Background: To determine whether DNA double-strand break (DSB) repair is coupled to transcription, we analyzed DSB repair at the active and inactive genes.
Results: Our results reveal that DSB repair at the active gene is faster than that at the inactive gene.
Conclusion: These results demonstrate a preferential DSB repair at the active gene.
Significance: This study supports the existence of transcription-coupled DSB repair.
Keywords: DNA Damage, DNA Repair, DNA Transcription, Gene Regulation, Genomic Instability, ADH1, DNA Repair, DSB, MAT Active Gene
Abstract
Previous studies have demonstrated transcription-coupled nucleotide/base excision repair. We report here for the first time that DNA double-strand break (DSB) repair is also coupled to transcription. We generated a yeast strain by introducing a homing (Ho) endonuclease cut site followed by a nucleotide sequence for multiple Myc epitopes at the 3′ end of the coding sequence of a highly active gene, ADH1. This yeast strain also contains the Ho cut site at the nearly silent or poorly active mating type α (MATα) locus and expresses Ho endonuclease under the galactose-inducible GAL1 promoter. Using this strain, DSBs were generated at the ADH1 and MATα loci in galactose-containing growth medium that induced HO expression. Subsequently, yeast cells were transferred to dextrose-containing growth medium to stop HO expression, and the DSB repair was monitored at the ADH1 and MATα loci by PCR, using the primer pairs flanking the Ho cut sites. Our results revealed a faster DSB repair at the highly active ADH1 than that at the nearly silent MATα locus, hence implicating a transcription-coupled DSB repair at the active gene in vivo. Subsequently, we extended this study to another gene, PHO5 (carrying the Ho cut site at its coding sequence), under transcriptionally active and inactive growth conditions. We found a fast DSB repair at the active PHO5 gene in comparison to its inactive state. Collectively, our results demonstrate a preferential DSB repair at the active gene, thus supporting transcription-coupled DSB repair in living cells.
Introduction
Cellular DNA is continuously attacked by both endo- and exogenous factors causing DNA damage (1–3). The most versatile cellular pathway for dealing with a large variety of structurally unrelated DNA lesions is nucleotide excision repair (NER),2 which mainly removes helix-distorting lesions, including UV-induced cyclobutane pyrimidine dimers, 6-4 photoproducts, and 4-nitroquinoline-1-oxide-induced bulky chemical adducts. The other frequent damages, like oxidative lesions and small base alterations, are processed by base excision repair (4). The very toxic DNA DSBs induced by ionizing radiation are repaired via homologous recombination (HR) or non-homologous endjoining (5–8). Further, the occurrence of DNA lesions triggers checkpoints at the key stages in the cell cycle. Checkpoints monitor the progression of cell cycle post-DNA damage and maintain the proper order of events (3, 9–11). The up-regulation activity of checkpoint proteins in response to DNA damage will impose a temporary arrest of cell cycle progression to allow DNA repair or to induce apoptosis (cell death).
Although DSB is a threat to the genomic integrity of a cell, it occurs during normal DNA metabolism such as replication, meiosis, and immune system development. Programmed DSBs in meiosis are found to promote several major events beyond recombination and synaptonemal complex formation (5, 12). Cells can take advantage of DSB-induced recombination to generate genetic diversity in physiological processes such as meiosis and generation of antibodies by V(D)J recombination in lymphocytes.
An extremely cytotoxic ramification of DNA damage is when lesions in the actively transcribed coding sequence cause stalling of the transcription machinery (13, 14). Persistent transcriptional arrest interferes with cellular function or triggers apoptosis (15, 16), and thus the efficient removal of lesions from the coding regions of active genes is essential for proper cellular function. A specific repair mode, referred to as transcription-coupled repair, removes lesions from the coding sequences of active genes in both prokaryotes and eukaryotes (17–24). In prokaryotes, the transcription repair coupling factor displaces DNA damage-stalled RNA polymerase, which facilitates the recruitment of the DNA repair machinery to the lesion in the active gene. Eukaryotic transcription-coupled repair is considerably more complex and is not well understood, although the phenomenon of transcription-coupled repair in eukaryotes was reported more than 20 years ago (19, 21, 22).
Transcription-coupled repair is one of the two subpathways of NER. The other one is the global genome repair (GGR or GG-NER) that is responsible for the removal of DNA lesions throughout the genome. The basic steps of NER are: 1) recognition of the DNA lesion, 2) dual incisions of the DNA strand carrying the lesion to form a 24–32-nucleotide oligomer, 3) release of the excised oligomer bracketing the lesion, 4) repair synthesis to fill in the resulting gap using the undamaged strand as template, and, finally, 5) ligation. DNA damage recognition differs between GG-NER and TC-NER, but the subsequent steps are shared. The damage recognition step makes TC-NER a faster process than GG-NER. In TC-NER, recognition of the lesion is tied to RNA polymerase stalling at the DNA damage site and involves Rad26p in yeast or Cockayne syndrome group B protein (CSB) in humans (25–34). Like TC-NER, base excision repair is also coupled to transcription (35–37). However, it is yet to be determined whether DSB repair is coupled to transcription. Here, we report a fast DSB repair at the transcriptionally active gene as compared with the inactive gene, thus implying transcription-coupled DSB repair.
EXPERIMENTAL PROCEDURES
Plasmids
The plasmid pFA6a-13Myc-KanMX6 (38) was used for genomic myc epitope tagging of Ino80p at the C-terminal. The same plasmid was also used to insert the Ho cut site just before multiple myc epitope tags at the ADH1 and PHO5 coding sequences. The plasmid pFA6a-3HA-His3MX6 (38) was used for genomic HA epitope tagging of Rad50p and Ku70p at their C-terminals. The plasmid pRS406 (39) was used to knock out RAD52 and DNL4.
Strains
The yeast (Saccharomyces cerevisiae) strain JKM179 contains a Ho cut site at the MATα locus and expresses HO under the GAL1 promoter in galactose-containing growth medium. JKM179 was obtained from the Haber laboratory (40). The genotype of JKM179 is hoΔ MATα hmlΔ::ADE1 hmrΔ::ADE1 ade1–100 leu2–3,112lys5 trp1::hisG' ura3–52 ade3::GAL::HO. The Ho cut site followed by a nucleotide sequence encoding multiple myc epitope tags was added before the stop codons at the original chromosomal loci of ADH1 and PHO5 in JKM179 to generate the PCY23 and RSY33 strains, respectively. Multiple myc epitope tags were added to the chromosomal locus of INO80 in JKM179 to generate the ASY32 strain. Likewise, multiple HA epitope tags were added to the chromosomal loci of RAD50 and KU70 in PCY23 to generate the RSY30 and RSY31 strains, respectively. The RAD52 and DNL4 genes were knocked out in PCY23 to generate the PCY36 and PCY37a strains, respectively.
Growth Media
For induction of the GAL1 promoter to express HO, yeast cells were grown in galactose-containing medium (YPG) (yeast extract, peptone plus 2% galactose). To stop the expression of HO, yeast cells were grown in raffinose-containing (YPR) (yeast extract, peptone plus 2% raffinose) or dextrose-containing (YPD) (yeast extract, peptone plus 2% dextrose) medium. To study the DSB repair at the Ho cut sites at ADH1 and MATα loci, yeast cells were initially grown in YPG and subsequently switched to YPD as described in the legend for Fig. 3B. To study DSB repair at the Ho cut site at PHO5, yeast cells (RSY33) were initially grown in YPG without inorganic phosphate (Pi) and then switched to YPD (transcriptionally inactive condition) or YPD-Pi (transcriptionally active condition), as described in the legend for Fig. 6, B and C.
