Abstract
The changes in fast dynamics of HP35 with a double CN vibrational dynamics label (HP35-P2) as a function of the extent of denaturation by urea were investigated with two-dimensional infrared (2D IR) vibrational echo spectroscopy. Cyanophenylalanine (PheCN) replaces the native phenylalanine at two residues in the hydrophobic core of HP35, providing vibrational probes. NMR data show that HP35-P2 maintains the native folded structure similar to wild type, and that both PheCN residues share essentially the same environment within the peptide. A series of time-dependent 2D IR vibrational echo spectra were obtained for the folded peptide and the increasingly unfolded peptide. Analysis of the time dependence of the 2D spectra yields the system’s spectral diffusion, which is caused by the sampling of accessible structures of the peptide under thermal equilibrium conditions. The structural dynamics become faster as the degree of unfolding is increased.
Keywords: fast peptide dynamics, 2D IR experiments, HP35 urea denaturation, CN labeled HP35
I. Introduction
Chicken villin headpiece 35 (HP35), a small F-actin binding motif domain in actin bundling proteins, has an important place in biophysics as the fastest folding peptide with the folding time on the microsecond timescale.1,2 Because it folds so fast, it is a convenient system for detailed computational studies. The 35-amino acid peptide folds into a globular bundle of three alpha helices with three phenylalanine residues in the hydrophobic core.3–5 Its simplicity and robustness also made it amenable to many types of experiments as well, such as x-ray crystallography,6 NMR,7,8 fluorescence,9,10 and triplet-triplet excitation transfer.11,12 In many respects, HP35 is an excellent model system for exploring protein structure, folding, and stability.
Recently, there have been reported a number of infrared studies of HP35, accelerated in part by the use of nitrile (CN) group as a vibrational dynamics label (VDL), such as temperature-jump spectroscopy13 and 2D IR vibrational echo spectroscopy. 2D IR spectroscopy takes advantage of ultrafast IR laser system’s time resolution to probe protein and peptide fast structural dynamics on the subpicosecond and longer time scales.14,15 The role of the VDL is critical in a 2D IR experiment as its response to structural fluctuations is the experimental observable. CN is a useful group as a VDL in biological 2D IR experiments as it has a relatively small perturbation to protein system, it is sensitive to the electric field, and it absorbs in an isolated spectral region of a peptide IR spectrum.16,17 CN-functionalized HP35 has been subjected to previous 2D IR studies that examined its structure,18 guanidinium (GuHCl)-induced unfolding and the effect of solvent viscosity on its dynamics,19 and stability.20
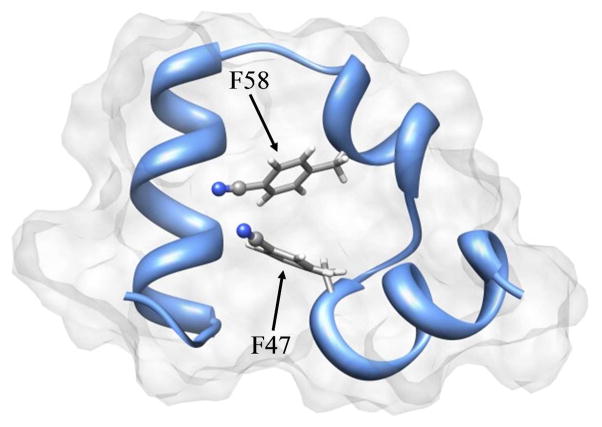
In this work, the urea-induced unfolding of HP35 was studied in detail by 2D IR and other spectroscopic techniques. The experimental design is similar to the previous GuHCl-unfolding study, where the fast dynamics of folded and unfolded HP35-P2 and the effect of solvent viscosity were investigated.19 In that study only the full denatured state (high GuHCl concentration) was compared to the folded state (no GuHCl). Here, the dynamics at intermediate concentrations of urea were studied. Although urea and GuHCl are both commonly used chemical denaturants, there are differences in their mechanism by which they unfold proteins.21 HP35-P2, a variant of HP35 (Phe47PheCN/Phe58PheCN) that contains two CN groups to act as the VDLs, was used. The structural representation, obtained from molecular dynamics simulation based on a modified x-ray structure (PDB 1YRF) is shown in Figure 1. The nature of the HP35-P2 was explored by NMR spectroscopy, which provides strong evidence that it maintains a folded, native-like structure similar to wild type HP35, and that the two CN’s share essentially the same environment within the hydrophobic core of the folded peptide. In addition, previous experiments show that the dynamics of the folded peptide are the same with one PheCN or two PheCNs, which suggests that the CN label does not significantly perturb the folded state.20 The 2D IR experiments show that as the degree of unfolding is increased with higher urea concentrations, the dynamics become significantly faster, suggesting an overall loss of the well-defined tight structure of the folded peptide.
Figure 1.
Structure of HP35-P2 (modified from PDB 1YRF with molecular dynamics simulations). CN labels are shown on F58 and F47 residues.
II. Experimental Methods
A. Sample Preparation
HP35-P2 with the amino acid sequence L42SDEDFCNKAVFGMTRSAFCNANPLWKQ-QNLKKEKGLF76 was synthesized by Stanford University School of Medicine Protein and Nucleic Acid Facility using Fmoc-based solid state synthesis, where FCN denotes para-cyano-L-phenylalanine (PheCN). The peptide was purified by HPLC, and integrity and purity were checked with electrospray-mass spectrometry. PheCN for the optical experiments was obtained from Peptech and used without further purification.
For FT-IR and 2D IR experiments, HP35-P2 and PheCN were dissolved in 50 mM sodium acetate buffer (pH 5.0) at various urea concentrations. The concentration of the peptide and PheCN were approximately 15 mM and 75 mM, respectively. The sample was sandwiched between two 3 mm CaF2 windows with a 50 μm Teflon spacer, resulting in an absorbance of 0.012 for the peptide and 0.03 for PheCN. All experiments were carried out at 25 °C.
To obtain good signal-to-noise ratios in the 2D IR experiments at the longest times, it is necessary to use samples with high peptide concentration. The samples are 15 mM. A concentration study was conducted in previous experiments on HP35-P2 to determine if aggregation occurs at 15 mM.19 It was found that when the concentration exceeds 16 mM, a second peak appears in the FT-IR spectrum of the CN absorption. A temperature dependent studied showed that this second peak goes away when the sample is mildly heated and HP35-P2 becomes more soluble. This result indicates that the second peak is due to aggregation. In addition at 16 mM, the sample would form visible aggregates after standing at room temperature for ~4 days. No such visible aggregation was observed at 15 mM regardless of the length of time over which the sample was observed. These results indicate that the sample is free of aggregation at the concentration of 15 mM. HP35 would become even less likely to be aggregated as urea generally disfavors aggregation. Therefore, the sample was kept at 15 mM and checked with FT-IR for the additional aggregation peak before and after each experiment.
