Abstract
Background
Plant mitochondrial genome has unique features such as large size, frequent recombination and incorporation of foreign DNA. Cytoplasmic male sterility (CMS) is caused by rearrangement of the mitochondrial genome, and a novel chimeric open reading frame (ORF) created by shuffling of endogenous sequences is often responsible for CMS. The Ogura-type male-sterile cytoplasm is one of the most extensively studied cytoplasms in Brassicaceae. Although the gene orf138 has been isolated as a determinant of Ogura-type CMS, no homologous sequence to orf138 has been found in public databases. Therefore, how orf138 sequence was created is a mystery. In this study, we determined the complete nucleotide sequence of two radish mitochondrial genomes, namely, Ogura- and normal-type genomes, and analyzed them to reveal the origin of the gene orf138.
Results
Ogura- and normal-type mitochondrial genomes were assembled to 258,426-bp and 244,036-bp circular sequences, respectively. Normal-type mitochondrial genome contained 33 protein-coding and three rRNA genes, which are well conserved with the reported mitochondrial genome of rapeseed. Ogura-type genomes contained same genes and additional atp9. As for tRNA, normal-type contained 17 tRNAs, while Ogura-type contained 17 tRNAs and one additional trnfM. The gene orf138 was specific to Ogura-type mitochondrial genome, and no sequence homologous to it was found in normal-type genome. Comparative analysis of the two genomes revealed that radish mitochondrial genome consists of 11 syntenic regions (length >3 kb, similarity >99.9%). It was shown that short repeats and overlapped repeats present in the edge of syntenic regions were involved in recombination events during evolution to interconvert two types of mitochondrial genome. Ogura-type mitochondrial genome has four unique regions (2,803 bp, 1,601 bp, 451 bp and 15,255 bp in size) that are non-syntenic to normal-type genome, and the gene orf138 was found to be located at the edge of the largest unique region. Blast analysis performed to assign the unique regions showed that about 80% of the region was covered by short homologous sequences to the mitochondrial sequences of normal-type radish or other reported Brassicaceae species, although no homology was found for the remaining 20% of sequences.
Conclusions
Ogura-type mitochondrial genome was highly rearranged compared with the normal-type genome by recombination through one large repeat and multiple short repeats. The rearrangement has produced four unique regions in Ogura-type mitochondrial genome, and most of the unique regions are composed of known Brassicaceae mitochondrial sequences. This suggests that the regions unique to the Ogura-type genome were generated by integration and shuffling of pre-existing mitochondrial sequences during the evolution of Brassicaceae, and novel genes such as orf138 could have been created by the shuffling process of mitochondrial genome.
Background
Compared with animal mitochondrial genomes that are compact and relatively uniform in size, plant mitochondrial genomes are extremely complex and have characteristic features, including large genome size, frequent reorganization and incorporation of foreign DNA [1]. Plant mitochondrial genome varies in size from 208 kb in Brassica hirta to 11.3 Mb in Silene conica[2,3], due to the expansion of non-coding sequence and duplication of a large segment. It is also well known that plant mitochondrial genome evolves rapidly in structure and slowly in sequence. Plant mitochondria contain a number of repeats. The recombination via repeat sequences is believed to be responsible for extensive structural change. Incorporation of foreign DNA fragments that originated from plastid and nuclear genomes also can be found in plant mitochondrial genome. Because of frequent recombination and incorporation of foreign DNA, extensive genome reorganization and gene-order shuffling may occur. These changes in mitochondrial genome can produce novel open reading frames, some of which result in cytoplasmic male sterility (CMS) [4].
CMS is a maternally inherited trait in which a plant is unable to produce functional pollen. CMS is an economically important trait for F1 hybrid seed production in many crops. CMS has been widely observed in higher plants, and many reports show that CMS is caused by alteration of the mitochondrial genome [5]. These alterations often create a novel open reading frame (ORF), which consists of a chimeric sequence generated by gene shuffling or by fusion between a native gene(s) and/or an unknown sequence(s).
The Ogura male-sterile cytoplasm, which was originally found in an unknown variety of Japanese radish, is the most widely studied cytoplasm in Brassicaceae[6]. This cytoplasm is used in F1 seed production of European, Chinese and Japanese radishes. The cytoplasm was introduced into Brassica crops by intergeneric hybridization and somatic cell fusion, and has been utilized for F1 seed production in Brassica crops such as rapeseed worldwide [7,8].
A mitochondrial gene, orf138, is responsible for Ogura male sterility and specifically present in the mitochondrial genome of various radishes with Ogura-type cytoplasm [9]. The accumulation of ORF138 protein is observed in plants expressing the CMS phenotype, and the association of ORF138 with the inner mitochondrial membrane of male-sterile plants was reported [10]. Although it appeared that expression of the nuclear chalcone synthase gene, which is related to flavonoid biosynthesis, is inhibited in the anther of Ogura radish [11], the molecular mechanism underlying Ogura CMS by ORF138 protein is not known. The sequence of the gene orf138 itself is also interesting; there is no evidence that orf138 is a chimeric gene consisting of native mitochondrial genes or sequence. No sequence homologous to orf138 is found in public databases; therefore, the origin of orf138 is a mystery. In addition, it has been shown that the mitochondrial genome of Ogura-type cytoplasm is highly rearranged compared with that of normal cytoplasm in radish. These rearrangements occurred in the loci atp1atp6cox1 and most notably orf138-atp8[12-17]. The gene orf138 is specifically located in the 5’ upstream region of the gene atp8 in Ogura radish, whereas the region is occupied by the gene cox1 in normal radish [16]. In the evolution of Raphanus, how and when these extensive rearrangements occurred between two types of mitochondrial genome are not known.
In this study, in order to obtain a better understanding of the origin of orf138 and Ogura-type mitochondrial genome in radish, we determined the complete nucleotide sequences of two mitochondrial genomes of radish, that is, Ogura- and normal-type mitochondrial genomes, and compared their structures. Gene contents and unique regions for each genome were analyzed in detail at the sequence level. Furthermore, we focused on short repeated sequences that may be involved in the recombination, leading to extensive rearrangements of the two genomes and to generation of the gene orf138.