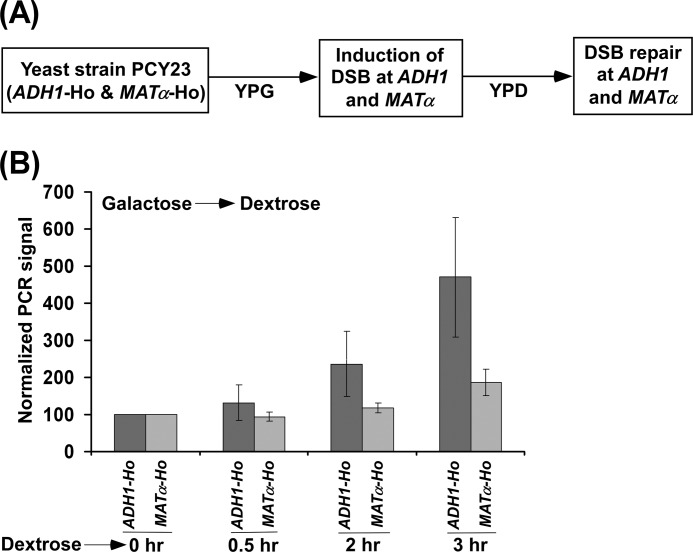
FIGURE 3.
Analysis of DSB repair at the ADH1 and MATα loci. A, schematic diagram for the experimental strategy. B, the yeast strain PCY23 was initially grown in YPG up to an A600 of 0.2 and then switched to YPD for 0.5, 2, and 3 h. Genomic DNAs from these yeast cultures were analyzed by PCR for DSBs at the ADH1 and MATα loci. The PCR signals at the 0 h time point for both the ADH1 and MATα loci were set to 100, and the signals at other time points were normalized with respect to 100. Normalized PCR signals are plotted in the form of a histogram.
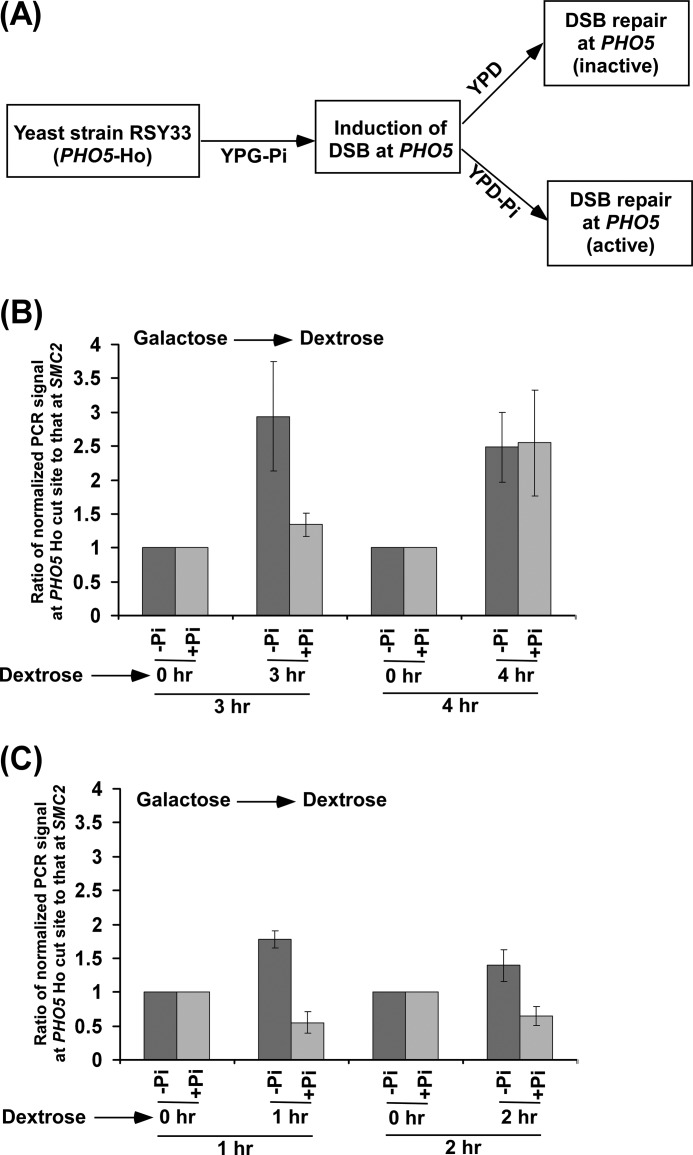
FIGURE 6.
Analysis of DSB repair at the PHO5 gene under transcriptionally inactive and active conditions. A, schematic diagram for the experimental strategy. Inactive and active refer to DSB repairs under transcriptionally inactive and active conditions, respectively. B and C, the yeast strain RSY33 was initially grown in YPG-Pi up to an A600 of 0.4 and then switched to YPD (denoted as +Pi) or YPD-Pi (denoted as -Pi) for 1, 2, 3, and 4 h. Genomic DNAs from these yeast cultures were prepared and analyzed by PCR for DSBs at the PHO5 locus using the primer pair flanking the Ho cut site. A specific region within SMC2 was amplified as control using the same genomic DNAs. The PCR signals at the 0 h time point for the PHO5 and SMC2 loci were set to 100, and the PCR signals at 1, 2, 3, and 4 h were normalized with respect to 100. The ratios of normalized PCR signals at 1, 2, 3 and 4 h at the PHO5 Ho cut site to those at SMC2 are plotted in the form of a histogram. A ratio that is greater than 1 would indicate the DSB repair.
Genomic DNA Preparation
The genomic DNA was extracted from 5 ml of yeast culture. Briefly, the harvested cells were suspended in 200 μl of lysis buffer (50 mm HEPES (pH 7.5), 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, and 0.1% Na-deoxycholate) with 200 μl of volume-equivalent of glass beads and then vortexed for 30 min at 4 °C using a Tomy vortexer (MT-360). The whole-cell extract was collected by punching a hole at the bottom of the Eppendorf tube and then partitioned with 200 μl of phenol:chloroform:isoamylalcohol. The aqueous phase following phenol:chloroform extraction was treated with ethanol to precipitate genomic DNA.
Analysis of DSB and Its Repair
The genomic DNA was analyzed for Ho-induced DSB and repair at the ADH1, PHO5, and MATα loci using the primer pairs flanking the Ho cut sites at the ADH1, PHO5, and MATα loci. The primer pair used to analyze DSB at the Ho cut site at ADH1 was as follows: 5′-CTGGTTACACCCACGACGGTTCTT-3′ and 5′-CCGAGATTCATCAACTCATTGCTGG-3′. The primer pair used to analyze DSB at the Ho cut site at PHO5 was 5′-ACCTCTAATTCTAAGAGATGTCATGAC-3′ and 5′-GAATTCGAGCTCGTTTAAAC-3′. The primer pair used to analyze the DSB at the Ho cut site at the MATα locus was 5′-AGTATGCTGGATTTAAACTCATCTGTGATTTGTGG-3′ and 5′-GATGCTAAGAATTGATTGTTTGCTTGAG-3′.