B. FT-IR and CD Spectroscopy
Absorption spectra were collected with Nicolet 6700 FT-IR spectrometer (ThermoFisher Scientific) with 1 cm−1 resolution. The nitrile absorption bands from HP35-P2 and PheCN are at ~2235 cm−1, on top of a broad water bending and libration combination mode. The spectra were background-corrected by subtracting the spectra of the matching solvent. CD spectra of HP35-P2 at various urea concentrations were measured in the far UV region (200–250 nm) at 0.5 nm resolution (Jasco J810). The concentration of the sample was 50 μM in a 1-mm quartz cell. The unfolding of the peptide was monitored at 222 nm.
C. NMR Spectroscopy
NMR experiments were carried out at 25 °C with 10 mM peptide in 50 mM sodium acetate buffer (D2O, pD 5.0). 1D 1H, 2D correlation (COSY), 1H/13C heteronuclear single-quantum correlation (HSQC), and 1H/13C heteronuclear multiple-bond correlation (HMBC) spectra were acquired on a 600 MHz Varian Inova NMR spectrometer with a Varian triple resonance {H,C,N} z-gradient probe. The 1H spectrum was acquired with 4 scans, an acquisition time of 4 seconds and a recycle delay of 0.5 seconds. 1D 13C in natural abundance was acquired on a 500 MHz Varian Inova NMR spectrometer with a Varian Switchable {X,H} z-gradient probe. The spectrum was acquired with 11,000 scans with an acquisition time of 2 seconds, a recycle delay of 3 seconds, and a 5 ms (40°) pulse length.
D. 2D IR Vibrational Echo Spectroscopy
The experimental setup and methods of 2D IR vibrational echo spectroscopy are described in detail in previous work.22,23 A Ti:Sapphire oscillator/regenerative amplifier pumped an optical parametric amplifier (OPA). The OPA produced 160 fs, 6 μJ pulses at a 1 kHz repetition rate centered at 2235 cm−1. The OPA output was then split into four beams: three excitation pulses that were focused onto the sample, and a local oscillator pulse (LO) for heterodyne detection. The average energy of each excitation pulse was 1.2 μJ, and the spot size (standard deviation of the electric field) at the sample was ~75 μm.
In the 2D IR experiment, three pulses were sequentially incident on the sample; the time between pulses 1 and 2 is τ (coherence time), and the time between pulses 2 and 3 is Tw (population time). The interaction of the time ordered pulses with the molecular oscillators in the sample generated the vibrational echo pulse propagating in the unique phase-matched direction with its peak at time t ≤ τ.
To obtain the phase information from the vibrational echo signal, optical heterodyne detection was used. The echo wave packet was spatially and temporally overlapped with the LO pulse. Scanning of τ, which causes the echo wave packet to move in time relative to the fixed LO pulse, createed a temporal interferogram between the vibrational echo signal and the LO. The interferogram provided the phase information. The combined vibrational echo-LO signal was then frequency resolved by a diffraction grating in a monochromator used as a spectrograph, and recorded by a liquid nitrogen-cooled 32-element HgCdTe detector. This spectrum, which was one of the two Fourier transforms of the echo wave packet, provided the vertical axis of the 2D IR spectrum, ωm. The horizontal axis ωτ was obtained by numerically Fourier transforming the interferogram at each ωm. For a single 2D IR spectrum, Tw was fixed and τ was scanned. The results provided one 2D spectrum. A series of such spectra were collected as a function of Tw.
The time-dependent dynamic information on the peptide dynamics was obtained from the change in the 2D spectrum shape as Tw was changed. The change in shape is related to spectral diffusion, which occurs because the frequency of the VDL changes due to the structural fluctuation of the peptide.
Qualitatively, the experiment works as follows. The first and second laser pulses “label” the initial structures of the peptides by “tagging” their initial frequencies. After the second spectral diffusion occurs for the time Tw, during which the “labeled” peptide molecules experience structural evolution which changes their frequencies. Finally, the third pulse and the subsequent vibration echo emission ends the period during which structural evolution occurs and reads out the information about the final structure of the labeled species by their echo emission frequencies. If there is no structural evolution during the spectral diffusion period, Tw,, then the read out frequencies would be identical to the initial labeled frequencies. On the other hand, if there is structural evolution during the spectral diffusion period, some of the correlation between the initially labeled and final frequencies would be lost. This is manifest in 2D spectra as a change in the shape of the 2D IR spectrum as Tw is increased. At short Tw, the 2D spectrum is elongated along the diagonal because the excitation frequencies (ωτ, horizontal axis) and the detection frequencies (ωm, vertical axis) are correlated. As Tw increases, structural changes of the peptides cause the VDL frequency to change, so the correlation between the initial and final frequencies is lost. At sufficiently long Tw, all structures are sampled, and the 2D spectrum will become round. Therefore, the rates of peptide structural fluctuations, which cause spectral diffusion, are manifested as the rate of the shape change in 2D spectra.
The rate of the shape change is quantified by the center line slope (CLS) method, which measures the slope of the line created by the maxima of slices cut along the ωm axis. CLS has been shown to be mathematically equivalent to the normalized frequency-frequency correlation function (FFCF), which is the connection between the experimental observables and the underlying dynamics of the peptide.24,25 The combination of the CLS data and the IR absorption spectrum permits the determination of the full FFCF.
The FFCF is modeled with a multiexponential function,
| (1) |
where Δi and τi are the frequency fluctuation amplitude and the time constant associated with the ith component, respectively. This form has been successfully used in many 2D IR studies of proteins.26,27 The multiexponential form allows the determination of Δi and τi. For structural dynamics that are sufficiently slow that they fall outside the experimental time window (determined by the vibrational lifetime of the VDL), the τi cannot be determined, but the corresponding Δi can still be obtained. If Δτ < 1, the component is motionally narrowed. Then only the product Δ2τ = 1/T2*can be obtained. T2* is known as the pure dephasing time, and can be calculated from the relationship
| (2) |
where dephasing time T2 is determined from the CLS and linear absorption spectrum and vibrational lifetime T1 is obtained from independent IR pump-probe experiments (4.5 ps for CN in HP35-P2). The pure dephasing line width is given by Γ*= 1/πT2*.
