Abstract
Nitration of tryptophan residues is a novel post-translational modification. In the present study, we examined whether NO2Trp (nitrotryptophan)-containing proteins are produced in the hippocampus and cerebellum of the adult rat under physiological conditions in vivo. Using Western blot analysis with anti-6-NO2Trp-specific antibody, we found many similar immunoreactive spots in the protein extracts from both regions. These spots were subsequently subjected to trypsin digestion and LC-ESI-MS/MS (LC-electrospray ionization-tandem MS) analysis. We identified several cytoskeletal proteins and glycolytic enzymes as NO2Trp-containing proteins and determined the position of nitrated tryptophan residues with significant ion score levels (P<0.05) in several proteins in both regions. We also observed that the total amount of NO2Trp-containing proteins in the cerebellum was significantly greater than that in the hippocampus (P<0.05). Moreover, IP (immunoprecipitation) assays using anti-aldolase C antibody showed that the relative intensity of immunostaining for NO2Trp over aldolase C was much higher in cerebellum than in hippocampus. The amounts of nNOS (neuronal nitric oxide synthase) and eNOS (endothelial nitric oxide synthase) were much greater in cerebellum than in hippocampus. This is the first evidence of several specific sites of nitrated tryptophan in proteins under physiological conditions in vivo.
Keywords: cerebellum, hippocampus, 6-nitrotryptophan, proteome, tandem MS analysis, tryptophan residue
Abbreviations: AD, Alzheimer's disease; ALP, alkaline phosphatase; DTT, dithiothreitol; eNOS, endothelial nitric oxide synthase; IEF, isoelectric focusing; iNOS, inducible nitric oxide synthase; IP, immunoprecipitation; IPG, immobilized pH gradient; LC-ESI-MS/MS, LC-electrospray ionization-tandem MS; nNOS, neuronal nitric oxide synthase; NO2Trp, nitrotyptophan; PDCE2, dihydrolipoly(lysine) residue acetyltransferase component of pyruvate dehydrogenase complex; RNS, reactive nitrogen species; SCOT, succinyl-CoA 3-ketoacid coenzyme A transferase; TBST, Tris-buffered saline with Tween 20
INTRODUCTION
Protein nitration is a post-translational modification that is induced by RNS (reactive nitrogen species) such as peroxynitrite (ONOO−) and nitrogen dioxide (NO2). ONOO− is produced by the very fast reaction between NO and superoxide radical in vivo. NO2 is formed by the reaction between H2O2 and nitrite, which is catalysed with myeloperoxidase and also by decomposition of ONOO−. These RNS are known to have the potential to cause oxidation or nitration of various biomolecules. One of the modifications by RNS is the nitration of tyrosine residues in proteins [1–3]. It has been reported that nitrotyrosine-containing proteins were observed in neurons of AD (Alzheimer's disease) [4,5], Lewy bodies of Parkinson's disease [6] and aged rat cerebellum [7]. Therefore nitrotyrosine has been used as a marker of oxidative and nitrative stress in brain. On the other hand, nitrotyrosine-containing proteins were detected endogenously in the brain of adult mice (9 weeks old) by using proteomic analysis [8]. Moreover, it has been shown that protein tyrosine nitration may regulate cell differentiation [9] and control microtubule dynamics [10]. Thus, protein tyrosine nitration also occurs under physiological conditions in vivo.
We have found another type of protein nitration as a novel post-translational modification, namely, formation of NO2Trp (nitrotryptophan) in proteins. Although several types of nitrated tryptophan, such as 5- and 6-NO2Trp, have been detected through in vitro and in vivo studies [11,12], we have demonstrated that 6-NO2Trp is the major nitrated products [12,13]. Previously, Ishii et al. [14] demonstrated the presence of 27–33 nmol of 6-NO2Trp per mol of tryptophan in protease-digested liver of mice after acetaminophen administration by using a quantitative method with LC-ESI-MS/MS (LC-electrospray ionization-tandem MS). Therefore we have developed monoclonal and polyclonal anti-6-NO2Trp-specific antibodies and constructed a method to detect 6-NO2Trp residue-containing proteins in ONOO− modified PC12 cells or their cell lysates by using these antibodies and LC-ESI-MS/MS analyses [15,16]. We identified several glycolytic enzymes and functional proteins as 6-NO2Trp residue-containing proteins and determined the positions of 6-NO2Trp in the amino acid sequences [16]. In addition, we found changes of 6-NO2Trp-containing proteins in the differentiation process from PC12 cells to neuron-like cells induced by NGF (nerve growth factor). We also successfully identified several specific sites of nitrated tryptophan in these proteins [17]. These findings raise the possibility that 6-NO2Trp-containing proteins could be detected in physiological conditions in vivo. We have chosen rat cerebellum and hippocampus as targets for the formation of 6-NO2Trp-containing proteins in physiological processes. Cerebellum contains all three isoforms of NOS (nitric oxide synthase) and the highest levels of nNOS (neuronal NOS), which generate NO as a component of ONOO−, compared with other brain regions [18]. Hippocampus CA1 and cerebellar granule cells are known to be more sensitive to oxidative stress than other brain regions under basal conditions [19].
In the present study, we found 12 NO2Trp-containing proteins from hippocampus and seven proteins from cerebellum and determined the positions of the nitrated tryptophan residues in these amino acid sequences. We also observed that the total amount of 6-NO2Trp-containing proteins in the cerebellum was significantly greater than that in hippocampus (P<0.05). Moreover, we suggested that an increase in nitration rate of the proteins might contribute to the abundance of 6-NO2Trp-containing proteins in cerebellum, at least partially, by focusing on aldolase C as a typical example. This is the first evidence of several specific sites of nitrated tryptophan in proteins under physiological conditions in vivo.
MATERIALS AND METHODS
Animals
The institutional ethics review committee of Juntendo University approved all experimental protocols. Adult male Fischer 344/N rats (6 months of age, n=5) were obtained from Japan SLC Inc. The rats were housed under a 12 h light/12 h dark in an environmentally controlled room (23±1°C room temperature, 55±5% relative humidity) and were provided with food and water ad libitum.
Sample preparation
The rats were deeply anaesthetized with sodium pentobarbital and their brains were removed. Hippocampus and cerebellum were dissected from the brain and frozen in isopentane cooled to approximately −80°C. The frozen hippocampus and cerebellum were stored at −80°C until use. The frozen hippocampus and cerebellum were homogenized in a lysis buffer containing 40 mM Tris/HCl, 8 M urea, 4% CHAPS, 65 mM DTT (dithiothreitol), 1 mM EDTA, and Complete protease inhibitor (Roche). The homogenates were then centrifuged at 15000 g for 15 min, and the middle layer, containing the proteins, was carefully withdrawn. Protein concentrations were determined by the Bradford method (Protein assay kit; Bio-Rad).
Two-dimensional electrophoresis and SDS/PAGE
Three samples from each hippocampus and cerebellum were used for proteomic analysis. A portion [200 μg (Western blotting) and 300 μg (Sypro Ruby staining)] of protein was dissolved in the rehydration buffer containing 8 M urea, 4% CHAPS, 40 mM DTT, 0.5% IPG (immobilized pH gradient) buffer and Bromophenol Blue. Then, this solution was applied to an IPG strip (Immobiline™ DryStrip pH 3–10 NL, 7 cm; GE Healthcare) and the IPG strip was rehydrated for at least 10 h at 20°C. Next, IEF (isoelectric focusing) was performed at 20°C on an Ettan IPGphor 3 apparatus (GE Healthcare) using a protocol provided by the supplier. After IEF, IPG strips were equilibrated for 15 min in a solution containing 50 mM Tris/HCl (pH 6.8), 6 M urea, 30% glycerol, 2% SDS, 1% DTT and Bromophenol Blue. Two-dimensional SDS/PAGE was carried out using 10% polyacrylamide gels. Subsequently, Western blotting and Sypro Ruby staining were performed. For semi-quantitative analyses of the proteins and enzymes, SDS/PAGE and Western blotting were carried out.