The disappearance of the PCR signal would indicate the presence of a DSB. A specific region of SMC2 was amplified as a control. SMC2 is not damaged by Ho. The primer pair for amplification of a specific region of SMC2 was 5′-GACGACCTTGTAACAGTCCAGACAG-3′ and 5′-GGCGAATTCCATCACATTATACTAACTACGG-3′.
ChIP Assay
The ChIP assay was performed as described previously (41–44). Briefly, yeast cells were treated with 1% formaldehyde, collected, and resuspended in lysis buffer. Following sonication, cell lysate (400 μl of lysate from 50 ml of yeast culture) was precleared by centrifugation, and then 100 μl of lysate was used for each immunoprecipitation. Immunoprecipitated protein-DNA complexes were treated with proteinase K, the cross-links were reversed, and DNA was purified. Immunoprecipitated DNA was dissolved in 20 μl of TE 8.0 (10 mm Tris HCl (pH 8.0) and 1 mm EDTA), and 1 μl of immunoprecipitated DNA was analyzed by PCR. PCR reactions contained [α-32P]dATP (2.5 μCi for a 25-μl reaction), and the PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, “input” DNA was isolated from 5 μl of lysate without going through the immunoprecipitation step, and dissolved in 100 μl of TE 8.0. To compare PCR signal arising from the immunoprecipitated DNA with the input DNA, 1 μl of input DNA was used in the PCR analysis.
For analysis of recruitment of Ino80p, Ku70p, and Rad50p, the above ChIP protocol was modified as described previously (25). Briefly, a total of 800 μl of lysate was prepared from 100 ml of yeast culture. Following sonication, 400 μl of lysate was used for each immunoprecipitation (using 10 μl of anti-HA or anti-myc antibody and 100 μl of protein A/G plus agarose beads from Santa Cruz Biotechnology, Inc.), and an immunoprecipitated DNA sample was dissolved in 10 μl of TE 8.0, of which 1 μl was used for the PCR analysis. In parallel, the PCR analysis for input DNA was performed using 1 μl of DNA that was prepared by dissolving purified DNA from 5 μl of lysate in 100 μl of TE 8.0. The primer pairs used for PCR analysis were as follows: MATα-Ho, 5′-GGTTTTGTAGAGTGGTTGACGAAT-3′ and 5′-GCTATACTGACAACATTCAGTACTCG-3′; ADH1-ORF, 5′-CGGTAACAGAGCTGACACCAGAGA-3′ and 5′-ACGTATCTACCAACGATTTGACCC-3′; ADH1-UAS, 5′-GAGTTTCCGGGTGTACAATATGG-3′ and 5′-CTATTGTATATCTCCCCTCCGC-3′; ADH1-Core, 5′-GGTATACGGCCTTCCTTCCAGTTAC-3′ and 5′-GAACGAGAACAATGACGAGGAAACAAAAG-3′; PHO5-ORF, 5′-ACCTCTAATTCTAAGAGATGTCATGAC-3′ and 5′-ACAATGTCATCATTGGCATCGTAGTC-3′; and Chr-V, 5′-GGCTGTCAGAATATGGGGCCGTAGTA-3′ and 5′-CACCCCGAAGCTGCTTTCACAATAC-3′. UAS, upstream activating sequence; core, core promoter; and Chr-V, Chromosome-V.
Autoradiograms were scanned and quantitated by the National Institutes of Health image 1.62 program. Immunoprecipitated DNAs were quantitated as the ratio of immunoprecipitate to input in the autoradiogram.
Total RNA Preparation
Total RNA was prepared from yeast cell culture as described by Peterson et al. (45). Briefly, 10 ml of yeast culture was harvested and then suspended in 100 μl of RNA preparation buffer (500 mm NaCl, 200 mm Tris-HCl, 100 mm Na2EDTA, and 1% SDS) along with 100 μl of phenol/chloroform/isoamyl alcohol and 100 μl of volume-equivalent of glass beads (acid-washed, Sigma). Subsequently, the yeast cell suspension was vortexed with a maximum speed (10 in a VWR mini-vortexer, catalog no. 58816-121) five times (30 s each). The cell suspension was put in ice for 30 s between pulses. After vortexing, 150 μl of RNA preparation buffer and 150 μl of phenol/chloroform/isoamyl alcohol were added to the yeast cell suspension followed by vortexing for 15 s with a maximum speed on a VWR mini-vortexer. The aqueous phase was collected following 5 min of centrifugation at maximum speed in a microcentrifuge machine. The total RNA was isolated from the aqueous phase by ethanol precipitation.
RT-PCR Analysis
RT-PCR analysis was performed according to the standard protocols (46). Briefly, total RNA was prepared from 10 ml of yeast culture. Ten micrograms of total RNA was used in the reverse transcription assay. RNA was treated with RNase-free DNase (M610A, Promega) and then reverse-transcribed into cDNA using oligo(dT) as described in the protocol supplied by Promega (A3800, Promega). PCR was performed using synthesized first strand as template and the primer pairs targeted to the ADH1 and MATα ORFs. RT-PCR products were separated by 2.2% agarose gel electrophoresis and visualized by ethidium bromide staining. The primer pairs used in the PCR analysis were as follows: ADH1, 5′-CGGTAACAGAGCTGACACCAGAGA-3′ and 5′-ACGTATCTACCAACGATTTGACCC-3′ and MATα, 5′-GGTTTTGTAGAGTGGTTGACGAAT-3′ and 5′-GCTATACTGACAACATTCAGTACTCG-3′.
