15N vs air = 0.64–2.02). Because nitrogenase enzymes typically function under low oxygen conditions, it is possible that bacteria fix nitrogen on the seed coat during early seedling expansion before photosynthetic activities commence (Dakora et al. 2008) or during dark periods when ROS secretion may be lower. Alternatively some bacteria may adapt to presence of ROS by formation of resistant biofilms or production of ROS scavengers to shelter nitrogenase enzymes (Watson and Schubert 1969). Surface disinfection of seeds reduced incorporation of 15N into seedlings but it did not eliminate it (Table 2). Johnston-Monje and Raizada (2011) reported that Pantoea agglomerans is endophytic in numerous ancestral and modern lines of corn; and Feng et al. (2006) reported that this bacterium is endophytic in rice. Because of these reports, it seems possible that this bacterium may become endophytic in tall fescue. However, we did not visualize it within seedling tissues. It is also possible that some surface bacteria may escape the surface disinfection procedure by embedding in the seed coat or forming dry biofilms in layers under or within paleas and lemmas. Continued presence of some diazotrophic bacteria on or within tall fescue seedlings could account for the 15N delta over air of 17.05 to 214.09 in seedlings from surface sterilized seeds, which suggests significant nitrogen fixation over that seen in the non-15N enriched seed and seedling controls (delta over air = 0.64 to 2.02).
Factors affecting hydrogen peroxide secretion by seedlings
Our experiments suggest that secretion of H2O2 by tall fescue seedlings may be a regulated process. In experiments summarized in Table 1, we observed that seedlings generated from surface disinfected seeds with reduced levels of external seed coat bacteria appeared to secrete less H2O2 from roots than seedlings from seeds that were not surface disinfected. This outcome suggests that plants may be responding to the presence of bacteria by secreting ROS proportional to the amount of bacteria on plants. This conclusion is further supported by ROS activity visualization on seedling shoot tissues where we observed staining for ROS activity only at sites of bacterial oxidation (Fig. 1e).
Factors such as light and nutrient environment of seedlings may also impact H2O2 secretion by seedlings. We observed that seedlings grown under dark conditions produced less H2O2 than seedlings maintained under light conditions. Reduction of ROS secretion under dark conditions might permit bacteria to more efficiently employ oxygen-sensitive nitrogenase enzymes to fix atmospheric nitrogen at night while high ROS secretion in light may result in greater harvesting of the fixed nitrogen. In another experiment (Table 1), we observed that the amount of ROS secreted by seedlings could be reduced or stopped completely by inclusion 0.1 % of organic forms of nitrogen (e.g., amino acids or yeast extract) into the media around seedlings, suggesting that H2O2 secretion may show feedback control where seedling tissues with sufficient organic nitrogen discontinue ROS secretion. Inorganic forms of nitrogen, even at very low levels (0.01 %), tended to reduce the number of seedlings showing H2O2 secretion (Table 1). This may be an indication that inorganic nitrogen application to plants may negatively impact the proposed oxidative system for acquisition of organic nitrogen from microbes. In experiments where we disinfected seeds to decrease bacterial populations, yeast extract and amino acids reduced H2O2 secretion from roots. The full significance of these observations will require additional experimentation using inorganic and organic sources of nitrogen. However, taken collectively, the results summarized in Table 1 support the hypothesis that the function of H2O2 secretion by seedlings relates to organic nitrogen acquisition.
ROS effects on bacterial cells
Visualization of bacterial oxidation on plant tissues suggests that oxidation does not immediately involve complete oxidation to inorganic forms. Instead, a partial oxidation of bacteria and their constituent compounds occurs, likely denaturing cell membranes and walls. Disarticulation of larger organic compounds, like proteins and nucleic acids, into smaller fragments follows. This phenomenon may be evidenced by bulls-eye rings around oxidizing bacteria when the nucleic acid probe SYTO9® (Fig. 2) is applied. The bulls-eye rings may result from diffusion of nucleic acid fragments and their adsorption to plant cell walls. More intense secretion of H2O2 in the vicinity of the bacterial cell may result in clearing of nucleic acids nearest the original site of the bacterium resulting in the bulls-eye-ring effect.