III. Results and Discussions
A. Steady-state Experimental Results
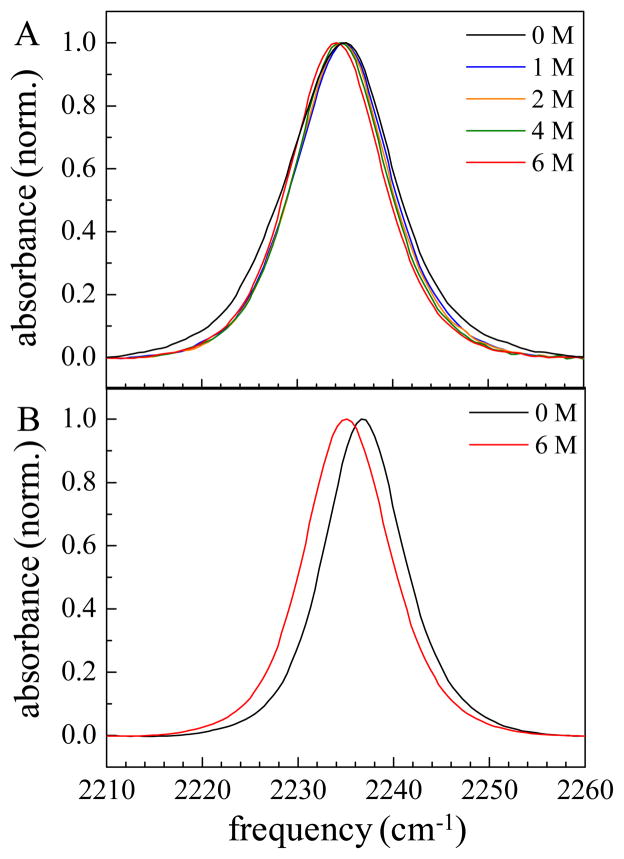
The stretching mode of CN on PheCN of HP35-P2 absorbs near 2235 cm−1, which is within the frequency range of other aromatic nitriles.17 Figure 2 shows the FT-IR spectra of HP35-P2 (A) and PheCN (B) for various urea concentrations. As was reported previously,19 a single IR absorption band is observed for the each peptide sample in water and various concentrations of urea despite the presence of two CN groups in each peptide.
Figure 2.
Normalized FT-IR absorption spectra of the CN stretching mode of HP35-P2 (A) and PheCN (B) at various urea concentrations.
The FT-IR spectra of HP35-P2 nitrile mode at different urea concentration shows very small incremental red shifts as the concentration is increased, from 2234.5 cm−1 in 0 M urea to 2234.1 cm−1 in 6 M (see Table 1). The full width at half maximum (FWHM) of the band decreases with the initial addition of urea, but the linewidth remains unchanged with increased urea concentration. It is notable that there is a significant reduction in FWHM between 0 M and 1 M band, while further addition of urea does not produce additional changes. This indicates that an important change of structure occurs at 1 M urea. Addition of more urea does not produce structural changes that affect the absorption spectrum. Further changes in structure are manifested by changes in the CD spectrum and in the peptide dynamics discussed below. Similar trends in both absorption frequency and width were observed with guanidinium hydrochloride (GuHCl) dependent CN absorption spectra of HP35-P2.19 For comparison, FT-IR of the amino acid PheCN in 0 M and 6 M urea were also taken. The CN stretch absorption of PheCN shows a minute increase in width and a red shift similar that observed for HP35-P2.
Table 1.
FT-IR
| center (cm−1) | FWHM (cm−1) | |
|---|---|---|
| 0 M | 2234.5 ± 0.2 | 13.5 ± 0.2 |
| 1 M | 2234.7 ± 0.2 | 12.4 ± 0.2 |
| 2 M | 2234.6 ± 0.2 | 12.2 ± 0.2 |
| 4 M | 2234.5 ± 0.2 | 11.9 ± 0.2 |
| 6 M | 2234.1 ± 0.2 | 12.0 ± 0.2 |
| PheCN 0 M | 2236.9 ± 0.2 | 10.5 ± 0.2 |
| PheCN 6 M | 2235.2 ± 0.2 | 11.1 ± 0.2 |
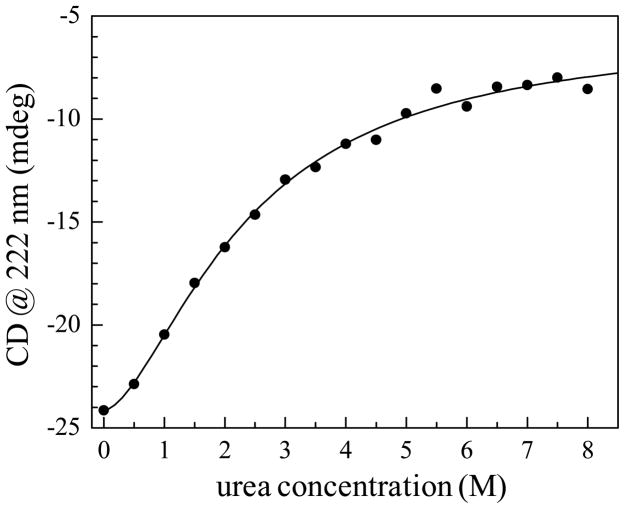
The equilibrium urea-induced changes in secondary structure of HP35-P2 were monitored by plotting CD data at 222 nm, which reflects alpha helical content, as a function of urea concentration (Figure 3). The midpoint of unfolding occurs at approximately 2 M. The CD unfolding curve shows a sigmoidal profile, indicating a transition that can be described using a two-state model, which is consistent with most previous work on HP35. It is possible that there is a mixture of populations of the native species and a single unfolded species are present in the intermediate urea concentrations, between 1 M and 4 M. This is not evident in the FT-IR spectra, in contrast to what is observed for thermal unfolding experiments on CO-bound cytochrome c.41 For cyt c, the absorption spectra corresponding to the native and unfolded states were clearly resolved, and at the intermediate temperatures the FT-IR spectrum of CO was the sum of two distinct absorption bands. Further, it was clear that the average structure of unfolded cyt c changed continuously with temperature as its peak position and linewidth change with temperature. These results along with the 2D IR data demonstrated that thermal unfolding of cyt c was more complex than a simple two-state transition although the CD data could be fit with the two-state model. For HP35-P2, however, the CN absorption spectra do not show two peaks, one for the folded and one for the unfolded states. If the splitting between an unfolded and folded band is very small, two peaks would not be observed, but there would most likely be an increase of linewidth caused by the two overlapping displaced bands. Such a broadening is not observed (see Table 1). It is possible that the spectrum of HP35-P2 at intermediate urea concentration is a sum of folded and unfolded spectra, as would be the case in the two-state model, or there could be an ensemble of partially unfolded structures that changes at each intermediate concentration, in the similar manner as has been clearly observed for cytochrome c.28 In either case, the infrared experimental results are not necessarily in contradiction to the widely accepted two-state model. However, ensemble averaged observables can display what appears to be two-state behavior even if the underlying process is much more complex.29
Figure 3.
