Background: Most reported ADAM17 inhibitors are zinc-binding and not selective.
Results: Novel selective ADAM17 inhibitors were discovered and characterized.
Conclusion: Novel ADAM17 inhibitors act via a non-zinc-binding mechanism.
Significance: Selective non-zinc-binding inhibitors of ADAM proteases can be useful research and therapeutic tools.
Keywords: ADAM, ADAMTS, Allosteric Regulation, Cell Surface Enzymes, Chemical Biology, Enzyme Inhibitors, ADAM Protease, Combinatorial Library, Exosite Inhibitor, Exosite Substrate, Glycoproteolysis
Abstract
A disintegrin and metalloprotease (ADAM) proteases are implicated in multiple diseases, but no drugs based on ADAM inhibition exist. Most of the ADAM inhibitors developed to date feature zinc-binding moieties that target the active site zinc, which leads to a lack of selectivity and off-target toxicity. We hypothesized that secondary binding site (exosite) inhibitors should provide a viable alternative to active site inhibitors. Potential exosites in ADAM structures have been reported, but no studies describing substrate features necessary for exosite interactions exist. Analysis of ADAM cognate substrates revealed that glycosylation is often present in the vicinity of the scissile bond. To study whether glycosylation plays a role in modulating ADAM activity, a tumor necrosis factor α (TNFα) substrate with and without a glycan moiety attached was synthesized and characterized. Glycosylation enhanced ADAM8 and -17 activities and decreased ADAM10 activity. Metalloprotease (MMP) activity was unaffected by TNFα substrate glycosylation. High throughput screening assays were developed using glycosylated and non-glycosylated substrate, and positional scanning was conducted. A novel chemotype of ADAM17-selective probes was discovered from the TPIMS library (Houghten, R. A., Pinilla, C., Giulianotti, M. A., Appel, J. R., Dooley, C. T., Nefzi, A., Ostresh, J. M., Yu, Y., Maggiora, G. M., Medina-Franco, J. L., Brunner, D., and Schneider, J. (2008) Strategies for the use of mixture-based synthetic combinatorial libraries. Scaffold ranking, direct testing in vivo, and enhanced deconvolution by computational methods. J. Comb. Chem. 10, 3–19; Pinilla, C., Appel, J. R., Borràs, E., and Houghten, R. A. (2003) Advances in the use of synthetic combinatorial chemistry. Mixture-based libraries. Nat. Med. 9, 118–122) that preferentially inhibited glycosylated substrate hydrolysis and spared ADAM10, MMP-8, and MMP-14. Kinetic studies revealed that ADAM17 inhibition occurred via a non-zinc-binding mechanism. Thus, modulation of proteolysis via glycosylation may be used for identifying novel, potentially exosite binding compounds. The newly described ADAM17 inhibitors represent research tools to investigate the role of ADAM17 in the progression of various diseases.
Introduction
A disintegrin and metalloprotease (ADAM)2 proteases are implicated in multiple diseases, including but not limited to several types of cancer (3, 4), rheumatoid arthritis (5), and Alzheimer disease (6). Multiple clinical trials of small molecule inhibitors of the most studied ADAM protease, ADAM17, were discontinued due to toxicity (5, 7) as a result of the lack of selectivity of these inhibitors. Most of the ADAM inhibitors developed to date feature zinc-binding moieties that target the active site zinc (8). There are ∼70 known human metalloproteases (ADAM, ADAMTS, and MMP) (9) that have zinc in their active site, which explains off-target toxicities of zinc-binding inhibitors (10).
Secondary binding site (exosite) inhibitors provide a viable alternative to active site inhibitors (11–16). Selective exosite inhibitors of MMP-13 that were discovered by several groups (17–21), including ours, are currently being investigated for arthritis therapy. Exosites are defined as sites outside of the active site that participate in substrate recognition and binding (11).