Western blot analysis
After two-dimensional electrophoresis and standard SDS/PAGE, proteins in gels were transferred on to a PVDF membrane (Immobilon-P, 0.45 μm; Millipore). Non-specific binding sites were blocked for 1 h at room temperature with Block Ace (DS Pharma Biomedical) and an additional 1 h with non-fat dried skimmed milk powder in TBST (Tris-buffered saline with Tween 20), pH 7.6. The membranes were incubated overnight at 4°C with primary antibodies followed by 1 h incubation at room temperature with ALP (alkaline phosphatase)-conjugated secondary antibodies. The signals were visualized by chemiluminescence using Immunstar-AP substrate (Bio-Rad). The primary antibodies used in the present study were as follows: rabbit and mouse anti-6-NO2Trp antibody developed in our laboratory [15], mouse anti-nNOS, eNOS (endothelial nitric oxide synthase), iNOS (inducible NOS) antibodies (BD Biosciences), goat anti-aldolase C antibody (Santa Cruz Biotechnology), and mouse anti-β-actin antibody (Sigma–Aldrich). The secondary antibodies used in the present study were as follows: ALP-conjugated anti-mouse IgG Fc-specific (1:50000, Sigma–Aldrich), ALP-conjugated anti-rabbit IgG Fc-specific (1:50000, Thermo Scientific–Pierce), ALP-conjugated anti-goat IgG (1:50000, Millipore Chemicon). For the control experiment for the specificity of the anti-6-NO2Trp antibodies, we used a modified method, which was originally reported in a previous study [20] to reduce nitrated proteins. After the transfer, the membranes were exposed to a boiled 0.1 M PBS (pH 7.2) containing 10 mM DTT and 25 μM bovine Hb (haemoglobin) (Sigma) or boiled 0.1 M PBS without both DTT and Hb for 3 min. Then, the membranes were washed with TBST, non-specific binding was blocked, and Western blotting was performed. Band densities were determined using Image J software.
Nano ESI-MS/MS
Sypro Ruby-stained gel spots, which were observed as anti-6-NO2Trp-positive signals in the Western blotting, were cut and digested with trypsin. The tryptic peptides were subjected to LC-ESI-MS/MS analysis using a Thermo Fisher Scientific LXQ mass spectrometer with nano-LC (AMR). The LXQ mass spectrometer system consists of a nano-ESI apparatus and an ion trap mass spectrometer. Samples were introduced into the mass spectrometer at 500 nl/min. Typical ESI conditions were as follows: ion spray voltage 1.8 kV, heated capillary temperature 200°C, capillary voltage 40 V and tube lens 115 V. Collision-induced dissociation-MS/MS experiments were performed on mass-selected precursor ions using standard isolation and excitation procedures (activation q value 0.25, activation time 30 ms). The collision energy used was 35 (arbitrary units). The conditions of nano-LC were as follows: Magic C18 column (0.2 mm internal diameter×150 mm) and elution with 0.1% formic acid in 2% CH3CN (solvent A) and 0.1% formic acid in 90% CH3CN (solvent B) using a programme of 5% solvent B for 10 min equilibration and a gradient at 2% solvent B/min for 30 min with a flow rate of 500 nl/min. A database search of Swiss-Prot was performed using the MASCOT search engine (Matrix Science). For identification of 6-NO2Trp residues, a modification of 44.99 Da was applied on each of the tryptophan residues. In addition, the following modifications were accounted for during the search: oxidation of Met (+16 Da), methylation of His (+14 Da) and formylation of lysine residue (+28 Da).
IP (immunoprecipitation)
For IP, 60 μl of Protein G–Sepharose 4 Fast Flow beads (GE Healthcare) was washed with IP buffer containing 10 mM Tris/HCl, pH 7.5, 150 mM NaCl, 100 mM EDTA, 1% Nonidet P-40 and Complete protease inhibitor (Roche). The beads were suspended in l ml of IP buffer containing 500 μg of protein from the homogenates of hippocampus or cerebellum and rotated at 4°C for 1 h to remove proteins which are non-specifically bound to the beads. After rotation, the beads were centrifuged and the supernatants were placed in new microcentrifuge tubes. Subsequently, new beads and 2 μg of goat anti-aldolase C antibody were added to the tubes, and rotated overnight at 4°C. The protein-bound beads were collected by centrifugation and washed three times with IP buffer. The bound proteins were solubilized with sample buffer and incubated at room temperature for 2 h. After incubation, the protein samples were loaded on to 10% polyacrylamide gels. Then, Western blotting using mouse anti-6-NO2Trp and goat anti-aldolase C antibodies was performed.
Statistical analysis
Results are presented as means±S.D. A paired Student's t test was used to analyse the differences between hippocampus and cerebellum. Statistical significance was set at P<0.05.
