Abstract
Brown fat expresses two PGC-1α isoforms (PGC-1α and NT-PGC-1α) and both play a central role in the regulation of cellular energy metabolism and adaptive thermogenesis by interacting with a wide range of transcription factors including PPARγ, PPARα, ERRα, and NRF1. PGC-1α consists of 797 amino acids, whereas alternative splicing of the PGC-1 α gene produces a shorter protein called NT-PGC-1α (aa 1–270). We report in this paper that transcriptional activity of PGC-1α and NT-PGC-1α is differently affected by the transcriptional regulator, Twist-1. Twist-1 suppresses PGC-1α but not NT-PGC-1α. The inhibition of PGC-1α activity by Twist-1 is mediated by direct interaction through the C-terminal region of PGC-1α (aa 353–797). Thus, the absence of the corresponding C-terminal domain in NT-PGC-1α allows NT-PGC-1α to be free from Twist-1-mediated inhibition. Overexpression of Twist-1 in brown adipocytes suppresses transcription of a subset of PGC-1α-target genes involved in mitochondrial fatty acid oxidation and uncoupling (CPT1β, UCP1, and ERRα). In contrast, NT-PGC-1α-mediated induction of these genes is unaffected by Twist-1. These findings show that differences in inhibitory protein-protein interactions of PGC-1α and NT-PGC-1α with Twist-1 lead to differential regulation of their function by Twist-1.
1. Introduction
The transcriptional coactivator PGC-1α was first identified as a coactivator of PPARγ in brown adipose tissue and is now known to interact with a broad range of nuclear receptors and transcription factors to regulate mitochondrial biogenesis in most tissues but also control adaptive thermogenesis, fatty acid/glucose metabolism, ROS metabolism, and muscle fiber type switching in a tissue-specific manner [1–7]. The function of PGC-1α among tissues is regulated by signaling inputs that increase transcription of PGC-1α and modulate the transcribed protein through tissue-specific posttranslational modifications [8–14]. This allows PGC-1α to function as a key regulator to link nutritional and environmental stimuli to the tissue-specific transcriptional programs.
Alternative splicing of PGC-1α produces an additional transcript that encodes a shorter isoform called NT-PGC-1α (aa 1–270) [15]. NT-PGC-1α is coexpressed with PGC-1α in metabolically active tissues and its expression is coregulated by the nutritional and environmental cues which activate the gene [15–17]. PGC-1α is a short-lived nuclear protein containing 797 amino acids. A variety of post-translational modifications enhance the stability and activity of PGC-1α by decreasing its targeting to the proteosome. In contrast, NT-PGC-1α is relatively stable since it is less effectively targeted to the proteosome due to lack of the C-terminal domain involved in proteosomal targeting [15]. Constitutive activation of target genes by NT-PGC-1α is effectively limited by a mechanism that sequesters NT-PGC-1α to the cytoplasm in a CRM1-dependent manner [16]. NT-PGC-1α activity is primarily modulated by increased translocation to the nucleus. PKA-dependent phosphorylation of NT-PGC-1α increases its nuclear retention and subsequent recruitment to the transcriptional complexes [16].
Another layer of regulation of PGC-1α function is mediated by direct interaction with other regulatory proteins. Previous studies have shown that p160MBP, RIP140, and Twist-1 bind to PGC-1α and repress its transcriptional activity. p160 myb binding protein was originally identified as a protein that interacts with the regulatory domain of PGC-1α (aa 200–400) in C2C12 myoblasts [18]. The docking of p160MBP on PGC-1α inhibits transcription of PGC-1α target genes [9, 18]. RIP140 is a transcriptional corepressor for a number of nuclear receptors in adipose tissue and skeletal muscle where it represses many PGC-1α target genes [19]. Mechanistically, RIP140 directly interacts with PGC-1α (aa 184–797) and suppresses its activity [20]. Recently, the transcription factor Twist-1 was also identified as a negative regulator of PGC-1α in brown adipose tissue. Twist-1 is a helix-loop-helix (HLH)-containing transcription factor involved in early development, apoptosis, cancer, and osteoblast differentiation [21–23]. A recent study reported that Twist-1 is recruited to the PGC-1α target genes by docking to the C-terminal domain of PGC-1α (aa 350–797) to negatively modulate oxidative metabolism and UCP1-dependent uncoupling in brown adipose tissue [24]. Interestingly, all of these negative regulators bind to the central to C-terminal region of PGC-1α, suggesting that these regulators would have little or no inhibitory effect on NT-PGC-1α function in the nucleus.
