Abstract
Snapshots of three consecutive steps in the proteolytic reaction of HIV-1 protease (PR) were obtained in crystal structures at resolutions of 1.2 to 1.4 Å. Structures of wild type protease and two mutants (PRV32I and PRI47V) with substitutions of V32I and I47V, which are common in drug resistance, reveal the gem-diol tetrahedral intermediate, the separating N- and C-terminal products, and the C-terminal product of an autoproteolytic peptide. These structures represent three stages in the reaction pathway and shed light on the reaction mechanism. The near-atomic resolution geometric details include a short hydrogen bond between the intermediate and the outer carboxylate oxygen of one catalytic Asp25 that is conserved in all three structures. The two products in the complex with mutant PRI47V have 2.2 Å separation of the amide and carboxyl carbon of the adjacent ends suggesting partial cleavage prior to product release. The complex of mutant PRV32I with a single C-terminal product shows density for water molecules in the other half of the binding site, including a partial occupancy water molecule interacting with the product carboxylate end and the carbonyl oxygen of one conformation of Gly27, which suggests a potential role of Gly27 in recycling from the product complex to the ligand-free enzyme. These structural details at near atomic resolution enhance our understanding of the reaction pathway and will assist in design of mechanism-based inhibitors as antiviral agents.
Keywords: aspartic protease, hydrolase, drug resistance, catalytic mechanism
INTRODUCTION
The human immunodeficiency virus type 1 protease (HIV-1 PR) acts as a dimer of two identical 99 amino-acid subunits to process the viral Gag and Gag-Pol polyproteins into functional proteins (Figure 1A).1 The indispensable function of PR in replication of infectious virus makes it an important target for antiretroviral therapy. However, the efficacy of PR inhibitors decreases over time due to the evolution of drug resistance, primarily by mutations in the PR.2 Even treatment with highly active antiretroviral therapy (HAART) does not completely eliminate resistant virus. So there is continuing need for new PR inhibitors to combat drug resistance. Improved knowledge of the PR reaction intermediates will help in the design of novel mechanism-based inhibitors.
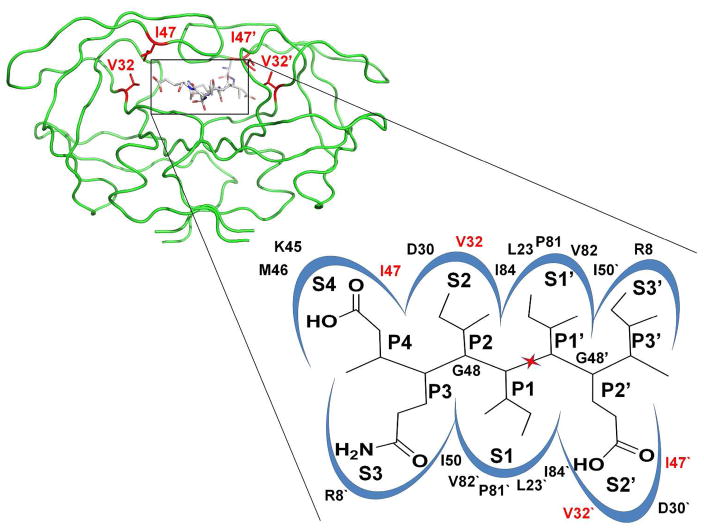
Figure 1.
A) Structure of HIV-1 PR dimer in a green backbone representation. The sites of mutations Val32 and Ile47 are shown as red stick for the side chain atoms in both subunits, with the prime indicating the “second” subunit. The TI peptide is shown in sticks colored by atom type. B) Schematic illustration of the substrate binding site of HIV-1 PR. The peptide DQIIxIEI (P4-P3′) is shown in the S4-S3′ subsites of the PR dimer. The scissile peptide bond is indicated by the red star. PR residues contributing to the binding site are indicated.
The aspartyl protease family is widely distributed in a variety of organisms and its members participate in diverse biological functions, however, the detailed proteolytic mechanism is not fully understood.3 Several experimental or theoretical studies have addressed the reaction mechanism of peptide cleavage by aspartyl proteases. 18O-exchange mass spectrometry experiments with HIV-1 PR suggest that the peptide hydrolysis reaction proceeds via the formation of a reversible and metastable gem-diol reaction intermediate.4 Recent analysis by neutron crystallography has provided critical information on the location of hydrogen atoms in the active site of HIV-1 PR complexed with an inhibitor, which does not contain the gem-diol reaction intermediate.5 X-ray crystallographic analysis of reaction intermediates trapped in the enzyme structure has proved to be a powerful tool to probe catalytic mechanisms.6 Veerapandian et al. used the aspartic protease endothiapepsin complexed with a renin inhibitor mimicking both hydroxyls in the putative intermediate to study the proteolytic mechanism.7, 8 X-ray structures were reported for N-terminal and C-terminal peptide products bound to PRs from HIV-1 and the closely related Simian immunodeficiency virus.9 Other studies soaked tethered HIV-1 PR crystals with a substrate peptide to trap different stages of the reaction, including the tetrahedral intermediate.10, 11 Our group has reported the highest resolution of 1.5 Å for the structures of a tetrahedral reaction intermediate in the wild type and mutant HIV-1 PR.12 In these examples, however, the structural information is limited by the resolution and the disorder observed frequently for peptide intermediates.
