Abstract
Acetylcholinesterase (AChE) is a crucial enzyme in the cholinergic nervous system that hydrolyses neurotransmitter acetylcholine (ACh) and terminates synaptic signals. The catalytic serine of AChE can be phosphonylated by soman, one of the most potent nerve agents, and subsequently undergo an aging reaction. This phosphonylation and aging process leads to irreversible AChE inhibition, results in accumulation of excess ACh at the synaptic clefts and causes neuromuscular paralysis. By employing Born-Oppenheimer ab initio QM/MM molecular dynamics simulations with umbrella sampling, a state-of-the-art approach to simulate enzyme reactions, we have characterized the aging mechanism of soman phosphonylated AChE and determined its free energy profile. This aging reaction starts with the scission of the O2-Cα bond, which is followed by methyl migration, and results in a tertiary carbenium intermediate. At the transition state, the scissile O2-Cα bond is already cleaved with an average O-C distance of 3.2 ± 0.3 Å and the migrating methyl group is shared between Cα and Cβ carbons with C-C distances of 1.9 ± 0.1 and 1.8 ± 0.1 Å, respectively. The negatively charged phosphonate group is stabilized by a salt bridge with the imidazole ring of the catalytic histidine. A major product of aging, 2,3-dimehtyl-2-butanol can be formed swiftly by the reaction of a water molecule. Our characterized mechanism and simulation results provide new detailed insights into this important biochemical process.
Keywords: nerve agents, irreversible inhibition, dealkylation, ab initio QM/MM molecular dynamics simulation, free energy and umbrella sampling
1. INTRODUCTION
Acetylcholinesterase (AChE) is a key serine hydrolase in the neuromuscular and cholinergic nervous system. It terminates synaptic signals by rapidly hydrolyzing neurotransmitter acetylcholine (ACh) with a rate close to the diffusion-controlled limit1–3. The catalytic site of AChE is buried at the bottom of a 20 Å deep, narrow and highly electronegative catalytic gorge4–6 and its catalytic triad (Ser200, His440 and Glu327) as well as its oxyanion hole (peptidic NH groups of Gly118, Gly119, and Ala201) are typical of serine hydrolyses. (Torpedo californica AChE residue numbering is used throughout this paper.) AChE catalytic mechanism of ACh hydrolysis proceeds through successive acylation and deacylation reaction steps2,7–11 and the overall catalytic process is initiated by a nucleophilic attack on carbonyl carbon of the bound ACh by Ser200 with His440 serving as a general base. Catalytic Ser200, which is crucial in ACh hydrolysis, is also reactive towards an array of covalent inhibitors such as pesticides and more dangerously nerve agents12.
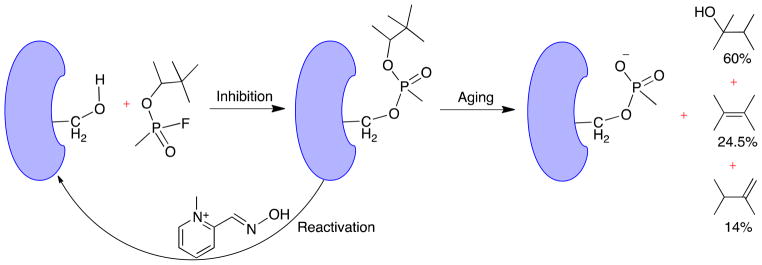
Nerve agents are a military subclass of organophosphate (OP) compounds and are responsible for ~300,000 fatalities per year worldwide13,14. Nerve agents have been used as terrorist agents in multiple occasions15–18 and have been classified as weapons of mass destruction by the United Nations in 1993 (UN Resolution 687). Furthermore, less lethal OP pesticides pose a public health challenge, in case of accidental and/or repeated exposure19–22. At a biochemical level, nerve agents exert their toxicity by phosphonylating the catalytic Ser200 of AChE and effectively blocking ACh hydrolysis. Phosphonylated adducts can either be reactivated to a limited extent by oximes such as 2-PAM (2-pyridine aldoxime methyl chloride), HI-6 (asoxime chloride), TMB-4 (trimedoxime bromide) and toxogonine23–26 or undergo a very fast secondary aging/dealkylation reaction resulting in an “aged” enzyme which cannot be reactivated27 (Figure 1). Phosphonylation and subsequent aging reactions of AChE result in accumulation of excess ACh at the synapse, which causes neuromuscular paralysis from overstimulation with fatal consequences. The lethality of nerve agents depend on how fast the phosphonylated adducts can undergo aging reaction, which varies with the bound inhibitors’ alkoxyl groups28,29.
Figure 1.
Schematic illustration of AChE phosphonylation by soman, re-activation by pralidoxime (2-PAM) and subsequent irreversible aging.