The present study was designed to investigate the effect of known PGC-1α repressors on NT-PGC-1α function and found that Twist-1 plays a differential role in the regulation of PGC-1α and NT-PGC-1α activity in brown adipocytes. Twist-1 significantly suppressed PGC-1α-mediated activation of CPT1β, UCP1, and ERRα by docking to the C-terminal region of PGC-1α. In contrast, NT-PGC-1α-dependent induction of these genes was not affected by Twist-1 due to lack of interaction with Twist-1.
2. Materials and Methods
2.1. Cell Cultures and Brown Adipocyte Differentiation
COS-1 cells were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin (Invitrogen). Immortalized PGC-1α-deficient mouse brown preadipocyte cell lines expressing empty vector, PGC-1α, or NT-PGC-1α [16] were maintained in DMEM supplemented with 10% FBS, 1% glutamine, and 1% penicillin/streptomycin. Preadipocytes were grown to confluence in culture medium supplemented with 20 nM insulin and 1 nM T3 (differentiation medium). Differentiation of brown adipocytes was induced (day 1) by incubating the cells in differentiation medium supplemented with 0.5 mM isobutylmethylxanthine (IBMX), 0.5 μM Dexamethasone, and 0.125 mM indomethacin for 48 hours. Thereafter, the cells were maintained in differentiation medium until day 7, followed by treatment with dibutyryl cAMP for 4 h.
2.2. Luciferase Reporter Assay
COS-1 cells were transiently transfected using Fugene6 (Roche) with following plasmids. For a transcriptional repression assay with RIP140, GAL4-responsive luciferase reporter (pGK), GAL4-DBD-fused mouse ERRα-LBD, and plasmids expressing PGC-1α-HA, NT-PGC-1α-HA, and RIP140 were used. For a transcriptional repression assay with Twist-1, (PPRE)3-TK-luc, pSV sport-PPARγ1, and plasmids expressing PGC-1α-HA, NT-PGC-1α-HA, and Twist-1 were used. pRL-SV40 control plasmid expressing Renilla luciferase was used for normalization. Cells were harvested for luciferase assay 48 h after transfection, and luciferase activity was determined using a Promega Dual Luciferase assay kit (Promega). The firefly luciferase activity was normalized with Renilla luciferase activity. Data represent mean ± SEM of at least four independent experiments.
2.3. Immunoprecipitation and Western Blot
COS-1 cells were transfected with pcDNA3.1-Flag-Twist-1 and pcDNA3.1-PGC-1α-HA or pcDNA3.1-NT-PGC-1α-HA. Cells were harvested 48 h after transfection, washed with ice-cold PBS, and lysed in IP buffer (20 mM HEPES-NaOH, pH 7.0, 150 mM NaCl, 0.2% NP-40) supplemented with a protease inhibitor cocktail (Roche). Lysates were precleared with protein G-agarose beads and immunoprecipitated with IgG and anti-Flag antibody (Sigma) overnight at 4°C. After washings, immunoprecipitates were subjected to Western blot analysis using anti-PGC-1α antibody directed against the N-terminus of PGC-1α (aa 1–200) and anti-Flag antibody.
2.4. Retroviral Infection
GP-293 cells were cotransfected with pVSV-G and pBABE-zeo or pBABE-zeo-Twist-1 using Profection transfection system (Promega). Following transfection, the cells were incubated at 32°C to increase viral titer. Virus-containing medium was collected, filtered through the 0.45 μm filter, and used to infect target cells. Immortalized PGC-1α-deficient mouse brown preadipocyte cells that ectopically express empty vector, PGC-1α, or NT-PGC-1α [16] were infected with the viral supernatant supplemented with 8 μg/mL polybrene. The medium was aspirated after 2 h and replaced with fresh viral supernatant, and the procedure was repeated. After 8 h of infection, the cells were replaced with fresh DMEM medium supplemented with 10% FBS, 1% glutamine, and 1% penicillin/streptomycin. Selection was initiated with zeocin (Invitrogen) 48 h after infection.