Investigations into the effects of ROS on bacterial cells are consistent with our observations and hypotheses (Cabiscol et al. 2000). In a review on ROS damage to bacterial cells, Cabiscol et al. (2000) report that nucleic acids, proteins and lipids are impacted by ROS. H2O2 produces highly reactive hydroxyl radicals via the Fenton reaction that occurs in the presence of metal ions. Lipids in bacterial walls and membranes are major targets where polyunsaturated fatty acids are peroxidized and degraded. The decomposition of the cell walls of bacteria, composed of lipids and proteins, result in loss of the cell shape. In nucleic acids ROS attacks both the nitrogenous base and sugar moieties, producing single- and double-strand breaks in the nucleic acid backbone. Proteins are denatured with alterations to amino acids and frequently show peptide fragmentation (Cabiscol et al. 2000). ROS denatured proteins show enhanced susceptibility to protease degradation (Galek et al. 1990). Van der Valk and Van Loon (1988) demonstrated that cell walls of oat leaves possess extracellular acidic proteases and Godlewski and Adamczyk (2007) demonstrated that plants secrete proteases from their roots. Similar extracellular proteases in tall fescue seedlings could contribute to the process of protein degradation on seedling roots and shoots. This process could occur regardless of whether bacteria are epiphytic on plant tissues or are endophytes within tissues.
Role of ROS in oxidation/digestion of microbes and their proteins for defense and nutrition
Generally, scientists consider plant-secreted ROS, such as H2O2, to be destructive metabolic byproducts of cellular respiration and photosynthesis that plants must manage internally to prevent cellular damage. Certain ROS moieties also function as signaling molecules, mediating defensive or other plant reactions (D’Autréaux and Toledano 2007). Foreman et al. (2003) demonstrated that H2O2 is produced by plasmalema NADPH oxidases and may be secreted into apoplastic spaces in planta (see also Dunand et al. 2007). Frahry and Schopfer (1998) demonstrated that H2O2 is secreted from roots of soybean (Glycine max). The proposal that ROS plays a role in oxidation of plant microbes or their protein products to extract nitrogen is a new concept in plant biology that will undoubtedly require additional confirmation. However, in animals, ROS are known to be involved in the killing and degradation action of animal phagocytic leukocytes in oxygen-dependent defense (Robinson 2008). In phagocytic leukocytes the killing and degradation action of H2O2 results from its capacity to form more potent ROS moieties, including hydroxyl radicals, singlet oxygen, ozone (Robinson 2008). A similar process could also be occurring on or within plant tissues. It is known that plants also secrete H2O2 defensively in ‘oxidative bursts’ at sites of pathogen colonization (Lamb and Dixon 1997). This defensive ROS is known to have a direct deleterious effect in killing microbial pathogens. ROS has additionally been shown to be involved in nutritive phagocytosis in starfish and other simple animals (Coteur et al. 2002). In carnivorous pitcher plants, plant-secreted ROS plays a nutritional role in digestion of insect proteins (Chia et al. 2004). Galek et al. (1990) similarly reported that the ‘venus flytrap’ (Dionaea muscipula) employs ROS to pre-digest insect proteins prior to protease action on the denatured peptide fragments.
We hypothesize that the oxidation process occurring on seedlings is comparable to that seen in some forms of phagocytosis in animals and nutritive digestion as seen in carnivorous plants. We suggest that ROS secreted from seedling tissues function as ‘digestive agents’ that play a role in degradation of microbes and their protein products to obtain nitrogen for the plant host. Proteases secreted by plant roots (Godlewski and Adamczyk 2007) or maintained in plant cell walls (Van der Valk and Van Loon 1988) may contribute to the process of microbial protein degradation. Amino acid or oligopeptide transporters in plant roots may transport products into root cells (Kielland 1994; Jamtgard et al. 2008). Even without the action of protease enzymes, ROS may be effective in oxidizing proteins to smaller oligopeptides or amino acids that may be assimilated by the seedlings (Hill et al. 2011). Investigations by Kocha et al. (1997) demonstrated that hydrogen peroxide-mediated protein degradation occurs without the presence of proteases or other catalyzing enzymes. Through H2O2-mediated non-specific degradation of proteins, smaller peptide fragments and eventually nitrates (Palenik and Morel 1990) may form on seedling tissue surfaces. Bacterial proteins may provide a significant amount of the nitrogen needed by developing grass seedlings. When it is considered that rapidly growing seedlings have a high requirement for nitrogen and that their roots are not yet fully developed for efficient nitrogen absorption from soils, it is not unexpected that some plants would evolve a mechanism to scavenge nitrogen from the diazotrophic bacteria that rapidly colonize seedlings during germination. To describe the process whereby plants may oxidize microbes or their proteins to obtain nitrogen we propose the term ‘oxidative nitrogen scavenging’ (ONS).