Enzymes from multiple families and classes have been shown to contain exosites: serine, cysteine, and metalloproteases (22–25), metallo-β-lactamases (26), kinases (27), oxidoreductases (28), nucleases (29), and deacetylases (30). Moreover, non-zinc-binding exosite inhibitors were reported for MMP and ADAMTS proteases (17, 18, 22), which are closely related to ADAM proteases. Therefore, although currently there are only a few reports of potential exosites in ADAM protease structures (31, 32), it is likely that ADAM proteases possess exploitable exosites. A recent report by Tape et al. (33) demonstrated that it is possible to achieve selective binding to the ADAM17 ectodomain by an antibody that exploits exosites.
Substrate recognition by ADAM proteases is a largely unexplored area. Substrate specificity of closely related proteases from ADAMTS and MMP families was shown to be due to a combination of sequence features and substrate topology (34–37). Although cleavage site sequence specificity was addressed for several members of the ADAM family (38–40), there are no studies of the effects of secondary structure on substrate recognition by ADAM proteases.
Similarly, it is not known whether other substrate features, such as glycosylation, play a role in ADAM substrate specificity. Glycosylation was shown to cause peptides to assume a repertoire of different conformations (41, 42) due either to stabilization or destabilization of glycosylated structure as compared with a non-modified peptide (43, 44). Additionally, it was shown that the rate of enzymatic hydrolysis of glycosylated peptides was dependent on the distance of the glycosylation site from the scissile bond (45). This suggests the possibility of glycosylation serving as specific cleavage signal or, alternatively, an effect of different peptide conformations on enzyme hydrolytic activity.
ADAM substrates exhibit various degrees of glycosylation, whereas distances of glycosylation sites from respective scissile bonds also vary significantly. For example, the cleavage site of TNFα by ADAM17 is only four residues away from a glycosylated residue (46), whereas glycosylation occurs 14 residues away from the TGFα cleavage site (47) and more than 200 residues away from the L-selectin cleavage site (48).
In this work, we have investigated the role of glycosylation in the specificity of ADAM-catalyzed reactions using TNFα as a model substrate. Enzyme-substrate interactions based on glycosylation were subsequently utilized to identify novel, potentially exosite-binding ADAM17 inhibitors.
EXPERIMENTAL PROCEDURES
Substrate Synthesis, Purification, and Characterization
Experimental details are listed in the supplemental materials. Briefly, substrate synthesis was performed on a Protein Technology PS3 peptide synthesizer using Fmoc (N-(9-fluorenyl)methoxycarbonyl) solid-phase peptide chemistry methodology. Substrates were purified using reversed-phase HPLC. Fractions were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and by analytical reversed-phase HPLC.
Circular Dichroism Spectroscopy
CD spectra were recorded over the range λ = 190–250 nm with a Jasco J-810 spectropolarimeter using a 0.2-cm path length quartz cell. Substrates solutions were prepared in deionized water. Raw CD data (millidegrees) was normalized for respective substrate concentrations to obtain molar ellipticity (θ) to allow for direct comparison of CD signatures of different substrates.
Enzyme Kinetics
ADAM and MMP enzymes were purchased from R&D Systems (Minneapolis, MN) with the exception of MMP-14, which was purchased from Millipore (Temecula, CA). ADAM8 and -12 were activated as per the manufacturer's instructions. Active enzyme concentrations were determined as described elsewhere (40). Substrate stock solutions were prepared at various concentrations in R&D Systems recommended assay buffers. Assays were conducted by incubating a range of substrate concentrations (2–100 μm) with various ADAM enzyme concentrations at 25 °C. Fluorescence was measured on a multimode microplate reader Synergy H4 (Biotek Instruments, Winooski, VT) using λex = 360 nm and λem = 460 nm. Rates of hydrolysis were obtained from plots of fluorescence versus time, using data points from only the linear portion of the hydrolysis curve. The slope from these plots was divided by the fluorescence change corresponding to complete hydrolysis and then multiplied by the substrate concentration to obtain rates of hydrolysis in units of μm/s. Kinetic parameters were calculated by non-linear regression analysis using the GraphPad Prism version 5.01 suite of programs. ADAM and MMP substrate cleavage sites were established by MALDI-TOF MS.