The urea-dependent unfolding curve of HP35-P2 monitored by CD at 222 nm. The midpoint of unfolding occurs near 2 M urea.
B. The Structure of HP35-P2
In HP35-P2, two CN groups are inserted into the hydrophobic core of HP35 to serve as the vibrational reporter of the peptide’s structural dynamics. Two important issues are 1) whether the peptide modified with the CNs in the interior of the peptide still folds into a structure similar to the native state of the wild type HP35, and 2) whether the two CNs are reporting on the same dynamics of the peptide. Some insights can be gained from previous IR experiments.20 In the previous study, the linear absorption spectra and the dynamics of HP35 obtained with 2D IR spectroscopy were compared for HP35 labeled with one PheCN (HP35-P, with Phe58PheCN mutation) and with HP35-P2. The FT-IR absorption spectra of the two HP35 variants were found to be identical. In addition, the dynamical observables, the CLS, are identical within experimental error. Since there is no change in going from one CN insertion to two, then these observations suggest that the perturbation in going from the wild type peptide to the peptides with PheCNs is small. Indeed, CN is a very small group expected to cause much smaller perturbation than other types of labels such as fluorescence tags30 and there have been other proteins,31,32 as well as HP35,10,18 in which the introduction of CN resulted in no major disruption of the structure.
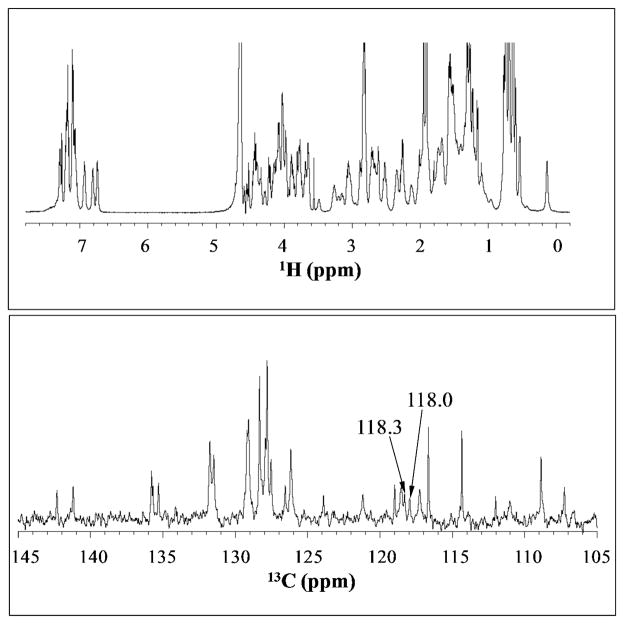
NMR can provide more sensitive data about the structure of the peptide and the environment around the CNs. The 1HNMR of HP35-P2 is shown in Figure 4 (top panel). The spectrum shows sharp and well-dispersed peaks, indicating a native-like state rather than unfolded or molten globule structures.33–35 In addition, the spectrum bears a close resemblance to that of the wild type HP35.35 Shown also in Figure 4 (bottom panel) is the aromatic region of natural abundance 13CNMR spectrum. The CN carbons were assigned, based on 1H/13C HSQC and 1H/13C HMBC (see supporting information), to the peaks at 118.0 and 118.3 ppm. The NMR relaxation times, T2*, are 35 ms and 24 ms, respectively, which are within the expected range for amino acids not exposed to solvent The similarity of the chemical shifts and relaxation times suggest that the two CNs are in very similar environments.
Figure 4.
1H NMR (top) and the 13C NMR spectra (bottom) of HP35-P2.
While the FT-IR and NMR data and previous 2D IR experiments20 support the hypothesis that both CN labels experience essentially identical environments in the native state, it is possible that this may not be the case when the peptide unfolds. However, at each urea concentration, only one absorption band is observed, and the linewidths are identical within experimental error once some urea is added (see Table 1). If the CN labels are exposed to significantly different environments when unfolded, a splitting of the peak might be observed. If the difference in environments is not sufficient to produce two distinct peaks, there would be a broadening of the total absorption bandwidth. Therefore, it is reasonable to assume that even as the peptide becomes increasingly unfolded with higher urea concentrations, the environments experienced by the two nitrile probes are very similar.
C. Dynamics of Urea-denatured HP35-P2
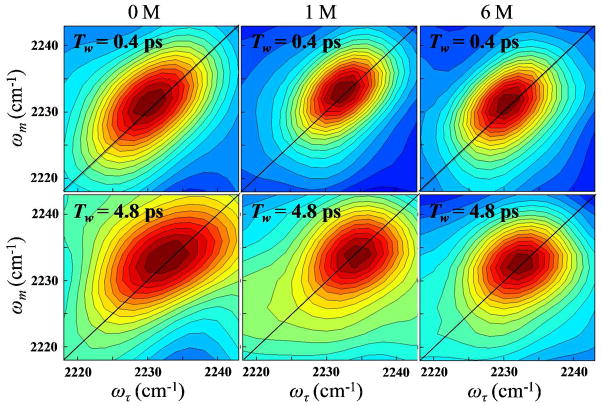
The 2D IR vibrational echo spectra of the CN stretching mode of HP35-P2 at 0 M, 1 M, and 6 M urea are shown in Figure 5. The top row displays spectra at early waiting time, Tw = 0.4 ps and the bottom row shows spectra at a later waiting time, Tw = 4.8 ps. Only the positive bands corresponding to the 0–1 transition were analyzed. The shape of the 2D IR spectrum changes with increasing Tw; it becomes less elongated with increasing time because of spectral diffusion. This shape change is quantified by calculating the CLS, which is directly related to the FFCF as briefly outline above.24
Figure 5.