Results
Radish mitochondrial genome
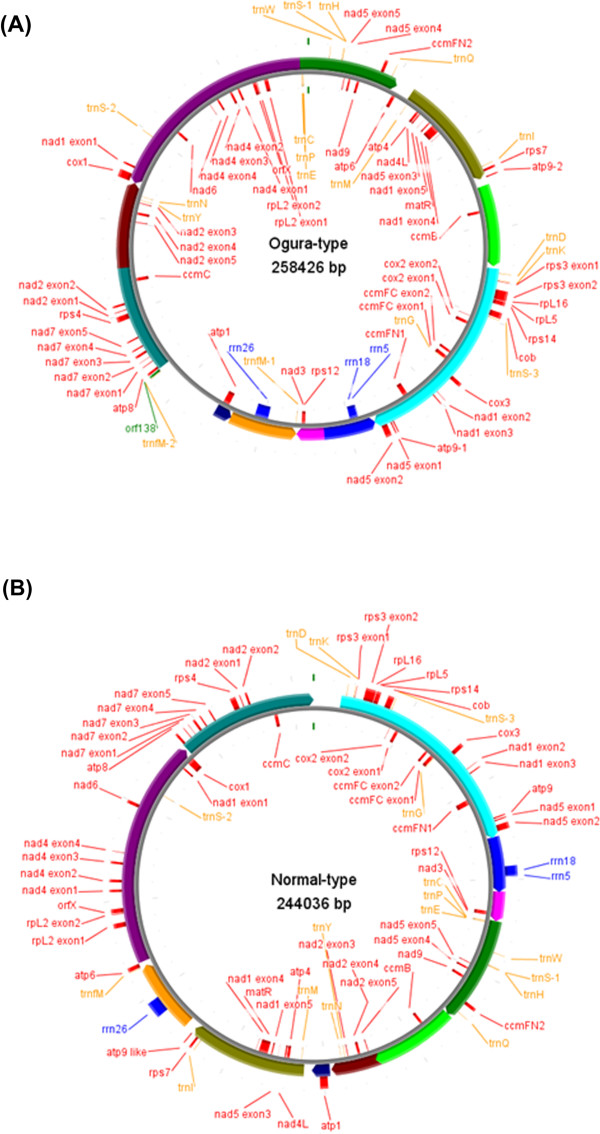
The Ogura and normal mitochondrial genomes are assembled as 258,426-bp and 244,036-bp circular molecules, respectively (Figure 1). The overall GC content of both mtDNAs is 45.2%, being comparable to those of other mtDNAs of Brassicaceae. About 7% of Ogura and 3% of normal sequences are unique to each mitochondrial genome, and 80% of Ogura and 84% of normal sequences show high similarity to the sequence of rapeseed (Brassica napus L.) mitochondrial genome (GenBank: AP006444.1) [18].
Figure 1.
The organization of Ogura-type (A) and normal-type (B) mitochondrial genomes represented as a "master circle". Features on forward and reverse strands are drawn on the outside and inside of the circles, respectively. Protein-coding genes are shown in red, rRNAs in blue, tRNAs in orange and orf138 in lime green. The arcs in the same colors indicate syntenic regions between Ogura- and normal-type genomes (refer to Figure 2). Genome maps were made with CGviewer [49].
Gene organization of radish mitochondrial genome
Normal-type mitochondrial genome contained 33 protein-coding and three rRNA genes, while Ogura-type genomes contained same genes and one additional atp9 (atp9-2). Gene content is identically conserved between the two radish mitochondrial genomes and it is also identical to that of rapeseed, except atp9-2 in Ogura-type genome. Normal-type genome contains similar sequence to atp9-2, but 3’ part of this sequence lacks similarity to atp9-2. The atp9-2 like sequence in normal-type genome encodes 70 amino acid protein, which is lacking atp9 domain partially. Eighteen of the conserved 33 protein-coding genes produce components of the electron transport chain and ATP synthase: nine subunits of complex I(nad1nad2nad3nad4nad4Lnad5nad6nad7 and nad9), apocytochrome b (cob) of complex III, three subunits of complex IV (cox1cox2 and cox3) and five subunits of complex V (atp1atp4atp6atp8 and atp9). Five additional proteins are involved in the biogenesis of cytochrome c (ccmBccmCccmFN1/ccmFN2 and ccmFC). Another eight genes encode ribosomal proteins (rpl2rpl5 and rpl16, and rps3rps4rps7rps12 and rps14). The two remaining genes encode maturase (matR) and orfX. The gene ccmFN is divided into two reading frames in mitochondrial genomes of radish like in those of rapeseed and Arabidopsis[18,19]. Three rRNA genes (rrn5rrn18 and rrn26) are highly conserved between the two mitochondrial genomes of radish.
The 5’ region of the gene atp6 is highly diverged between the two radishes because of recombination that occurred in the locus [20]. The 3’ end of Ogura-type orfX also lacks similarity to that of normal-type. This is caused by a 48-bp repeated sequence that is regarded as a minisatellite. Eleven SNPs were identified in the nine genes: four were synonymous and seven were non-synonymous (Table 1). As for tRNA, both Ogura and normal mitochondrial genomes contain 17 tRNAs, whereas Ogura genome has an additional trnfM (trnfM-2). The trnfM-2, which is located close to the gene orf138 (Figure 1), has a specific SNP compared with other sequenced mitochondrial trnfMs in Brassicaceae.
Table 1.
Difference in genes coding protein between normal-type and Ogura-type
| Gene | Nucleic acid | Amino acidb |
|---|---|---|
|
nad4 |
1010 T-C |
Non-synonymous (337 L-P) |
|
cox1 |
117 G-T |
Synonymous |
|
atp6 |
no homology in 5′ terminal |
|
|
atp8 |
370 A-C |
Non-synonymous (124 I-L) |
| |
444 T-C |
Non-synonymous (150 V-A) |
|
atp9 |
65 A-G |
Non-synonymous (22 I-V) |
|
rpL2 |
840 C-T |
Synonymous |
|
rpL16 |
492 G-A |
Synonymous |
|
rps4 |
776 C-T |
Non-synonymous (259 S-F) |
|
ccmC |
351 G-A |
Synonymous |
|
ccmFC |
605 A-G |
Non-synonymous(202 E-G) |
| |
606 G-T |
|
| orfX | no homology in 3′ terminal |
a Location of SNP (Normal-Ogura).
b Location of amino acid mutation (Normal-Ogura).
We searched for a sequence homologous to the orf138 in Ogura and normal mtDNA sequences. Only a 67-bp sequence, which includes the 3’ terminal of the orf138 sequence and its 3’ UTR, was detected. This sequence is homologous to 5’ flanking region of ccmFN1 in reported Brassicaceae genomes. However, except for this short sequence, no homology was found between the main part of the gene orf138 and other parts of the Brassicaceae mitochondrial genome.