2.5. Real-Time PCR Analysis
Total RNA was isolated from brown adipocytes using Tri-Reagent (Molecular Research Center) and RNeasy kits (Qiagen). For quantitative RT-PCR analysis, 2 μg of RNA samples were reverse transcribed using oligo dT primers and M-MLV reverse transcriptase (Promega), and 4 ng of cDNA were used in quantitative PCR reactions in the presence of a fluorescent dye (Cybergreen, Takara) on Applied Biosystems 7900 (Applied Biosystems). Relative abundance of mRNA was normalized to that of cyclophilin mRNA. The primers for Twist-1, UCP1, CPT1β, Cox7a1, ERRα, VLCAD, PPARα, and cyclophilin were previously described [15, 16, 24].
3. Results
3.1. Twist-1 Negatively Regulates PGC-1α but Not NT-PGC-1α
Brown adipose tissue expresses two PGC-1α isoforms (PGC-1α and NT-PGC-1α), both of which regulate transcription of mitochondrial and thermogenic genes by promoting the activity of several nuclear receptors including PPARs [15–17]. RIP140 and Twist-1, which have been shown to negatively regulate PGC-1α activity in adipose tissue, were tested for their effect on regulation of NT-PGC-1α function. NT-PGC-1α (aa 1–270) partially contains a docking region for RIP140 that binds to the wide region of PGC-1α (aa 186–797), whereas the C-terminal Twist-1-binding domain (aa 353–797) is missing in NT-PGC-1α (Figure 1(a)). To assess the ability of these transcriptional regulators to modulate NT-PGC-1α activity, we carried out transient co-transfection and luciferase reporter assays as described in Materials and Methods. For a transcriptional repression assay with RIP140, Gal4-DBD-fused ERRα-LBD was used since RIP140 in part decreases transcriptional activity of full length ERRα [25]. The transcriptional activity of Gal4-ERRα-LBD was not affected by RIP140 (data not shown). Co-expression of RIP140 with PGC-1α significantly inhibited the ability of PGC-1α to increase Gal4-ERRα-LBD-mediated transcription of the reporter gene (Figure 1(b)). Similarly, RIP140 suppressed NT-PGC-1α-mediated induction of the reporter gene (Figure 1(b)), indicating that amino acids 186–270 in NT-PGC-1α are sufficient for RIP140 binding and repression. A transcriptional repression assay with Twist-1 showed that Twist-1 largely suppressed the ability of PGC-1α to increase PPARγ-mediated transcription, whereas NT-PGC-1α-dependent increase of reporter gene expression was not affected by Twist-1 (Figure 1(c)).