RESULTS AND DISCUSSION
Induction of DSBs at the Highly Active ADH1 and Nearly Silent MATα Genes
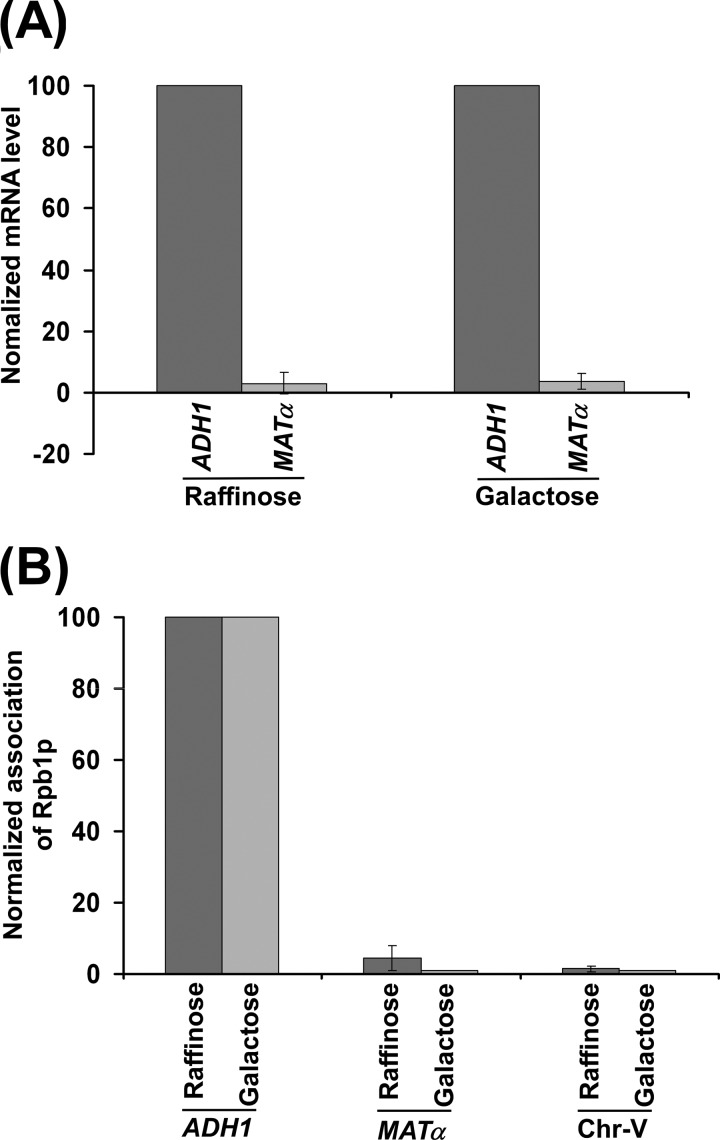
Cells in S-phase have a fast kinetics of DNA DSB repair post-irradiation as compared with cells in G1 or G2 phase (47). The fast repair kinetics of DSBs in S phase is attributed to functionally active DNA polymerases. To determine whether RNA polymerases have any influence on the repair of DNA DSB, we analyzed the effect of RNA polymerase II-dependent transcription on DSB repair by introducing the Ho cut site at the ADH1 coding sequence and MATα locus as described below. Our RT-PCR analysis revealed that ADH1 is a highly active gene as compared with the MATα locus (Fig. 1A). MATα appeared to be a poorly active or nearly silent gene, consistent with previous studies (48). These observations were further substantiated by the analysis of the association of RNA polymerase II with these two loci. We found a robust association of RNA polymerase II with ADH1 as compared with the MATα locus (Fig. 1B). A primer pair targeted to the transcriptionally inactive region of chromosome V was also used in the PCR analysis as a nonspecific DNA control (49). Collectively, these results demonstrate that ADH1 is a highly active gene, consistent with previous studies (48, 50), and MATα is a nearly silent gene as compared with ADH1. However, the coding sequences of both genes have a similar nucleosomal density (51).
FIGURE 1.
MATα is a poorly active gene in comparison to a highly active ADH1 gene. A, RT-PCR analysis of ADH1 and MATα mRNAs. Yeast cells were grown in YPR as well as YPG. Total RNA was prepared from harvested culture and analyzed for ADH1 and MATα mRNAs. The level of ADH1 mRNA was set to 100, and MATα mRNA was normalized with respect to 100. Normalized mRNA levels are plotted in the form of a histogram. B, analysis of RNA polymerase II association with the ADH1 and MATα loci. Yeast cells were grown in YPR as well as YPG and cross-linked by formaldehyde. Immunoprecipitation was performed as described previously (41–44) using a mouse monoclonal antibody 8WG16 (Covance) against the carboxyl terminal domain of the largest subunit (Rpb1p) of RNA polymerase II. Immunoprecipitated DNAs were analyzed by PCR using the primer pairs targeted to ADH1 and MATα. The ratio of the PCR signal of immunoprecipitated DNA to that of input DNA was determined and referred to as the ChIP signal. The ChIP signal at ADH1 was set to 100, and the ChIP signals at MATα and Chr-V were normalized with respect to 100. The normalized ChIP signals (represented as normalized occupancy) are plotted in the form of a histogram.
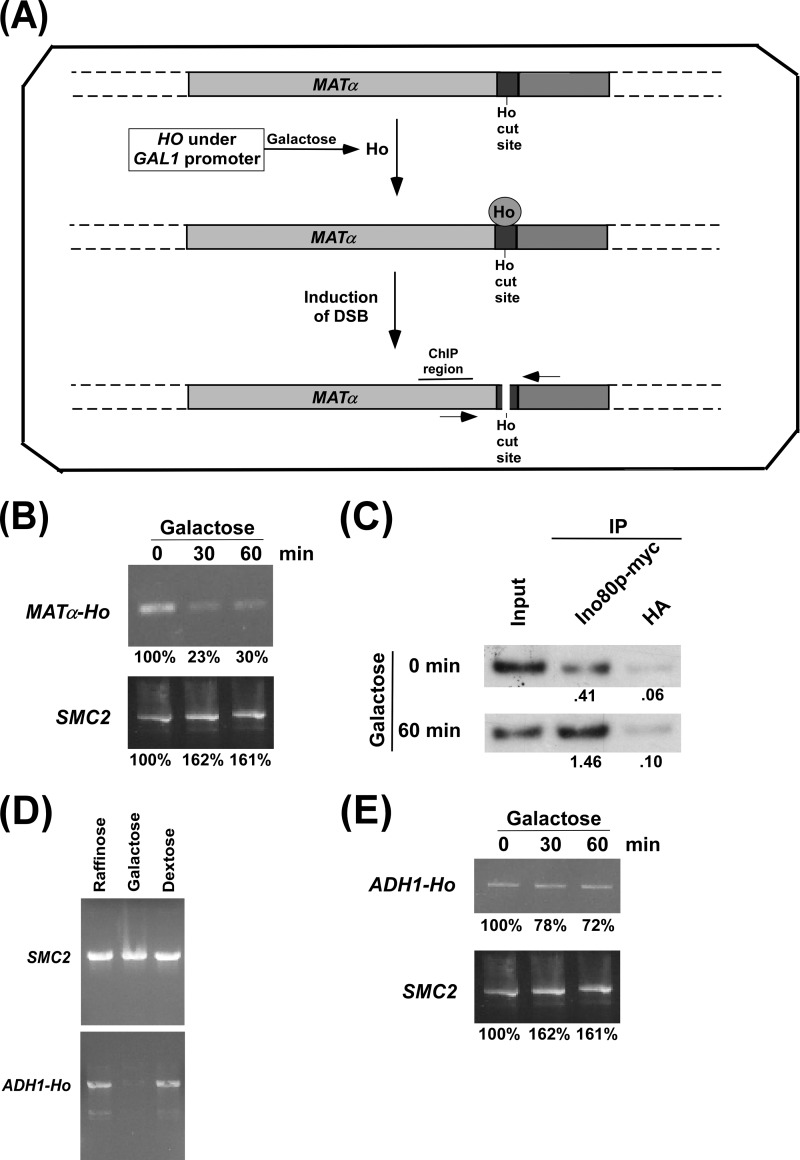
To determine whether DSB repair is coupled to transcription, we first analyzed the DSB at the nearly silent MATα locus using the yeast strain that has a Ho cut site at the MATα locus and expresses HO under the control of the GAL1 promoter in galactose-containing growth medium (Fig. 2A). The yeast strain was initially grown in raffinose-containing medium up to an A600 of 0.9 and then switched to galactose-containing growth medium for 30 and 60 min. In raffinose-containing growth medium, HO was not expressed, as the GAL1 promoter is not induced in the presence of raffinose (44, 52). Therefore, the Ho cut site at the MATα locus was intact in raffinose-containing growth medium (Fig. 2B). Upon switching to galactose-containing growth medium, HO was expressed as the GAL1 promoter is induced in the presence of galactose (44, 52). Such an expression of HO would induce a DSB at the Ho cut site at the MATα locus. The generation of a DSB at the MATα locus was monitored by PCR using the primer pair flanking the Ho cut site (Fig. 2A). The disappearance of the PCR signal would indicate the DSB at the MATα locus. Our PCR analysis revealed that the PCR signal was significantly reduced upon switching the growth medium from raffinose to galactose for 30 and 60 min (Fig. 2B). As a control, we amplified a specific region of SMC2 that is not damaged by Ho. We find that like the MATα locus, the PCR signal at SMC2 did not decrease upon switching the growth media from raffinose to galactose for 30 and 60 min (Fig. 2B). These results support that the expression of HO in galactose-containing growth medium induced DSB at the MATα locus.