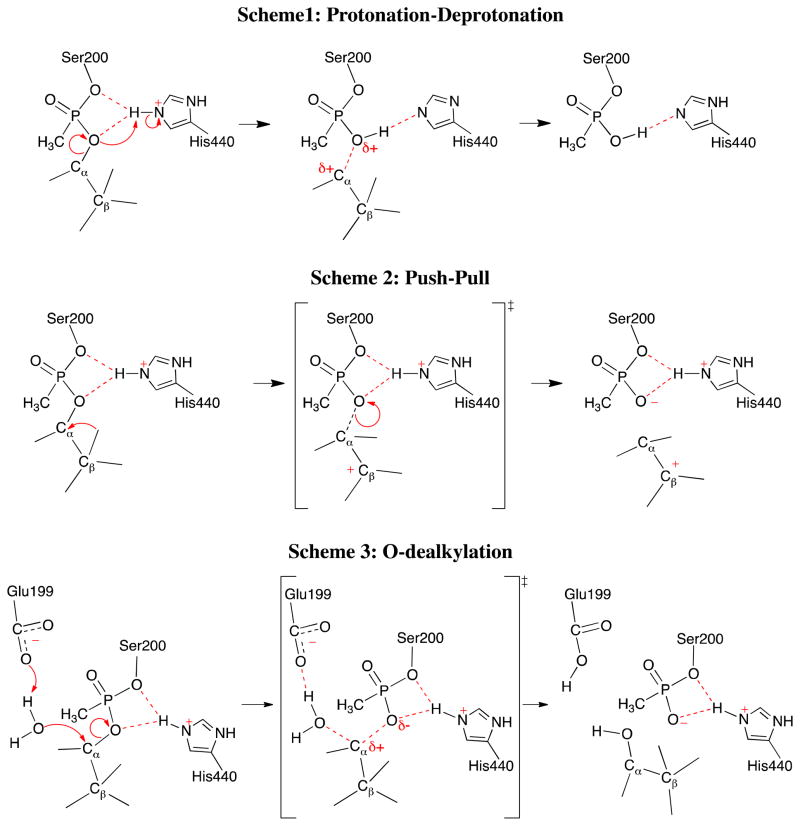
Soman (3,3-dimethylbutan-2-yl methylphosphonofluoridate) has a highly branched alkoxyl group and is one of the most lethal nerve agents with an aging half-life of 4.62 min and 1.87 min at pH 7 when bound with TcAChE30 and HuBChE31 respectively. Soman has two chiral centers, namely the P and Cα atoms and the stereochemistry of the P atom is inverted once a covalent adduct is formed between32. The crystal structure of non-aged soman in complex with TcAChE was recently obtained by Wiek et al.33, in which soman is in the PSCαR stereoisomer configuration (Figure 2). In spite of extensive biochemical and structural studies29–31,33–39, mechanistic details of AChE-soman aging remain to be elusive. Several mechanisms were suggested for the aging of soman inhibited AChE as shown in Figure 3. One is the protonation-deprotonation mechanism, in which protonation of the pinacolyl oxygen by the positively charged imidazole ring of catalytic His440+ is suggested to initiate the dealkylation reaction (Scheme 1 of Figure 3)27,31,34. The second proposed scheme is known as the push-pull mechanism, in which a methyl migration from overpopulated Cβ to Cα of the pinacolyl group is suggested to be the initial rate-determining step (Scheme 2 of Figure 3)30,34,39. For this scheme, Glu199 and Trp84 were proposed to act as “pusher” driving methyl rearrangement. But, it has already been suggested that Glu199 cannot act as one of the pushers of the push-pull scheme since its side chain is closer to Cα than to the Cβ carbon. Nevertheless, the experimentally determined low molecular weight products of aging provide evidence for the methyl rearrangement during the aging reaction27,38. For nerve agents with unbranched alky groups such as tabun, O-dealkylation mechanism of aging has been proposed, in which Glu199 of the active site acts as a base to induce hydrolytic scission of O2-Cα covalent bond (Scheme 3 of Figure 3)40. In all these previously proposed aging mechanisms, Glu199 has been assumed to be deprotonated.
Figure 2.
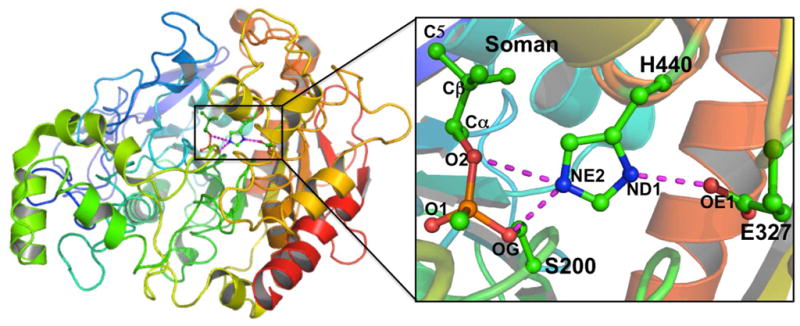
Illustration of overall secondary structure of the non-aged soman inhibited AChE (PDB ID: 2WFZ)33. The catalytic triad and the covalently bound soman are shown in ball-stick representation and are enlarged to the right panel.
Figure 3.
Previously suggested aging reaction mechanisms for soman inhibited AChE.
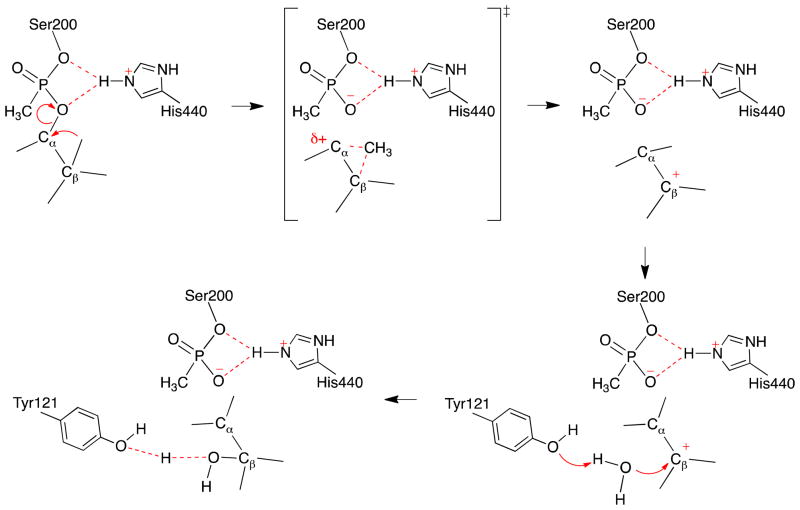
Although a variety of computational mechanistic studies of AChE catalysis8,41–44, covalent phosphonylation/inhibition45–48 and dephosphorylation/reactivation12,49 have been carried out, the aging/dealkylation mechanism of soman inhibited AChE has not been studied computationally. In the present work, we filled this gap and characterized the aging mechanism of soman phosphonylated AChE by employing Born Oppenheimer ab initio QM/MM molecular dynamics simulations with umbrella sampling method, an approach that has been extensively applied to study enzyme reactions8,50–59. The characterized aging mechanism is schematically illustrated in Figure 4. Mechanistic details revealed by this work would facilitate the design of novel compounds capable of slowing the aging reactions.
Figure 4.
Illustration of the characterized aging reaction mechanism of soman-phosphonylated AChE.