Figure 1.
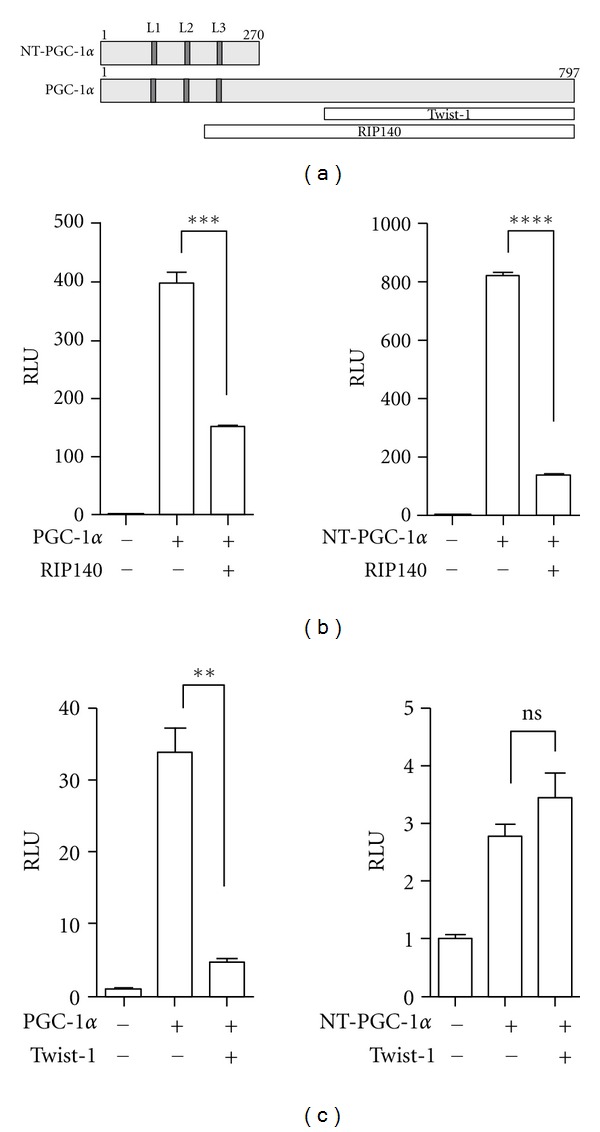
Effects of transcriptional regulators on PGC-1α and NT-PGC-1α function. (a) Schematic diagram of PGC-1α and NT-PGC-1α proteins. L1, L2, and L3 represent leucine-rich motifs, one of which (L2) bears the conserved LXXLL sequence that interacts with nuclear receptors. Two white boxes represent the interaction regions of PGC-1α with transcriptional regulators, Twist-1, and RIP140, respectively. (b), (c) Transcriptional repression assay using a luciferase reporter. pcDNA3.1, PGC-1α, and NT-PGC-1α were cotransfected in COS-1 cells with Gal4-ERRα-LBD and RIP140 (b) or PPARγ and Twist-1 (c). Luciferase activity was determined 48 h after transfection and the relative luciferase units were calculated as described in Materials and Methods. Data represent mean ± SEM of at least three independent experiments. Significant difference is determined by student t-test (**P < 0.01; ***P < 0.001; ****P < 0.0001; ns: not significant).
3.2. Twist-1 Does Not Interact with NT-PGC-1α
Twist-1 suppresses PGC-1α activity by docking to the C-terminal domain of PGC-1α (Figure 1) [24]. To test that no repression of NT-PGC-1α activity by Twist-1 is due to lack of interaction between two proteins, Flag-Twist-1 was co-expressed with PGC-1α-HA or NT-PGC-1α-HA in COS-1 cells and immunoprecipitated with IgG and anti-Flag antibody. PGC-1α was efficiently coprecipitated with Twist-1 but not with IgG control (Figure 2(a)). In contrast, NT-PGC-1α was not coimmunoprecipitated with Twist-1 (Figure 2(b)), suggesting that Twist-1 is not recruited to NT- PGC-1α target genes.
Figure 2.
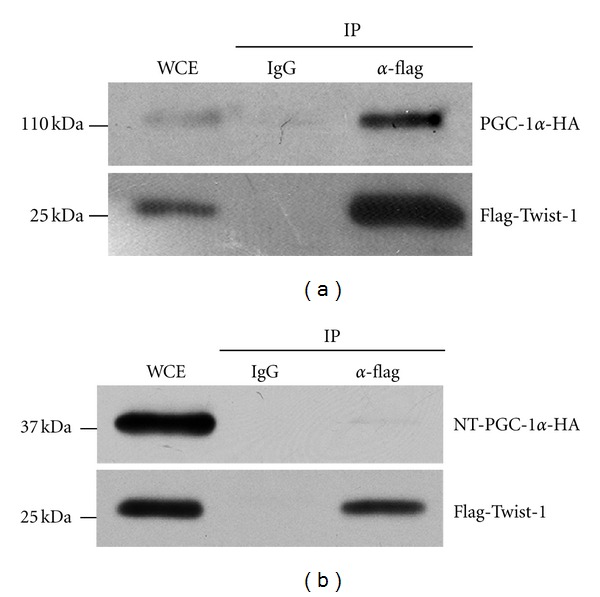
Interactions of Twist-1 with PGC-1α but not with NT-PGC-1α. PGC-1α-HA (a) and NT-PGC-1α-HA (b) were co-expressed with Flag-Twist-1 in COS-1 cells. Immunoprecipitates with IgG or anti-Flag antibody were separated by SDS-PAGE and immunoblotted with anti- PGC-1α (a, top panel) or HA antibody (b, top panel). Expression of Flag-Twist-1 was confirmed by probing with anti-Flag antibody (a and b, bottom panels).