FIGURE 2.
Analysis of DSB at the MATα and ADH1 loci. A, schematic diagram showing the generation of DSB at the MATα locus following the induction of HO expression. The primers flanking the Ho cut site are marked by two arrow-headed lines and were used in PCR for analysis of DSB. B, PCR analysis of DSB at the MATα locus. The yeast strain that contains a Ho cut site at the MATα locus and expresses HO under the GAL1 promoter was initially grown in YPR up to an A600 of 0.9 (log phase) and then switched to YPG for 30 and 60 min (an A600 of 1.0 is ∼3 × 107 yeast cells/ml; 46). Genomic DNA was analyzed by PCR to determine DSB using the primer pair flanking the DSB site. The PCR signal at 0 min was set to 100. The PCR signals at 30 or 60 min in YPG were normalized with respect to the 0 min time point in YPG. C, the ChIP assay for analyzing the recruitment of Ino80p to the DSB site. The yeast strain (ASY32) was grown as in B prior to cross-linking. Immunoprecipitation (IP) was performed using an anti-myc antibody (9E10, Santa Cruz Biotechnology, Inc.) against myc-tagged Ino80p. An anti-HA was used as a nonspecific antibody control. The ratio of immunoprecipitate over the input in the autoradiogram is indicated below the immunoprecipitated band. D, analysis of DSB at ADH1 in YPR, YPG, and YPD media. The yeast strain PCY23 was grown in YPR, YPG, or YPD up to an A600 of 0.2 at 30 °C and harvested. Genomic DNA was analyzed for DSB by PCR using the primer pair flanking the HO cut site at ADH1. E, induction of DSB at the ADH1 locus. The yeast strain PCY23 was grown as in B.
To further support the induction of DSB at the MATα locus, we analyzed the recruitment of the INO80 complex at the Ho cut site because previous studies (53, 54) have demonstrated the recruitment of INO80 to DSB. INO80 is an ATP-dependent chromatin remodeling complex and is required to promote DSB repair (53–55). To analyze the recruitment of the INO80 complex at the MATα-Ho locus, we tagged the Ino80p component of INO80 by myc epitope in the JKM179 strain that has a Ho cut site at the MATα locus and expresses HO under the GAL1 promoter. Subsequently, the ChIP assay was performed following the switch of the growth medium from raffinose to galactose for 60 min. Immunoprecipitated chromatin was analyzed by PCR at the site proximal (represented as “ChIP region” in Fig. 2A) to the DSB. Consistent with previous studies (53, 54), we observed an enhanced recruitment of INO80 to the ChIP region in galactose-containing growth medium (Fig. 2C), thus supporting the presence of DSB at the MATα locus.
Next, we analyzed the DSB at the highly active ADH1 gene. In this direction, we introduced a Ho cut site followed by a nucleotide sequence for multiple myc epitope tags just before the translational stop codon of ADH1 in the JKM179 strain. To confirm the presence of the Ho cut site at the ADH1 locus in the generated strain (PCY23), the DSB was analyzed in raffinose, dextrose, or galactose-containing growth medium. In the raffinose- or dextrose-containing growth media, the GAL1 promoter is not induced (44, 52), and thus, HO would not be expressed. Hence, DSB would not be observed at the ADH1 locus in raffinose- or dextrose-containing growth medium. The DSB at the ADH1 locus was analyzed by PCR using the primer pair flanking the Ho cut site. The PCR signal at the ADH1 locus disappeared in galactose-containing growth medium but not raffinose- or dextrose-containing growth medium (Fig. 2D). As a control, we amplified a specific region at the SMC2 locus and found that the PCR signal at SMC2 did not disappear in galactose-containing growth medium (Fig. 2D). These results demonstrate the presence of DSB (and hence the Ho cut site) at the ADH1 locus in galactose-containing growth medium.
We next analyzed the induction of DSB at the ADH1 locus following the switch of the growth medium from raffinose to galactose for 30 and 60 min. In this direction, we inoculated the yeast strain in raffinose-containing medium, and grown up to an A600 of 0.9 at 30 °C. Subsequently, yeast cells were transferred to galactose-containing growth medium for 30 and 60 min, and DSB was analyzed at the ADH1 locus by PCR. We find a modest decrease in the PCR signals after 30 and 60 min in galactose-containing growth medium (Fig. 2E). The PCR signal at the control SMC2 gene did not decrease after 30 and 60 min in galactose-containing growth medium (Fig. 2E). Thus, there is a modest level of DSB at ADH1 following the switch of the growth medium from raffinose to galactose for 30 and 60 min (Fig. 2E). On the other hand, we observed a dramatic decrease in the PCR signal at the Ho cut site at the MATα locus (Fig. 2B). These observations indicate that the nearly silent MATα locus has significantly more DNA DSB than the highly active ADH1 gene.
Fast DSB Repair at ADH1 in Comparison to MATα
Why is the extent of DSB at ADH1 less than that at the MATα locus? When HO is expressed in galactose-containing growth medium, both the generation and repair of DSB occur simultaneously, as Ho is constantly present in galactose-containing growth medium. Therefore, we do not observe 100% DSB. However, less DSB would be observed if DSB repair occurs faster than its generation in galactose-containing growth medium. Intriguingly, we observed less DSB at the highly active ADH1 gene than the nearly silent MATα locus. Thus, these observations indicate that there might be a fast DSB repair at ADH1 as compared with the MATα locus. To confirm that the DSB at the highly active ADH1 gene is repaired fast in comparison to the nearly silent MATα locus, we analyzed DSB repair at the ADH1 and MATa loci. In this direction, we grew the yeast cells in galactose-containing growth medium up to an A600 of 0.2 to induce maximal DSB at ADH1 (as done in Fig. 2D) and then switched to dextrose-containing growth medium for 0.5, 2, and 3 h to allow the cells to repair the DSB as shown schematically in Fig. 3A. We isolated genomic DNAs from these yeast cultures and analyzed DSB repair by PCR using the primer pairs flanking the Ho cut sites at the ADH1 and MATα loci. The increase in PCR signal would indicate the progression of DSB repair. Intriguingly, we find that the PCR signal at ADH1 increased more than that at the MATα locus (Fig. 3B). These observations support that the DSB at the highly active ADH1 is repaired fast as compared with the nearly silent MATα locus. Therefore, we detected less DSB at ADH1 than at the MATα locus (Fig. 2, B and E).