More recently, our studies have focused on understanding the influence on the reaction intermediates of selected mutations found in drug resistance. Moreover, we identified a 1.2 Å resolution structure of the wild type PR with a tetrahedral intermediate. PR recognizes peptide substrates of at least 6 residues binding in subsites S3–S3′ within the active site cavity (Figure 1B).1 Mutations of PR residues Val32 and Ile47 were selected since they contribute hydrophobic interactions with substrates or inhibitors (Figure 1A) and mutations at these sites are common in drug resistance.13 Multi-drug-resistant mutation V32I appears in about 20% of patients treated with amprenavir14 and is associated with high levels of drug resistance to lopinavir/ritonavir.13 Drug resistant mutation I47V is located in the flexible flap and interacts with inhibitor. Mutation I47V is associated with resistance to darunavir, lopinavir, tipranavir and ritonavir in therapy.13, 15
We describe crystal structures of PR and its mutants with different reaction intermediates and the implications for the proteolytic mechanism. The crystal structures of wild type HIV-1 PR (PRWT) and its mutants containing the single substitutions of I47V (PRI47V) and V32I (PRV32I) were refined at the near-atomic resolutions of 1.2 to 1.4 Å and, by serendipity, illustrate three different steps in the hydrolytic reaction. Peptide products and the reaction intermediate corresponding to an autoproteolytic cleavage site were observed in the structures, as described previously.12 These near atomic resolution crystal structures provide more accurate information for the catalytic mechanism and the design of next generation anti-viral inhibitors.
EXPERIMENTAL PROCEDURES
Protein Preparation and Crystallization of HIV-1 PRWT, PRV32I, and PRI47V
The mutants were constructed, expressed in bacteria and the protein purified as described previously.12 The crystallization trials employed the hanging drop method using equal volumes of enzyme-inhibitor and reservoir solution. PR and mutant proteins at 2.2mg/mL concentration were mixed with inhibitor or peptide (dissolved in DMSO) in a molar ratio of 1:5 and incubated on ice for 30 min prior to centrifugation to remove any insoluble material. PRWT was crystallized from 0.1 M sodium acetate buffer at pH 4.8, 0.41 M potassium chloride, and an investigational inhibitor. PRV32I was crystallized from 0.06 M sodium acetate buffer at pH 5.6, 0.67 M sodium chloride, and a synthetic peptide. PRI47V crystals were grown from 0.05 M sodium acetate buffer at pH 5.0, 1.2 M sodium formate, and 2.5% PEG8000.
X-ray Data Collection and Refinement
Single crystals were mounted on fiber loops with 25 % (v/v) glycerol as cryoprotectant in the reservoir solution. X-ray diffraction data were collected at the SER-CAT beamline of the Advanced Photon Source, Argonne National Laboratories. Diffraction data were integrated, scaled, and merged using the HKL2000 package.16 PRWT, PRV32I and PRI47V were solved by the molecular replacement program Molrep17 using structures 3B7V, 1FG6, and 2F8G as the respective starting models.12, 18, 19 Refinement was carried out using SHELX-97.20 No electron density was seen for the inhibitor used in the crystallization solution for PRWT, however, the gem-diol intermediate of autoproteolysis of PR residues 59–67 (YDQIIxIEIA) fitted the observed density. The synthetic peptide sequence did not fit the electron density in PRV32I and was replaced by the single C-terminal product of residues 59–63 (YDQII). Tyr59 at P5 was refined as Ala due to the poor electron density for its side chain atoms in PRWT and PRV32I. PRI47V was refined with both N-terminal and C-terminal products extending in opposing directions (residues 60–63 with 64–65 in one orientation and residues 61–63 with 64–66 in the other). The structures were refined with anisotropic atomic displacement parameters (B factors). Hydrogen atoms were added at the final stages of the refinement. The molecular graphics program COOT was used for map display and model building.21 Structural figures were made using PyMol.22 The structures were compared by superimposing their Cα atoms and using HIVAGENT23 to calculate the distance between two atoms. The cut-off distances for different types of interactions were as described in 24.
RESULTS AND DISCUSSION
Crystallographic Analysis
The crystal structures of PRWT, PRV32I and PRI47V were determined in the same space group P21212 and the crystallographic statistics are summarized in Table 1. The asymmetric units include one PR homodimer, and the residues in the two subunits are labeled 1–99 and 1′–99′. The diffraction data extend to a resolution of 1.2 Å for PRWT, 1.3 Å for PRI47V and 1.4 Å for PRV32I and the structures were refined to R factors of 14.4 to 17.5 %. The majority of protein residues and solvent molecules showed clear electron density in all the structures. The tetrahedral intermediate and cleavage products of PR residues 59–67 (YDQII*IEIA, where * indicates the cleavage site between P1 and P1′ residues) matched the electron density in the active site cavities of the PRWT, PRI47V, and PRV32I structures. These peptides are assumed to derive from very slow autoproteolysis, since the L63I substitution almost eliminates a site of autoproteolytic cleavage.25 The PRWT dimer included two partial occupancy gem-diol intermediates: residues 59–65 (designated P5-P2′) were fitted in one conformation and 61–67 (P3-P4′) for the conformation in the opposite orientation with relative occupancies of 0.4 and 0.5, respectively. The two mutants trapped the product peptides. A single C-terminal product containing residues 59–63 (P5-P1) was seen in PRV32I. Ala was refined instead of Tyr59 at P5 due to weak electron density for the side chain in PRWT and PRV32I. PRI47V was refined with both N-terminal and C-terminal products in two alternate conformations comprising residues 60–63 (P4-P1) with 64–65 (P1′-P2′) at 0.5 occupancy and residues 61–63 (P3-P1) with 64–66 (P1′-P3′) at 0.4 occupancy. The side chains of P4 Asp and P3 Glu were not visible in the electron density. The solvent, consisting of water molecules, sodium ions, chloride ions and glycerol, was modeled with 204, 181 and 140 molecules in PRWT, PRI47V and PRV32I, respectively. Alternate conformations were refined for 26 residues in PRWT, 13 residues in PRI47V and 7 residues in PRV32I. Generally, alternate conformations were seen for the longer side chains on surface residues. A few internal residues showed alternate conformations for the side chain or main chain: Ile84 Leu97, Ile33′ and Ile84′ of PRWT, Gly27, Ile84, and Ile33′ of PRV32I, Val47, Ile64, Ile84, Ile33′, and Ile84′ of PRI47V. The backbone structures were almost identical for the three dimers with low pairwise RMS deviations of 0.15–0.17 Å for all Cα atoms.