The 0–1 transition region of the 2D IR spectra of HP35-P2. First column is the peptide at 0 M, second column is at 1 M, and third column is at 6 M urea. Top row shows each spectra at a short Tw = 0.4 ps and the bottom row at a long Tw = 4.8 ps.
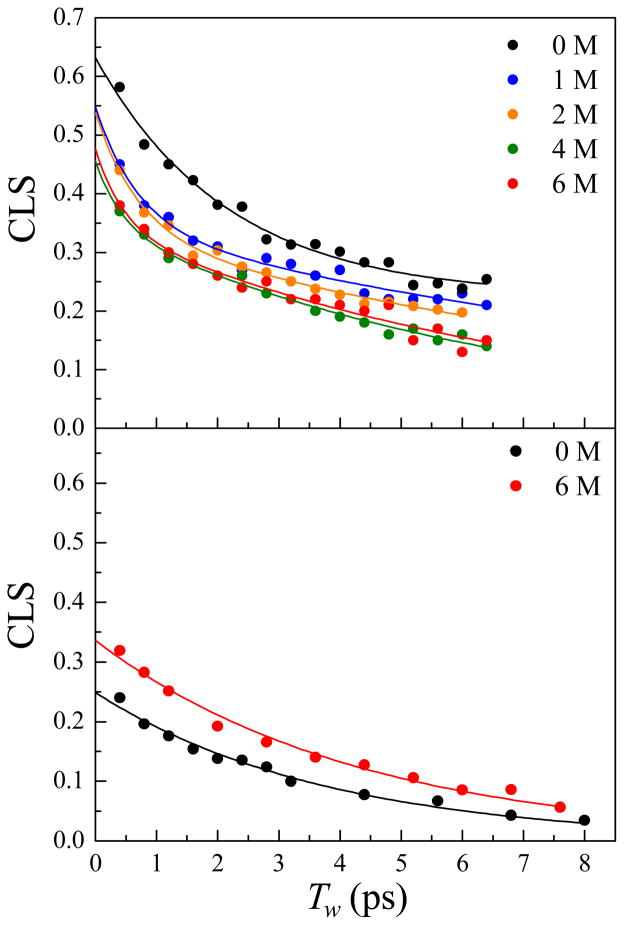
The CLS data for HP35-P2 (top panel) and PheCN (bottom panel) at various urea concentrations are shown in Figure 6. The 1 M and 2 M data are very different from the folded peptide data (0 M), showing faster decays. However, the 1M and 2 M data are quite similar to each other. The 4 M and 6 M data decay even faster, but these two decays are virtually identical. The data are fit to the multiexponential form shown in Equation (1). For HP35-P2 in 0 M urea, one exponential term plus a static offset was sufficient to fit data, while for other urea concentrations two exponential decay terms were necessary (no static offset). Detailed statistical analysis using Akaike information criterion (AIC),36 which is a method that compare models, was employed to determine the most appropriate fitting function. The method accounts for the number of fitting parameters in selecting between models. It is evident from the CLS data that the overall dynamics of the peptide become faster at higher urea concentrations. The results show that there is more rapid conformational sampling occurring as the peptide is increasingly unfolded within the ~7 ps experimental time window. The difference between 1 and the intercepts of the data on the vertical axis at Tw = 0 is a measure of the homogeneous component. The larger the difference is, the greater the homogeneous (pure dephasing) component. As the urea concentration increases, the pure dephasing component of the dynamics increases.
Figure 6.
CLS data and fits for HP35-P2 (top panel) and PheCN (bottom panel) at various urea concentrations.
Table 2 shows the FFCF parameters obtained from the fits of the CLS data and absorption spectra. In 0 M urea, the native state HP35-P2 has a 2.2 ps spectral diffusion component and a static component, that is, a component that is too slow to measure within the experimental time window. Upon the introduction of urea, there are two significant changes that occur for all concentrations. First, the timescale of the slow component is no longer too slow to measure, and second, the timescale of the fast component becomes much faster (< 1 ps). Even at the lowest urea concentration studied, 1 M, the dynamics become much faster. For both 1 M and 2 M urea samples, the fast component (τ1) is ~0.7 ps, and for 4 M and 6 M urea samples it is ~0.45 ps. The slower spectral diffusion component (τ2) for 1 M and 2 M samples is ~12 ps, while for the 4 M and 6 M samples it is ~7 ps. The homogeneous component, expressed as T2*, the pure dephasing time. or Γ*, the homogeneous linewidth, also changes with increasing urea concentration. For the folded peptide (0 M), T2* = 2.8 ps. For both the 1 M and 2 M samples, T2* = 2.4 ps, and for the 4 M and 6 M samples, T2* = 2.0 ps. Thus the dephasing time becomes faster with increasing extend of urea denaturation.
Table 2.
FFCF parameters
| sample | Γ* (cm−1) | T2* (ps) | Δ1 (cm−1) | τ1 (ps) | Δ2 (cm−1) | τ2 (ps) |
|---|---|---|---|---|---|---|
| 0 M | 3.8 ± 0.3 | 2.8 ± 0.3 | 3.7 ± 0.1 | 2.2 ± 0.3 | 2.7 ± 0.1 | > 20 |
| 1 M | 4.4 ± 0.5 | 2.4 ± 0.2 | 2.4 ± 0.2 | 0.70 ± 0.29 | 3.1 ± 0.1 | 13 ± 3 |
| 2 M | 4.4 ± 0.4 | 2.4 ± 0.2 | 2.4 ± 0.2 | 0.71 ± 0.21 | 3.0 ± 0.1 | 11 ± 2 |
| 4 M | 5.2 ± 0.5 | 2.0 ± 0.2 | 1.7 ± 0.4 | 0.43 ± 0.30 | 3.0 ± 0.1 | 7.0 ± 0.5 |
| 6 M | 5.0 ± 0.5 | 2.1 ± 0.3 | 1.8 ± 0.5 | 0.45 ± 0.43 | 3.0 ± 0.1 | 7.5 ± 1.0 |
| PheCN 0 M | 6.7 ± 0.1 | 1.6 ± 0.1 | -- | -- | 2.2 ± 0.1 | 3.8 ± 0.2 |
| PheCN 6 M | 6.1 ± 0.1 | 1.7 ± 0.1 | -- | -- | 2.7 ± 0.1 | 4.3 ± 0.2 |
These trends in FFCF parameters show that in general, structural interconversion between conformations of the peptide becomes more rapid as the extent of unfolding is increased. Although the experiments can be carried out only to ~7 ps, the data in this time window are influenced by structural fluctuations that occur out to a factor of ~3 longer time scale. For the folded peptide, the slowest component is to slow to measure, but it has a relatively large amplitude (Δ2), showing that there are significant dynamics much slower than ~20 ps. In contrast, the urea denatured peptides show slow components of 7–13 ps, with the timescale becoming faster with increasing urea concentration. The frequency fluctuation amplitudes associated with the slowest dynamics are the same for all urea denatured samples. Having the same amplitudes may indicate that the slowest component reflects dynamics among a similar distribution of structural peptide states for all urea concentrations,. However, the rate of interconversion between the states becomes faster, moving into the experimental time window with the introduction of 1M urea, and becoming increasingly fast with increasing urea concentration.