RESULTS
Western blotting with the 6-NO2Trp antibody
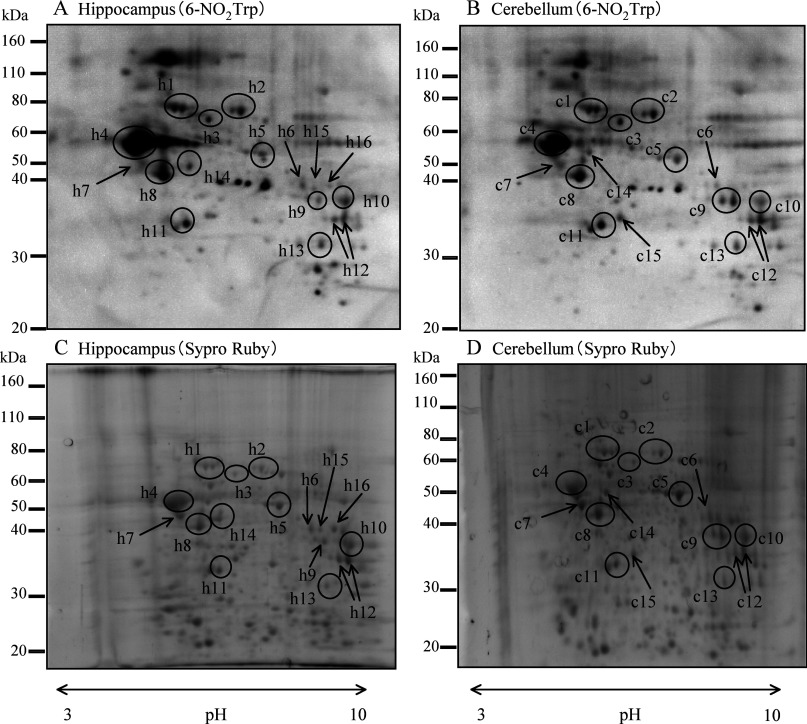
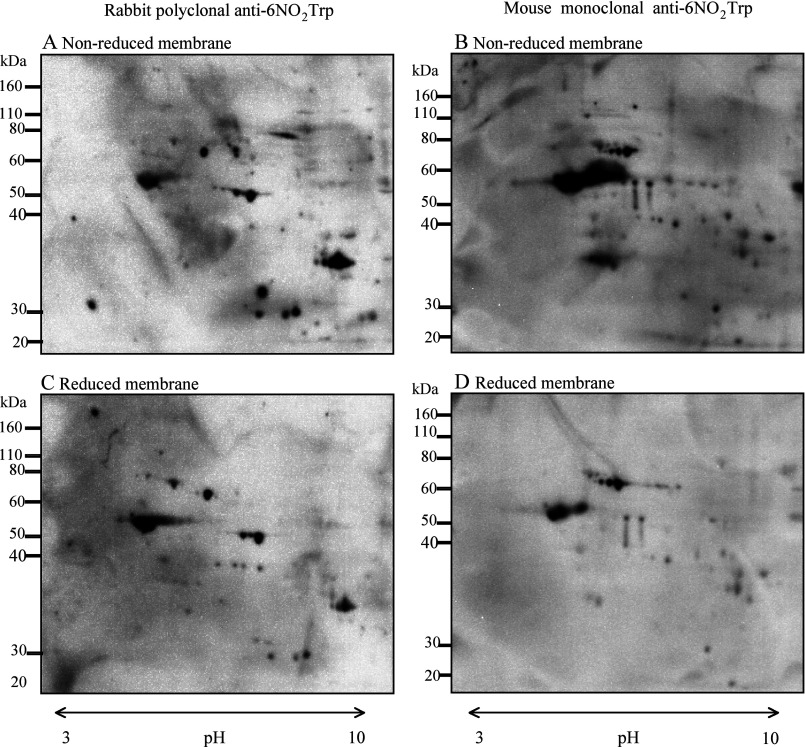
Figure 1 shows representative results of Western blotting with the anti-6-NO2Trp antibody (Figures 1A and 1B) and Sypro Ruby staining gels (Figures 1C and 1D) after two-dimensional PAGE of hippocampus (Figures 1A and 1C) and cerebellum (Figures 1B and 1D). Many immunoreactive spots of anti-6-NO2Trp antibody were observed in both of the brain regions from the 6-month-old adult rats. Although several immunoreactive spots were detected only in hippocampus or cerebellum, most of the immunoreactive spots were observed in both. In the control experiment, NO2Trp was reduced (Figures 2C and 2D) by the method described in the text. The results of the same treatment without reducing agents are shown in Figures 2(A) and 2(B). The immunoreactive spots of rabbit polyclonal and mouse monoclonal anti-6-NO2Trp antibody became weak and several spots disappeared by the reduction. Although complete disappearance of the spots had not been attained by this treatment, it has been reported that reduction can be problematic and sometimes may not fully reduce all of the nitrated protein present [21]. Therefore these results indicate the specificity of both anti-6-NO2Trp antibodies is considerably high.
Figure 1. Representative results of Western blotting with an anti-6-NO2Trp antibody and Sypro Ruby-stained gel after two-dimensional electrophoresis.
(A) and (C) are Western blotting and Sypro Ruby gel staining of hippocampus respectively. (B) and (D) are those of cerebellum. Open circled spots and spots indicated by arrows in hippocampus (h1–h16) and cerebellum (c1–c15) were subjected to LC-ESI-MS/MS analysis.
Figure 2. Representative non-reduced and reduced membranes.
The samples in the membranes were treated without reducing agents (A and B) and with reducing agents (C and D) as described in the text. Immunoreactive spots in (A) and (C) are detected using rabbit polyclonal anti-6-NO2Trp antibody. Immunoreactive spots in (B) and (D) are detected using mouse monoclonal anti-6-NO2Trp antibody.
LC-ESI-MS/MS analysis of 6-NO2Trp-antibody positive spots
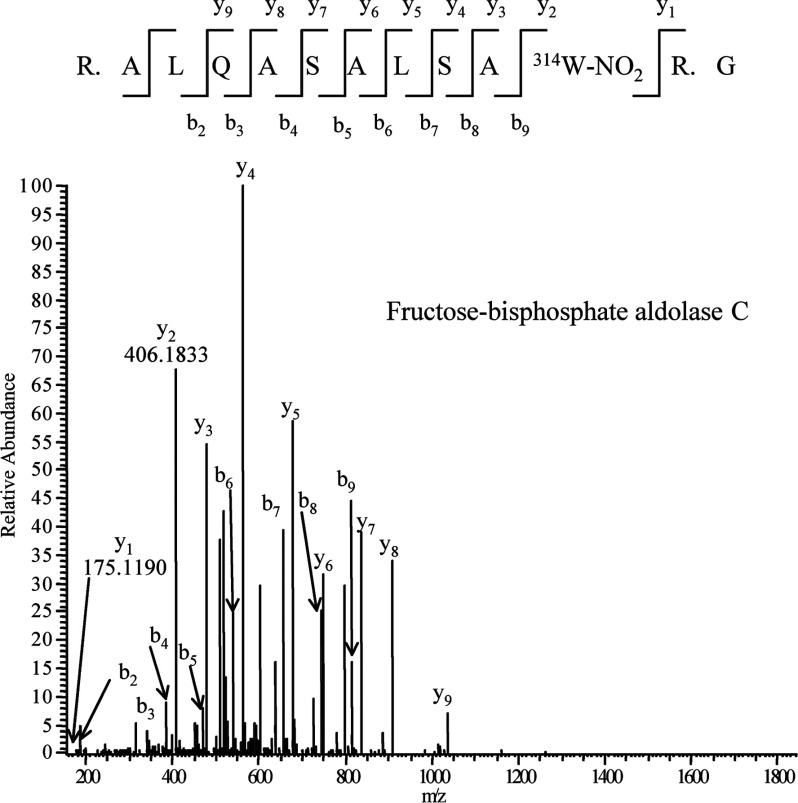
A total of 16 immunoreactive spots and 15 spots on the membrane for the samples from hippocampus and cerebellum respectively were cut out from Sypro Ruby-stained gels and digested with trypsin. The digested samples were analysed with LC-ESI-MS/MS. Putative 6-NO2Trp-containing proteins from both regions are shown in Table 1. Although four proteins including mitochondrial electron transfer flavoprotein subunit α and creatine kinases were identified only in hippocampus and three proteins including voltage-dependent anion-selective channel protein 1 were identified only in cerebellum as putative 6-NO2Trp-containing proteins (Table 1), most of the other spots included the same 12 proteins, such as α-enolase, tubulins and fructose-bisphosphate aldolases (Table 1). We successfully determined the positions of the nitrated tryptophan residues in amino acid sequences of 12 proteins from hippocampus (Table 2) and seven proteins from cerebellum (Table 3). These proteins in both regions included cytoskeletal proteins and glycolytic enzymes. Figure 3 shows the MS/MS spectrum of fructose-bisphosphate aldolase C as a typical example of the MS/MS analyses.
Table 1. Putative NO2Trp-containing proteins in the hippocampus and cerebellum.