2. METHODS SECTION
Preparation of the Reactant Structure
The initial reactant structure of AChE-soman was based on the crystal structure of non-aged soman phosphonylated TcAChE33 (pdb code: 2WFZ). Protonation states were determined by using H++ program60 and examining the local hydrogen bond network. One ambiguity is the protonation state of Glu199 and the pKa calculations were not conclusive to determine it. Hence, we prepared three models including unprotonated Glu199, protonated Glh199 and Glu199Gln mutant. Non-standard soman-serine residue partial charges were fitted with HF/6-31G(d) calculations and the restrained electrostatic potential (RESP) protocol implemented by the Antechamber module in Amber1161 package (see Table S1). Other force field parameters were adapted from the standard GAFF force field by the Antechamber module.
The Amber1161 package with Amber99SB force field61–64 was used to perform all the classic MD simulations. The prepared systems were neutralized by adding Na+ ions. Then, each system was solvated in a rectangular box of TIP3P water molecules65 with a minimum solute-wall distance of 10 Å. Each resultant system, which consisted of ~65,700 atoms, was first geometry optimized followed by a series of equilibrations and a final 20 ns molecular dynamics production run for the NPT ensemble was carried out. The time step was 1 fs and periodic boundary condition was used. The system was maintained at 1 atm and coupled with isotropic position scaling, while the temperature was controlled at 300 K with Berendsen thermostat method66. Long-range electrostatic interactions were treated with Particle Mash Ewald (PME) method and 12 Å cutoff was used for both PME and van deer Waals (vdW) interaction.
QM/MM Simulations
A randomly chosen MD snapshot, selected after an initial equilibration period of 1 ns, was used as the starting structure for the QM/MM simulations. The chosen structure was first examined for hydrogen bonding between the catalytic triad residues. The QM/MM model was prepared by removing ions and waters beyond 30 Å from the hydroxyl oxygen of Ser200, which consists of ~14,000 atoms. The QM subsystem consists of those fragments that are potentially involved in the bond forming/breaking process, as illustrated in Figure S1: for the dealkylation step, it includes side chains of soman modified Ser200, His440, Glu327 and Glu199 as well as a potentially reactive water molecule in the active site; for the second hydration step, it contains side chains of modified Ser200, His440, Glu327 and Tyr121 as well as the tertiary carbenium ion and a potentially reactive water molecule located between Tyr121 and the tertiary carbenium.
The QM/MM boundary was described by the pseudobond approach67–72. The QM subsystem was described using B3LYP functional with 6-31G* basis set while MM subsystem was described using Amber99SB force field and TIP3P water model. The spherical boundary condition was employed and only atoms within 20 Å of the reaction site were allowed to move. 18 and 12 Å cutoffs were used for electrostatics and VdW interactions respectively. No cutoff was employed for the electrostatic interactions between the QM and MM regions. Beeman algorithm73 was used to integrate Newton equations of motions with time step of 1 fs. Berendsen thermostat method66 was employed to control system temperature at 300 K. All QM/MM calculations were run using modified versions Q-Chem74 and Tinker75 packages.
The QM/MM system was first geometry optimized and subsequently several reaction paths have been calculated using the reaction coordinate driving (RCD) method71,72 until the minimum energy path was found for each suggested aging schemes. For each structure along the determined reaction path, a 500 ps MD simulation with a MM force field was performed to equilibrate the MM subsystem while the QM subsystem was kept frozen. The final snapshot from MM equilibration was used as the starting structure for B3LYP(6-31G*) QM/MM MD simulations with umbrella sampling method76–79, in which the ab initio QM/MM total energy of the system is biased with a harmonic potential to sample successive regions along the reaction coordinate. For each MD simulation (windows) along the reaction coordinate, configurations for the last 15 ps were collected for data analysis after a 5 ps equilibration period. The probability distributions along the reaction coordinates were determined for each window and pieced together using the weighted histogram analysis method (WHAM)80–82 to calculate the free energy profile. More details and formulations regarding ab initio QM/MM and umbrella sampling can be found in Ref. 79.
3. RESULTS AND DISCUSSION
3.1 Examination of previously suggested aging mechanisms
In all previously suggested aging mechanisms for soman inhibited AChE, the glutamate residue (Glu199 in Torpedo California AChE) vicinal to the catalytic gorge has been assumed to be deprotonated. Thus in our first prepared soman-AChE model, Glu199 is deprotonated. For each previously suggested mechanism, as shown in Figure 3, we have employed B3LYP(6-31G*) QM/MM simulations to determine its reaction energy profiles by choosing appropriate reaction coordinates.
First, we examined the proposed protonation-deprotonation reaction scheme, in which His440+ is suggested to donate its HE2 proton to soman O2 oxygen prior to the dealkylation of the alkyl chain as schematically shown in Scheme 1 of Figure 3. Our calculated potential energy barrier for the proton transfer from His440+ NE2 nitrogen to soman is ~40 kcal/mol as shown in Figure S2. The protonated product is not stable and collapses back to the reactant state during unrestrained QM/MM MD simulations. These results indicate that this previously suggested scheme is energetically unfeasible. In addition, we have also tested a concerted scheme in which protonation of soman occurs in concert with dealkylation as shown in Figure S3. Although the calculated activation energy for this scheme is about 20 kcal/mol, the proton remains on His440+ while the O2-Cα bond, connecting the alkyl chain to the phosphonate moiety, is cleaved. Furthermore, once the O2-Cα distance is elongated to ~3 Å, the resultant secondary carbocation is not stable: in some windows, one of the methyl groups from Cβ carbon migrates spontaneously to Cα carbon resulting in a stable 2,3-dimehtylbutyl carbenium ion, which is essentially a methyl-migration mechanism; whereas in some other windows, waters included in the QM subsystem forms a covalent bond with the positively charged Cα carbon, which is similar to the O-dealkylation scheme. Thus in summary, our simulation results do not support the protonation-deprotonation mechanism.