The fast component of the dynamics of folded HP35-P2 (2.2 ps) also becomes considerably faster with urea denaturation (0.4–0.7 ps). It is important to note that all of the time constants should be taken as reflective of the time scales associated with various structural dynamics as opposed to being linked to a single process that occurs with the specific time constant. In addition to changes in the timescales of the dynamics, the amplitudes of frequency fluctuation associated with the faster dynamics (Δ1) decrease with increasing concentration of urea. This decrease indicates that not only do the dynamics become faster with increasing extent of denaturation, but the distribution of frequencies, or by proxy the distribution of peptide states, sampled on the faster timescale is reduced. One possible explanation for the observed reduction in Δ1 associated with increasing urea denaturation is that some of peptide dynamics become so fast upon denaturation that they become part of the homogeneous dynamics. This possibility is consistent with a the observed change in T2*; however, because Δ and τ cannot be separated for the homogeneous dynamics, this interpretation cannot be verified.
Inspection of Table 2 reveals that the FFCF parameters do not appear to change continuously with increasing urea concentration, in contrast to the CD data. Although it should be noted that the error bars of many of the parameters overlap, in general the FFCF parameters obtained for the 1 M and 2 M urea solutions and those for the 4 M and 6 M urea solutions are very similar. The shape of the CLS decay curve changes from the native state peptide in 0 M urea to partially unfolded state in 1 M urea, as was observed in FT-IR. However, there is no significant change in the CLS when the concentration of urea is increased to 2 M, although the CD data (Figure 3) indicates that there is continuous change between 1 and 2 M urea. The CLS curve then changes at 4 M urea concentration, but again it remains the same at 6 M urea. The CD signal is nonspecific and reports on the global change of the peptide structure, in particular, the alpha helical content. The 2D IR vibrational echo measurement reports on the influence on the VDL of fluctuating electric fields produced by the structural dynamics.37–41 The VDL is sensitive to the global fluctuations of the peptide including the solvent, but can also be sensitive to the more local environmental influences.42 The changes in the dynamics with urea concentration reported by the 2D IR experiments indicate that some loss of alpha helical content seen by CD does not have a significant impact on the peptides dynamics sensed by the VDL. These observations may be associated with specific structural alterations in the vicinity of the VDL, which effectively communicate the associated modified dynamics to the VDL. For instance, based on the urea concentration-dependent CLS, one might conjecture that significant modifications of the hydrophobic core occur between 0 and 1 M urea, and then between 2 and 4 M urea to produce faster dynamics. The 2D IR results are amenable to molecular dynamics simulations, which should be particularly tractable given the short time range of the experiments.
The increasingly faster equilibrium conformational dynamics in denatured states may be associated with the expansion of the peptide into a less restricted structure. A previous 2D IR study of HP35 found significantly slower dynamics of the HP35 mutant, Lys65Nle/Lys70Nle mutant (HP35-P NleNle).20 This mutant has improved stability resulting from the replacement in the hydrophobic core of two charged lysine residues with norleucines.4 It was posited that the mutation resulted in a better hydrophobic packing and more inflexible structure of the core, and consequently, reduced conformational dynamics. Then, the increased dynamics of HP35-P2 upon the addition of urea can be surmised to be a result of the opposite effect: a reduction in rigid structure of the peptide.
The center frequencies of the absorption bands in the FT-IR spectra of the PheCN residues in HP35-P2 do not change dramatically with variation in the urea concentration, and further, in 6 M urea the frequency of the PheCN of HP35-P2 does not equal the frequency of the single PheCN amino acid. The spectral differences suggest that urea-denatured HP35-P2 does not adopt a random coil configuration where the PheCN side chains are completely exposed to solvent. The bottom panel of Figure 6 displays the CLS data for the amino acid PheCN in 0 M and 6 M urea; the FFCF parameters are listed in Table 2. In going from 0 M to 6 M, there is only a mild change in the dynamics. As can be seen from Figure 6 and Table 2, the dynamics of the HP35-P2 and those of PheCN in 6 M urea are very different. The urea-denatured peptide shows dynamics that are of a different function form and has a slower component than observed for the free amino acid. The peptide dynamics likely arise from fairly complex motions among structural states of the unfolded peptide ensemble. This comparison shows that the dynamics of the peptide reported by the VDL in 6 M is not strictly dominated by the solvent interacting with solvent exposed CNs on the labeled PheCNs.
It is interesting to compare the present results to the results of previous experiments where fast dynamics of HP35-P2 were measured as a function of the concentration of the denaturant GuHCl.19 In contrast to urea, which is neutral, Gu is charged. This difference can affect the nature of the unfolded state. With either urea or GuHCl, increasing the denaturant concentration resulted in fast structural fluctuations of the peptide. However, at the highest GuHCl concentration (6 M) the HP35-P2 dynamics were the same as those of the single labeled amino acid PheCN in 6M GuHCl. This result was interpreted as indicating that the unfolded peptide has its CN labeled phenylalanines completely solvent exposed, and the observed dynamics are dominated by solvent fluctuations. In contrast, at the highest concentration of urea (6 M), the peptides structural fluctuations are still significantly slower than those of single amino acid PheCN in 6 M urea. This result suggests that the peptide’s PheCNs are not completely solvent exposed when the peptide is unfolded in urea. The difference may arise because of the different electrostatic interactions of the two denaturants with the peptide.