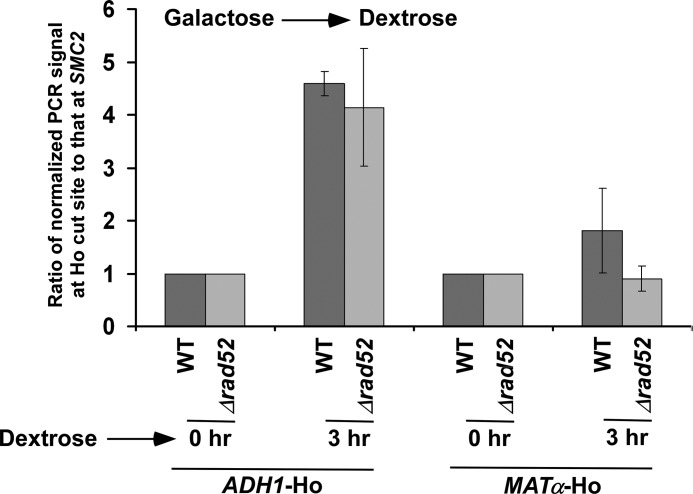
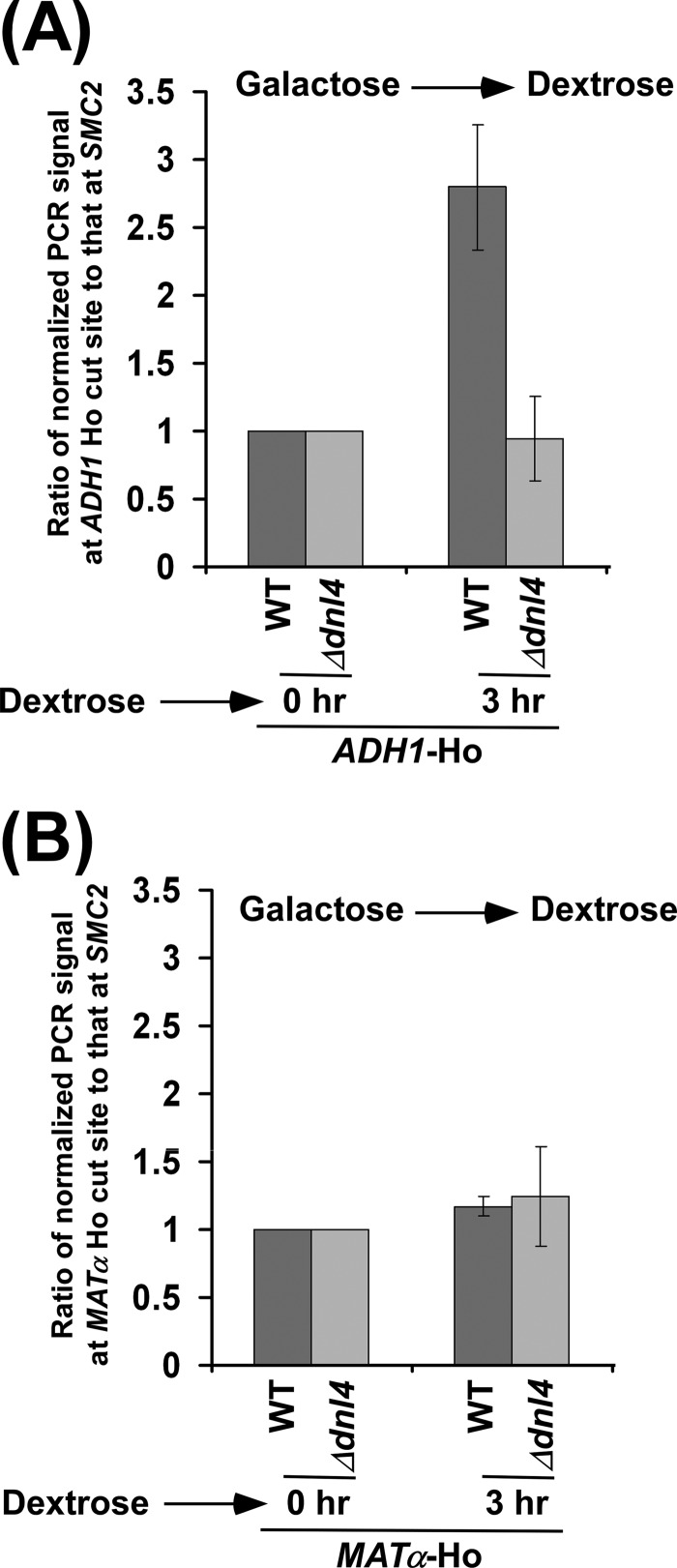
HR-dependent DSB repair is absent at the MATα locus because of the deletion of the donor in the parent JKM179 strain (40). Thus, DSB repair at the MATα locus occurs via non-homologous end joining. An enhanced DSB repair at ADH1 as compared with MATα could be due to the presence of HR at ADH1. To test this possibility, we analyzed DSB repair at ADH1 in the presence and absence of Rad52p that is essential for HR-dependent DSB repair (56). We find that the absence of Rad52p did not significantly alter the DSB repair at ADH1 (Fig. 4). Thus, DSB repair at ADH1 does not appear to be promoted via HR but rather non-homologous end joining. Such DSB repair is impaired in the absence of DNA ligase Dnl4p (Figs. 5, A and B).
FIGURE 4.
Analysis of DSB repair at the ADH1 and MATα loci in the presence and absence of Rad52p. Both the wild-type (PCY23) and Δrad52 (PCY36) strains were grown as in Fig. 3B. Genomic DNAs from these yeast cultures were prepared and analyzed by PCR for DSBs at the ADH1 and MATα loci. A specific region within SMC2 was amplified as control, using the same genomic DNAs. The PCR signals at 0 h time point for the ADH1, MATα and SMC2 loci were set to 100, and the PCR signals at 3 h were normalized with respect to 100. The ratios of normalized PCR signals at 3 h at the ADH1 and MATα Ho cut sites to that at SMC2 are plotted in the form of a histogram. A ratio that is greater than 1 would indicate the DSB repair.
FIGURE 5.
Analysis of DSB repair at ADH1 in the presence and absence of Dnl4p. Both the wild-type (PCY23) and Δdnl4 (PCY37a) strains were grown as in Fig. 3B. The DSB repairs at the ADH1 (A) and MATα (B) loci were analyzed as in Fig. 4.
How is the DSB repair facilitated at the active gene? We hypothesize that RNA polymerase II promotes the recruitment of the repair factors to the DSB site at the active gene (and hence stimulates DSB repair), analogous to the fact that RNA polymerase II facilitates the recruitment of TC-NER-specific factor Rad26p to the DNA lesion at the active gene to promote NER (25). In support of this hypothesis, a recent study (57) implicated the interaction of RNA polymerase II with RPA (replication protein A complex), which is involved in DSB repair. Further, the INO80 chromatin remodeling complex that promotes DSB repair is recruited to the gene in a transcription-dependent manner (58, 59). Similarly, the recruitment of the MRX (Mre11p-Rad50p-Xrs2p) complex and Ku proteins to the DSB site might be promoted by RNA polymerase II or transcription machinery to stimulate DSB repair. To test this, we generated yeast strains carrying HA epitope tags at the C-terminals of Rad50p (Mre11p-Rad50p-Xrs2p) and Ku70p (Ku proteins) and then performed the ChIP experiments to analyze their association with the active ADH1 gene. Our ChIP analysis revealed that Rad50p and Ku70p did not associate with ADH1 (supplemental Fig. S1, A and B). Thus, RNA polymerase II or active transcription machinery does not appear to promote the targeting of Mre11p-Rad50p-Xrs2p or Ku70p to DSB. However, like the association of RPA and INO80 with the active gene (57–59), we have demonstrated recently that TC-NER factor Rad26p associates with active genes (25, 60) and facilitates chromatin disassembly (60, 61). Such chromatin regulatory function of Rad26p might be enhancing DSB repair at the active gene. Further, DSB repair at the active coding sequence is likely to be promoted by a transcription-coupled open chromatin structure or chromatin remodeling/modifying factors. These possibilities remain to be investigated. Nonetheless, this study demonstrates for the first time that DSB repair at the active gene occurs faster than that at the inactive gene, thus supporting transcription-coupled DSB repair.