All proteins cleared P<0.05 of the individual ion scores by the Mascot search. Accession numbers are from the Swiss-Prot database. Sequence coverage indicates an average of the samples in each region. ‘h’ and ‘c’ under sequence coverage indicate ‘hippocampus’ and ‘cerebellum’ respectively.
| Sequence coverage (%) | ||||||
|---|---|---|---|---|---|---|
| Spot number | Protein | Nominal mass (Da) | Calculated pI value | h | c | Accession number |
| h1, c1 | Heat-shock cognate 71 kDa protein | 71055 | 5.37 | 66 | 76 | P63018 |
| h2, c2 | Serum albumin | 70682 | 6.09 | 39 | 64 | P02770 |
| h3, c3 | PDCE2, mitochondria | 67637 | 8.76 | 35 | 55 | P08461 |
| h4, c4 | Tubulin β-2A chain | 50274 | 4.78 | 57 | 77 | P85108 |
| Tubulin β-2B chain | 50377 | 4.78 | 57 | 77 | Q3KRE8 | |
| Tubulin β-2C chain | 50225 | 4.79 | 54 | 69 | Q6P9T8 | |
| Tubulin β-5 chain | 50095 | 4.78 | 53 | 75 | P69897 | |
| h5, c5 | α-Enolase | 47440 | 6.61 | 69 | 83 | P04764 |
| h6, c6 | Glutamine synthetase | 42982 | 6.64 | 37 | 61 | P09606 |
| h7, c7 | γ-Enolase | 47510 | 5.03 | 83 | 92 | P07323 |
| h8, c8 | Actin, cytoplasmic 2 | 42381 | 5.23 | 69 | 86 | P63259 |
| h9, c9 | Fructose-bisphosphate aldolase C | 39658 | 6.67 | 87 | 89 | P09117 |
| h10, c10 | Fructose-bisphosphate aldolase A | 39783 | 8.31 | 70 | 91 | P05065 |
| h11, c11 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β1 | 38151 | 5.6 | 49 | 69 | P54311 |
| h12, c12 | GAPDH | 36090 | 8.14 | 58 | 72 | P04797 |
| h13 | Electron transfer flavoprotein subunit α, mitochondria | 35272 | 8.62 | 44 | − | P13803 |
| h14 | Creatine kinase B-type | 42983 | 5.56 | 55 | − | P07335 |
| h15 | Phosphoglycerate kinase 1 | 44909 | 8.02 | 67 | − | P16617 |
| h16 | Creatine kinase U-type, mitochondrial | 47398 | 8.02 | 47 | − | P25809 |
| c13 | Voltage-dependent anion-selective channel protein 1 | 30851 | 8.62 | − | 51 | Q9Z2L0 |
| c14 | Glial fibrillary acidic protein | 49984 | 5.35 | − | 94 | P47819 |
| c15 | L-Lactate dehydrogenase B chain | 36874 | 5.70 | − | 64 | P42123 |
Table 2. Nitrotryptophan-containing trypsin-digested peptide sequnces identified from hippocampus.
Boldface W indicates nitrated tryptophan residue. All peptides cleared P<0.05 of the ion scores by the Mascot search. Supplementary Figures S4—S16 are available online at http://www.bioscirep.org/bsr/032/bsr0320521add.htm.
| Protein | Peptide sequence | MS/MS spectrum |
|---|---|---|
| PDCEZ, mitochondrial | K475.VPEANSSW483MDTVIR.Q490 | Supplementary Figure S4 |
| R606.VVDGAVGAQW616LAEFKK.Y623 | Supplementary Figure S5 | |
| Tubulin β-2A, -2B, -2C and -5 chains | R77.SGPFGQIFRPDNFVFGQSGAGNNW101AK.G104* | Supplementary Figure S6 |
| α-Enolase | K358.LAQSNGW365GVMVSHR.S373 | Supplementary Figure S7 |
| Glutamine synthetase | K25.IQLMYIW32VDGTGEGLR.C42 | Supplementary Figure S8 |
| γ-Enolase | K358.LAQENGW365GVMVSHR.S373 | Supplementary Figure S9 |
| Actin, cytoplasmic 2† | K68.YPIEHGIVTNW79DDMEK.I85 | Supplementary Figure S10 |
| Fructose-bisphosphate aldolase C | R304.ALQASALSAW314R.G315 | Figure 2 |
| Fructose-bisphosphate aldolase A | R304.ALQASALKAW314GGK K318 | Supplementary Figure S11 |
| GAPDH | K84. W85GDAGAEYVVESTGVFTTMEK.A106 | Supplementary Figures S12 |
| K307.LISW311YDNEYGYSNR.V322 | Supplementary Figures S13 | |
| Electron transfer flavoprotein subunit α, mitochondrial | K187.APSSSSAGISEW199LDQK.L204 | Supplementary Figure S14 |
| Phosphoglycerate kinase 1‡ | K382. W383NTEDKVSHVSTGGGASLELLEGK.V407 | Supplementary Figure S15 |
| Creatine kinase U-type, mitochondrial | K257.SFLIW262VNEEDHTR. V271 | Supplementary Figure S16 |
*This peptide sequence is identical among four isoforms of tubulin β identified in the present study, and tubulin β was counted as one protein.
†Methylation of histidine residue (+14 Da) is added for the identification of nitrated tryptophan residue.
‡Formylation of lysine residue (+28 Da) is added for the identification of nitrated tryptophan residue.
Table 3. NO2Trp-containing trypsin-digested peptide sequences identified from cerebellum.
Boldface W indicates nitrated tryptophan residue. All peptides cleared P<0.05 of the individual ion scores by the Mascot search. Supplementary Figures S17–S23 are available online at http://www.bioscirep.org/bsr/032/bsr0320521add.htm.
| Protein | Peptide sequence | MS/MS spectrum |
|---|---|---|
| Tubulin β-2A, -2B, -2C and -5 chains | R77.SGPFGQIFRPDNFVFGQSGAGNN101WAK. G104* | Supplementary Figure S17 |
| α-Enolase | K358.LAQSNGW365GVMVSHR.S373 | Supplementary Figure S18 |
| γ-Enolase | K358.LAQENGW365GVMVSHR.S373 | Supplementary Figure S19 |
| Actin, cytoplasmic 2† | K68.YPIEHGIVTNW79DDMEK.I85 | Supplementary Figure S20 |
| Fructose-bisphosphate aldolase C | R304.ALQASALSAW314R.G315 | Supplementary Figure S21 |
| GAPDH | K307.LISW311YDNEYGYSNR.V322 | Supplementary Figure S22 |
| Voltage-dependent anion-selective channel protein 1 | K61.YRW64TEYGLTFTEK.W75 | Supplementary Figure S23 |
*This peptide sequence is identical among four isoforms of tubulin β identified in the present study, and tubulin β was counted as one protein.
†Methylation of His (+14 Da) is added for the identification of nitrated tryptophan residue.
Figure 3. MS/MS spectrum of the tryptic peptide 304R. ALQASALSA314W-NO2R. G315 from fructose-bisphosphate aldolase C.
W-NO2 indicates NO2Trp residue.