The faster dynamics of the unfolded state observed for HP35-P2 is in contrast to that of cytochrome c (cyt c), which was also studied with 2D IR to examine its equilibrium temperature dependent unfolding.28 At relatively low temperature, unfolded cyt c has slower dynamics in the ~100 ps experimental time window, as well as a substantial increase in the long time component outside of the experimental time window, compared to the folded protein. However, as the temperature is increase further, the dynamics become faster, but do not become as fast as those observed for the folded protein. The 2D IR studies along with FT-IR experiments show that the unfolded cyt c has a broader range of structures, but there are higher barriers separating distinct structures relative to the ensemble of native structures.43,44 The differences between cyt c and HP35-P2 may be due to the fact that cyt c is a larger protein and is known to retain residual structures even under severely denaturing conditions.45,46
In studying the influence of the addition of urea to the solutions on the peptide dynamics, it is necessary to consider the effect of the viscosity change of the solvent. A 2D IR study of HP35-P2 found that increasing the viscosity of solvent by a factor of 2.7 resulted in significant slowing of the dynamics observed via the nitrile VDLs in the native state peptide.19 The viscosity of 6 M urea solution is 1.4 cP,47 reflecting a 40% increase in viscosity from water. Again, the bottom panel of Figure 6 shows the CLS data of PheCN in 0 M and 6 M urea, and the calculated FFCF parameters are listed in Table 2. The dynamics of PheCN in 6 M urea are only slightly slower than in 0 M, showing that a 40% change in viscosity has a weak influence on the dynamics. The slowing is probably due to the combined effect of increased viscosity and disruption in hydrogen bonding networks.48 In HP35-P2, however, the dynamics become substantially faster in 6 M urea compared to 0 M. This observation indicates that the viscosity effect is weak, and is overcome by the influence of the structural changes of the denatured peptide.
IV. Concluding Remarks
In this work, the fast dynamics of CN labeled HP35, a widely studied peptide for simulations and experiments, were examined at various stages of unfolding using the denaturant urea under equilibrium conditions using 2D IR vibrational echo experiments. NMR studies were also carried out to ascertain that the structure of double CN-labeled HP35, HP35-P2, formed a native-like structure and that both CNs in the peptide sensed the essentially the same environments inside the hydrophobic core. The results of 2D IR experiments demonstrate that the overall dynamics of HP35-P2 become increasingly fast as the concentration of urea is increased. A comparison can be made to a recent study where a mutant of HP35 (HP35-P NleNle),20 which is more stable in part due to improved hydrophobic core packing,4 displayed substantially slower dynamics compared to the wild type. The comparison suggests that in HP35, the unfolding by urea causes the peptide to become less tightly structured with increasingly rapid structural sampling particularly involving the hydrophobic core. Although the concentration of 15 mM used here due to the low absorbance of nitrile VDL can be too high for routine application to other protein and peptide systems, improvements in laser and detection technology may make nitrile group more attractive as VDL. Recent developments in 2D IR vibrational echo experiments using pulse shaping techniques are greatly reducing the time required to acquire a 2D spectrum.49–51 These methods also allow phase cycling techniques that improve signal-to-noise ratios (s/n). Increased rate of data collection means that more time can be spent averaging to pull out weak signals. For large biological molecules, that do not undergo rapid orientational relaxation, additional large increases in s/n can be achieved using particular polarizations of the beams. Therefore, it is likely that in the near future, measurements on low concentration samples with weakly absorbing VDLs will become routine.
As a protein or peptide folds, it can pass through many intermediate partially folded structures. The folding of HP35 in particular has been studied by simulation studies, which showed short-lived intermediate and partially unfolded structures on the path to the completely folded peptide.2,5 As protein folding is relatively slow, microseconds at the fastest, each intermediate structure will have sufficient time for picosecond time scale sampling of multiple conformations associated with intermediate structures. This sampling can lead to the path to the next intermediate in a progression that ultimately results in the folded structure. In this study, each of these intermediates is in equilibrium because of the fixed urea concentration. The fast dynamics measured by 2D IR are the ensemble average of the equilibrium structures at each given urea concentration. While the relationship between the equilibrium structures at various points of unfolding using urea and kinetic unfolding is unclear, it seems possible that these equilibrium intermediates and their dynamics may have a close resemblance to the non-equilibrium structures on the folding pathway. The results presented here are particularly amenable to molecular dynamics simulations, as HP35 is a widely studied system and conditions such as solvent and timescale are well suited to computational methods. Simulations of these HP35-P2 experiments may provide insights into the structures and dynamics of folding intermediates.
Supplementary Material
Acknowledgments
We are grateful to Ruth Sommese for assistance in CD data collection. We would like to thank Kyle Beauchamp for providing the material for Figure 1, which came from MD simulations he performed. This work was supported by the National Institute of Health (2-R01-GM061137-09).