Grass species that show ONS by seedlings
In our survey (Table 3) of grass seedlings to identify additional species that show ONS, the phenomenon was most notable in several grass species of subfamily Pooideae, including Bromus tectorum, Festuca arundinaceae, Festuca ovina, Lolium perenne, Poa pratensis and Poa annua. Phallaris arundinacea, also subfamily Pooideae, did not show H2O2 secretion. The highly selected and long cultivated grasses wheat, rye, sorghum and corn did not show evidence of H2O2 secretion (Table 3). In species where we report the phenomenon of seedling H2O2 secretion and bacterial oxidation (Table 3), seeds possess adherent plant tissues (dry paleas and lemmas) that disperse with the seeds. It is possible that seeds are colonized by bacteria on their paleas and lemmas during the period of maturation prior to their release from plants. Thus, seed coat adherent paleas and lemmas may function to vector bacteria to seedlings. Phallaris arundinacea, wheat, rye, sorghum and corn possess seeds that are free of adherent paleas and lemmas and thus may not be adapted to vector bacteria of their seed coats. It is possible that ancestral forms of wheat, rye, sorghum and corn possessed seed-transmitted diazotrophic microbes but that these have been lost in cultivation. However, if ancestors of crop species possessed the capacity for ONS it may be possible to improve modern crops by re-establishing these symbioses to reduce the need for external nitrogen inputs.
Many of the grasses that we have identified as showing enhanced H2O2 secretion from roots are known for their competitive abilities and have come to be used as forage, conservation or turf grasses. These species include the fescues (Festuca arundinaceae and F. ovina), perennial ryegrass (Lolium perenne), annual bluegrass (Poa annua) and Kentucky bluegrass (Poa pratensis). Poa annua and Bromus tectorum are both generally aggressive and often ‘invasive’ (Sperry et al. 2006; Chwedorzewska 2008). It is possible that the competitive abilities of these grass species may be, at least in part, due to their capacity to extract nutrients from bacteria. Further, oxidative systems to extract nutrients from microbes may not be limited to grasses. Many other plants are known to thrive under conditions where nitrogen is difficult or impossible to extract from soils. Future work will need to evaluate the existence and importance of oxidative nitrogen scavenging systems in enabling plants in diverse families to grow in nitrogen limited habitats.
Conclusions
The proposal that some plants possess a system to extract nitrogen from microbes through oxidation is new, but not unreasonable given that plants possess the basic metabolic machinery (e.g., NADPH oxidases for ROS generation, proteases, oligopeptide and amino acid transporters) needed for such a process (Kielland 1994; Godlewski and Adamczyk 2007; Jamtgard et al. 2008). Further, many plants are known to be rich sources of diazotrophic microbes that inhabit much of their exterior and interior surfaces (Arnold and Lutzoni 2007; Reinhold-Hurek and Hurek 2011). A capacity of some plants to directly extract nitrogen from symbiotic bacteria would alter our longstanding view of the role of these ubiquitous microbes in and on those plants. Rather than being casual or opportunistic inhabitants of plants, diazotrophic microbes may represent a reservoir of fixed nitrogen that plants cultivate when available soil nitrogen is abundant and harvest through oxidation in circumstances when nitrogen cannot be extracted from the soil. Definitive validation of ONS in plants would also alter our view of how plants gain nutrients in demonstrating ‘microbivory’ as an option for nutrient acquisition. This is a nutritional mode where typically small animals or protists consume bacteria as a nutrient source (Mikola 1998). We are not the first investigators to suggest that plants may show microbivory. Paungfoo-Lonhienne et al. (2010) presented evidence that some plants appear to phagocytize bacteria as a nutrient source. Confirmation of the functioning and importance of the proposed ONS mechanism will require additional experimentation to track the flow of nitrogen into cells and tissues and evaluate the precise roles of ROS, plant proteases and plant cell transporters in the degradation and absorption process.