Second, we studied the proposed push-pull mechanism of aging. The free energy profile is smooth and continuous with activation energy of 20 kcal/mol, shown as the red curve in Figure 5. Key structures along this reaction scheme are shown in Figure 6a. The 2,3-dimethyl-3-butyl carbenium ion intermediate formed from this reaction mechanism can yield products consistent with experimental results38. However, unlike the proposed push-pull mechanism, our simulations suggest that methyl rearrangement occurs only after the O2-Cα covalent bond is cleaved as schematically outlined in Figure 4. Thus our simulations do not support that Glu199 plays the role of a pusher in the soman-aging reaction and our computationally characterized reaction scheme is essentially a methyl-migration mechanism.
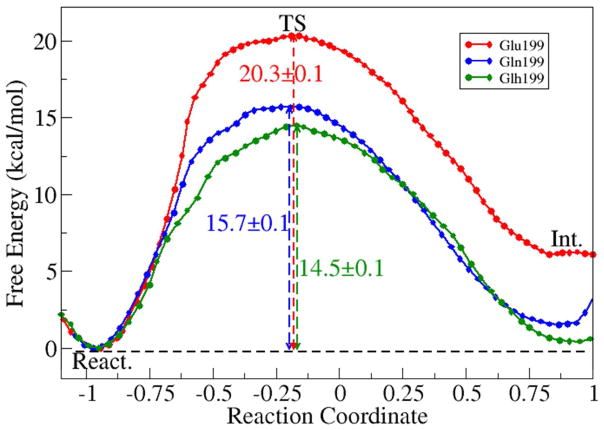
Figure 5.
Free energy profiles for the methyl migration step of aging reaction for the unprotonated Glu199 (red), Glu199Gln mutant (blue) and the protonated Glu199 (green) AChE models. The reaction coordinate was chosen as the difference between two bond lengths: RC = d(somanCβ…somanC5) − d(somanCα…somanC5). The total length of ab initio QM/MM MD simulations for this reaction path is 1.04 ns (Unprotonated Glu199: 20 windows * 20 ps each; Glu199Gln mutant: 17 windows * 20 ps each; protonated Glh199: 16 windows * 20 ps each).
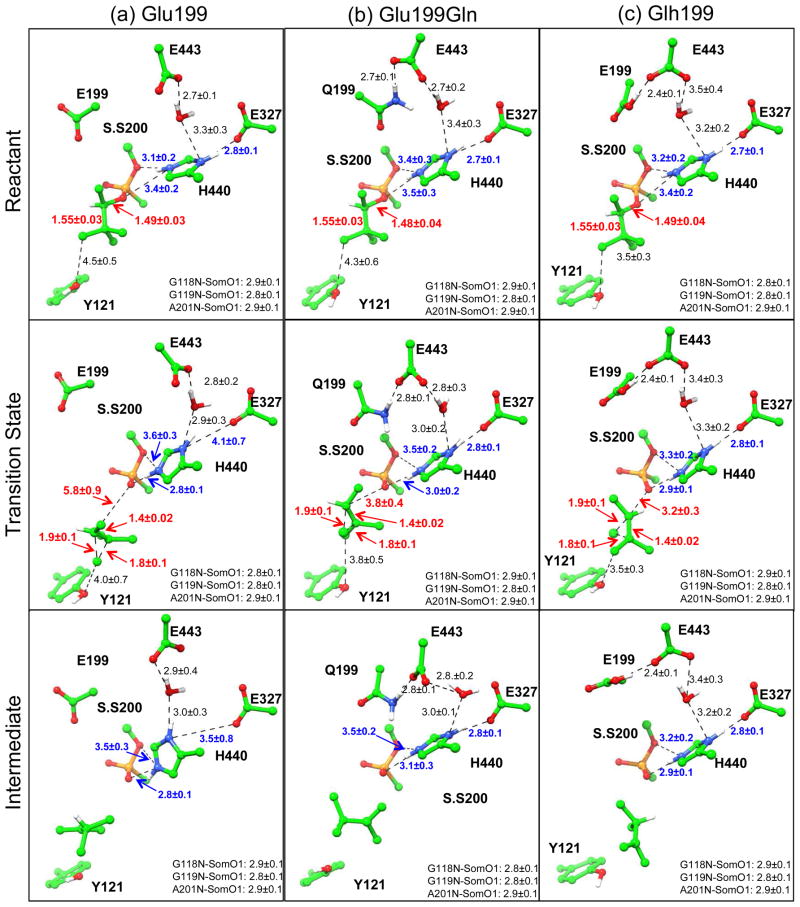
Figure 6.
Illustration of key structures along the methyl migration step of aging reaction: (a) unprotonated Glu199, (b) Glu199Gln mutant and (c) protonated Glh199 models. S.S200 is the soman modified serine residue. The labeled distances (A) are averages and standard deviations calculated along the last 15 ps of 20 ps QM/MM MD simulations.
Lastly, the O-dealkylation mechanism of aging was also studied, considering that some of our ab initio QM/MM MD simulations suggest that this might be feasible. In this scheme, the crystal water (Wat1206) hydrogen bonded to the carboxyl side chain of Glu199 acts as a hydrolytic agent and induce dealkylation of the bound soman. The calculated activation energy for this O-dealkylation reaction is 21 kcal/mol (Figure S4), comparable to the above methyl-migration scheme. By comparing the red curve in Figure 5 and Figure S4b, we can see that 3,3-dimehtyl-2-butanol product from the O-dealkylation is energetically more stable than the 2,3-dimehtyl-butyl carbenium ion intermediate from the methyl migration scheme. These results suggest that for the unprotonated Glu199 model, O-dealkylation scheme should be one of the major aging pathways, however its low molecular weight product 3,3-dimehtyl-2-butanol has not been observed experimentally.