Footnotes
Supporting Information Available
Additional information is available in Supporting Information. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.McKnight CJ, Matsudaira PT, Kim PS. Nat Struc Bio. 1997;4:180–184. doi: 10.1038/nsb0397-180. [DOI] [PubMed] [Google Scholar]
- 2.Duan YK, Peter A. Science. 1998;282:740. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 3.Frank BS, Vardar D, Buckley DA, McKnight CJ. Protein Sci. 2002;11:680–687. doi: 10.1110/ps.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubelka J, Chiu TK, Davies DR, Eaton WA, Hofrichter J. J Mol Biol. 2006;359:546–553. doi: 10.1016/j.jmb.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Zagrovic B, Pande VS. J Am Chem Soc. 2006;128:11742–11743. doi: 10.1021/ja0640694. [DOI] [PubMed] [Google Scholar]
- 6.Chiu TK, Kubelka J, Herbst-Irmer R, Eaton WA, Hofrichter J, Davies DR. Nat Acad Sci USA. 2005;102:7517–7522. doi: 10.1073/pnas.0502495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang YF, Goger MJ, Raleigh DP. Biochemistry. 2006;45:6940–6946. doi: 10.1021/bi052484n. [DOI] [PubMed] [Google Scholar]
- 8.Grey MJ, Tang YF, Alexov E, McKnight CJ, Raleigh DP, Palmer AG. J Mol Biol. 2006;355:1078–1094. doi: 10.1016/j.jmb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Kubelka J, Eaton WA, Hofrichter J. J Mol Biol. 2003;329:625–630. doi: 10.1016/s0022-2836(03)00519-9. [DOI] [PubMed] [Google Scholar]
- 10.Glasscock JM, Zhu YJ, Chowdhury P, Tang J, Gai F. Biochemistry. 2008;47:11070–11076. doi: 10.1021/bi8012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiner A. J Pept Sci. 2011;17:413–419. doi: 10.1002/psc.1353. [DOI] [PubMed] [Google Scholar]
- 12.Reiner A, Henklein P, Kiefhaber T. Nat Acad Sci USA. 2010;107:4955–4960. doi: 10.1073/pnas.0910001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brewer SH, Song BB, Raleigh DP, Dyer RB. Biochemistry. 2007;46:3279–3285. doi: 10.1021/bi602372y. [DOI] [PubMed] [Google Scholar]
- 14.Finkelstein IJ, Zheng J, Ishikawa H, Kim S, Kwak K, Fayer MD. Phys Chem Chem Phys. 2007;9:1533–1549. doi: 10.1039/b618158a. [DOI] [PubMed] [Google Scholar]
- 15.Thielges MC, Fayer MD. Acc Chem Research. 2012 doi: 10.1021/ar200275k. ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suydam IT, Boxer SG. Biochemistry. 2003;42:12050–12055. doi: 10.1021/bi0352926. [DOI] [PubMed] [Google Scholar]
- 17.Waegele MM, Culik RM, Gai F. J Phys Chem Lett. 2011;2:2598–2609. doi: 10.1021/jz201161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urbanek Diana C, DYV, Serrano Arnaldo L, Gai Feng, Hochstrasser Robin M. J Phys Chem Lett. 2010;1:3311–3315. doi: 10.1021/jz101367d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung JK, Thielges MC, Fayer MD. Nat Acad Sci USA. 2011;108:3578–3583. doi: 10.1073/pnas.1100587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung JK, Thielges MC, Fayer MD. J Am Chem Soc. 2012;134:12118–12124. doi: 10.1021/ja303017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanzi F, Madan B, Sharp K. J Am Chem Soc. 1998;120:10748–10753. [Google Scholar]
- 22.Park S, Kwak K, Fayer MD. Laser Phys Lett. 2007;4:704–718. [Google Scholar]
- 23.Zheng J, Kwak K, Fayer MD. Acc of Chem Res. 2007;40:75–83. doi: 10.1021/ar068010d. [DOI] [PubMed] [Google Scholar]
- 24.Kwak K, Park S, Finkelstein IJ, Fayer MD. J Chem Phys. 2007;127:124503. doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 25.Kwak K, Rosenfeld DE, Fayer MD. J Chem Phys. 2008;128:204505. doi: 10.1063/1.2927906. [DOI] [PubMed] [Google Scholar]
- 26.Thielges MC, Chung JK, Fayer MD. J Am Chem Soc. 2011;133:3995–4004. doi: 10.1021/ja109168h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finkelstein IJ, Ishikawa H, Kim S, Massari AM, Fayer MD. Proc Natl Acad Sci USA. 2007;104:2637–2642. doi: 10.1073/pnas.0610027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung JK, Thielges MC, Bowman SEJ, Bren KL, Fayer MD. J Am Chem Soc. 2011;133:6681–6691. doi: 10.1021/ja111009s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DILL KA, SHORTLE D. Annu Rev Biochem. 1991;60:795–825. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
- 30.Lindquist BA, Furse KE, Corcelli SA. PCCP. 2009;11:8119–8132. doi: 10.1039/b908588b. [DOI] [PubMed] [Google Scholar]
- 31.Fafarman AT, Boxer SG. J Phys Chem B. 2010;114:13536–13544. doi: 10.1021/jp106406p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fafarman AT, Webb LJ, Chuang JI, Boxer SG. J Am Chem Soc. 2006;128:13356–13357. doi: 10.1021/ja0650403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baum J, Dobson CM, Evans PA, Hanley C. Biochemistry. 1989;28:7–13. doi: 10.1021/bi00427a002. [DOI] [PubMed] [Google Scholar]
- 34.Kiefhaber T, Labhardt AM, Baldwin RL. Nature. 1995;375:513–515. doi: 10.1038/375513a0. [DOI] [PubMed] [Google Scholar]
- 35.Gao JM, Kelly JW. Protein Sci. 2008;17:1096–1101. doi: 10.1110/ps.083439708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akaike H. IEEE Trans Automat Contr. 1974;AC19:716–723. [Google Scholar]
- 37.Williams RB, Loring RF, Fayer MD. J Phys Chem B. 2001;105:4068–4071. [Google Scholar]
- 38.Merchant KA, Noid WG, Akiyama R, Finkelstein I, Goun A, McClain BL, Loring RF, Fayer MD. J Am Chem Soc. 2003;125:13804–13818. doi: 10.1021/ja035654x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merchant KA, Noid WG, Thompson DE, Akiyama R, Loring RF, Fayer MD. J Phys Chem B. 2003;107:4–7. [Google Scholar]
- 40.Bagchi S, Nebgen BT, Loring RF, Fayer MD. J Am Chem Soc. 2010;132:18367–18376. doi: 10.1021/ja108491t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagchi S, Boxer SG, Fayer MD. J Phys Chem B. 2012;116:4034–4042. doi: 10.1021/jp2122856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finkelstein IJ, Goj A, McClain BL, Massari AM, Merchant KA, Loring RF, Fayer MD. J Phys Chem B. 2005;109:16959–16966. doi: 10.1021/jp0517201. [DOI] [PubMed] [Google Scholar]
- 43.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 44.Onuchic JN, LutheySchulten Z, Wolynes PG. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 45.Russell BS, Melenkivitz R, Bren KL. Proc Natl Acad Sci USA. 2000;97:8312–8317. doi: 10.1073/pnas.150239397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segel DJ, Fink AL, Hodgson KO, Doniach S. Biochemistry. 1998;37:12443–12451. doi: 10.1021/bi980535t. [DOI] [PubMed] [Google Scholar]
- 47.Kawahara K, Tanford C. J Biological Chem. 1966;241:3228–3232. [PubMed] [Google Scholar]
- 48.Park S, Fayer MD. Nat Acad Sci USA. 2007;104:16731–16738. doi: 10.1073/pnas.0707824104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shim SH, Strasfeld D, Fulmer E, Zanni M. Opt Lett. 2006;31:838–878. doi: 10.1364/ol.31.000838. [DOI] [PubMed] [Google Scholar]
- 50.Shim SH, Strasfeld DB, Zanni MT. Opt Express. 2006;14:13120–13130. doi: 10.1364/oe.14.013120. [DOI] [PubMed] [Google Scholar]
- 51.Shim SH, Zanni MT. Phys Chem Chem Phys. 2009;11:748–761. doi: 10.1039/b813817f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.