Acknowledgements
We are grateful to the New Jersey Agricultural Experiment Station and the Rutgers University Turf Science Center for resources and financial support.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- Álvarez-Loayza P, White JF, Jr, Torres MS, Balslev H, Kristiansen T. Light converts endosymbiotic fungus to pathogen, influencing seedling survival and niche-space filling of a common tropical tree, Iriartea deltoidea. PLoS One. 2011;6(1):e16386. doi: 10.1371/journal.pone.0016386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology. 2007;88:541–549. doi: 10.1890/05-1459. [DOI] [PubMed] [Google Scholar]
- Bacon CW, Hinton DM. In planta reduction of maize seedling stalk lesions by the bacterial endophyte Bacillus mojavensis. Can J Microbiol. 2011;57:1–8. doi: 10.1139/w11-031. [DOI] [PubMed] [Google Scholar]
- Bacon CW, White JF Jr (1994) Stains, media, and procedures for analyzing endophytes. In: Bacon CW, White JF Jr (eds) Biotechnology of endophytic fungi of grasses, CRC Press, pp. 47–56.
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J. Microb. Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Bashan Y, Singh CS, Levanony H. Contribution of Azospirillum brasilense Cd to growth of tomato seedlings is not through nitrogen fixation. Can. J. Botany. 1989;67:2429–2434. doi: 10.1139/b89-312. [DOI] [Google Scholar]
- Behrendt U, Ulrich A, Schumann P. Fluorescent pseudomonads associated with the phyllosphere of grasses; Pseudomonas trivialis sp. nov., Pseudomonas poae sp. nov. and Pseudomonas congelans sp. nov. Int J Syst Evol Microbiol. 2003;53:1461–1469. doi: 10.1099/ijs.0.02567-0. [DOI] [PubMed] [Google Scholar]
- Bremer E, Janzen HH, Gilbertson C. Evidence against associative N2 fixation as a significant N source in long-term wheat plots. Plant Soil. 1995;175:13–19. doi: 10.1007/BF02413006. [DOI] [Google Scholar]
- Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. International Microbiol. 2000;3:3–8. [PubMed] [Google Scholar]
- Chia TF, Aung HH, Osipov A, Goh NK, Chia LS. Carnivorous pitcher plant uses free radicals in the digestion of prey. Redox Report. 2004;9:255–261. doi: 10.1179/135100004225006029. [DOI] [PubMed] [Google Scholar]
- Chwedorzewska KJ. Poa annua L. in Antarctica: searching for the source of introduction. Polar Biol. 2008;87:603–615. [Google Scholar]
- Clarke BB, White JF, Jr, Hurley RH, Torres MS, Sun S, Huff DR. Endophyte-mediated suppression of dollar spot disease in fine fescue. Plant Dis. 2006;90:994–998. doi: 10.1094/PD-90-0994. [DOI] [PubMed] [Google Scholar]
- Clay K, Holah J, Rudgers JA. Herbivores cause a rapid increase in hereditary symbiosis and alter plant community composition. PNAS. 2005;102:12465–12470. doi: 10.1073/pnas.0503059102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coteur G, Wamau M, Jangoux M, Dubois P. Reactive oxygen species (ROS) production by amoebocytes of Asterias rubens (Echinodermata) Fish Shellfish Immunol. 2002;12:187–200. doi: 10.1006/fsim.2001.0366. [DOI] [PubMed] [Google Scholar]
- D’Autréaux B, Toledano MB. ROS as signaling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- Dakora FD, Chimphango SBM, Valentine AJ, Wlmerich C, Newton WE (2008) Biological nitrogen fixation: Towards poverty alleviation through sustainable agriculture. Proceedings of the 15th International Nitrogen Fixation Congress and the 12th International Conference of the African Association for Biological Nitrogen Fixation, 21–26 January 2007, Cape town, South Africa. Current plant sciences and biotechnology in agriculture 42. Springer, New York.