Semi-quantitative studies of 6-NO2Trp-containing enzyme and NOS by Western blotting
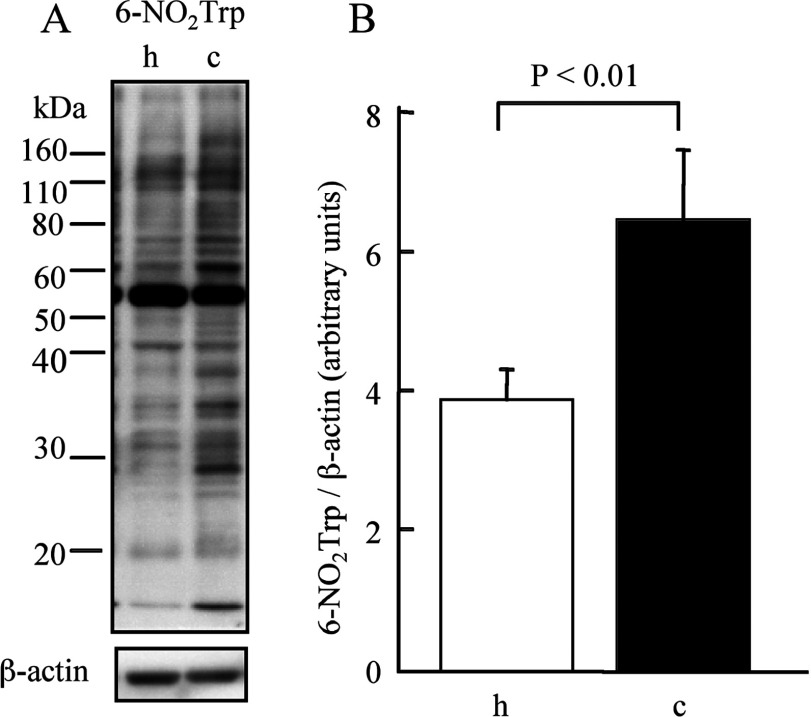
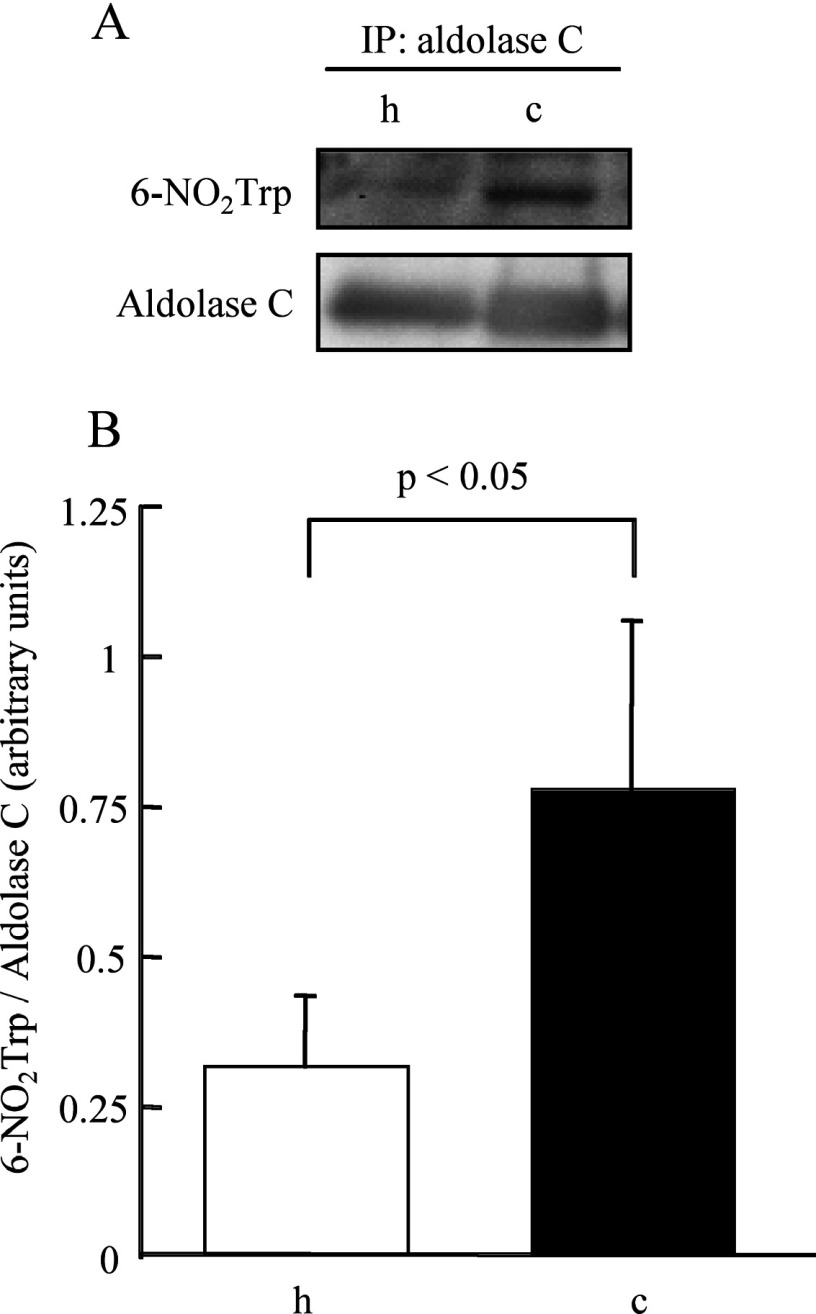
In order to compare gross amounts of 6-NO2Trp-containing proteins in hippocampus and cerebellum quantitatively, we measured total intensity of the bands on the membrane by Western blot analysis using β-actin as a loading control. Figure 4(A) shows the difference in 6-NO2Trp-containing protein bands between hippocampus and cerebellum. The total intensity of 6-NO2Trp-containing proteins in the cerebellum was significantly greater than that in hippocampus (P<0.01, Figure 4B). We observed the appearance of several new bands and the increased intensity of several bands for the cerebellum (Figure 4A). In order to clarify whether there is a protein with a higher rate of nitration in the cerebellum, we chose spots c9 and h9, which have been identified as aldolase C, as a target protein (Table 1), as the intensity of spot c9 in the Western blotting of two-dimensional electrophoresis was also clearly higher than that of h9 (Figures 1A and 1B). We isolated aldolase C by IP using anti-aldolase C antibody from the extracts of cerebellum and hippocampus and applied the isolated samples to SDS/PAGE and Western blotting. The relative intensity of immunostaining for 6-NO2Trp over aldolase C was much higher in cerebellum than in hippocampus (Figure 5). Therefore the increase in the nitration rate of the proteins, as shown in aldolase C, contribute, at least in part, to the increase in gross amounts of 6-NO2Trp-containing proteins shown in Figure 4.
Figure 4. Semi-quantitative studies of 6-NO2Trp-containing proteins.
(A) Representative result of Western blotting with an anti-6-NO2Trp antibody of hippocampus and cerebellum. (B) The difference in the relative amounts of NO2Trp -containing proteins between hippocampus and cerebellum measured by densitometry. h, hippocampus; c, cerebellum.
Figure 5. Comparison of the degree of tryptophan nitration of fructose-bisphosphate aldolase C between the hippocampus and cerebellum.
(A) Representative results of Western blotting for anti-6-NO2Trp antibody and anti-aldolase C antibody after IP with anti-aldolase C antibody. (B) The difference in the degree of tryptophan nitration between hippocampus and cerebellum measured by densitometry. h, hippocampus; c, cerebellum.
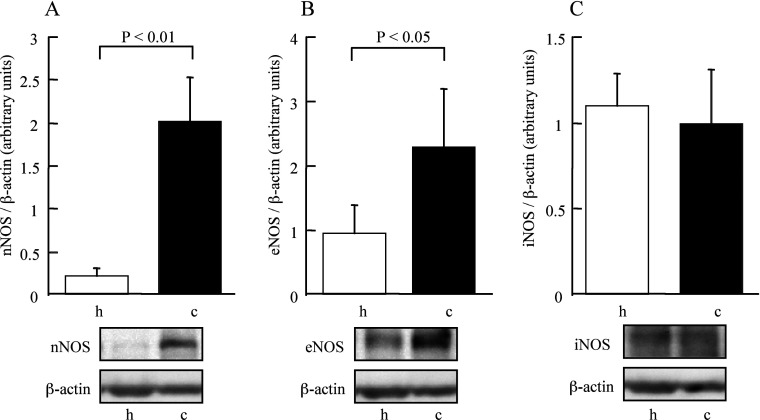
Finally, we compared relative amounts of NOS proteins in the extracts from cerebellum and hippocampus. Figure 6 shows the results of Western blot analysis using anti-nNOS, -eNOS and -iNOS antibodies. The amounts of nNOS and eNOS were significantly larger in cerebellum than in hippocampus. We did not observe any difference in protein expression of iNOS between these regions.