Reorganization between two types of radish mitochondrial genome
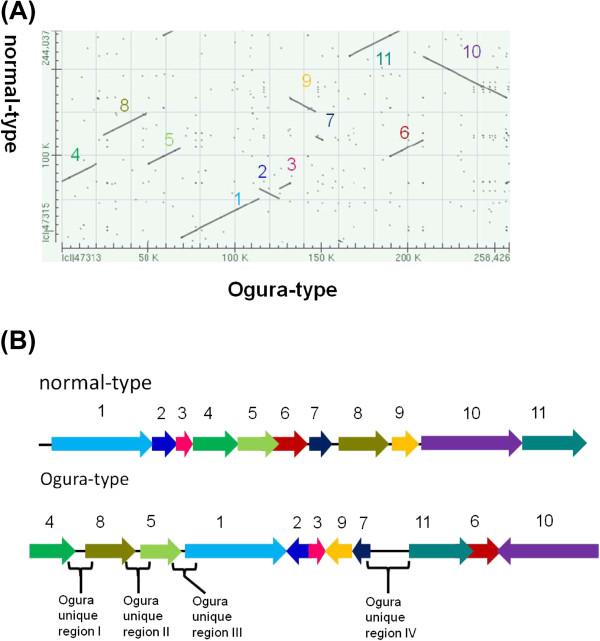
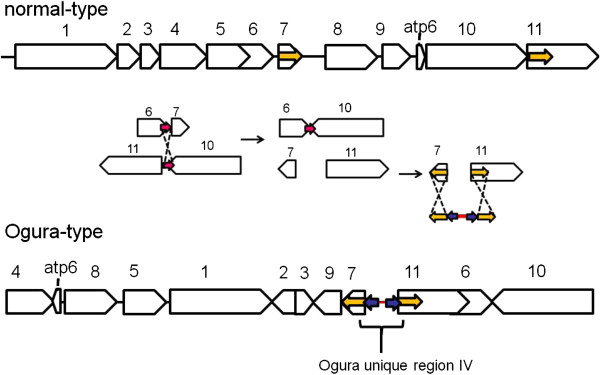
The bl2seq analysis suggested that the radish mitochondrial genome consists of 11 syntenic regions (length >3 kb) (Figure 2). The sequence in the Ogura-type mitochondrial genome shows quite high homology (99.9%) to that of normal mitochondrial genome within these syntenic regions. This analysis revealed that Ogura-type mitochondrial genome has a large unique region (15,255 bp) between syntenic regions 7 and 11. This unique region corresponds to the “region A” that was reported by Makaroff et al. [12]. The gene orf138 is located at one of the terminals of this region (Figure 1).
Figure 2.
Comparison of the location and orientation of syntenic regions between Ogura- and normal-type mitochondrial genomes. (A) Ogura-type genome on the X-axis, plotted against normal-type genome on the Y-axis. The numbers for the syntenic regions correspond to those indicated in (B). (B) Schematic illustration of the eleven syntenic regions in radish mitochondrial genomes.
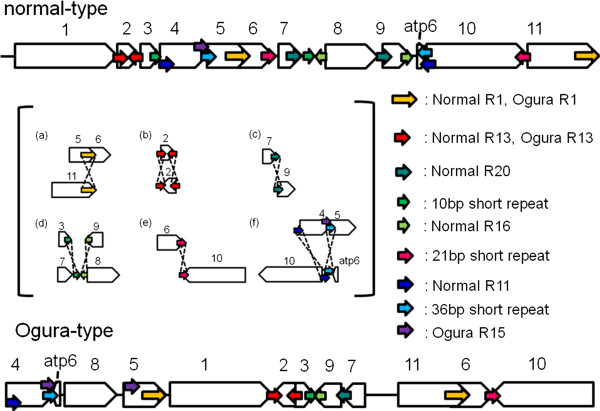
Repeats in two mitochondrial genomes have been investigated. If a pair of sequences has over 90% similarity, the pair is defined as a repeat. A pair of large repeats of 9,732 bp was found in Ogura-type mitochondrial genome, while a pair of large repeats of 5,530 bp was identified in normal-type. Even though the length of large repeats is different, sequence of the large repeats are highly conserved within 5,530 bp. In Ogura-type mitochondrial genome, the large repeats are located in syntenic regions 5 and 11/6 (Figure 3, Ogura R1). A 9,732-bp sequence, corresponding to Ogura-type repeat, is conserved in syntenic region 5/6 of the normal-type genome, but it is truncated in syntenic region 11 because normal-type genome has a specific sequence between syntenic regions 1 and 11. In rapeseed mitochondrial genome, a 2,427-bp sequence that includes the gene cox2 was reported as a large direct repeat. Large repeated sequences found in Raphanus are completely different from those reported in rapeseed [18]. These large repeats are considered to be involved in the formation of a tripartite structure; each radish mitochondrial genome could recombine into two sub-genomic circles. Two expected sub-genomic circles are 130,760 bp and 127,666 bp in Ogura-type, while they are 139,398 bp and 104,638 bp in normal-type. In addition, these repeats are also related to reorganization among syntenic regions 5, 6 and 11 (Figure 3a).
Figure 3.
The possible rearrangements that had occurred between two Ogura- and normal-type genomes via short repeated sequences. Large blank arrows indicate each syntenic region. Small arrows in the same color show the position and orientation of repeats. The length and location of the repeats are summarized in Additional file 1 and Additional file 2.
We defined 22 and 23 short repeats in Ogura- and normal-type genomes, respectively (Additional file 1 and Additional file 2). These repeats can account for reorganization of mitochondrial genome via homologous recombination. We have identified repeats that can be involved in reorganization from normal- to Ogura-type mitochondrial genomes, although the direction of changes cannot be fixed. The pair of inverted repeats (Normal R13) is located on either side of syntenic region 2 in normal-type genome. These inverted repeats may change the orientation of syntenic region 2 in Ogura-type genome (Figure 3b). In normal-type mitochondrial genome, a pair of direct repeats (Normal R20) is located at the edge of syntenic regions 7 and 9. These repeats can be related to generation of a linkage between 7 and 9 in Ogura-type genome (Figure 3c). In normal-type genome, Normal R16 is present at the edge of syntenic region 9 and in the unique sequence located between syntenic regions 7 and 8 in the inverted orientation. A 10-bp short repeat is also present in this unique sequence, and it exists at the edge of syntenic region 3 in the direct orientation. These repeats can be associated with the generation of a linkage between syntenic regions 3 and 9 in Ogura-type genome (Figure 3d). Syntenic regions 6 and 10 in normal-type genome have a 21-bp short inverted repeat. This inverted repeat can be involved in inversion and recombination between syntenic regions 6 and 10 in Ogura-type genome (Figure 3e). Finally, in normal-type genome, syntenic region 4 has Normal R11 and a short 36-bp repeat at each edge. Normal R11 is also present at one edge of syntenic region 10, being overlapped with a 36-bp repeat. In Ogura-type genome, a linkage between syntenic regions 10 and 4 is generated via these two sequences. This recombination event also changes the location of the gene atp6 (Figure 3f). Moreover, in Ogura-type genome, two copies of a short direct repeat (Ogura R15) are present at the edge of syntenic regions 4 and 5, between which syntenic region 8 is inserted with a novel sequence (Figure 3f).
Open reading frames that are unique to each mitochondrial genome type
A comprehensive comparison of predicted ORFs was conducted between Ogura- and normal-type mitochondrial genomes. We picked up unique ORFs that can encode over 100 amino acids and were generated by recombination. As a result, six ORFs including orf138 were specifically detected in Ogura-type mitochondrial genome (Table 2). The five Ogura-type ORFs were located between syntenic regions 7 and 11, whereas orf344 was in syntenic region 3. With the exception of orf138, the nucleotide sequences of the five ORFs showed high similarity to the reported sequences in plant mitochondrial genomes of other species. As for the normal-type genome, three unique ORFs were detected. The orf322 has strong similarity to the orf344 in Ogura-type. The extension of orf344 was caused by a new initiation codon that was created in 5’ region of orf322 through a recombination between syntenic regions 3 and 9. The two remaining ORFs were located within the region unique to the normal-type genome located between syntenic regions 11 and 1. Protein Blast analysis was conducted to characterize unique ORFs. Even though homologous amino acid sequences were not found for orf150 in normal-type and for orf122 and orf138 in Ogura-type, other unique ORFs encoded similar proteins that have been reported in the mitochondrial genome of rapeseed or Arabidopsis.