Library Screening
Mixture libraries (1, 2) were solubilized in 3% DMSO/H2O and added to polypropylene 384-well plates (Greiner catalog no. 781280). ADAM10 and -17 glycosylated and non-glycosylated substrate assays followed the same general protocol. 5 μl of 3× enzyme solution (30 nm) in assay buffer (10 mm Hepes, 0.001% Brij-35, pH 7.5) were added to solid bottom white 384-well low volume plates (Nunc, catalog no. 264706). Next, 5 μl of test compounds or pharmacological controls were added to corresponding wells. After a 30-min incubation at room temperature, the reactions were started by the addition of 5 μl of 3× solutions of the respective substrates (30 μm). Fluorescence was measured every 30 min for 2 h using the multimode microplate reader Synergy H4 (Biotek Instruments, Winooski, VT) using λex = 360 nm and λem = 460 nm. Rates of hydrolysis were obtained from plots of fluorescence versus time, and inhibition was calculated using rates obtained from wells containing substrates only (100% inhibition) and substrates with enzyme (0% inhibition). Three parameters were calculated on a per plate basis: (a) the signal-to-background ratio; (b) the coefficient for variation (equal to the (S.D./mean) × 100) for all compound test wells); and (c) the Z′-factor (18). The IC50 value of the pharmacological control (N-hydroxy-1-(4-methoxyphenyl)sulfonyl-4-(4-biphenylcarbonyl)piperazine-2-carboxamide; Calbiochem catalog no. 444252) was also calculated to ascertain the assay robustness. Each mixture library was screened in triplicate at 0.33 mg/ml.
Inhibition Kinetics
Glycosylated substrate and ADAM17 working solutions were prepared in buffer containing 10 mm Hepes, 0.001% Brij-35, pH 7.5. All reactions were conducted in 384-white polystyrene well plates (Nunc, catalog no. 264706). Determinations of inhibition constants and modalities were conducted by incubating the range of glycosylated substrate concentrations (1–50 μm) with 2.5 nm ADAM17 at room temperature in the presence of varying concentrations of inhibitors. Fluorescence was measured as described above. Rates of hydrolysis were obtained from plots of fluorescence versus time, using data points from only the linear portion of the hydrolysis curve.
All kinetic parameters were calculated using GraphPad Prism version 5.01 (GraphPad Software, Inc., La Jolla, CA). All Ki and K′i values were determined by non-linear regression (hyperbolic equation) analysis using the mixed inhibition model, which allows for simultaneous determination of mechanism of inhibition (13). The mechanism of inhibition was determined using the “alpha” parameter derived from a mixed model inhibition by GraphPad Prism. The mechanism of inhibition was additionally confirmed by Lineweaver-Burk plots.
Dual Inhibition Kinetics
A matrix of two different inhibitor combinations was created in 384-white polystyrene well plates (Nunc, catalog no. 264706) by serially diluting inhibitors in buffer containing 10 mm Hepes, 0.001% Brij-35, pH 7.5. ADAM17 ectodomain and glycosylated substrate were then added resulting in 2.5 nm and 10 μm final assay concentrations, respectively. Fluorescence was measured by a Synergy H4 (Biotek Instruments, Winooski, VT) using λex = 360 nm and λem = 460 nm. Rates of hydrolysis were obtained from plots of fluorescence versus time, using data points from only the linear portion of the hydrolysis curve.
Initial rates of glycosylated substrate hydrolysis in the presence of two inhibitors were arranged as Yonetani-Theorell plots with 1/vij plotted as a function of [I] in the presence of varying concentrations of inhibitor J. In Yonetani-Theorell plots, the intersecting lines indicate simultaneous (i.e. mutually non-exclusive) binding of both inhibitors to the enzyme (49, 50).