Table 1.
Crystallographic Data Collection and Refinement Statistics
| PRWT | PRI47V | PRV32I | |
|---|---|---|---|
| Space group | P21212 | P21212 | P21212 |
| Unit cell dimensions: (Å) | |||
| A | 58.41 | 58.08 | 58.06 |
| B | 86.20 | 86.30 | 86.14 |
| C | 46.36 | 46.35 | 46.30 |
| Resolution range (Å) | 10-1.2 | 10-1.31 | 50-1.4 |
| Unique reflections | 66188 | 55879 | 45833 |
| Rmerge (%) overall (final shell) | 9.7 (39) | 6.3 (51) | 7.2 (43) |
| I/σ(I) overall (final shell) | 15.03 (2.2) | 27.8 (2.1) | 18.3 (2.1) |
| Completeness (%) overall (final shell) | 94.3 (56.8) | 98.8 (90.6) | 93.0 (98.7) |
| Data range for refinement (Å) | 10-1.2 | 10-1.31 | 10-1.4 |
| R (%) | 0.14 | 0.15 | 0.17 |
| Rfree (%) | 0.18 | 0.18 | 0.23 |
| No. of solvent atoms (total occupancies) | 206 (191) | 181 (169.5) | 140 (134) |
| RMS deviation from ideality | |||
| Bonds (Å) | 0.015 | 0.012 | 0.010 |
| Angle distance (Å) | 0.034 | 0.031 | 0.029 |
| Average B-factors (Å2) | |||
| Main-chain atoms | 19.2 | 17.1 | 16.8 |
| Side-chain atoms | 24.9 | 20.4 | 23.3 |
| Peptide Intermediate | 54.1 | 53.5 | 32.2 |
| Solvent | 36.4 | 33.3 | 30.5 |
| Intermediate Peptide | (Y)a DQIIxIE | (D)(Q)II+IE | (Y)DQII |
| Peptide relative occupancy | 0.5/0.4 | 0.5/0.4 | 1.0 |
Parentheses indicate peptide residues refined as alanine due to poor electron density for the longer side chain.
Structural changes around the mutated residues
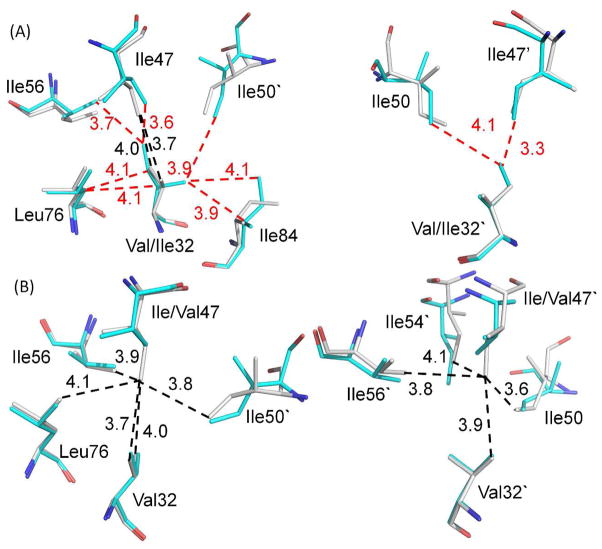
The two drug resistant mutations are conservative substitutions of hydrophobic residues in the substrate binding cavity. The side chain of Val32 in the PRWT structure forms van der Waals contacts with the internal hydrophobic cluster comprising residues Ile50′, Ile47, Ile56, Leu76, Thr80 and Ile84, and similar interactions were seen in the other subunit. The longer Ile32 side chain in the PRV32I mutant has the potential to form new van der Waals contacts within the cluster, as described for the PRV32I complex with amprenavir.26 In one subunit, the Cδ1 methyl of Ile32 side chain is directed toward the hydrophobic cluster providing new van der Waals contacts with several hydrophobic side chains. In the other subunit, however, the Cδ1 methyl of Ile32′ rotates to form new hydrophobic contacts only with Ile47′ and Ile50 (Figure 2A). The 3.3Å distance between the Cδ1 methyl groups of Ile32′ and Ile47′ is unusually short for a C-H…H-C interaction, which may indicate a destabilizing interaction.27 Ile32 forms a van der Waals contact with Ile at the P2 position in the product peptide similar to the contact seen for Val32 in PRWT, while no product is seen in the other subunit.