Table 2.
Mitotype specific open-reading frames caused by recombination
| ORF | Similarity of predicted protein | Location | |
|---|---|---|---|
| Ogura-type specific |
orf122 |
No similarity |
between syntenic regions 7 and 11 |
| |
orf154 |
AAG51754.1 reverse transcriptase, putative[Arabidopsis thaliana] |
between syntenic regions 7 and 11 |
| |
orf117 |
YP_717156.1 hypothetical protein BrnapMp059 [Brassica napus] |
between syntenic regions 7 and 11 |
| |
orf138 |
No similarity |
between syntenic regions 7 and 11 |
| |
orf102 |
NP_085574.1 hypothetical protein ArthMp044 [Arabidopsis thaliana] |
between syntenic regions 7 and 11 |
| |
orf344 |
YP_717145.1 hypothetical protein BrnapMp048 [Brassica napus] |
the edge of syntenic region 3 |
| normal-type specific |
|
|
|
| |
orf322 |
YP_717145.1 hypothetical protein BrnapMp048 [Brassica napus] |
the edge of syntenic region 3 |
| |
orf150 |
No similarity |
between syntenic regions 1 and 10 |
| orf145 | NP_085575.1 hypothetical protein ArthMp101 [Arabidopsis thaliana] | between syntenic regions 1 and 10 |
Regions unique to the Ogura-type mitochondrial genome
Ogura-type mitochondrial genome has four unique regions that are non-syntenic to normal-type mitochondrial genome. Unique region I is 2,802 bp, which is located between syntenic regions 4 and 8 (Figure 2B). Unique regions II and III are 1,601-bp and 451-bp sequences that are present between syntenic regions 8 and 5, and between syntenic regions 5 and 1, respectively. The largest unique region IV is located between syntenic regions 7 and 11 (Figure 2B). Total length of the unique regions in Ogura-type genome is 20,110 bp, which occupies 7% of the whole Ogura-type mitochondrial genome. The largest unique region contains five unique ORFs including orf138.
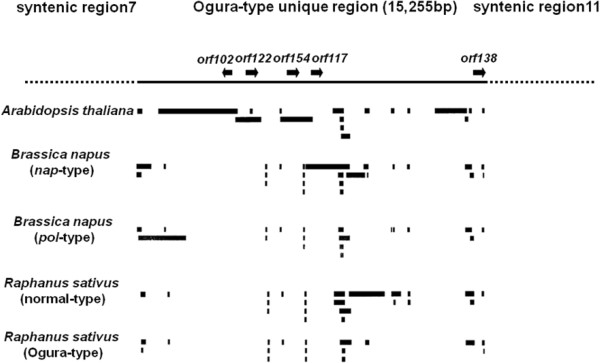
To obtain more information on the sequence unique to Ogura-type genome, nucleotide Blast search was conducted to assign the sequences. The search showed that over 80% of these regions consist of sequences homologous to those reported for Brassicaceae or radish mitochondrial genomes. Bl2seq analyses were further conducted between the sequence unique to Ogura-type and each of five mitochondrial genomes in Brassicaceae (normal and Ogura-type radish, A. thaliana and two types of Brassica napus). Seventy-two percent of unique region I (2,044 bp in a total of 2,802 bp) and 100% of unique region II (1,601 bp) were occupied by five mitochondrial sequences of Brassicaceae (Additional file 3). As for the smallest unique region III, 97% of the sequence (440 bp in a total of 451 bp) was shown to have similarity to rapeseed mtDNAs. As for the largest unique region IV, sequences homologous to the five Brassicaceae mitochondrial genomes covered 80% of this region (12,187 bp in a total of 15,255 bp), even though orf138 sequence itself had no similarity. Homologous sequences detected by bl2seq were plotted on unique region IV in Ogura-type mitochondrial genome (Figure 4). Homologous sequences were scattered throughout this region. Insertion of plastid or nuclear sequence has not been detected in this region. It seems, therefore, that the region unique to Ogura-type mitochondrial genome was created by frequent recombination and shuffling of mitochondrial DNA sequences during the evolution of Brassicaceae.
Figure 4.
The sequences homologous to Brassicaceae mitochondrial genome plotted on Ogura-type specific region IV. A black line indicates the region specific to the Ogura-type mitochondrial genome (15,255 bp in size) located between syntenic regions 7 and 11. The gene orf138 is located at the edge of this region. Black boxes indicate the sequence homologous to radish and/or other Brassicaceae mitochondrial genomes.
Discussion
Comparative sequence analysis of Ogura- and normal-type mitochondrial genomes
Ogura-type mitochondrial genome was 14 kb larger than that of normal type. Region IV that is unique to Ogura-type mitochondrial genome mainly contributes to this size difference. Comparative analysis has been performed between mitochondrial genomes of CMS and normal lines in other higher plant species. In sugar beet, Owen-type CMS line has a significantly larger mitochondrial genome than fertile line because the Owen-type mitochondrial genome contains one 86-kb large repeat [21]. In wheat, the mitochondrial genome of the CMS line Ks3 was reported to be 192 kb larger than that of common wheat [22]. This difference is also caused by additional large repeats, including a 98-kb repeat. In contrast, a rice (Oryza sativa) LD-CMS line has a smaller mitochondrial genome than fertile line, while CW-CMS derived from wild rice (O. rufipogon) has a larger genome than fertile line [23]. The difference in mitochondrial genome size within the Oryza genus is also attributable to large repeats. It should be noted that the size difference between two radish mitochondrial genomes mainly arises from a novel sequence of 15,255 bp, not from a large repeated sequence.
The reported Brassicaceae species including Brassica and Arabidopsis have small mitochondrial genomes among higher plants [24]. It is also known that Brassicaceae mitochondrial genomes reported so far do not contain much nuclear sequence. The number of large repeats (>1 kb) is less in reported Brassicaceae (Arabidopsis and Brassica) than in other plants species [25]. While mitochondrial genomes of rice or wheat have more than ten large repeats, Brassica and Raphanus have only two large repeats. In addition, sequences derived from nuclear or plastid genomes were rarely observed. This is one of the reasons why mitochondrial genomes of Brassica and Raphanus are small. Because of the small genome size and few large repeats, we can consider Brassica and Raphanus as species that have the “simple” mitochondrial genomes in higher plants. An understanding of the remarkable reorganization events that occurred in the simple radish genome will provide us with a clear picture of mitochondrial genome evolution, such as insertion of a 15-kb large novel sequence in Ogura-type.