Figure 6. Comparison of the protein amounts of NOS isoforms between the hippocampus and cerebellum.
(A) nNOS, (B) eNOS, (C) iNOS. Upper part: the difference in the amounts of the proteins estimated by densitometry. h, hippocampus; c, cerebellum.
DISCUSSION
We have found nitration of tryptophan residues in cultured cells of a model for neuronal differentiation (PC12 cells) and suggested that tryptophan nitration may play a role in the cell-differentiation process in a previous study [17]. In the present study, we focused on rat brain as a target for in vivo study. This is the first study to identify several specific sites of nitrated tryptophan on proteins in vivo in a physiological state (Tables 2 and 3). Ishii et al. [14] reported the presence of 4- and 6-NO2Trp in the protease-digested liver homogenate from mice after administration of acetaminophen by using the LC-ESI-MS/MS method. They found 2.24–3.92 and 26.96–32.71 nmol/mol tryptophan for 4- and 6-NO2Trp respectively in the homogenate [14]. This evidence supports in vivo formation of NO2Trp, although it is from the oxidative stress conditions. On the other hand, formation of 5-hydroxy-6-NO2Trp by SCOT (succinyl-CoA 3-ketoacid coenzyme A transferase) has been reported in the heart [22] and kidney [23] of rats. We also found several peptides having an increase in mass of 61 (molecular mass corresponding to +NO2-H+OH-H), which may correspond to the formation of 5-hydroxy-6-NO2Trp, with a significant ion score in our system in the present study (results not shown). However, Wang et al. [24] did not find formation of 5-hydroxy-6-NO2Trp but found only nitrotyrosine in SCOT from a diabetic model mouse. Therefore formation of 5-hydroxy-6-NO2Trp might depend on the biological conditions.
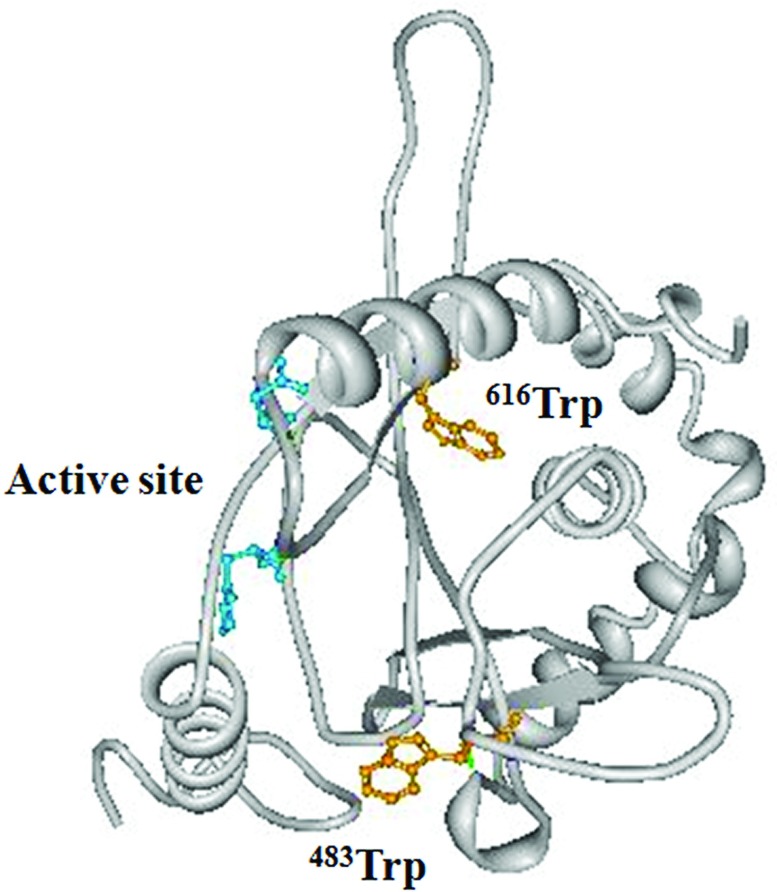
We have shown that the nitration of single tryptophan at position 32 in human recombinant Cu, Zn-SOD (copper/zinc superoxide dismutase) decreases the enzymatic activity [13,25] and the nitration of three tryptophan residues in egg-white lysozyme resulted in total loss of the activity in our previous study [26]. These observations indicate that the nitration of tryptophan residues in enzymes could affect the activity of enzymes. We have investigated the position of the modified tryptophan residues in the three-dimensional structure of the identified proteins in the present study. We found that five of the modified tryptophan residues are located near substrates or coenzyme-binding sites of the enzymes, namely, Trp616 and Trp483 in PDCE2 [dihydrolipoly(lysine)-residue acetyltransferase component of PDH (pyruvate dehydrogenase) complex] in mitochondria, Trp314 in fructose-bisphosphate aldolase C (Supplementary Figure S1 at http://www.bioscirep.org/bsr/032/bsr0320521add.htm), Trp85 and Trp311 in GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Supplementary Figure S2 at http://www.bioscirep.org/bsr/032/bsr0320521add.htm) and Trp383 in phosphoglycerate kinase 1 (Supplementary Figure S3 at http://www.bioscirep.org/bsr/032/bsr0320521add.htm). We show the three-dimensional structure of human PDCE2 in Figure 7 as a typical example. The amino acid sequences of PDCE2 are 93% identical between human and rat. The positions of the nitrated tryptophan residues are shown in black in Figure 7. Trp616 and Trp483 are located within the CoA-binding region. The addition of a nitro group to these tryptophan residues may cause steric hindrance and a different state on the indole π electron, consequently these changes may affect CoA binding and modulate the activity of the enzyme. However, further studies using isolated proteins are required to clarify the effects of the nitration of the tryptophan residues for each of the enzymes.
Figure 7. Three-dimensional models of active site region of human PDCE2.
The results for three-dimensioanl modelling were downloaded from the Protein Data Bank. The accession number of the protein is 3B8K. Both Trp616 and Trp483 (shown in yellow), which were nitrated in the present study are located within the CoA-binding region and near the catalytic amino acid residues (shown in blue).