3.3. Twist-1 Differentially Regulates a Subset of PGC-1α- and NT-PGC-1α-Target Genes in Brown Adipocytes
Twist-1 is selectively expressed in adipose tissue and its overexpression in brown adipocytes specifically suppresses PGC-1α-mediated activation of fatty acid oxidation and uncoupling genes [24]. Since NT-PGC-1α is co-expressed with PGC-1α in brown adipocytes and regulates many PGC-1α target genes [15–17], we hypothesized that PGC-1α and NT-PGC-1α are differently regulated by Twist-1 in brown adipocytes. To investigate a differential role of Twist-1 in PGC-1α- and NT-PGC-1α-target gene expression in brown adipocytes, the PGC-1α-deficient mouse brown preadipocyte cell lines expressing empty vector, PGC-1α, or NT-PGC-1α [16] were transduced with Twist-1 retrovirus to overexpress Twist-1. PPAR expression is not affected by PGC-1α or NT-PGC-1α [26]. In addition, Twist-1 does not change PPAR expression or activity [24]. After retroviral infection, the mRNA levels of Twist-1 were ~16- and ~12-fold increased in the PGC-1α-deficient brown preadipocytes expressing PGC-1α and NT-PGC-1α, respectively (Figure 3).
Figure 3.
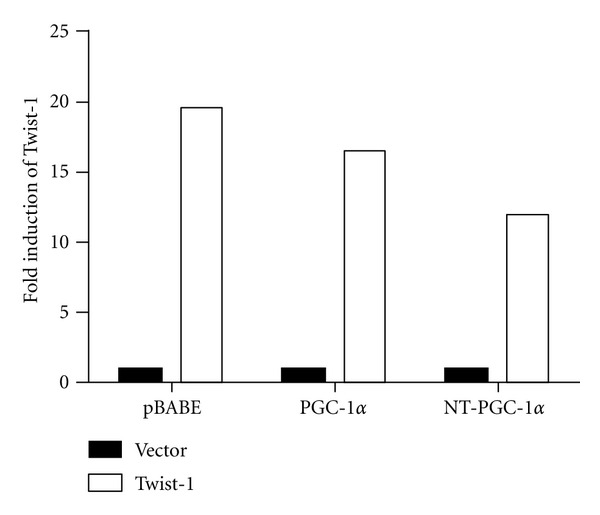
The elevated Twist-1 levels in brown preadipocytes by retroviral infection. Immortalized PGC-1α-deficient brown preadipocyte cells that express pBABE-neo vector, PGC-1α, or NT-PGC-1α were transduced with pBABE-zeo or Twist-1 retrovirus. Zeocine-resistant cells were pooled and Twist-1 expression was assessed by real time-PCR analysis. Relative abundance of Twist-1 mRNA was normalized to that of cyclophilin mRNA.
These brown preadipocyte cell lines were then differentiated for 7 days and treated with dibutyryl cAMP to maximize protein stability/activity of PGC-1α and increase nuclear retention of NT-PGC-1α [15, 16]. In response to cAMP-induced signaling, p38 MAPK increases stabilization and activation of PGC-1α protein by preventing its proteosomal targeting [8], whereas cAMP-activated PKA phosphorylates NT-PGC-1α, leading to inhibition of CRM1-mediated nuclear export [16]. Expression of aP2, a marker of adipocyte differentiation, was relatively comparable among the cell lines (Figure 4(a)). In agreement with previous findings [24], overexpression of Twist-1 in differentiated brown adipocytes significantly suppressed PGC-1α-mediated induction of CPT1β, UCP1, and ERRα, which are implicated in mitochondrial fatty acid oxidation and uncoupling (Figure 4(b)). In contrast, Twist-1 had no suppressive effect on NT-PGC-1α-dependent induction of CPT1β, UCP1, and ERRα (Figure 4(b)), indicating that Twist-1 differently regulates PGC-1α and NT-PGC-1α activity in brown adipocytes. With no addition of dibutyryl cAMP, PGC-1α and NT-PGC-1α are able to activate their target gene expression although their transcriptional activity is reduced. In the absence of cAMP signaling activation, cAMP-dependent increase of basal UCP1 levels was ~80% reduced, leading to large fold changes in PGC-1α- and NT-PGC-1α-mediated induction of UCP1 (12.2-fold and 10.3-fold, respectively) (Figure S1, available online at doi: 10.1155/2012/320454). Similarly, Twist-1 significantly suppressed PGC-1α-mediated induction of UCP1, whereas NT-PGC-1α-mediated increase of UCP1 gene expression was not affected by Twist-1 (Figure S1, available online at doi: 10.1155/2012/320454). Expression of many mitochondrial genes and nuclear receptors is also regulated by PGC-1α and NT-PGC-1α [15–17]. However, neither PGC-1α- nor NT-PGC-1α-dependent induction of Cox7a1, PPARα, and VLCAD (Figure 4(c)) and MCAD, Atp5b (not shown) was affected by Twist-1. This suggests that Twist-1 negatively regulates only a subset of PGC-1α target genes in brown adipocytes.