As expected, gene contents and their sequences were highly conserved between the two mitochondrial genomes in radish. The 5’ sequence of the atp6 in Ogura-type mitochondrial genome is, however, different from that of normal-type. Recombination had occurred at the site of syntenic region 4 and atp6, which produced a new initiation codon for atp6 in Ogura-type genome. According to the genome sequence, Ogura-type atp6 encodes a different polypeptide from that of normal-type atp6. However, the N-terminal of ATP6 polypeptide of Ogura-type is processed to produce a protein identical to normal-type ATP6 [20]. The 3’ end of orfX also lacks similarity to the sequence of normal-type. This is caused by a 48-bp repeated sequence regarded as a minisatellite. This minisatellite will be a useful marker to detect polymorphism among radish mitochondrial genomes including Ogura and normal types [26]. A total of 11 SNPs were detected in nine protein-coding genes between Ogura and normal types. The number of SNPs in radish was similar to that in sugar beet; the sugar beet Owen-CMS line contains 24 SNPs in 11 protein-coding genes compared with the fertile lines [21]. As for tRNA, 17 tRNAs were highly conserved between two radish mitochondrial genome types, but Ogura-type contains one additional trnfM (trnfM-2). This additional trnfM-2 is located near orf138, and it is interesting to note that this trnfM-2 has a unique SNP that has not been found in other reported trnfMs in Brassicaceae. As for unique ORFs, Ogura-type has six unique ORFs including orf138. These ORFs can be related to unique biological characteristics in Ogura-type cytoplasm. In particular, transcription of four ORFs (orf122orf154orf117 and orf102) that are specific to Ogura-type genome should be investigated in future study.
The origin of Ogura-type unique sequence
CMS-related ORFs have been isolated in various plant species, such as T-urf13 in maize [27], orf522 in sunflower [28], pcf in petunia [29], orf456 in pepper [30], orf107 in sorghum [31] and orf79 in rice [32]. In Brassicaceae crops, in addition to orf138 in radish, three CMS-related ORFs have been reported: orf224 and orf222 in rapeseed [33], and orf267 in Brassica tournefortii[34]. With the exception of orf138, all these CMS-related ORFs have a chimeric structure that contains the sequence of known mitochondrial genes. The orf138, however, does not contain any conserved mitochondrial sequence. The sequence of orf138 is totally novel, and is not present in normal-type mitochondrial genome of radish. It is quite interesting to reveal how the orf138 sequence was created during the evolution of radish mitochondrial genome. However, it now appears difficult to infer the origin of the gene orf138, since no sequence homologous to the main part of orf138 is present in either Ogura- or normal-type mitochondrial genome.
In this study, we have determined the nucleotide sequence for unique region IV (15,255 bp in size) in Ogura-type mitochondrial genome. The gene orf138 is located in this region. Blast analysis using the sequence in region IV as a query showed that 80% of this region has similarity to the mitochondrial sequences in other Brassicaceae plants, including rapeseed and Arabidopsis. The analysis also showed that the homologous sequences are scattered throughout unique region IV (Figure 4). Therefore, region IV is a mosaic of mitochondrial sequences from Brassicaceae, suggesting that the sequence was generated through shuffling and fusion of pre-existing mitochondrial sequences in ancestral Brassicaceae. The sequence of which the gene orf138 consists may be one of the products of this shuffling process. It is unlikely that the sequence of orf138 is derived from foreign DNA, such as a nuclear or plastid sequence, because no sequence with clear homology to foreign DNA is found within this unique region. However the possibility remains that they may be derived from horizontal gene transfer from unidentified organism.
This composition of novel sequence in radish contrasts to that reported for sugar beet. In sugar beet mitochondrial genome, 7.6% of the unique regions showed significant homology to previously determined mitochondrial sequences, 17.9% to nuclear DNA, 4.6% to mitochondrial episome and 0.1% to plastid DNA [35].
The emergence of Ogura-type cytoplasm
The sequence of Ogura-type unique region IV can be derived from a mixture of sequences in the reported Brassicaceae mitochondrial genome. The next question relates to how this unique region IV was generated or integrated into the radish mitochondrial genome. We searched for repeated sequences around this region. Ogura-type unique region IV was shown to be sandwiched between syntenic regions 7 and 11 (Figure 5). There were identical 176-bp inverted repeats at the edges of regions 7 and 11. On the insides of these repeated sequences, another pair of 28-bp repeated sequences was found in inverted orientation. These 28-bp sequences are specific to the Ogura-type mitochondrial genome, and not found in normal-type genome. This characteristic arrangement of repeats indicates that unique region IV was integrated into the radish mitochondrial genome when recombination between syntenic regions 7 and 11 occurred.
Figure 5.
A possible model for integration of the sequence (unique region IV) in Ogura-type mitochondrial genome. Yellow small arrows indicate identical 176-bp inverted repeats at the edges of regions 7 and 11. On the insides of these repeated sequences, another pair of 28-bp inverted repeats is present (purple small arrows). Unique region IV had been integrated into radish mitochondria genome via these repeats when recombination between syntenic regions 7 and 11 occurred.
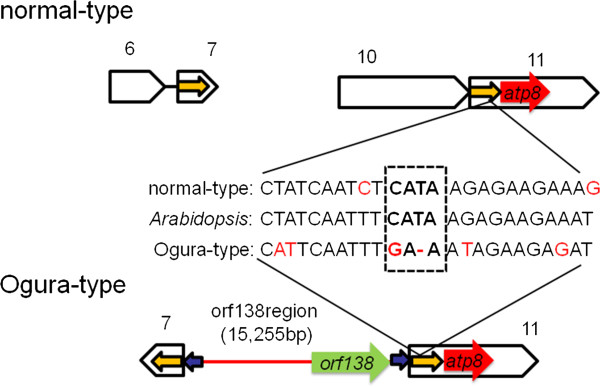
Previously, we conducted sequence analysis on the atp8 locus in normal- and Ogura-type cytoplasm from wild and cultivated radishes [36]. While plants with normal-type cytoplasm contained several types of sequence, plants with Ogura-type cytoplasm had only one type of sequence. These results suggested that a mutational event, which created linkage of atp8 and orf138, occurred only once in the history of radish [36]. The present data indicate that this event probably occurred at the same time as when recombination between syntenic regions 7 and 11 occurred via 176-bp repeats and the unique region IV was integrated into the Ogura-type mitochondrial genome (Figure 5). Interestingly, the radish mitochondrial genome may have lost one putative promoter region for atp8 by this recombination (Figure 6). In Arabidopsis, the promoters of mitochondrial genes have been determined [37]. A promoter sequence of atp8 (CTATCAATCTCATAAGAGAAGAAAT) is almost conserved at the edge of syntenic region 11 of normal radish. The position of the promoter exactly matched the size of atp8 transcript (ca. 600 bp in size) [16]. The recombination of syntenic regions 7 and 11 would destroy the promoter sequence. Therefore, recombination between syntenic regions 7 and 11 in normal radish can lead to loss of biological function in mitochondria. A reasonable hypothesis to explain the generation of Ogura-type mitochondrial genome is that the mitochondrial genome rearrangement that could compensate for the loss of biological function had been selected among many rearranged genomes. Integration of unique region IV can provide a new promoter for the gene atp8. Actually, in Ogura-type genome, atp8 is co-transcribed with orf138 and trnfM, while in normal-type, atp8 is transcribed from its own promoter.