The above results raise some doubts regarding whether Glu199 should be deprotonated during the AChE-soman aging process, which has been widely assumed to be the case based on the experimental mutation result that its mutation to glutamine reduced the experimental rate of aging by 150–700 fold within physiological pH range30. Thus we were motivated to computationally examine the AChE-soman aging process for the Glu199Gln mutant. By mutating the Glu199 to glutamine, the base to induce hydrolytic cleavage of soman’s alkyl chain in the O-dealkylation scheme is removed and thus we only studied the methyl-migration aging scheme for the Glu199Gln model. As shown in Figure 5 (blue), our calculated activation energy for the Glu199Gln model is 15.7 kcal/mol, which is 4.6 kcal/mol smaller than the unprotonated Glu199 model. This does not agree with the experimental result that Glu199Gln mutant should increase the overall activation energy by ~3 kcal/mol30. These results further indicate that Glu199 may be protonated in the aging process of soman phosphonylated AChE.
3.2 Glu199 is protonated in soman inhibited AChE
It should be noted that during the hydrolysis of the neurotransmitter acetylcholine (ACh), pKa calculations, EVB simulations and state-of-the-art ab initio QM/MM studies have all suggested that Glu199 is only negatively charged in the acylation stage which is essential in binding the positively charged substrate, but protonated during the deacylation stage after the release of the positively charged choline from the active site8,43. Since the soman inhibited AChE consists of a phosphonate group with a highly branched alkyl chain in the active site, the pKa calculations with H++ suggested that Glu199 is protonated in the soman phosphonylated AChE complex60. Hence, we prepared the third model for the soman inhibited AChE in which Glu199 is protonated (Glh199) and examined its aging mechanism.
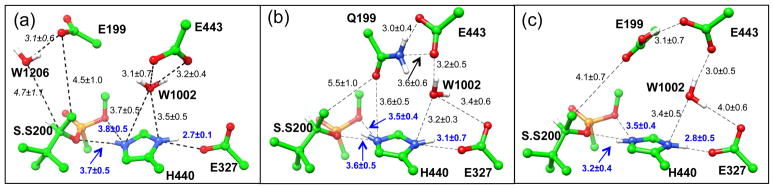
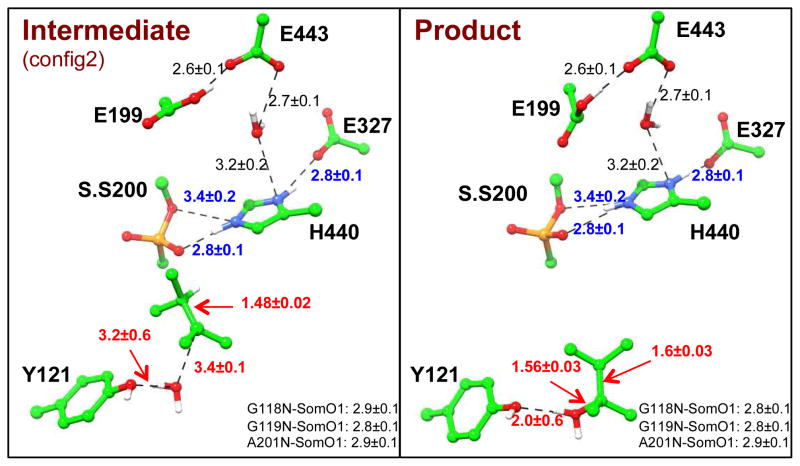
As shown in Figure 7, in comparison with the unprotonated Glu199 model as well as the Glu199Gln model, His440+ side chain is less mobile when Glu199 is protonated and the structural integrity of the catalytic site is better maintained as indicated by stronger hydrogen bonding between the imidazole ring nitrogen atoms and the soman modified Ser200 and Glu327 oxygen atoms. Also, in the protonated Glh199 model, His440+ χ1 and χ2 angles are less mobile especially in comparison to the unprotonated Glu199 model at the transition state (Figure S6). For the more stable protonated Glu199 model, aging though methyl migration mechanism was studied by using the same computational protocol as for the other two models. The determined free energy profile is smooth and continuous, as shown in Figure 5 (green). The calculated activation energy for aging reaction is 14.5 kcal/mol, which is lower than that of either the unprotonated Glu199 or Glu199Gln mutant models with the same computational protocols. This value can be compared with the estimated experimental activation barrier of 21.1 kcal/mol at pH 7 using transition state theory83. From Figure 6c, we can see that in the protonated Glu199 model system, aging of AChE-soman is initiated with scission of O2-Cα covalent bond and subsequent methyl migration from overpopulated Cβ carbon to the positively charged Cα carbon. At the transition state, scission of O2-Cα bond has already occurred (3.2 ± 0.3 Å) and the migrating methyl group is shared between Cα and Cβ carbons with C-C distance of 1.9 ± 0.1 Å and 1.8 ± 0.1 Å respectively. Furthermore, upon scission of O2-Cα bond, the histidine ring re-orients a little bit which enables the formation of a stronger salt bridge to stabilize the transition state: the average N-O distance between His440+ NE2 and soman O2 decreases from 3.4 ± 0.2 Å to 2.9 ± 0.1 Å at the transition state and remains so throughout the reaction. The observed His440+ NE2 shift is 1.08 ± 0.32 Å on average and is in agreement with X-ray structure displacement of ~0.8 Å33. The His440+ - Glu327 salt bridge is stable throughout the aging reaction in the protonated Glu199 model (reactant NO distance is 2.8 ± 0.1 Å) while it breaks down when Glu199 is unprotonated. Meanwhile, the phosphonyl O1 oxygen of soman is stabilized by three hydrogen bonds with the oxyanion hole, made up of backbone NH groups of Ala201, Gly118, and Gly119, with initial reactant N-O distances of 2.9 ± 0.1 Å, 2.8 ± 0.1 Å and 2.8 ± 0.1 Å respectively. Methyl rearrangement results in the 3,2-dimethyl-butyl carbenium ion intermediate (Int.), where water molecule migrates within the vicinity of Cβ carbon and bridges it to the hydroxyl group of Tyr121.
Figure 7.
Hydrogen bond network of non-aged soman inhibited AChE active site (a) unprotonated Glu199, (b) Glu199Gln mutant and (c) the protonated Glh199 models. Shown configurations correspond to the 5 ns snapshot from atomistic MD simulation. S.S200 refers the soman modified serine residue. The labeled distances (A) are averages and standard deviations calculated along 20 ns atomistic MD simulations. W1206 in panel (a) is only stable for ~10 ns after which it moves further away from the vicinity of the active site. Hence the average reported in italics for W1206 distance is only for the first 10ns.