- Döbereiner J. History and new perspectives of diazotrophs in association with non-leguminous plants. Symbiosis. 1992;13:1–13. [Google Scholar]
- Döbereiner J, Baldani VLD, Olivares FL, Reis VM. Endophytic diazotrophs: The key to BNF in gramineous plants. In: Hegasi NA, Fayez M, Monib M, editors. Nitrogen fixation with non-legumes. Egypt: The American University in Cairo Press; 1994. pp. 395–408. [Google Scholar]
- Dunand C, Crèvecoeur M, Penel C. Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol. 2007;174:332–341. doi: 10.1111/j.1469-8137.2007.01995.x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Shen D, Song W. Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J Appl Microbiol. 2006;100:938–945. doi: 10.1111/j.1365-2672.2006.02843.x. [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, Davies JM, Dolan L. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- Frahry G, Schopfer P. Hydrogen peroxide production by roots and its stimulation by exogenous NADH. Physiol Plant. 1998;103:395–404. doi: 10.1034/j.1399-3054.1998.1030313.x. [DOI] [Google Scholar]
- Galek H, Osswald WF, Elstner EF. Oxidative protein modification as predigestive mechanism of the carnivorous plant Dionaea muscipula: an hypothesis based on in vitro experiments. Free Radic Biol Med. 1990;9:427–434. doi: 10.1016/0891-5849(90)90020-J. [DOI] [PubMed] [Google Scholar]
- Glick BR. The enhancement of plant growth by free-living bacteria. Can J Microbiol. 1995;41:109–117. doi: 10.1139/m95-015. [DOI] [Google Scholar]
- Godlewski M, Adamczyk B. The ability of plants to secrete proteases by roots. Plant Physiol. Biochem. 2007;45:657–664. doi: 10.1016/j.plaphy.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Hamilton CE, Gundel PE, Helander M, Saikkonen K. Endophytic mediation of reactive oxygen species and antioxidant activity in plants: a review. Fungal Diversity. 2012;54:1–10. doi: 10.1007/s13225-012-0158-9. [DOI] [Google Scholar]
- Hill S, Postgate JR. Failure of putative nitrogen fixing bacteria to fix nitrogen. J Gen Microbiol. 1969;58:277–285. doi: 10.1099/00221287-58-2-277. [DOI] [PubMed] [Google Scholar]
- Hill PW, Quilliam RS, DeLuca TH, Farrar J, Farrell M, et al. Acquisition and assimilation of nitrogen as peptide-bound and d-enantiomers of amino acids by wheat. PLoS One. 2011;6(4):e19220. doi: 10.1371/journal.pone.0019220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurek T, Reinhold-Hurek B, Kellenberger, Grimm B, Fendrik J, Neimann EG. Occurrence of effective nitrogen scavenging bacteria in the rhizosphere of kallar grass. Plant Soil. 1988;110:339–348. doi: 10.1007/BF02226814. [DOI] [Google Scholar]
- Hurek T, Reinhold-Hurek B, Kellenberger E, Van Montagu M. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J Bacteriol. 1994;176:1913–1923. doi: 10.1128/jb.176.7.1913-1923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James EK. Nitrogen fixation in endophytic and associative symbiosis. Field Crops Res. 2000;65:197–209. doi: 10.1016/S0378-4290(99)00087-8. [DOI] [Google Scholar]
- James EK, Olivares FL. Infection and colonization of sugar cane and other graminaceous plants by endophytic diazotrophs. Crit Rev Plant Sci. 1998;17:77–119. doi: 10.1016/S0735-2689(98)00357-8. [DOI] [Google Scholar]
- James EK, Reis VM, Olivares FL, Baldani JI, Dobereiner J. Infection of sugar cane by the nitrogen-fixing bacterium Acetobacter diazotrophicus. J Exp Bot. 1994;45:757–766. doi: 10.1093/jxb/45.6.757. [DOI] [Google Scholar]
- Jamtgard S, Nasholm T, Huss-Danell K. Characteristics of amino acid uptake in barley. Plant Soil. 2008;302:221–231. doi: 10.1007/s11104-007-9473-4. [DOI] [Google Scholar]
- Jayani RS, Saxena S, Gupta R. Microbial pectinolytic enzymes: a review. Process Biochem. 2005;40:2931–2944. doi: 10.1016/j.procbio.2005.03.026. [DOI] [Google Scholar]
- Johnston-Monje D, Raizada MN. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS One. 2011;6(6):e20396. doi: 10.1371/journal.pone.0020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielland K. Amino acid absorption by arctic plants: implications for plant nutrition and nitrogen cycling. Ecology. 1994;75:2373–2383. doi: 10.2307/1940891. [DOI] [Google Scholar]