Figure 2.
The mutations alter internal hydrophobic contacts. Residues are shown from superimposed structures of PRWT with A) PRV32I and B) PRI47V. The two subunits are shown in the left and right panels, respectively. The PRWT residues are colored gray for carbon atoms, while the mutants are colored cyan. The van der Waals interactions are indicated by dashed lines in black for PRWT and in red for the mutants with interatomic distances in Å. The mutation of I47V to a smaller side chain in PRI47V eliminates hydrophobic contacts seen for Ile47 in PRWT. The opposite effect occurs with substitution of the large side chain in PRV32I.
The hydrophobic side chain of Ile47 forms internal hydrophobic contacts and interacts with P2 and P2′ residues of the reaction intermediate in both subunits of the PRWT complex. These hydrophobic interactions are retained in the PRI47V mutant. In PRWT, the Cδ1 of Ile47 interacts with Ile50′, Val32, Val56 and Leu76, while the Cδ1 of Ile47′ shows van der Waals interactions with the corresponding residues in the other subunit and with Ile54′. Mutation I47V substitutes the shorter Val side chain and eliminates van der Waals contacts with adjacent residues, thus probably decreasing the stability of the hydrophobic cluster in PRI47V (Figure 2B).
PRWT –TI interaction
The tetrahedral intermediate (TI) of residues Y59DQII*IEIA67 (where * indicates the gem-diol group) was observed in the Fo-Fc electron density map in PRWT in two alternate conformations extending in opposite directions (Figures 3A, S1A and S2A), as found frequently in PR complexes with peptide analogs.19, 28 Residues P3-P4′ were fitted in one conformation and residues P5-P2′ in the opposite orientation, with occupancies refined to 0.5 and 0.4, respectively. The omit electron density (Fo–Fc) map for the gem-diol structure clearly indicates four hydroxyl oxygens (Figures 3B and S1B). The two alternate conformations of the TI peptide formed essentially identical interactions with PRWT. Interactions with the catalytic Asp25 and 25′ are described later. The main chain atoms of the TI peptide formed hydrogen bond interactions with residues Gly27, Asp29, Gly48, Gly27′, Asp29′, Asp30′, and Gly48′ (Figure 4A). The side chain of Glu at P2′ showed hydrogen bond interactions with Asp29 and Asp30. A shorter interaction of 2.4 Å seen between the carboxylate side chains of P2 Glu and Asp30 is consistent with protonation of P2 Glu, as described in other crystal structures of PR with peptide analogs.19, 28 Additional stabilizing interactions include water mediated hydrogen bonds and hydrophobic contacts between PRWT and the peptide.
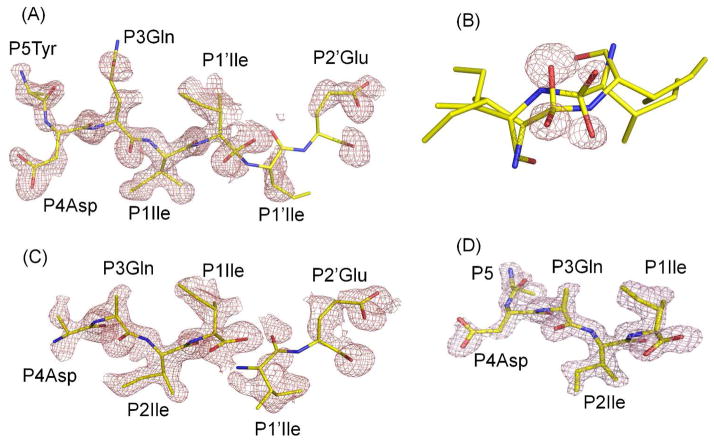
Figure 3.
Electron density maps for the peptide intermediates. A) Omit map (FO-FC) for major conformation of TI in the PRWT structure. Contour level is 2.0σ. B) FO-FC omit map for the hydroxyl oxygen atoms of P1 Ile in the major and minor conformations contoured at 3.5σ. C) Omit electron density map (FO-FC) for major conformation of product peptides in the PRI47V structure. Contour level is 2.0σ. D) FO-FC omit map for the peptide C-terminal product of PRV32I contoured at 2.5σ. The corresponding stereo figures are S1A–D.
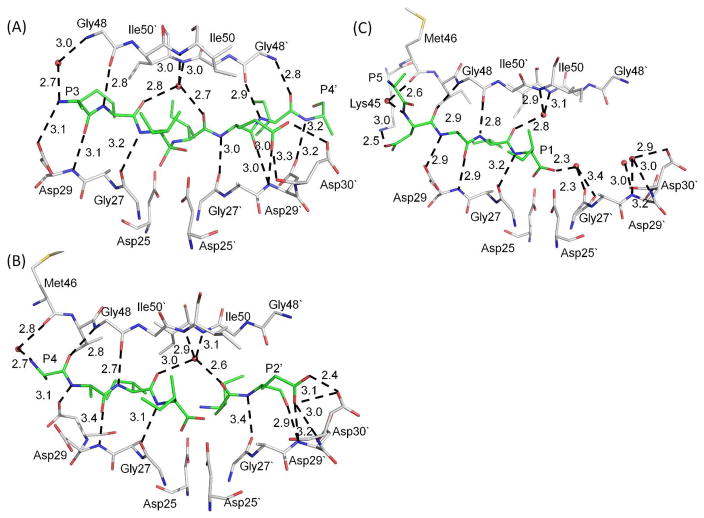
Figure 4.