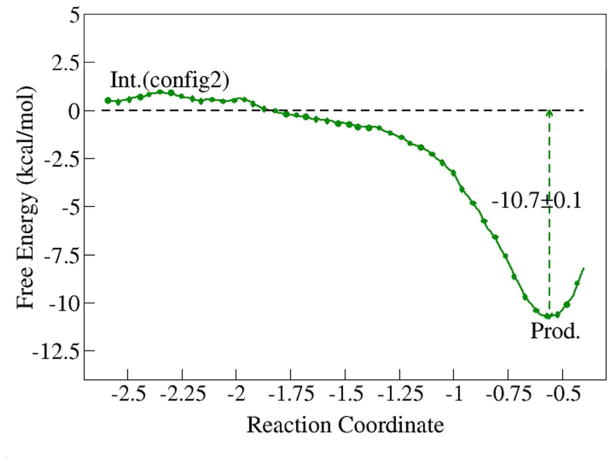
To examine how the carbenium intermediate can be hydrolyzed to yield one major final product, a second QM/MM system was prepared as shown in Figure S1b. The free energy profile for this hydration reaction is shown in Figure 8 and corresponding key structures are illustrated in Figure 9. The hydration of 2-3-dimehtylbutyl to 2-3-dimehtylbutanol by AChE is an exothermic and spontaneous process and yields the major product that has been experimentally observed38. The final aged AChE enzyme is ~10 kcal/mol more stable than the reactant, in agreement with the experimental results that the stability of aged AChE is significantly increased in comparison to its non-aged conjugate39,84–88.
Figure 8. Protonated Glh199 model.
Free energy profile for hydration of 2,3-dimethyl-butyl carbenium ion. The reaction coordinate was chosen as the difference between two bond lengths: RC = d(watO-watH) − d(somanCβ…watO). The total length of ab initio QM/MM MD simulations for this reaction path is 220 ps (11 windows * 20 ps each).
Figure 9. Protonated Glh199 model.
Illustration of key structures along hydration step of the aging reaction. S.S200 is the soman modified serine residue. The distances (A) are averages and standard deviations calculated along the last 15 ps of 20 ps QM/MM MD simulations.
4. CONCLUSIONS
By employing Born-Oppenheimer ab initio QM/MM molecular dynamics simulations with umbrella sampling, we have characterized the complete aging mechanism of soman inhibited AChE and determined its free energy profile. We find that aging is favored when Glu199, vicinal to the catalytic gorge, is protonated. The AChE-soman aging process begins with the scission of O2-Cα covalent bond, resulting in the 2,3-dimethylbutyl carbenium ion. At the transition state, the scissile O2-Cα covalent bond is cleaved and the migrating methyl group is shared between Cα and Cβ carbons. Immediately upon scission of O2-Cα covalent bond, the negative charge developed on the phosphonate group is stabilized by forming a strong salt bridge with the re-oriented imidazole ring of the catalytic His440+. Methyl migration, subsequent to the scission of O2-Cα bond, leads to the formation of the 2,3-dimethylbutyl carbenium intermediate. Finally, the tertiary carbenium can be hydrated swiftly by a water molecule facilitated by Try121 hydroxyl group to result in the 2,3-dimehtyl-2-butanol product. Overall, the aging of soman inhibited AChE is an exothermic reaction and results in a stable aged-AChE conjugate and 2,3-dimehtyl-2-butanol as the major product, in agreement with experimental work.
To provide such detailed mechanistic insights into this important biological process is not only of fundamental importance but also of significant interest for the development of novel strategies to reduce the toxic effect of soman. Currently, treatment against organophosphate poisoning involves a combination therapy, in which anti-cholinergic agents such as atropine is administered with reactivating oxime compounds. However, this treatment is not effective against high-exposure levels and the limited reactivating capacity of oxime compounds make it necessary to continue the search to improve oxime potency89,90 and develop novel approaches that are either capable of slowing the aging reaction or reactivating aged AChEs91.
Supplementary Material
Acknowledgments
This work was supported by NSF-IGERT and NIH (R01- GM079223). We thank NSF-XSEDE and NYU-ITS for providing computational resources.
Footnotes
Supporting Information Available: Figures S1–S6 and Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rosenberry TL. Adv Enzymol Relat Areas Mol Biol. John Wiley & Sons, Inc; 2006. pp. 103–218. [DOI] [PubMed] [Google Scholar]
- 2.Quinn DM. Chem Rev. 1987;87:955–979. [Google Scholar]
- 3.Silman I, Sussman JL. Curr Opin Pharmacol. 2005;5:293–302. doi: 10.1016/j.coph.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 5.Goncalves AS, Franca TCC, Wilter A, Figueroa-Villar JD. J Braz Chem Soc. 2006;17:968–975. [Google Scholar]
- 6.Matos KS, da Cunha EFF, da Silva Goncalves A, Wilter A, Kuča K, Franca TCC, Ramalho TC. J Biomol Struct Dyn. 2012;30:546–558. doi: 10.1080/07391102.2012.687521. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Kua J, McCammon JA. J Am Chem Soc. 2002;124:10572–10577. doi: 10.1021/ja020243m. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, Wang S, Zhang Y. J Phys Chem B. 2010;114:8817–8825. doi: 10.1021/jp104258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harel M, Quinn DM, Nair HK, Silman I, Sussman JL. J Am Chem Soc. 1996;118:2340–2346. [Google Scholar]
- 10.Pryor AN, Selwood T, Leu LS, Andracki MA, Lee BH, Rao M, Rosenberry T, Doctor BP, Silman I, Quinn DM. J Am Chem Soc. 1992;114:3896–3900. [Google Scholar]
- 11.Malany S, Sawai M, Sikorski RS, Seravalli J, Quinn DM, Radic Z, Taylor P, Kronman C, Velan B, Shafferman A. J Am Chem Soc. 2000;122:2981–2987. [Google Scholar]