Fast DSB Repair at PHO5 under Transcriptionally Active Conditions in Comparison with the Inactive State
To extend our results to another set of transcriptionally active and inactive genes, we introduced a Ho cut site followed by a nucleotide sequence for multiple myc epitope tags just before the translational stop codon of an inducible PHO5 gene in the aforesaid yeast (JKM179) strain that had a Ho cut site at the MATα locus and expressed HO under the GAL1 promoter. The PHO5 gene is transcriptionally induced in the absence of Pi and repressed in the presence of Pi (62–65). Thus, transcription of PHO5 would be repressed in YPD (which contains Pi) and induced in the YPD-Pi (YPD without Pi) medium. Using this generated strain, we induced DSB at the PHO5 coding sequence in YPG-Pi (supplemental Fig. S2) and then analyzed DSB repair under transcriptionally active (YPD-Pi) and inactive (YPD) conditions, as shown schematically in Fig. 6A. We find that DSB repair at the PHO5 coding sequence occurs fast under transcriptionally active condition as compared with the inactive state (Fig. 6, B and C). However, at the later time point (4 h), DSB repairs at the active and inactive PHO5 were the same (Fig. 6B). Therefore, our data support a fast DSB repair at the transcriptionally active PHO5 gene as compared with inactive PHO5, hence implying transcription-coupled DSB repair.
CONCLUSION
Previous studies (17–24, 35–37) have demonstrated transcription-coupled nucleotide/base excision repairs. However, it remained unknown whether transcription-coupled DSB repair exists. Here, we have developed experimental systems to analyze the induction and repair of DSBs at the active and inactive genes. Using such systems, we demonstrate that DSB repair at the active gene occurs faster than that at the inactive genomic locus. Therefore, our results support for the first time the existence of transcription-coupled DSB repair, hence providing a new regulatory process of genome repair in vivo.
Acknowledgments
We thank James E. Haber for the yeast strain and Geetha Durairaj, Shivani Malik, Abhijit Shukla, and Shruti Bagla for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R15GM088798-01 (to S. R. B.), American Heart Association Grant-in-Aid 10GRNT4300059 (Greater Midwest Affiliate), a Mallinckrodt Foundation grant, and Excellence in Academic Medicine (EAM) grants of the Southern Illinois University School of Medicine. This work was also supported by Grants R01 CA123232, R01 CA129537, and R01 CA154320 (to T. K. P.).

This article contains supplemental Figs. S1 and S2.
- NER
- nucleotide excision repair
- DSB
- double-strand break
- HR
- homologous recombination
- TC-NER
- transcription-coupled NER
- GG-NER
- global genome NER
- YPG
- yeast extract, peptone plus 2% galactose
- YPR
- yeast extract, peptone plus 2% raffinose
- YPD
- yeast extract, peptone plus 2% dextrose
- Pi
- inorganic phosphate.
REFERENCES
- 1. Feuerhahn S., Egly J. M. (2008) Tools to study DNA repair. What's in the box? Trends Genet. 24, 467–474 [DOI] [PubMed] [Google Scholar]
- 2. Klaunig J. E., Kamendulis L. M., Hocevar B. A. (2010) Oxidative stress and oxidative damage in carcinogenesis. Toxicol. Pathol. 38, 96–109 [DOI] [PubMed] [Google Scholar]
- 3. Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 4. Seeberg E., Eide L., Bjoras M. (1995) The base excision repair pathway. Trends Biochem. Sci. 20, 391–397 [DOI] [PubMed] [Google Scholar]
- 5. Scott S. P., Pandita T. K. (2006) The cellular control of DNA double-strand breaks. J. Cell. Biochem. 99, 1463–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pandita T. K., Richardson C. (2009) Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 37, 1363–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanaar R., Hoeijmakers J. H., van Gent D. C. (1998) Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 8, 483–489 [DOI] [PubMed] [Google Scholar]
- 8. Cromie G. A., Connelly J. C., Leach D. R. (2001) Recombination at double-strand breaks and DNA ends. Conserved mechanisms from phage to humans. Mol. Cell 8, 1163–1174 [DOI] [PubMed] [Google Scholar]
- 9. Hartwell L. H., Weinert T. A. (1989) Checkpoints. Controls that ensure the order of cell cycle events. Science 246, 629–634 [DOI] [PubMed] [Google Scholar]
- 10. Hartwell L. H. (1992) Role of yeast in cancer research. Cancer, 69, 2615–2621 [DOI] [PubMed] [Google Scholar]
- 11. Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. (2002) Toward maintaining the genome. DNA damage and replication checkpoints. Annu. Rev. Genet. 36, 617–656 [DOI] [PubMed] [Google Scholar]
- 12. Richardson C., Horikoshi N., Pandita T. K. (2004) The role of the DNA double-strand break response network in meiosis. DNA Repair 3, 1149–1164 [DOI] [PubMed] [Google Scholar]
- 13. Conaway J. W., Shilatifard A., Dvir A., Conaway R. C. (2000) Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25, 375–380 [DOI] [PubMed] [Google Scholar]
- 14. Sims R. J., 3rd, Mandal S. S., Reinberg D. (2004) Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell Biol. 16, 263–271 [DOI] [PubMed] [Google Scholar]
- 15. Ljungman M., Zhang F. (1996) Blockage of RNA polymerase as a possible trigger for UV light-induced apoptosis. Oncogene 13, 823–831 [PubMed] [Google Scholar]
- 16. Yamaizumi M., Sugano T. (1994) UV-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene 9, 2775–2784 [PubMed] [Google Scholar]
- 17. Friedberg E. C., Aguilera A., Gellert M., Hanawalt P. C., Hays J. B., Lehmann A. R., Lindahl T., Lowndes N., Sarasin A., Wood R. D. (2006) DNA repair. From molecular mechanism to human disease. DNA Repair 5, 986–996 [DOI] [PubMed] [Google Scholar]
- 18. Hanawalt P. C., Spivak G. (2008) Transcription-coupled DNA repair. Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 [DOI] [PubMed] [Google Scholar]
- 19. Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. (1985) DNA repair in an active gene. Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40, 359–369 [DOI] [PubMed] [Google Scholar]
- 20. Leadon S. A. (2000) Transcription-coupled repair. A multifunctional signaling pathway. Cold Spring Harbor Symp. Quant. Biol. 65, 561–566 [DOI] [PubMed] [Google Scholar]
- 21. Mellon I., Spivak G., Hanawalt P. C. (1987) Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249 [DOI] [PubMed] [Google Scholar]
- 22. Smerdon M. J., Thoma F. (1990) Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell 61, 675–684 [DOI] [PubMed] [Google Scholar]
- 23. Sweder K. S., Hanawalt P. C. (1993) Transcription-coupled DNA repair. Science 262, 439–440 [DOI] [PubMed] [Google Scholar]