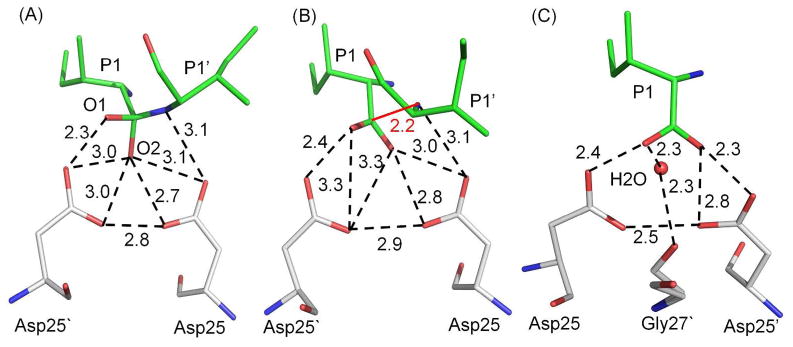
Hydrogen bond interactions: A) the tetrahedral intermediate with PRWT; B) the two products with PRI47V mutant, and C) the single product with PRV32I mutant. Only the major conformation is shown for peptide intermediate or products in A) and B). Hydrogen bond interactions are indicated by dashed lines with distances in Å. Interactions of Asp25/25′ are omitted for clarity. PR is shown in gray carbons, TI and product peptides are in green. The water and carbonyl oxygen of Gly27′ in C) were refined with partial occupancy of 0.45.
Interactions of PRI47V with Two Products
PRI47V structure revealed two alternate conformations of both the N- and C-terminal products extending in opposite directions (Figures 3C, S1C and S2B). The occupancy of residues P4-P1 and P1′-P2′ in one conformation was 0.5, and P3-P1 and P1′-P2′ residues were refined with 0.4 occupancy for the opposite orientation. The alternate conformations maintained similar interactions with the two subunits of PRI47V. Interactions with the catalytic Asp25 and 25′ are described later. Four hydrogen bond interactions connected the main chain atoms of product P4-P1 with residues Gly27, Asp29 and Gly48, and two hydrogen bond interactions linked main chain atoms of product P1′-P2′ with residues Gly27′ and Asp29′ (Figure 4B). The side chain carboxylate oxygen atoms of P2′ Glu interact with Asp29′ and Asp30′, as described for P2′ in the TI peptide.
Interactions of PRV32I with P5-P1 Product
The PRV32I structure showed clear electron density for the single peptide product of residues P5-P1, except for the side chain of P5 (Figures 3D and S1D). Individual water molecules were fitted in the other subunit since no peptide product was visible in the electron density. The polar interactions of the single product with PRV32I are shown in Figure 4C. Two waters near the P2′ position formed hydrogen bond interactions to the main chain of Asp29′ and main chain and side chain of Asp30′, resembling the interactions of the P2′ Glu in the other peptides. Another water molecule at 0.5 occupancy interacts with both alternate conformations of the carbonyl of Gly27′ and with the product carboxyl terminus. Hydrogen bond interactions connect the main chain of the product to PRV32I residues Gly27, Asp29, and Gly48, as observed in the other complexes. A new salt bridge was seen between the side chains of P4 Asp and Lys45, and the P4 main chain amide had a water-mediated interaction with the carbonyl oxygen of Met46. Hydrophobic interactions were also observed between the side chains of the product and side chains of PRV32I.
Interactions with catalytic residues
The interactions with the catalytic residues are essentially identical in the major and minor conformations of the TI and two product peptides as shown by the superposition in Figure S3. In the TI complex with PRWT, the gem-diol structure lies between P1 Ile and P1′ Ile (Figure 5A). The O1 hydroxyl of P1 Ile forms a very short 2.3Å hydrogen bond to the outer carboxylate Oδ2 atom of Asp25′, while the O2 hydroxyl interacted with all four carboxylate oxygens of Asp25 and 25′. The nitrogen of the gem-diol-amine moiety had a hydrogen bond interaction to Oδ2 of Asp25.
Figure 5.
Hydrogen bond interactions with the catalytic residues Asp25 and 25′. A) PRWT-TI complex, B) PRI47V with both products, and C) PRV32I with single product. Hydrogen bond interactions are indicated by dashed lines with distances in Å. The PR is shown in gray color, TI and products are in green color. The short separation of 2.2 with distances in Å between the N- and C-terminal products is indicated by the red line in B). Only one conformation (0.5 occupancy) of Gly27′ and interacting water is shown in C).
The PRWT-TI interactions are comparable to those described in the 1.0 Å resolution crystal structure of endothiapepsin, a fungal aspartic proteinase, complexed with a gem-diol analog.8 In endothiapepsin, one short hydrogen bond interaction was seen between the O1 hydroxyl and Oδ2 of Asp35 with distance of 2.5Å; the O2 hydroxyl formed two hydrogen bond interactions with Oδ1 and Oδ2 of Asp219 with distances of 2.6 Å and 3.0 Å, respectively; and one hydrogen bond interaction to Oδ2 of Asp35. Unlike PRWT, the hydrogen bond interaction between the amide nitrogen of the gem-diol analog and the catalytic aspartate was absent in the endothiapepsin complex.