- 12.Goncalves AS, Franca TCC, Figueroa-Villar JD, Pascutti PG. J Braz Chem Soc. 2011;22:155–165. [Google Scholar]
- 13.Gupta RC. Toxicology of organophosphate and carbamate compounds. Academic Press; 2006. [Google Scholar]
- 14.Gunnell D, Eddleston M, Phillips M, Konradsen F. BMC public health. 2007;7:357. doi: 10.1186/1471-2458-7-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbas F. Arch Belg. 1984:302–310. [PubMed] [Google Scholar]
- 16.Council, U. N. S. . Report of the Mission Dispatched by the Secretary-General to Investigate Allegations of the Use of Chemical Weapons in the Conflict Between the Islamic Republic of Iran and Iraq. United Nations Security Council; 1986. [Google Scholar]
- 17.Macilwain C. Nature. 1993;363:3. doi: 10.1038/363003b0. [DOI] [PubMed] [Google Scholar]
- 18.Nagao M, Takatori T, Matsuda Y, Nakajima M, Iwase H, Iwadate K. Toxicol Appl Pharmacol. 1997;144:198–203. doi: 10.1006/taap.1997.8110. [DOI] [PubMed] [Google Scholar]
- 19.Smart JK. Medical Aspects of Chemical and Biological Warfare. Washington, DC: Office of the Surgeon General; 1997. pp. 9–86. [Google Scholar]
- 20.Okudera H, Morita H, Iwashita T, Shibata T, Otagiri T, Kobayashi S, Yanagisawa N. Am J Emerg Med. 1997;15:527–528. doi: 10.1016/s0735-6757(97)90201-1. [DOI] [PubMed] [Google Scholar]
- 21.Holstege CP, Kirk M, Sidell FR. Crit Care Clin. 1997;13:923–942. doi: 10.1016/s0749-0704(05)70374-2. [DOI] [PubMed] [Google Scholar]
- 22.Hay A, Roberts G. J Am Med Assoc. 1990;263:1065–1066. [Google Scholar]
- 23.da Silva Goncalves A, Franca TCC, Figueroa-Villarb JD, Pascutti PG. J Braz Chem Soc. 2010;21:179–184. [Google Scholar]
- 24.Wilson IB, Meislich EK. J Am Chem Soc. 1953;75:4628–4629. [Google Scholar]
- 25.Worek F, Szinicz L, Thiermann H. Chem Biol Interact. 2005;157:349–352. doi: 10.1016/j.cbi.2005.10.101. [DOI] [PubMed] [Google Scholar]
- 26.Marrs TC. Pharmacology & therapeutics. 1993;58:51–66. doi: 10.1016/0163-7258(93)90066-m. [DOI] [PubMed] [Google Scholar]
- 27.Michel HO, Hackley BE, Berkowitz L, List G, Hackley EB, Gillilan W, Pankau M. Arch Biochem Biophys. 1967;121:29–34. doi: 10.1016/0003-9861(67)90006-9. [DOI] [PubMed] [Google Scholar]
- 28.Aurbek N, Thiermann H, Szinicz L, Eyer P, Worek F. Toxicology. 2006;224:91–99. doi: 10.1016/j.tox.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 29.Beauregard G, Lum J, Roufogalis BD. Biochem Pharmacol. 1981;30:2915–2920. doi: 10.1016/0006-2952(81)90252-5. [DOI] [PubMed] [Google Scholar]
- 30.Saxena A, Doctor BP, Maxwell DM, Lenz DE, Radic Z, Taylor P. Biochem Biophys Res Commun. 1993;197:343–349. doi: 10.1006/bbrc.1993.2481. [DOI] [PubMed] [Google Scholar]
- 31.Shafferman A, Ordentlich A, Barak D, Stein D, Ariel N, Velan B. Biochem J. 1996;318:833–840. doi: 10.1042/bj3180833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman HA, Decker MM. J Biol Chem. 1989;264:3951–3956. [PubMed] [Google Scholar]
- 33.Sanson B, Nachon F, Colletier J-P, Froment M-T, Toker L, Greenblatt HM, Sussman JL, Ashani Y, Masson P, Silman I, et al. J Med Chem. 2009;52:7593–7603. doi: 10.1021/jm900433t. [DOI] [PubMed] [Google Scholar]
- 34.Kovach IM. J Phys Org Chem. 2004;17:602–614. [Google Scholar]
- 35.Barak D, Ordentlich A, Segall Y, Velan B, Benschop HP, De Jong LPA, Shafferman A. J Am Chem Soc. 1997;119:3157–3158. [Google Scholar]
- 36.Masson P, Fortier PL, Albaret C, Froment MT, Bartels CF, Lockridge O. Biochem J. 1997;327:601–607. doi: 10.1042/bj3270601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massiah MA, Viragh C, Reddy PM, Kovach IM, Johnson J, Rosenberry TL, Mildvan AS. Biochemistry. 2001;40:5682–5690. doi: 10.1021/bi010243j. [DOI] [PubMed] [Google Scholar]
- 38.Viragh C, Kovach IM, Pannell L. Biochemistry. 1999;38:9557–9561. doi: 10.1021/bi991112+. [DOI] [PubMed] [Google Scholar]
- 39.Viragh C, Akhmetshin R, Kovach IM, Broomfield C. Biochemistry. 1997;36:8243–8252. doi: 10.1021/bi962764q. [DOI] [PubMed] [Google Scholar]
- 40.Carletti E, Li H, Li B, Ekstrom F, Nicolet Y, Loiodice M, Gillon E, Froment MT, Lockridge O, Schopfer LM. J Am Chem Soc. 2008;130:16011–16020. doi: 10.1021/ja804941z. [DOI] [PubMed] [Google Scholar]
- 41.Kua J, Zhang Y, McCammon JA. J Am Chem Soc. 2002;124:8260–8267. doi: 10.1021/ja020429l. [DOI] [PubMed] [Google Scholar]
- 42.Fuxreiter M, Warshel A. J Am Chem Soc. 1998;120:183–194. [Google Scholar]
- 43.Vagedes P, Rabenstein B, Aqvist J, Marelius J, Knapp EW. J Am Chem Soc. 2000;122:12254–12262. [Google Scholar]
- 44.Wlodek ST, Antosiewicz J, Briggs JM. J Am Chem Soc. 1997;119:8159–8165. [Google Scholar]
- 45.Kwasnieski Ol, Verdier L, Malacria M, Derat E. J Phys Chem B. 2009;113:10001–10007. doi: 10.1021/jp903843s. [DOI] [PubMed] [Google Scholar]