Figure 4.
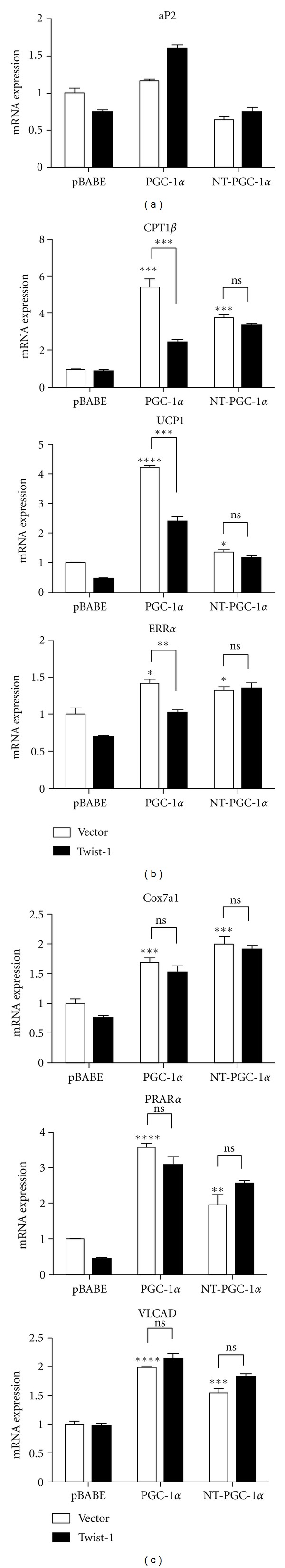
Differential effect of Twist-1 on PGC-1α and NT-PGC-1α target gene expression in brown adipocytes. (a), (b), (c) Quantitative real time PCR analysis of gene expression in differentiated PGC-1α-null brown adipocytes expressing empty vector (pBABE), PGC-1α, and NT-PGC-1α with empty vector (white bars) or Twist-1 (black bars). Relative abundance of mRNA levels was normalized to that of cyclophilin mRNA. Data represent mean ± SEM of at least four independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns: not significant.
4. Discussion
We previously reported that PGC-1α and NT-PGC-1α regulate a number of mitochondrial and thermogenic genes in brown adipose tissue [15–17]. Sympathetic stimulation of BAT by cold increases transcription of the PGC-1α gene by activating and recruiting cAMP-dependent transcription factors, ATF2 and CREB, to the PGC-1α promoter [1, 27]. Subsequent normal and alternative splicings of the transcribed RNA produce comparable mRNA levels of PGC-1α and NT-PGC-1α, respectively [15]. However, two transcripts produce structurally different proteins that possess fundamental differences in their protein size, stability, and localization. These different natures of two PGC-1α isoforms require different regulatory mechanisms to increase their transcriptional activity in response to the same signaling inputs. For example, cold/cAMP-activated p38 MAPK phosphorylation leads to stabilization and activation of PGC-1α protein by preventing its proteosomal targeting [8]. In contrast, cAMP-activated PKA phosphorylation increases nuclear accumulation of NT-PGC-1α and subsequent recruitment to the transcriptional complexes [16].