- 24. Tornaletti S. (2009) DNA repair in mammalian cells. Transcription-coupled DNA repair. Directing your effort where it's most needed. Cell Mol. Life Sci. 66, 1010–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malik S., Chaurasia P., Lahudkar S., Durairaj G., Shukla A., Bhaumik S. R. (2010) Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res. 38, 1461–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee S. K., Yu S. L., Prakash L., Prakash S. (2001) Requirement for yeast RAD26, a homolog of the human CSB gene, in elongation by RNA polymerase II. Mol. Cell Biol. 21, 8651–8656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee S. K., Yu S. L., Prakash L., Prakash S. (2002) Yeast RAD26, a homolog of the human CSB gene, functions independently of nucleotide excision repair and base excision repair in promoting transcription through damaged bases. Mol. Cell Biol. 22, 4383–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selby C. P., Sancar A. (1997) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 94, 11205–11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R. E., Hoeijmakers J. H., Vermeulen W. (2000) ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell Biol. 20, 7643–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bucheli M., Sweder K. (2004) In UV-irradiated Saccharomyces cerevisiae, overexpression of Swi2/Snf2 family member Rad26 increases transcription-coupled repair and repair of the non-transcribed strand. Mol. Microbiol. 52, 1653–1663 [DOI] [PubMed] [Google Scholar]
- 31. Malik S., Bagla S., Chaurasia P., Duan Z., Bhaumik S. R. (2008) Elongating RNA polymerase II is disassembled through specific degradation of its largest but not other subunits in response to DNA damage in vivo. J. Biol. Chem. 283, 6897–6905 [DOI] [PubMed] [Google Scholar]
- 32. Christians F. C., Hanawalt P. C. (1992) Inhibition of transcription and strand-specific DNA repair by α-amanitin in Chinese hamster ovary cells. Mutat. Res. 274, 93–101 [DOI] [PubMed] [Google Scholar]
- 33. Lee K. B., Wang D., Lippard S. J., Sharp P. A. (2002) Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc. Natl. Acad. Sci. U.S.A. 99, 4239–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sweder K. S., Hanawalt P. C. (1992) Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc. Natl. Acad. Sci. U.S.A. 89, 10696–10700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Larsen E., Kwon K., Coin F., Egly J. M., Klungland A. (2004) Transcription activities at 8-oxoG lesions in DNA. DNA Repair 3, 1457–1468 [DOI] [PubMed] [Google Scholar]
- 36. Banerjee D., Mandal S. M., Das A., Hegde M. L., Das S., Bhakat K. K., Boldogh I., Sarkar P. S., Mitra S., Hazra T. K. (2011) Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 286, 6006–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsutakawa S. E., Cooper P. K. (2000) Transcription-coupled repair of oxidative DNA damage in human cells. Mechanisms and consequences. Cold Spring Harbor Symp. Quant. Biol. 65, 201–215 [DOI] [PubMed] [Google Scholar]
- 38. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 39. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moore J. K., Haber J. E. (1996) Cell cycle and genetic requirements of two pathways of non-homologous endjoining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell Biol. 16, 2164–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bhaumik S. R., Green M. R. (2002) Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell Biol. 22, 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bhaumik S. R., Green M. R. (2003) Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 370, 445–454 [DOI] [PubMed] [Google Scholar]
- 43. Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S. R. (2006) Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell Biol. 26, 3339–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bhaumik S. R., Raha T., Aiello D. P., Green M. R. (2004) In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18, 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peterson C. L., Kruger W., Herskowitz I. (1991) A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64, 1135–1143 [DOI] [PubMed] [Google Scholar]
- 46. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Struhl K. (2001) Current Protocols in Molecular Biology, Wiley, New York [Google Scholar]
- 47. Pandita T. K., Hittelman W. N. (1992) The contribution of DNA and chromosome repair deficiencies to the radiosensitivity of ataxia-telangiectasia. Radiat Res. 131, 214–223 [PubMed] [Google Scholar]
- 48. Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., Young R. A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 [DOI] [PubMed] [Google Scholar]
- 49. Lee J. S., Shukla A., Schneider J., Swanson S. K., Washburn M. P., Florens L., Bhaumik S. R., Shilatifard A. (2007) Translating histone cross-talk between H2B monoubiquitination and H3 methylation by COMPASS and Dot1. Cell 131, 1084–1096 [DOI] [PubMed] [Google Scholar]
- 50. Li X. Y., Bhaumik S. R., Green M. R. (2000) Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288, 1242–1244 [DOI] [PubMed] [Google Scholar]
- 51. Jiang C., Pugh B. F. (2009) A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 10, R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bhaumik S. R., Green M. R. (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15, 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morrison A. J., Highland J., Krogan N. J., Arbel-Eden A., Greenblatt J. F., Haber J. E., Shen X. (2004) INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119, 767–775 [DOI] [PubMed] [Google Scholar]
- 54. van Attikum H., Fritsch O., Hohn B., Gasser S. M. (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788 [DOI] [PubMed] [Google Scholar]
- 55. Tsukuda T., Fleming A. B., Nickoloff J. A., Osley M. A. (2005) Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mortensen U. H., Lisby M., Rothstein R. (2009) Rad52. Curr. Biol. 19, R676–677 [DOI] [PubMed] [Google Scholar]
- 57. Sikorski T. W., Ficarro S. B., Holik J., Kim T., Rando O. J., Marto J. A., Buratowski S. (2011) Sub1 and RPA associate with RNA polymerase II at different stages of transcription. Mol. Cell, 44, 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Conaway R. C., Conaway J. W. (2009) The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem. Sci. 34, 71–77 [DOI] [PubMed] [Google Scholar]
- 59. Klopf E., Paskova L., Solé C., Mas G., Petryshyn A., Posas F., Wintersberger U., Ammerer G., Schüller C. (2009) Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 29, 4994–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Malik S., Chaurasia P., Lahudkar S., Uprety B., Bhaumik S. R. (2012) Rad26p regulates the occupancy of histone H2A-H2B dimer at the active genes in vivo. Nucleic Acids Research 40, 3348–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Malik S., Bhaumik S. R. (2012) Rad26p, a transcription-coupled repair factor, promotes the eviction and prevents the reassociation of histone H2A-H2B dimer during transcriptional elongation in vivo. Biochemistry, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. He Y., Swaminathan A., Lopes J. M. (2012) Transcription regulation of the Saccharomyces cerevisiae PHO5 gene by the Ino2p and Ino4p basic helix-loop-helix proteins. Mol. Microbiol. 83, 395–407 [DOI] [PubMed] [Google Scholar]
- 63. Barbaric S., Luckenbach T., Schmid A., Blaschke D., Hörz W., Korber P. (2007) Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 282, 27610–27621 [DOI] [PubMed] [Google Scholar]
- 64. Adkins M. W., Williams S. K., Linger J., Tyler J. K. (2007) Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Mol. Cell Biol. 27, 6372–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lau W. W., Schneider K. R., O'Shea E. K. (1998) A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae. Genetics 150, 1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]