The inhibitor KNI-272 has been used as a transition state mimic in neutron diffraction studies in order to locate important hydrogen atoms at the catalytic site of HIV PR.5 Although the hydroxymethylcarbonyl isostere of KNI-272 is not identical to the gem-diol of TI, the hydrogen bond interactions of O2 of KNI-272 are similar to those of the O2 of P1 Ile with both catalytic aspartates. Also, the O4 of KNI-272 resembles O1 of TI in forming a single hydrogen bond interaction to Oδ2 of Asp25, although the hydrogen bond is not particularly short.
The two product peptides bound in PRI47V are partially separated with 2.2 Å distance between the P1′ amide nitrogen and the P1 carbonyl carbon (Figure 5B), whereas the standard C-N separation of 1.3–1.4 Å occurs in the TI complex. The 2.2 Å distance may indicate that there is still some bonding interaction present between the N atom of the amino-product and the C atom of the carboxylate product. The N-terminus of P1′ Ile has one hydrogen bond interaction with Oδ2 of Asp25. One carboxylate oxygen of P1 Ile forms a short 2.4 Å-long hydrogen bond interaction to the outer Oδ2 of Asp25′, as seen for the tetrahedral intermediate. The other carboxylate oxygen of P1 has hydrogen bond interactions with the four aspartate carboxylate oxygens. This analysis suggests that the structure of PRI47V with two products represents the stage immediately after hydrolysis of the peptide bond.
In PRV32I, a partial (0.5) occupancy water (Figure 5C) in the catalytic site forms hydrogen bond interactions linking the carboxylate oxygen of P1 and the alternate conformations of the carbonyl oxygen of Gly27′. One carboxylate oxygen of P1 forms a short hydrogen bond with Oδ2 of Asp25 with distance of 2.4 Å, and the other carboxylate oxygen of the product forms two hydrogen bond interactions (2.3 and 2.8 Å) with Oδ1 and Oδ2 of Asp25′. These hydrogen bond interactions of the P1 carboxylate group are similar to those reported in other structures with synthetic peptide products.9 This complex appears to represent the reaction step after release of the N-terminal product from the catalytic site. The alternate positions of the carbonyl oxygen of Gly27′ may reflect conformational changes occurring on release of the N-terminal product, since this oxygen usually interacts with an amide of the bound substrate peptide, as shown in the PRWT–TI and PRI47V complex with two products.
Implications for the reaction pathway
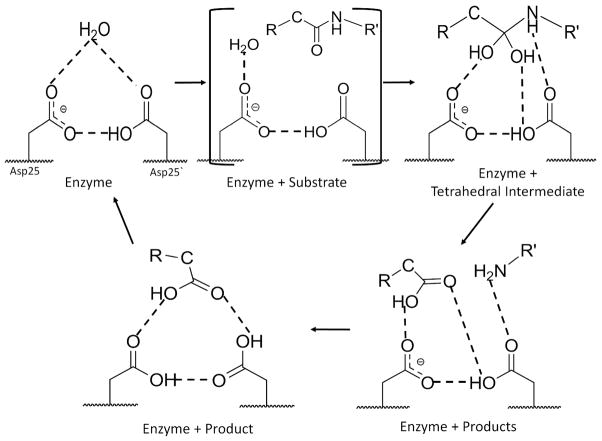
Reaction intermediates have been described in a variety of enzyme crystal structures, as reviewed in 29. Several metastable intermediates can sometimes exist along a reaction path indicating the catalytic process proceeds through a series of energy barriers. In the case of HIV PR, a number of intermediates can be trapped by simply crystallizing the protein in the presence of a peptide substrate, which suggests that the energy barriers the hydrolysis reaction has may be of similar height. Thus, it is entirely possible that the reaction pathway of the peptide hydrolysis by PR does not include a single rate-limiting step. The three new crystal structures represent three consecutive steps in the proteolytic reaction of HIV PR and provide improved geometric details due to the near atomic resolution X-ray data. The majority of the interactions with the bound peptides are conserved in the wild type enzyme and the mutants in agreement with the report that mutants share similar transition states to wild type PR.30 The interactions observed in the new structures have been mapped on the scheme for the reaction (Figure 6). The hydrogen atoms around the catalytic sites cannot be identified in X-ray structures at this resolution, however, their locations were deduced from the short interactions. One Asp25 is protonated and the other deprotonated in most states, in agreement with neutron diffraction studies on an inhibitor complex.5 In the absence of substrate, the active site cavity of PR generally contains water, or possibly a metal cation, interacting with the two catalytic aspartates.31 This water reacts with the bound peptide to form the gem-diol intermediate, represented by the PRWT-TI structure (Figure 5A). The tetrahedral intermediate dissociates into two products in the step shown in the PRI47V complex (Figure 5B). Then, the N-terminal product is released, in association with rotation of the carbonyl group of Gly27′, as shown in the PRV32I structure (Figure 5C). This state with a single product is shown with protonation of both Asp25 and 25′ to reflect the short interactions observed in the crystal structure, which suggests diprotonation in the simplest interpretation, as found in theoretical studies of PR with some inhibitors.32 Finally, the C-terminal product is released and the enzyme recycles to the first step. All three intermediate stages retain the short 2.3–2.4 Å hydrogen bond, which may be a low barrier hydrogen bond, of the hydroxyl group of the peptide intermediate with one of the catalytic aspartates, as reported in lower resolution crystal structures.9–11 Moreover, the amide of the cleaved bond in the gem-diol intermediate and the freed amino terminus of the product also form hydrogen bond interactions with the carboxylate of Asp25.