Here we show an additional regulatory mechanism that differently modulates transcriptional activity of PGC-1α and NT-PGC-1α in the nucleus of brown adipocytes. The mode of action is mediated by direct interaction of PGC-1α with a negative regulator Twist-1, which is abundantly expressed in brown adipocytes. Twist-1 is recruited to PGC-1α target genes by docking to the C-terminal region of PGC-1α and inhibits their expression by subsequently recruiting the histone deacetylase HDAC5 to the PGC-1α target gene promoters (e.g., UCP1 and CPT1) [24]. In contrast, Twist-1 has no inhibitory effect on NT-PGC-1α-mediated induction of NT-PGC-1α target genes since NT-PGC-1α does not recruit Twist-1 to its target gene promoters. Despite potential inhibition of all PGC-1α target genes by Twist-1, Twist-1 suppresses only a subset of PGC-1α target genes, including UCP1, CPT1β, and ERRα. Twist-1 is a basic helix-loop-helix (bHLH)-containing transcription factor that binds to the canonical E-box and the related sequences in the regulatory regions of target genes [28], thus raising a possibility that the presence of potential E-boxes on the PGC-1α target gene promoters further specifies a subset of PGC-1α/Twist-1 target genes. However, it seems unlikely that subsequent docking of Twist-1 to the potential E-boxes on the PGC-1α target gene promoters is required for its inhibitory effect since Twist-1-mediated suppression does not depend on its DNA-binding activity [24]. Instead, Twist-1 exerts its transcriptional repression on PGC-1α target genes by altering chromatin conformational states by recruitment of histone deacetylases (HDAC) [24]. Thus, it is likely that subsequent recruitment of additional regulators (e.g., histone deaceylases) to the PGC-1α target gene promoters by Twist-1 is required for its suppression of PGC-1α-mediated gene expression.
5. Conclusion
Our findings demonstrate a differential regulation of PGC-1α and NT-PGC-1α activity by Twist-1 in brown adipocytes. Twist-1 suppresses PGC-1α-mediated transcriptional activation of a subset of PGC-1α target genes, including UCP1, CPT1β, and ERRα. In contrast, NT-PGC-1α-mediated induction of these genes is not affected by Twist-1.
Supplementary Material
Figure S1. Differential effect of Twist-1 on PGC-1α- and NT-PGC-1α-dependent induction of UCP1.
Quantitative real time PCR analysis of gene expression in differentiated PGC-1α-null brown adipocytes expressing empty vector (pBABE), PGC-1α, and NT-PGC-1α in the absence (white bars) or presence of Twist-1 (black bars). Differentiated brown adipocytes were collected without treatment with dibutyryl cAMP. Relative abundance of mRNA levels was normalized to that of cyclophilin mRNA. Data represent mean ± SEM of at least four independent experiments. ∗∗∗∗,p < 0.0001; ns, not significant.
Acknowledgments
The authors thank Dr. Anthony Firulli for providing pcDNA3.1-Flag-Twist-1. The authors also thank Yagini Joshi for excellent technical contribution and Cindi Tramonte for outstanding administrative support. This work was supported by NIH R01 DK 074772 (TWG), 1R01 DK096311 (TWG), ADA1-12-BS-58 (TWG), and NIH-8 P20 GM103528 (TWG, JSC). This work was also supported by PBRC Nutrition Obesity Research Center (NORC) Pilot & Feasibility Grant 2P30 DK072476 (JSC). This work used Genomics core facility that is supported in part by COBRE (NIH-8 P20 GM103528) and NORC (NIH 2P30-DK072476) center Grants from the National Institutes of Health.
Abbreviations
- PGC-1α:
Peroxisome proliferator-activated receptor γ coactivator 1α
- ERRα:
Estrogen-related receptor α
- NRF1:
Nuclear respiratory factor 1
- CPT1β:
Carnitine palmitoyltransferase 1β
- UCP1:
Uncoupling proteins 1
- ROS:
Reactive oxygen species
- PKA:
Protein kinase A
- p160MBP:
p160 Myb binding protein
- RIP140:
Receptor-interacting protein 140
- MAPK:
Mitogen-activated protein kinase
- VLCAD:
Very long-chain acyl-CoA dehydrogenase
- MCAD:
Medium-chain acyl-CoA dehydrogenase
- ATF2:
Activating transcription factor 2
- CREB:
cAMP response element-binding.