- Klein DA, Salzwedel JL, Dazzo FB. Microbial colonization of plant roots. In: Nakas JP, Hagedorn C, editors. Biotechnology of plant-microbe interactions. New York: McGraw-Hill Publishing; 1990. pp. 189–218. [Google Scholar]
- Kloepper JW. Plant growth-promoting rhizobacteria as biological control agents. In: Metting FB Jr, editor. Soil microbial ecology: Applications in agricultural and environmental management. New York: Marcel Dekker; 1993. pp. 255–274. [Google Scholar]
- Kloepper JW. Plant growth-promoting rhizobacteria (other systems) In: Okon Y, editor. Azospirillum/Plant Associations. Boca Raton: CRC Press, Inc.; 1994. pp. 137–166. [Google Scholar]
- Kocha T, Yamaguchi M, Ohtaki H, Fukuda T, Aoyagi T. Hydrogen peroxide-mediated degradation of protein: different oxidation modes of copper- and iron-dependent hydroxyl radicals on the degradation of albumin. Biochim Biophys Acta. 1997;1337:319–326. doi: 10.1016/S0167-4838(96)00180-X. [DOI] [PubMed] [Google Scholar]
- Kuldau G, Bacon CW. Clavicipitaceous endophytes: their ability to enhance grass resistance to multiple stresses. Biol Control. 2008;46:57–71. doi: 10.1016/j.biocontrol.2008.01.023. [DOI] [Google Scholar]
- Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Mol. Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- Lethbridge G, Davidson MS. Root-associated nitrogen-fixing bacteria and their role in the nitrogen nutrition of wheat estimated by 15N isotope dilution. Soil Biol Biochem. 1983;15:365–374. doi: 10.1016/0038-0717(83)90085-8. [DOI] [Google Scholar]
- Magnani G, Didonet C, Cruz L, Picheth C, Pedrosa F, Souza E. Diversity of endophytic bacteria in Brazilian sugarcane. Genet Mol Res. 2010;9:258. doi: 10.4238/vol9-1gmr703. [DOI] [PubMed] [Google Scholar]
- Malinowski M, Belesky DP, Lewis GC. Abiotic stresses in endophytic grasses. In: Roberts C, West C, Spiers D, editors. Neotyphodium in cool season grasses. Ames: Blackwell Publishing; 2005. pp. 187–199. [Google Scholar]
- Mantelin S, Touraine B. Plant growth-promoting bacteria and nitrate availability: impacts on root development and nitrate uptake. J Exp Bot. 2004;55:27–34. doi: 10.1093/jxb/erh010. [DOI] [PubMed] [Google Scholar]
- Mikola J. Effects of microbivore species composition and basal resource enrichment on trophic-level biomasses in an experimental microbial-based soil food web. Oecologia. 1998;117:396–403. doi: 10.1007/s004420050673. [DOI] [PubMed] [Google Scholar]
- Munkres KD. Histochemical detection of superoxide radicals and hydrogen peroxide by Age-1 mutants of Neurospora. Fungal Genet Newsl. 1990;37:24–25. [Google Scholar]
- Ortíz-Castro R, Valencia-Cantero E, López-Bucio J. Plant growth promotion by Bacillus megaterium involves cytokinin signaling. Plant Signal Behav. 2008;3:263–265. doi: 10.4161/psb.3.4.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Morel FMM. Amino acid utilization by marine phytoplankton: a novel mechanism. Limnol Oceanogr. 1990;35:260–269. doi: 10.4319/lo.1990.35.2.0260. [DOI] [Google Scholar]
- Paungfoo-Lonhienne C, Rentsch D, Robatzrk S, Webb RI, Sagulenko E, Nasholm T, Schmidt S, Lonhienne TGA. Turning the table: plants consume microbes as a source of nutrients. PLoS One. 2010;5(7):e11915. doi: 10.1371/journal.pone.0011915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick E, Keisari Y. A simple colorimetric method for the measurement of hydrogen peroxide produced by cells in culture. J Microbiol Methods. 1980;38:161–170. doi: 10.1016/0022-1759(80)90340-3. [DOI] [PubMed] [Google Scholar]
- Puente ME, Bashan Y. The desert epiphyte Tillandsia recurvata harbours the nitrogen-fixing bacterium Pseudomonas stutzeri. Can J Bot. 1994;72:406–408. doi: 10.1139/b94-054. [DOI] [Google Scholar]
- Puente ME, Li C, Bashan Y. Rock-degrading endophytic bacteria in cacti. Environ Exp Bot. 2009;66:389–401. doi: 10.1016/j.envexpbot.2009.04.010. [DOI] [Google Scholar]