Figure 6.
An atp8 promoter sequence affected by rearrangement among syntenic regions. Alignment of nucleotide sequences shows the comparison of predicted promoters for the gene atp8 among three mitochondrial genomes including Arabidopsis, and Ogura and normal radish. In Ogura-type genome, a promoter core (in a dotted box) is lost. This nucleotide change can affect transcription of the gene atp8.
Another possibility to explain the emergence of Ogura-type mitochondrial genome is substoichiometric shift (SSS). SSS is a phenomenon by which copy number of the substoichiometric molecule dramatically increases over generations [38]. Research in some plants has shown that SSS is related to the occurrence of CMS [39,40]. Recently, it has been reported that two mitochondrial genome types, pol -type and nap-type, co-exist among many rapeseed cultivars, and SSS can explain the emergence of these CMS [41]. In radish with normal-type cytoplasm, orf138 was reported to be present in substoichiometric molecules with low copy number by genomic PCR [42]. However, our previous analyses failed to detect the gene orf138 in various normal radishes. The fact that no sequence homologous to orf138 was found in the current pyrosequencing data (20,646,046 bp in total) from a normal radish suggests that substoichiometric molecules containing orf138 are absent in normal radish. The discrepancy between our results and a previous study [42] can be attributed to the differences of materials used and to the conditions of PCR including primers, type of DNA polymerase and particularly amplification cycles (30 cycles [43,44] vs. 40 cycles in ref. [42]). However, our PCR conditions can amplify two different configurations of orf138 regarded as subliminal fragments in various radishes with Ogura-type cytoplasm. This indicates that, if there were orf138 sequences in normal radish, they would be at quite a low level, and the contribution of SSS as a possible mechanism to explain generation of the Ogura-type mitochondrial genome from normal-type genome remains to be elucidated.
Previously, we conducted a large-scale sequence analysis of orf138 in 107 Japanese wild radishes, 29 cultivated radishes and seven R. raphanistrum[43]. On the basis of the pattern of mutation and the distribution of orf138 variants, we inferred the mechanism behind the differentiation of Ogura-type cytoplasm. These studies showed that the original Ogura-type radish is derived from Japanese wild radish and that orf138 sequence of Japanese wild radish was introduced from R. raphanistrum[44-46]. However, it was still unclear when Ogura-type cytoplasm was initially generated. This is still an open question, and comparative sequence analysis among more mitochondrial genomes in Raphanus and molecular evolutionary studies of the data should provide clues to resolve this issue.
Conclusions
The complete nucleotide sequence of the mitochondrial genome from Ogura male-sterile radish reveals that the genome has been highly rearranged compared with that of normal radish. The radish mitochondrial genome consists of 11 syntenic regions. The reorganization of the genome occurred via a pair of large repeats and multiple pairs of short repeats, and produced four regions unique to the Ogura-type mitochondrial genome. Most of these unique regions consist of sequences homologous to known Brassicaceae mitochondrial genomes. Insertions of sequences derived from plastid or nuclear genome were not identified. This suggested that the unique regions were generated by integration and shuffling of pre-existing mitochondrial sequences during the evolution of Brassicaceae, and novel genes such as orf138 could have been created by the shuffling process.
Methods
Plant materials
A cultivar 'Uchiki-Gensuke' (UC-G) and its male-sterile line 'MS-Gensuke' (MS-G) were used in this study. These two lines are in the category of Japanese cultivated radish (Raphanus sativus var. hortensis) with a long white root. UC-G and MS-G have normal cytoplasm and Ogura-type cytoplasm, respectively. The MS-G was developed by introducing Ogura-type male-sterile cytoplasm into UC-G and maintained by repeated backcrosses.
Mitochondrial DNA extraction
Mitochondrial DNA was isolated from leaves of MS-G and UC-G, according to the method of Bonen and Gray [47] using discontinuous (1.15 M, 1.30 M and 1.45 M sucrose) density gradient centrifugation with minor modifications. DNaseI-treated mitochondria were collected from the interface between 1.30 M and 1.45 M sucrose. MtDNA was purified by EtBr/CsCl centrifugation and ethanol precipitation.
Sequence assembly
For normal-type mitochondrial genome
Pyrosequencing using the GS-FLX system (Roche) and de novo assembly were conducted by Hokkaido System Science (Sapporo, Japan). A nucleotide sequence of 20,646,046 bp in total was obtained. Sequences were assembled to 105 contigs (4,414 bp long on average). On the basis of the physical map for the normal-type mitochondrial genome, we connected contigs to develop a master circle. If contigs had similarity to plant plastid genome and their depth value was low, we ignored them. All linkages between contigs were confirmed by genomic PCR (Additional file 4 and Additional file 5). By sequencing genomic-PCR product, we determined the sequences between contigs. Sequences were assembled using the software Sequencher ver. 4.9 (Gene Codes Corporation, Ann Arbor, MI, USA), and deposited in a database (DDBJ) under accession No. AB694743.
For Ogura-type mitochondrial genome
Pyrosequencing was conducted using the GS-FLX system (Roche) by Takara-Bio (Ohtsu, Japan). A nucleotide sequence of 12,059,770 bp in total for Ogura-type was obtained. Sequences were assembled to 147 contigs (4,402 bp long on average), using GS De Novo Assembler version 2.0 (Roche). The average sequence depth, which was defined as the total nucleotide number used for assembly divided by the total length of contigs, was 36. On the basis of linkage information of the contigs and the physical map of radish mitochondria, we connected contigs to develop a master circle by a parsimonious method, so that each contig appeared at least once to construct the smallest genome [23]. If the depth of linkage was under 10, we did not use the linkages to make the master circle. When it was difficult to judge whether a given contig really appears in the master circle, we checked the existence of the contig in all possible regions by genomic PCR and sequencing (Additional file 6). Sequences were assembled using the software Sequencher ver. 4.9, and deposited in a database (DDBJ) under accession No. AB694744.
Sequence analysis
The genes encoding known mitochondrial proteins and those for rRNAs, as well as repeated sequences, were identified using the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A tRNA gene search was conducted with the tRNA scan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/) [48]. ORFs encoding hypothetical proteins were identified using Getorf software (http://emboss.dbcls.jp/). ORFs predicted to encode proteins longer than 100 amino acids were picked up in the current analysis.
Abbreviations
CMS = Cytoplasmic male sterility; mtDNA = Mitochondrial DNA; ORF = Open reading frame; SNP = Single nucleotide polymorphism; SSS = Substoichiometric shifting.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YT carried out the sequence data analysis, and drafted the manuscript. MT isolated mitocnodrial DNA from radish. KY participated in the sequence analysis. HY prepared plant materials. TT supervised the work and edited the manuscript. All authors read and approved the final manuscript.