Figure 6.
Scheme of reaction pathway. The interactions of the peptide intermediates with the catalytic residues are illustrated based on the new crystal structures. The minimal number of hydrogen atoms and hydrogen bonds is indicated based on the interaction distances in the crystal structures. Crystal structures have been described for 4 stages. Only the active enzyme with bound substrate has not been seen, as indicated by the large parentheses.
The conserved catalytic triplets of Asp25-Thr26-Gly27 are important for the activity and the dimerization of PR. Previous studies by X-ray and neutron diffraction have shown similar hydrogen bond interactions with the catalytic Asp25/25′ to those described in our new structures.5, 12 The position of hydrogen atoms, however, cannot be determined unambiguously in these X-ray structures and further studies by neutron crystallography will be important to determine the protonation state of the catalytic residues in the reaction steps. Our structures suggest a new role for rotation of Gly27 to facilitate release of product as well as its known role in binding the main chain amides of peptides.28 Analysis of these structures emphasizes the essential roles of the residues 25–30 and flap residues 48–50 in binding peptide substrates and products and provides details of the geometry around the catalytic site at near atomic resolution. The critical interactions can be targeted in the design of new antiviral inhibitors for resistant HIV.
Supplementary Material
Acknowledgments
Funding: This research was supported, in whole or in part, by the Intramural Research Program of the NIDDK, National Institutes of Health (NIH), Intramural AIDS-Targeted Antiviral Program of the Office of the Director, NIH, and grant GM062920 from the NIH. C.S. and X.Y. were supported in part by the Georgia State University Research Program Enhancement award in Bioinformatics and by Georgia State University Molecular Basis of Disease Fellowships.
Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22ID at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Abbreviations
- HIV-1
human immunodeficiency virus type 1
- PRWT
wild type HIV-1 protease
- PRV32I
protease with V32I mutation
- PRI47V
protease with I47V mutation
- TI
gem-diol-amine tetrahedral intermediate
- RMS
root mean square
Footnotes
Accession codes: The structure coordinates and factors have been deposited in the Protein Data Bank (www.pdb.org) with accession codes 4FL8 for PRWT, 4FM6 for PRV32I, and 4FLG for PRI47V
Supporting Information Available
Figures are available for the minor conformation of peptides and their comparison with the major conformations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Weber IT, Kovalevsky AY, Harrison RW. Frontiers in Drug Design & Discovery. Vol. 3. Bentham Science Publishers; Oak Park, IL: 2007. Structures of HIV Protease Guide Inhibitor Design to Overcome Drug Resistance; pp. 45–62. [Google Scholar]
- 2.Weber I, Agniswamy J. HIV-1 Protease: Structural Perspectives on Drug Resistance. Viruses. 2009;1:1110–1136. doi: 10.3390/v1031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brik A, Wong CH. HIV-1 protease: mechanism and drug discovery. Org Biomol Chem. 2003;1:5–14. doi: 10.1039/b208248a. [DOI] [PubMed] [Google Scholar]
- 4.Hyland LJ, Tomaszek TA, Jr, Roberts GD, Carr SA, Magaard VW, Bryan HL, Fakhoury SA, Moore ML, Minnich MD, Culp JS, et al. Human immunodeficiency virus-1 protease. 1 Initial velocity studies and kinetic characterization of reaction intermediates by 18O isotope exchange. Biochemistry. 1991;30:8441–8453. doi: 10.1021/bi00098a023. [DOI] [PubMed] [Google Scholar]
- 5.Adachi M, Ohhara T, Kurihara K, Tamada T, Honjo E, Okazaki N, Arai S, Shoyama Y, Kimura K, Matsumura H, Sugiyama S, Adachi H, Takano K, Mori Y, Hidaka K, Kimura T, Hayashi Y, Kiso Y, Kuroki R. Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography. Proc Natl Acad Sci U S A. 2009;106:4641–4646. doi: 10.1073/pnas.0809400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson AR, Owen RL. Combining X-ray crystallography and single-crystal spectroscopy to probe enzyme mechanisms. Biochem Soc Trans. 2009;37:378–381. doi: 10.1042/BST0370378. [DOI] [PubMed] [Google Scholar]
- 7.Veerapandian B, Cooper JB, Sali A, Blundell TL, Rosati RL, Dominy BW, Damon DB, Hoover DJ. Direct observation by X-ray analysis of the tetrahedral “intermediate” of aspartic proteinases. Protein Sci. 1992;1:322–328. doi: 10.1002/pro.5560010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coates L, Tuan HF, Tomanicek S, Kovalevsky A, Mustyakimov M, Erskine P, Cooper J. The catalytic mechanism of an aspartic proteinase explored with neutron and X-ray diffraction. J Am Chem Soc. 2008;130:7235–7237. doi: 10.1021/ja801269x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose RB, Craik CS, Douglas NL, Stroud RM. Three-dimensional structures of HIV-1 and SIV protease product complexes. Biochemistry. 1996;35:12933–12944. doi: 10.1021/bi9612733. [DOI] [PubMed] [Google Scholar]