We observed a very similar pattern of immunoreactive spots between hippocampus and cerebellum (Figure 1). However, there were fewer identified peptide sequences with nitrated tryptophan in cerebellum (Table 3) than in hippocampus (Table 2). This may be attributable to the difference in relative contents of each protein in the total protein between cerebellum and hippocampus. On the other hand, the relative amount of NO2Trp-containing proteins in cerebellum was higher than that of hippocampus upon analysis using SDS/PAGE (Figure 4). This apparent discrepancy can be explained in part by the evidence that proteins with a large molecular mass or extremely low or high pI are not taken into isoelectric gels in two-dimensional electrophoresis [27]. Therefore the amount of unloaded NO2Trp-containing proteins may be larger in cerebellum than in hippocampus. We showed that the nitration rate of tryptophan residues in aldolase C in cerebellum was significantly higher than that in hippocampus (Figure 5). This evidence may result in an increased degree of nitration of each of the tryptophan residues in aldolase C or in an increased percentage of nitrated-tryptophan residues in aldolase C. Therefore the larger nitration amount of the proteins in cerebellum is attributed to the larger modification rate of the proteins at least partly. This higher level of nitration of the proteins could be attributable to the increased concentration of RNS in cerebellum. In rat brain, the enzyme activity and expression of nNOS in cerebellum are higher than those in hippocampus [28,29]. In the present study, we have confirmed that cerebellum contains significantly larger amounts of nNOS and eNOS than hippocampus. Thus the expression levels of nNOS and eNOS may affect the difference of NO2Trp content between hippocampus and cerebellum. In other words, the expression level of NOS may regulate the nitration of tryptophan residues as a post-translational modification. In normal rat brain, using light and electron microscopy, nitroty-rosine immunoreactivity was shown to be higher in brain regions enriched in nNOS-containing neurons and/or near the neurons, indicating that the expression level of nNOS affects protein nitration [30]. This observation supports the above-mentioned hypothesis.
Finally, we describe the possible significance of the endogenous formation of 6-NO2Trp in cerebellum and hippocampus. Formation of 3-nitrotyrosine in proteins by RNS has been widely reported in a number of pathological states associated with inflammation [1–3]. We have identified several proteins that contain nitrated tryptophan from affected areas of a model mouse of an inflammatory disease (H. Kawasaki, M. Tominaga, K. Takamori, H. Ogawa, A. Shigenaga and F. Yamakura, unpublished results). This model mouse is known to have oxidative stress in the affected areas. Therefore nitration of tryptophan residues may be caused widely under oxidative stress conditions in vivo, which is the same as the case of nitrotyrosine formation. Previously, Sacksteder et al. [8] found 31 unique nitrotyrosine sites within 29 different proteins in a whole mouse brain under basal conditions using LC/LC-MS/MS analyses. They pointed out that more than half of the tyrosine-nitrated proteins are involved in Parkinson's disease or AD. Therefore they suggested that these endogenously nitrated proteins have tyrosine residues specifically sensitive to nitration under mild conditions of oxidative and nitrative stress as a possible concept. Similar to this situation, the first significance for the endogenous formation of NO2Trp could be at sites sensitive to mild oxidative and nitrative stress. Three of the NO2Trp-containing enzymes that we identified in the present study (creatine kinase, actin and tubulin β) are related to AD and Parkinson's diseases [8]. Although we used adult rats (6 months old), not aged rats, they could have a low level of oxidative/nitrative stress in brain under normal conditions. Since NOS levels are much higher in cerebellum than in hippocampus (Figure 6), these stresses may be higher in cerebellum (Figure 4). Nitrotyptophan could be used as a sensitive marker to detect mild oxidative/nitrative stress in vivo.
The additional significance of endogenous NO2Trp formation is that it could have some physiological functions. The formation of nitrotyrosine has been proposed not only as a marker of oxidative/nitrative damage but also as a functionally relevant post-translational modification of proteins under normal physiological conditions [31]. Three major effects on protein function, that is, no change in protein function, loss of function and gain of function, due to tyrosine nitration can be envisaged. We have found changes in the nitration of tryptophan residues in several proteins during the differentiation process from PC12 cells to neuron-like cells and proposed that tryptophan nitration may contribute somewhat to the differentiation process in the cells [17]. Since we have only limited information on the effect of tryptophan nitration on enzyme function, that is, reducing or abolishing the activity of the isolated enzyme [13,25], we could not clarify the mechanism of the functional change of the enzymes by tryptophan nitration. However, many tryptophan residues in enzymes are known to have specific function in the proteins. For instance, carbohydrates are known to bind preferentially to tryptophan residues in proteins [32] and tryptophan residues exhibit important roles for the binding to lipid bilayer membrane in some enzymes [33]. Nitration of the tryptophan residues may change the interaction of the enzymes with those components. Although further studies are required to clarify the mechanism in detail, the participation of tryptophan nitration in some physiological processes of cerebellum and hippocampus could be of significance. As the nitration of tryptophan residues affects enzyme functions by mechanisms different from the nitration of tyrosine residues, studies on the tyrosine nitration alone are insufficient to understand whole RNS functions in physiological processes. Therefore tryptophan nitration is a very important and unique target to be elucidated.
In conclusion, we were able to identify NO2Trp-containing cytoskeletal proteins and glycolytic enzymes in both hippocampus and cerebellum. This is the first proteomic study to identify several NO2Trp-containing proteins in physiological conditions in vivo. Furthermore, we showed that there is a difference in the amounts of NO2Trp-containing proteins between hippocampus and cerebellum, and this difference may result in the difference in the nitration rate of tryptophan residues in the proteins from both regions and the difference in the formation NO in physiological states. These findings suggest the possibility of regulating the function of proteins by the nitration of tryptophan residues in the proteins in vivo together with the possibility of a biomarker for oxidative and nitrative stress.
Online data
AUTHOR CONTRIBUTION
Munehiro Uda designed and performed experiments, analysed the data and wrote the paper. Hiroaki Kawasaki and Takeshi Baba contributed to the discussions and wrote the paper. Ayako Shigenaga contributed to the acquisition and analysing of the LC-MS/MS data. Fumiyuki Yamakura designed the research and wrote the paper. All authors approved the final paper.
FUNDING
This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science [grant numbers 22700701 (to M.U.) and 21500695 (to F.Y.)].