Supplementary Material
List of repeated sequences in normal-type genome.Repeated sequences(>80 bp, >90% homology) in normal-type genome were listed.
List of repeated sequences in Ogura-type genome.Repeated sequences(>80 bp, >90% homology) in Ogura-type genome were listed.
The sequences homologous to Brassicaceae mitochondrial genome plotted on Ogura-type specific region I -III. A black line indicates the region specific to the Ogura-type mitochondrial genome. Black boxes indicate the sequence homologous to radish and/or other Brassicaceae mitochondrial genomes.
Validation of contig linkage by PCR analysis. The primer information used for this PCR analysis is described in Additional file 5.
The primers used for the PCR analysis to validate the linkage between contigs of normal-type genome. Information on the primers used for the PCR analysis in Additional file 4.
The primers used for PCR analysis to validate the short repeats. Information on the primers used for PCR and sequencing, in order to confirm short repeats in Ogura-type genome.
Contributor Information
Yoshiyuki Tanaka, Email: k4715@cc.kyoto-su.ac.jp.
Mizue Tsuda, Email: i1055136@cc.kyoto-su.ac.jp.
Keita Yasumoto, Email: k4479@cc.kyoto-su.ac.jp.
Hiroshi Yamagishi, Email: hiyamagi@cc.kyoto-su.ac.jp.
Toru Terachi, Email: terachi@cc.kyoto-su.ac.jp.
Acknowledgement
This study was supported in part by Private University Strategic Research Foundation Support Program, Grants-in-Aid for Scientific Research, Scientific Research (B) (No. 22380008), and the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Indurstry (BRAIN), JAPAN.
References
- Kubo T, Newton KJ. Angiosperm mitochondrial genomes and mutations. Mitochondrion. 2008;8(1):5–14. doi: 10.1016/j.mito.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Herbo LA. Unicircular structure of the Brassica hirta mitochondrial genome. Curr Genet. 1987;11(6–7):565–570. doi: 10.1007/BF00384620. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR. Rapid Evolution of Enormous, Multichromosomal Genomes in Flowering Plant Mitochondria with Exceptionally High Mutation Rates. PLoS Biol. 2012;10(1):e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Wise RP. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3(5):175–180. doi: 10.1016/S1360-1385(98)01235-7. [DOI] [Google Scholar]
- Hanson MR, Bentolila S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004;16(SUPPL):S154–S169. doi: 10.1105/tpc.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura H. Studies on the new male sterility in Japanese radish, with special references to the utilization of this sterility towards the practical raising of hybrid seeds. Mem. 1968;6:39–78. [Google Scholar]
- Sakai T, Imamura J. Intergeneric transfer of cytoplasmic male sterility between Raphanus sativus (cms line) and Brassica napus through cytoplast-protoplast fusion. Theor Appl Genet. 1990;80(3):421–427. doi: 10.1007/BF00210083. [DOI] [PubMed] [Google Scholar]
- Bannerot H, Boulidard L, Chupeau Y. Unexpected difficulties met with the radish cytoplasm in Brassica oleracea. Eucarpia Cruciferae Newsletter. 1977;2:16. [Google Scholar]
- Krishnasamy S, Makaroff CA. Organ-specific reduction in the abundance of a mitochondrial protein accompanies fertility restoration in cytoplasmic male-sterile radish. Plant Mol Biol. 1994;26(3):935–946. doi: 10.1007/BF00028860. [DOI] [PubMed] [Google Scholar]
- Duroc Y, Gaillard C, Hiard S, Defrance MC, Pelletier G, Budar F. Biochemical and functional characterization of ORF138, a mitochondrial protein responsible for Ogura cytoplasmic male sterility in Brassiceae. Biochimie. 2005;87(12):1089–1100. doi: 10.1016/j.biochi.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Yang S, Terachi T, Yamagishi H. Inhibition of chalcone synthase expression in anthers of Raphanus sativus with ogura male sterile cytoplasm. Ann Bot. 2008;102(4):483–489. doi: 10.1093/aob/mcn116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makaroff CA, Palmer JD. Mitochondrial DNA rearrangements and transcriptional alterations in the male-sterile cytoplasm of Ogura radish. Mol Cell Biol. 1988;8(4):1474–1480. doi: 10.1128/mcb.8.4.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makaroff CA, Apel IJ, Palmer JD. The atp6 coding region has been disrupted and a novel reading frame generated in the mitochondrial genome of cytoplasmic male-sterile radish. J Biol Chem. 1989;264(20):11706–11713. [PubMed] [Google Scholar]
- Makaroff CA, Apel IJ, Palmer JD. Characterization of radish mitochondrial atpA: influence of nuclear background on transcription of atpA-associated sequences and relationship with male sterility. Plant Mol Biol. 1990;15(5):735–746. doi: 10.1007/BF00016123. [DOI] [PubMed] [Google Scholar]
- Makaroff CA, Apel IJ, Palmer JD. The role of coxI-associated repeated sequences in plant mitochondrial DNA rearrangements and radish cytoplasmic male sterility. Curr Genet. 1991;19(3):183–190. doi: 10.1007/BF00336485. [DOI] [PubMed] [Google Scholar]
- Krishnasamy S, Makaroff CA. Characterization of the radish mitochondrial orfB locus: Possible relationship with male sterility in Ogura radish. Curr Genet. 1993;24(1–2):156–163. doi: 10.1007/BF00324680. [DOI] [PubMed] [Google Scholar]
- Rankin CT, Cutright MT, Makaroff CA. Characterization of the radish mitochondrial nad3/rps12 locus: Analysis of recombination repeats and RNA editing. Curr Genet. 1996;29(6):564–571. doi: 10.1007/BF02426961. [DOI] [PubMed] [Google Scholar]
- Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): Comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31(20):5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 1997;15(1):57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- Krishnasamy S, Grant RA, Makaroff CA. Subunit 6 of the Fo-ATP synthase complex from cytoplasmic male-sterile radish: RNA editing and NH2-terminal protein sequencing. Plant Mol Biol. 1994;24(1):129–141. doi: 10.1007/BF00040580. [DOI] [PubMed] [Google Scholar]
- Satoh M, Kubo T, Nishizawa S, Estiati A, Itchoda N, Mikami T. The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol Genet Genomics. 2004;272(3):247–256. doi: 10.1007/s00438-004-1058-9. [DOI] [PubMed] [Google Scholar]
- Liu H, Cui P, Zhan K, Lin Q, Zhuo G, Guo X, Ding F, Yang W, Liu D, Hu S, Yu J, Zhang A. Comparative analysis of mitochondrial genomes between a wheat K-type cytoplasmic male sterility (CMS) line and its maintainer line. BMC Genomics. 2011;12:163. doi: 10.1186/1471-2164-12-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Kazama T, Yamada M, Toriyama K. Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. BMC Genomics. 2010;11:209. doi: 10.1186/1471-2164-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Herbon LA. Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28(1–2):87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD. The mitochondrial genome of the legume vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One. 2011;6(1):e16404. doi: 10.1371/journal.pone.0016404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma Y, Yoshida Y, Terachi T, Toriyama K, Mikami T, Kubo T. Polymorphic minisatellites in the mitochondrial DNAs of Oryza and Brassica. Curr Genet. 2011;57(4):261–270. doi: 10.1007/s00294-011-0345-3. [DOI] [PubMed] [Google Scholar]