- Radajweski S, Ineson P, Parekh N, Murrell JC. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- Redman RS, Sheehan KB, Stout RG, Rodriguez RJ, Henson JM. Thermotolerance generated by plant/fungal symbiosis. Science. 2002;298:1581. doi: 10.1126/science.1072191. [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Hurek T. Living inside plants: bacterial endophytes. Curr Opin Plant Biol. 2011;14:435–443. doi: 10.1016/j.pbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Hurek T, Gillis M, Hoste B, Vancanneyt M, De Kersters K, Ley J. Azoarcus gen. nov., a nitrogen fixing Proteobacteria associated with the roots of Kallar grass (Leptochloa fusca (L.) Kunth.), and description of two species Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int J Syst Bacteriol. 1993;43:574–588. doi: 10.1099/00207713-43-3-574. [DOI] [Google Scholar]
- Robinson JM. Reactive oxygen species in phagocytic leucocytes. Histochem. Cell Biol. 2008;130:281–297. doi: 10.1007/s00418-008-0461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RJ, Woodward C, Kim YO, Redman RS. Habitat-adapted symbiosis as a defense against abiotic and biotic stresses. In: White JF Jr, Torres MS, editors. Defensive mutualism in microbial symbiosis. Boca Raton: CRC Press; 2009. pp. 335–346. [Google Scholar]
- Selosse MA, Schardl CL. Fungal endophytes of grasses: hybrids rescued by vertical transmission? An evolutionary perspective. New Phytol. 2007;173:452–458. doi: 10.1111/j.1469-8137.2007.01978.x. [DOI] [PubMed] [Google Scholar]
- Sperry LJ, Belnap J, Evans RD. Bromus tectorum invasion alters nitrogen dynamics in an undisturbed arid grassland ecosystem. Ecology. 2006;87:603–615. doi: 10.1890/05-0836. [DOI] [PubMed] [Google Scholar]
- Steenhoudt O, Vanderleyden J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol Rev. 2000;24:487–506. doi: 10.1111/j.1574-6976.2000.tb00552.x. [DOI] [PubMed] [Google Scholar]
- Stone JK, Bacon CW, White JF., Jr . An overview of endophytic microbes: endophytism defined. In: Bacon CW, White JF Jr, editors. Microbial endophytes. New York: Marcel-Dekker; 2000. pp. 3–30. [Google Scholar]
- Torres MS, White JF, Zhang X, Hinton DM, Bacon CW. Endophyte-mediated adjustments in host morphology and physiology and effects on host fitness traits in grasses. Fungal Ecol. 2012;5:322–330. doi: 10.1016/j.funeco.2011.05.006. [DOI] [Google Scholar]
- Tsavkelova E, Cherdyntseva T, Netrusov A. Bacteria associated with the roots of epiphytic orchids. Mikrobiologiya. 2004;73:710–715. [PubMed] [Google Scholar]
- Van der Valk H, Van Loon L. Subcellular localization of proteases in developing leaves of oats (Avena sativa L.) Plant Physiol. 1988;87:536–541. doi: 10.1104/pp.87.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fisher M, Heier T, Huckelhoven R, Neumann C, Wettstein D, Franken P, Kogel KH. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. PNAS. 2005;102:13386–13391. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JA, Schubert J. Action of hydrogen peroxide on growth inhibition of Salmonella typhimurium. J Gen Microbiol. 1969;57:25–34. doi: 10.1099/00221287-57-1-25. [DOI] [PubMed] [Google Scholar]
- White JF, Torres MS. Is endophyte-mediated defensive mutualism oxidative stress protection? Physiol Plant. 2010;138:440–446. doi: 10.1111/j.1399-3054.2009.01332.x. [DOI] [PubMed] [Google Scholar]
- White JF, Jr, Sullivan R, Balady G, Gianfagna T, Yue Q, Meyer W, Cabral D. A fungal endosymbiont of the grass Bromus setifolius: distribution in some Andean populations, identification and examination of beneficial properties. Symbiosis. 2001;31:241–257. [Google Scholar]
- Yashiro E, Spear RN, McManus PS. Culture-dependent and culture-independent assessment of bacteria in the apple phyllosphere. J Appl Microbiol. 2011;110:1284–1296. doi: 10.1111/j.1365-2672.2011.04975.x. [DOI] [PubMed] [Google Scholar]
- Zogg GP, Zak DR, Pregitzer KS, Burtow AJ. Microbial immobilization and the retention of anthropogenic nitrate in a northern hardwood forest. Ecology. 2000;81:1858–1866. doi: 10.1890/0012-9658(2000)081[1858:MIATRO]2.0.CO;2. [DOI] [Google Scholar]