- 10.Kumar M, Prashar V, Mahale S, Hosur MV. Observation of a tetrahedral reaction intermediate in the HIV-1 protease-substrate complex. Biochem J. 2005;389:365–371. doi: 10.1042/BJ20041804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A, Mahale S, Prashar V, Bihani S, Ferrer JL, Hosur MV. X-ray snapshot of HIV-1 protease in action: observation of tetrahedral intermediate and short ionic hydrogen bond SIHB with catalytic aspartate. J Am Chem Soc. 2010;132:6366–6373. doi: 10.1021/ja100002b. [DOI] [PubMed] [Google Scholar]
- 12.Kovalevsky AY, Chumanevich AA, Liu F, Louis JM, Weber IT. Caught in the Act: the 1.5 A resolution crystal structures of the HIV-1 protease and the I54V mutant reveal a tetrahedral reaction intermediate. Biochemistry. 2007;46:14854–14864. doi: 10.1021/bi700822g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer R, Wensing AM, Richman DD. 2011 update of the drug resistance mutations in HIV-1. Top Antivir Med. 2011;19:156–164. [PMC free article] [PubMed] [Google Scholar]
- 14.Wu TD, Schiffer CA, Gonzales MJ, Taylor J, Kantor R, Chou S, Israelski D, Zolopa AR, Fessel WJ, Shafer RW. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J Virol. 2003;77:4836–4847. doi: 10.1128/JVI.77.8.4836-4847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pazhanisamy S, Stuver CM, Cullinan AB, Margolin N, Rao BG, Livingston DJ. Kinetic characterization of human immunodeficiency virus type-1 protease-resistant variants. J Biol Chem. 1996;271:17979–17985. doi: 10.1074/jbc.271.30.17979. [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;267:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 17.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr, Sec D: Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 18.Kovalevsky AY, Tie Y, Liu F, Boross PI, Wang YF, Leshchenko S, Ghosh AK, Harrison RW, Weber IT. Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M. J Med Chem. 2006;49:1379–1387. doi: 10.1021/jm050943c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahalingam B, Louis JM, Hung J, Harrison RW, Weber IT. Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins. 2001;43:455–464. doi: 10.1002/prot.1057. [DOI] [PubMed] [Google Scholar]
- 20.Sheldrick GM, Schneider TR. SHELXL: high-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr, Sec D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 23.Tie Y. Biology. Georgia State University; Atlanta: 2006. Crystallographic Analysis and Kinetic Studies of HIV-1 Protease and Drug-Resistant Mutants; pp. 131–141. [Google Scholar]
- 24.Kovalevsky AY, Liu F, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. Ultra-high resolution crystal structure of HIV-1 protease mutant reveals two binding sites for clinical inhibitor TMC114. J Mol Biol. 2006;363:161–173. doi: 10.1016/j.jmb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mildner AM, Rothrock DJ, Leone JW, Bannow CA, Lull JM, Reardon IM, Sarcich JL, Howe WJ, Tomich CS, Smith CW, et al. The HIV-1 protease as enzyme and substrate: mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry. 1994;33:9405–9413. doi: 10.1021/bi00198a005. [DOI] [PubMed] [Google Scholar]
- 26.Shen CH, Wang YF, Kovalevsky AY, Harrison RW, Weber IT. Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters. Febs J. 2010;277:3699–3714. doi: 10.1111/j.1742-4658.2010.07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowley RL, Pakkanen T. Determination of a methane intermolecular potential model for use in molecular simulations from ab initio calculations. J Chem Phys. 1999;110:3368–3377. [Google Scholar]
- 28.Tie Y, Boross PI, Wang YF, Gaddis L, Liu F, Chen X, Tozser J, Harrison RW, Weber IT. Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. Febs J. 2005;272:5265–5277. doi: 10.1111/j.1742-4658.2005.04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber IT, Agniswamy J, Fu G, Shen CH, Harrison RW. Reaction intermediates discovered in crystal structures of enzymes. Adv Protein Chem Struct Biol. 2012;87:57–86. doi: 10.1016/B978-0-12-398312-1.00003-2. [DOI] [PubMed] [Google Scholar]
- 30.Kipp DR, Hirschi JS, Wakata A, Goldstein H, Schramm VL. Transition states of native and drug-resistant HIV-1 protease are the same. Proc Natl Acad Sci U S A. 2012;109:6543–6548. doi: 10.1073/pnas.1202808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heaslet H, Rosenfeld R, Giffin M, Lin YC, Tam K, Torbett BE, Elder JH, McRee DE, Stout CD. Conformational flexibility in the flap domains of ligand-free HIV protease. Acta Crystallogr, Sec D: Biol Crystallogr. 2007;63:866–875. doi: 10.1107/S0907444907029125. [DOI] [PubMed] [Google Scholar]
- 32.Piana S, Sebastiani D, Carloni P, Parrinello M. Ab initio molecular dynamics-based assignment of the protonation state of pepstatin A/HIV-1 protease cleavage site. J Am Chem Soc. 2001;123:8730–8737. doi: 10.1021/ja003145e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.