- Rhoads DM, Levings Iii CS, Siedow JN. URF13, a ligand-gated, pore-forming receptor for T-toxin in the inner membrane of cms-T mitochondria. J Bioenerg Biomembr. 1995;27(4):437–445. doi: 10.1007/BF02110006. [DOI] [PubMed] [Google Scholar]
- Gagliardi D, Leaver CJ. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 1999;18(13):3757–3766. doi: 10.1093/emboj/18.13.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young EG, Hanson MR. A fused mitochondrial gene associated with cytoplasmic male sterility is developmentally regulated. Cell. 1987;50(1):41–49. doi: 10.1016/0092-8674(87)90660-X. [DOI] [PubMed] [Google Scholar]
- Kim DH, Kang JG, Kim BD. Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.) Plant Mol Biol. 2007;63(4):519–532. doi: 10.1007/s11103-006-9106-y. [DOI] [PubMed] [Google Scholar]
- Tang HV, Chen W, Pring DR. Mitochondrial orf107 transcription, editing, and nucleolytic cleavage conferred by the gene Rf3 are expressed in sorghum pollen. Sex Plant Reprod. 1999;12(1):53–59. doi: 10.1007/s004970050171. [DOI] [Google Scholar]
- Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D. et al. Cytoplasmic male sterility of rice with Boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;18(3):676–687. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Homme Y, Stahl RJ, Li XQ, Hameed A, Brown GG. Brassica nap cytoplasmic male sterility is associated with expression of a mtDNA region containing a chimeric gene similar to the pol CMS- associated orf224 gene. Curr Genet. 1997;31(4):325–335. doi: 10.1007/s002940050212. [DOI] [PubMed] [Google Scholar]
- Landgren M, Zetterstrand M, Sundberg E, Glimelius K. Alloplasmic male-sterile Brassica lines containing B. tournefortii mitochondria express an ORF 3′ of the atp6 gene and a 32 kDa protein. Plant Mol Biol. 1996;32(5):879–890. doi: 10.1007/BF00020485. [DOI] [PubMed] [Google Scholar]
- Satoh M, Kubo T, Mikami T. The Owen mitochondrial genome in sugar beet (Beta vulgaris L.): Possible mechanisms of extensive rearrangements and the origin of the mitotype-unique regions. Theor Appl Genet. 2006;113(3):477–484. doi: 10.1007/s00122-006-0312-4. [DOI] [PubMed] [Google Scholar]
- Terachi T, Yamaguchi K, Yamagishi H. Sequence analysis on the mitochondrial orfB locus in normal and Ogura male-sterile cytoplasms from wild and cultivated radishes. Curr Genet. 2001;40(4):276–281. doi: 10.1007/s00294-001-0256-9. [DOI] [PubMed] [Google Scholar]
- Kuhn K, Weihe A, Borner T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005;33(1):337–346. doi: 10.1093/nar/gki179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloszynska M. Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes-though this be madness, yet there's method in't. J Exp Bot. 2010;61(3):657–671. doi: 10.1093/jxb/erp361. [DOI] [PubMed] [Google Scholar]
- Janska H, Sarria R, Woloszynska M, Arrieta-Montiel M, Mackenzie SA. Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell. 1998;10(7):1163–1180. doi: 10.1105/tpc.10.7.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Kaur AP, MacKenzie SA, Dweikat IM. Substoichiometric shifting in the fertility reversion of cytoplasmic male sterile pearl millet. Theor Appl Genet. 2009;118(7):1361–1370. doi: 10.1007/s00122-009-0986-5. [DOI] [PubMed] [Google Scholar]
- Chen J, Guan R, Chang S, Du T, Zhang H, Xing H. Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus l. PLoS One. 2011;6(3): . doi: 10.1371/journal.pone.0017662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lim H, Park S, Cho KH, Sung SK, Oh DG, Kim KT. Identification of a novel mitochondrial genome type and development of molecular markers for cytoplasm classification in radish (Raphanus sativus L.) Theor Appl Genet. 2007;115(8):1137–1145. doi: 10.1007/s00122-007-0639-5. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Terachi T. Intra- and inter-specific variations in the mitochondrial gene orf138 of Ogura-type male-sterile cytoplasm from Raphanus sativus and Raphanus raphanistrum. Theor Appl Genet. 2001;103(5):725–732. doi: 10.1007/s001220100614. [DOI] [Google Scholar]
- Yamagishi H, Terachi T. Molecular and biological studies on male-sterile cytoplasm in the Cruciferae. IV. Ogura-type cytoplasm found in the wild radish, Raphanus raphanistrum. Plant Breeding. 1997;116(4):323–329. doi: 10.1111/j.1439-0523.1997.tb01006.x. [DOI] [Google Scholar]
- Yamagishi H, Terachi T. Molecular and biological studies on male-sterile cytoplasm in the Cruciferae. 1.The origin and distribution of Ogura male-sterile cytoplasm in Japanese wild radishes (Raphanus sativus L.) revealed by PCR-aided assay of their mitochondrial DNAs. Theor Appl Genet. 1994;87(8):996–1000. doi: 10.1007/BF00225794. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Terachi T. Molecular and biological studies on male-sterile cytoplasm in the cruciferae. III. Distribution of Ogura-type cytoplasm among Japanese wild radishes and Asian radish cultivars. Theor Appl Genet. 1996;93(3):325–332. doi: 10.1007/BF00223172. [DOI] [PubMed] [Google Scholar]
- Bonen L, Gray MW. Organization and expression of the mitochondrial genome of plants. I. The genes for wheat mitochondrial ribosomal and transfer RNA: Evidence for an unusual arrangement. Nucleic Acids Res. 1980;8(2):319–335. doi: 10.1093/nar/8.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21(4):537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of repeated sequences in normal-type genome.Repeated sequences(>80 bp, >90% homology) in normal-type genome were listed.
List of repeated sequences in Ogura-type genome.Repeated sequences(>80 bp, >90% homology) in Ogura-type genome were listed.
The sequences homologous to Brassicaceae mitochondrial genome plotted on Ogura-type specific region I -III. A black line indicates the region specific to the Ogura-type mitochondrial genome. Black boxes indicate the sequence homologous to radish and/or other Brassicaceae mitochondrial genomes.
Validation of contig linkage by PCR analysis. The primer information used for this PCR analysis is described in Additional file 5.
The primers used for the PCR analysis to validate the linkage between contigs of normal-type genome. Information on the primers used for the PCR analysis in Additional file 4.
The primers used for PCR analysis to validate the short repeats. Information on the primers used for PCR and sequencing, in order to confirm short repeats in Ogura-type genome.