- 46.Beck JM, Hadad CM. Chem Biol Interact. 2010;187:220–224. doi: 10.1016/j.cbi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Gu J, Leszczynski J. J Phys Chem B. 2006;110:7567–7573. doi: 10.1021/jp060370v. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Gu J, Leszczynski J. J Phys Chem B. 2008;112:3485–3494. doi: 10.1021/jp076974w. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Zhang Y, Zhan CG. J Phys Chem B. 2009;113:16226–16236. doi: 10.1021/jp9055335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Hu P, Zhang Y. J Phys Chem B. 2007;111:3758–3764. doi: 10.1021/jp067147i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ke Z, Wang S, Xie D, Zhang Y. J Phys Chem B. 2009;113:16705–16710. doi: 10.1021/jp9080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ke Z, Zhou Y, Hu P, Wang S, Xie D, Zhang Y. J Phys Chem B. 2009;113:12750–12758. doi: 10.1021/jp903173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu R, Wang S, Zhou N, Cao Z, Zhang Y. J Am Chem Soc. 2010;132:9471–9479. doi: 10.1021/ja103932d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ke Z, Smith GK, Zhang Y, Guo H. J Am Chem Soc. 2011;133:11103–11105. doi: 10.1021/ja204378q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu R, Lu Z, Cao Z, Zhang Y. J Am Chem Soc. 2011;133:6110–6113. doi: 10.1021/ja111104p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Zhang Y. Chem Commun. 2011;47:1577–1579. doi: 10.1039/c0cc04112b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu R, Gong W, Liu T, Zhang Y, Cao Z. J Phys Chem B. 2012;116:1984–1991. doi: 10.1021/jp211403j. [DOI] [PubMed] [Google Scholar]
- 58.Hu P, Wang S, Zhang Y. J Am Chem Soc. 2008;130:16721–16728. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu P, Wang S, Zhang Y. J Am Chem Soc. 2008;130:3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. Nucleic Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, et al. AMBER. Vol. 11. Univ of California; San Francisco: 2010. [Google Scholar]
- 62.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins: Struct, Funct, Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Cieplak P, Kollman PA. J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 64.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 65.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 66.Berendsen HJC, Postma JPM, Van Gunsteren WF, DiNola A, Haak J. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 67.Zhang Y. Theor Chem Acc. 2006;116:43–50. [Google Scholar]
- 68.Zhang Y. J Chem Phys. 2005;122:024114–024120. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y, Lee TS, Yang W. J Chem Phys. 1999;110:46–54. [Google Scholar]
- 70.Liu H, Zhang Y, Yang W. J Am Chem Soc. 2000;122:6560–6570. [Google Scholar]
- 71.Zhang Y, Liu H, Yang W. J Chem Phys. 2000;112:3483–3492. [Google Scholar]
- 72.Hu H, Yang W. Annu Rev Phys Chem. 2008;59:573–601. doi: 10.1146/annurev.physchem.59.032607.093618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beeman D. J Comput Phys. 1976;20:130–136. [Google Scholar]
- 74.Shao Y, Molnar LF, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert AT, Slipchenko LV, Levchenko SV, O’Neill DP, et al. Q-Chem, Inc; Pittsburgh, PA: 2006. [Google Scholar]
- 75.Ponder J. TINKER: Software Tools for Molecular Design, Version 4.2. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benoit R. Comput Phys Commun. 1995;91:275–282. [Google Scholar]
- 77.Boczko EM, Brooks CL. J Phys Chem. 1993;97:4509–4513. [Google Scholar]
- 78.Patey G, Valleau J. J Chem Phys. 1975;63:2334–2339. 17. [Google Scholar]
- 79.Zheng H, Wang S, Zhang Y. J Comput Chem. 2009;30:2706–2711. doi: 10.1002/jcc.21296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Souaille M, Roux B. Comput Phys Commun. 2001;135:40–57. [Google Scholar]
- 81.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 82.Ferrenberg AM, Swendsen RH. Phys Rev Lett. 1988;61:2635–2638. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- 83.Evans M, Polanyi M. Trans Faraday Soc. 1935;31:875–894. [Google Scholar]
- 84.Gabel F, Masson P, Froment MT, Doctor B, Saxena A, Silman I, Zaccai G, Weik M. Biophys J. 2009;96:1489–1494. doi: 10.1016/j.bpj.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Masson P, Clery C, Guerra P, Redslob A, Albaret C, Fortier P. Biochem J. 1999;343:361–369. [PMC free article] [PubMed] [Google Scholar]
- 86.Masson P, Gouet P, Clery C. J Mol Biol. 1994;238:466–478. doi: 10.1006/jmbi.1994.1305. [DOI] [PubMed] [Google Scholar]
- 87.Masson P, Goasdoue JL. BBA - Protein Struct M. 1986;869:304–313. doi: 10.1016/0167-4838(86)90070-1. [DOI] [PubMed] [Google Scholar]
- 88.Ashani Y, Gentry MK, Doctor BP. Biochemistry. 1990;29:2456–2463. doi: 10.1021/bi00462a004. [DOI] [PubMed] [Google Scholar]
- 89.Worek F, Wille T, Koller M, Thiermann H. Biochem Pharmacol. 2012:1700–1706. doi: 10.1016/j.bcp.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 90.Mercey G, Verdelet T, Renou J, Kliachyna M, Baati R, Nachon F, Jean L, Renard PY. Acc Chem Res. 2012:756–766. doi: 10.1021/ar2002864. [DOI] [PubMed] [Google Scholar]
- 91.Masson P, Nachon F, Lockridge O. Chem Biol Interact. 2010;187:157–162. doi: 10.1016/j.cbi.2010.03.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.