References
- 1.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 2.Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 3.Yoon JC, Puigserver P, Chen G, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413(6852):131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 4.Rhee J, Inoue Y, Yoon JC, et al. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 6.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Molecular and Cellular Biology. 2000;20(5):1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St-Pierre J, Drori S, Uldry M, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127(2):397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 8.Puigserver P, Rhee J, Lin J, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Molecular Cell. 2001;8(5):971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 9.Knutti D, Kressler D, Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(17):9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jäer S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α . Proceedings of the National Academy of Sciences of the United States of America. 2007;104(29):12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes and Development. 2005;19(12):1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature. 2007;447(7147):1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 13.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 14.Herzig S, Long F, Jhala US, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413(6852):179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Huypens P, Adamson AW, et al. Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1α . The Journal of Biological Chemistry. 2009;284(47):32813–32826. doi: 10.1074/jbc.M109.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang JS, Huypens P, Zhang Y, Black C, Kralli A, Gettys TW. Regulation of NT-PGC-1αsubcellular localization and function by protein kinase A-dependent modulation of nuclear export by CRM1. The Journal of Biological Chemistry. 2010;285(23):18039–18050. doi: 10.1074/jbc.M109.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang JS, Fernand V, Zhang Y, et al. NT-PGC-1α protein is sufficient to link β3-adrenergic receptor activation to transcriptional and physiological components of adaptive thermogenesis. The Journal of Biological Chemistry. 2012;287(12):9100–9111. doi: 10.1074/jbc.M111.320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan M, Rhee J, St-Pierre J, et al. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: modulation by p38 MAPK. Genes and Development. 2004;18(3):278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White R, Morganstein D, Christian M, Seth A, Herzog B, Parker MG. Role of RIP140 in metabolic tissues: connections to disease. FEBS Letters. 2008;582(1):39–45. doi: 10.1016/j.febslet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 20.Hallberg M, Morganstein DL, Kiskinis E, et al. A functional interaction between RIP140 and PGC-1α regulates the expression of the lipid droplet protein CIDEA. Molecular and Cellular Biology. 2008;28(22):6785–6795. doi: 10.1128/MCB.00504-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes and Development. 1995;9(6):686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 22.Šošić D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-κB activity. Cell. 2003;112(2):169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 23.Bialek P, Kern B, Yang X, et al. A twist code determines the onset of osteoblast differentiation. Developmental Cell. 2004;6(3):423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 24.Pan D, Fujimoto M, Lopes A, Wang YX. Twist-1 is a PPARδ-inducible, negative-feedback regulator of PGC-1α in brown fat metabolism. Cell. 2009;137(1):73–86. doi: 10.1016/j.cell.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castet A, Herledan A, Bonnet S, Jalaguier S, Vanacker JM, Cavaillès V. Receptor-interacting protein 140 differentially regulates estrogen receptor-related receptor transactivation depending on target genes. Molecular Endocrinology. 2006;20(5):1035–1047. doi: 10.1210/me.2005-0227. [DOI] [PubMed] [Google Scholar]
- 26.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metabolism. 2006;3(5):333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Cao W, Daniel KW, Robidoux J, et al. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Molecular and Cellular Biology. 2004;24(7):3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes RM, Firulli AB. A twist of insight—the role of twist-family bHLH factors in development. International Journal of Developmental Biology. 2009;53(7):909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Differential effect of Twist-1 on PGC-1α- and NT-PGC-1α-dependent induction of UCP1.
Quantitative real time PCR analysis of gene expression in differentiated PGC-1α-null brown adipocytes expressing empty vector (pBABE), PGC-1α, and NT-PGC-1α in the absence (white bars) or presence of Twist-1 (black bars). Differentiated brown adipocytes were collected without treatment with dibutyryl cAMP. Relative abundance of mRNA levels was normalized to that of cyclophilin mRNA. Data represent mean ± SEM of at least four independent experiments. ∗∗∗∗,p < 0.0001; ns, not significant.