References
- 1.Beckman J. S., Koppenol W. H. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am. J. Physiol. Cell Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 2.Greenacre S. A., Ischiropoulos H. Tyrosine nitration: localisation, quantification, consequences for protein function and signal transduction. Free Radical Res. 2001;34:541–581. doi: 10.1080/10715760100300471. [DOI] [PubMed] [Google Scholar]
- 3.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophys. Res. Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 4.Smith M. A., Richey Harris P. L., Sayre L. M., Beckman J. S., Perry G. Widespread peroxynitrite-mediated damage in Alzheimer's disease. J. Neurosci. 1997;17:2653–2657. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Good P. F., Werner P., Hsu A., Olanow C. W., Perl D. P. Evidence of neuronal oxidative damage in Alzheimer's disease. Am. J. Pathol. 1996;149:21–28. [PMC free article] [PubMed] [Google Scholar]
- 6.Good P. F., Hsu A., Werner P., Perl D. P., Olanow C. W. Protein nitration in Parkinson's disease. J. Neuropathol. Exp. Neurol. 1998;57:338–342. doi: 10.1097/00005072-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Gokulrangan G., Zaidi A., Michaelis M. L., Schoneich C. Proteomic analysis of protein nitration in rat cerebellum: effect of biological aging. J. Neurochem. 2007;100:1494–1504. doi: 10.1111/j.1471-4159.2006.04334.x. [DOI] [PubMed] [Google Scholar]
- 8.Sacksteder C. A., Qian W. J., Knyushko T. V., Wang H., Chin M. H., Lacan G., Melega W. P., Camp D. G., II, Smith R. D., Smith D. J., et al. Endogenously nitrated proteins in mouse brain: links to neurodegenerative disease. Biochemistry. 2006;45:8009–8022. doi: 10.1021/bi060474w. [DOI] [PubMed] [Google Scholar]
- 9.Cappelletti G., Maggioni M. G., Tedeschi G., Maci R. Protein tyrosine nitration is triggered by nerve growth factor during neuronal differentiation of PC12 cells. Exp. Cell Res. 2003;288:9–20. doi: 10.1016/s0014-4827(03)00209-x. [DOI] [PubMed] [Google Scholar]
- 10.Rayala S. K., Martin E., Sharina I. G., Molli P. R., Wang X., Jacobson R., Murad F., Kumar R. Dynamic interplay between nitration and phosphorylation of tubulin cofactor B in the control of microtubule dynamics. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19470–19475. doi: 10.1073/pnas.0705149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuriel T., Hansler A., Gross S. S. Protein nitrotryptophan: formation, significance and identification. J. Proteomics. 2011;74:2300–2312. doi: 10.1016/j.jprot.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamakura F., Ikeda K. Modification of tryptophan and tryptophan residues in proteins by reactive nitrogen species. Nitric Oxide. 2006;14:152–161. doi: 10.1016/j.niox.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Yamakura F., Matsumoto T., Ikeda K., Taka H., Fujimura T., Murayama K., Watanabe E., Tamaki M., Imai T., Takamori K. Nitrated and oxidized products of a single tryptophan residue in human Cu,Zn-superoxide dismutase treated with either peroxynitrite-carbon dioxide or myeloperoxidase-hydrogen peroxide-nitrite. J. Biochem. 2005;138:57–69. doi: 10.1093/jb/mvi095. [DOI] [PubMed] [Google Scholar]
- 14.Ishii Y., Ogara A., Katsumata T., Umemura T., Nishikawa A., Iwasaki Y., Ito R., Saito K., Hirose M., Nakazawa H. Quantification of nitrated tryptophan in proteins and tissues by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J. Pharm. Biomed. Anal. 2007;44:150–159. doi: 10.1016/j.jpba.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda K., Yukihiro Hiraoka B., Iwai H., Matsumoto T., Mineki R., Taka H., Takamori K., Ogawa H., Yamakura F. Detection of 6-nitrotryptophan in proteins by Western blot analysis and its application for peroxynitrite-treated PC12 cells. Nitric Oxide. 2007;16:18–28. doi: 10.1016/j.niox.2006.04.263. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki H., Ikeda K., Shigenaga A., Baba T., Takamori K., Ogawa H., Yamakura F. Mass spectrometric identification of tryptophan nitration sites on proteins in peroxynitrite-treated lysates from PC12 cells. Free Radical Biol. Med. 2011;50:419–427. doi: 10.1016/j.freeradbiomed.2010.10.688. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki H., Shigenaga A., Uda M., Baba T., Ogawa H., Takamori K., Yamakura F. Nitration of tryptophan in ribosomal proteins is a novel post-translational modification of differentiated and naive PC12 cells. Nitric Oxide. 2011;25:176–182. doi: 10.1016/j.niox.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Siles E., Martinez-Lara E., Canuelo A., Sanchez M., Hernandez R., Lopez-Ramos J. C., Del Moral M. L., Esteban F. J., Blanco S., Pedrosa J. A., et al. Age-related changes of the nitric oxide system in the rat brain. Brain Res. 2002;956:385–392. doi: 10.1016/s0006-8993(02)03575-8. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Zaidi A., Pal R., Garrett A. S., Braceras R., Chen X. W., Michaelis M. L., Michaelis E. K. Genomic and biochemical approaches in the discovery of mechanisms for selective neuronal vulnerability to oxidative stress. BMC Neurosci. 2009;10:12. doi: 10.1186/1471-2202-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balabanli B., Kamisaki Y., Martin E., Murad F. Requirements for heme and thiols for the nonenzymatic modification of nitrotyrosine. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13136–13141. doi: 10.1073/pnas.96.23.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y. Z., Strong M., Huang Z. Q., Beckman J. S. Antibodies that recognize nitrotyrosine. Methods Enzymol. 1996;269:201–209. doi: 10.1016/s0076-6879(96)69022-3. [DOI] [PubMed] [Google Scholar]
- 22.Rebrin I., Bregere C., Kamzalov S., Gallaher T. K., Sohal R. S. Nitration of tryptophan 372 in succinyl-CoA:3-ketoacid CoA transferase during aging in rat heart mitochondria. Biochemistry. 2007;46:10130–10144. doi: 10.1021/bi7001482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bregere C., Rebrin I., Gallaher T. K., Sohal R. S. Effects of age and calorie restriction on tryptophan nitration, protein content, and activity of succinyl-CoA:3-ketoacid CoA transferase in rat kidney mitochondria. Free Radical Biol. Med. 2010;48:609–618. doi: 10.1016/j.freeradbiomed.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Peng F., Tong W., Sun H., Xu N., Liu S. The nitrated proteome in heart mitochondria of the db/db mouse model: characterization of nitrated tyrosine residues in SCOT. J. Proteome Res. 2010;9:4254–4263. doi: 10.1021/pr100349g. [DOI] [PubMed] [Google Scholar]
- 25.Yamakura F., Matsumoto T., Fujimura T., Taka H., Murayama K., Imai T., Uchida K. Modification of a single tryptophan residue in human Cu,Zn-superoxide dismutase by peroxynitrite in the presence of bicarbonate. Biochim. Biophys. Acta. 2001;1548:38–46. doi: 10.1016/s0167-4838(01)00212-6. [DOI] [PubMed] [Google Scholar]
- 26.Yamakura F., Ikeda K., Matsumoto T., Taka H., Kaga N. Formation of 6-nitrotryptophan in purified proteins by reactive nitrogen species: a possible new biomarker. Int. Cong. Ser. 2007;130:22–32. [Google Scholar]
- 27.Becker M., Schindler J., Nothwang H. G. Neuroproteomics – the tasks lying ahead. Electrophoresis. 2006;27:2819–2829. doi: 10.1002/elps.200500892. [DOI] [PubMed] [Google Scholar]
- 28.Forstermann U., Gorsky L. D., Pollock J. S., Schmidt H. H., Heller M., Murad F. Regional distribution of EDRF/NO-synthesizing enzyme(s) in rat brain. Biochem. Biophys. Res. Commun. 1990;168:727–732. doi: 10.1016/0006-291x(90)92382-a. [DOI] [PubMed] [Google Scholar]
- 29.Necchi D., Virgili M., Monti B., Contestabile A., Scherini E. Regional alterations of the NO/NOS system in the aging brain: a biochemical, histochemical and immunochemical study in the rat. Brain Res. 2002;933:31–41. doi: 10.1016/s0006-8993(02)02302-8. [DOI] [PubMed] [Google Scholar]
- 30.Bolan E. A., Gracy K. N., Chan J., Trifiletti R. R., Pickel V. M. Ultrastructural localization of nitrotyrosine within the caudate-putamen nucleus and the globus pallidus of normal rat brain. J. Neurosci. 2000;20:4798–4808. doi: 10.1523/JNEUROSCI.20-13-04798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webster R. P., Roberts V. H., Myatt L. Protein nitration in placenta – functional significance. Placenta. 2008;29:985–994. doi: 10.1016/j.placenta.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik A., Ahmad S. Sequence and structural features of carbohydrate binding in proteins and assessment of predictability using a neural network. BMC Struct Biol. 2007;7:1. doi: 10.1186/1472-6807-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge C., Georgiev A., Ohman A., Wieslander A., Kelly A. A. Tryptophan residues promote membrane association for a plant lipid glycosyltransferase involved in phosphate stress. J. Biol. Chem. 2011;286:6669–6684. doi: 10.1074/jbc.M110.138495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.