RESULTS AND DISCUSSION
Database and Literature Searches
To determine which structural features of ADAM substrates can play a role in enzyme recognition and specificity, we examined the cognate human substrates of ADAM proteases. Overall, more than 30 ADAM substrates were analyzed. The analysis revealed that several confirmed physiological substrates of ADAM proteases are post-translationally modified in the vicinity of the scissile bond. For example, pro-TNFα was reported to be O-glycosylated at Ser80 in human B-cell lymphoblastic leukemia cells (BALL-1) (46). The mucin type 2,3- and 2,6-sialylated and non-sialylated core 1 structure (β-Gal-(1→3)-GalNAc), also known as T- or TF-antigen, were identified. The physiological cleavage site by ADAM17 is at the Ala76↓Val bond, only 4 residues away from the modified Ser residue (Table 1). IL6-R (interleukin-6 receptor), which is cleaved by both ADAM10 and -17 (38, 51), was reported to be N-glycosylated by N-acetylglucosamine (GlcNAc) at four different positions (52, 53). Glycosylated Asn350 is only 7 residues away from the Gln357↓Asp scissile bond (Table 1).
TABLE 1.
ADAM cognate human substrates and their cleavage and glycosylation sites
| UniProt no. | Substrate | Cleavage site | Glycosylated residue | Distance from scissile bond (no. of residues) | Type of glycosylation | ADAM |
|---|---|---|---|---|---|---|
| P35070 | Pro-BTC | CVVA31↓DGNS | Asn34 | 3 | N-Linked (GlcNAc) | 10 |
| P01375 | Pro-TNFα | LAQA76↓VRSS | Ser80 | 4 | O-Linked (GalNAc) | 17 |
| P02786 | TRP1 | ECER100↓LAGT | Thr104, Asn251, Asn317, Asn727 | 4a | O-Linked (GalNAc) | 17 |
| P08887 | IL6-R | LPVQ357↓DSSV | Asn55, Asn93, Asn221, Asn350 | 7a | N-Linked (GlcNAc) | 10/17 |
| O14944 | Pro-EPR | DNPR59↓VAQV | Asn47 | 12 | N-Linked (GlcNAc) | 17 |
| Q99075 | Pro-HB-EGF | RKVR62↓DLQE | Thr75, Thr85 | 13a | O-Linked (GalNAc) | 10/17 |
| P01135 | Pro-TGFα | VAAA39↓VVSH | Asn25 | 14 | N-Linked (GlcNAc) | 17 |
a Distance from the closest glycosylated site is indicated in cases of multiple glycosylations.
We hypothesized that the proximity of the carbohydrate moiety to the scissile bond of ADAM protease cognate substrates can have an effect on ADAM activity and, therefore, on production of soluble forms of cell surface molecules. To test this hypothesis, we synthesized substrates based on the cleavage site within TNFα (PLAQA76↓VRSSS) with and without TF-antigen (Fig. 1A) and characterized these substrates by circular dichroism spectroscopy and in enzymatic assays.
FIGURE 1.
Glycosylated substrate and substrate characterization by circular dichroism spectroscopy. A, structure of glycosylated substrate (Glu(EDANS)-Pro-Leu-Ala-Gln-Ala-Val-Arg-Ser-Ser(TF)-Ser-Lys(DABCYL)). Shown are circular dichroism spectra of non-glycosylated (B) and glycosylated substrates (C). D, molar ellipticity of non-glycosylated and glycosylated substrates at λ = 222 nm as a function of TFE concentration. TF, Thomsen-Freidenreich.
Circular Dichroism Spectroscopic Analysis
In order to determine the effect of glycosylation on substrate conformation and stability, CD spectra of non-glycosylated and glycosylated substrates in water and varying concentrations of TFE were recorded. CD spectra of 100% aqueous solution (0% TFE) and 25% TFE solutions of both non-glycosylated and glycosylated substrates exhibited characteristics of random coils featuring a single large negative peak at around λ = 198 nm (Fig. 1, B and C). In the presence of high TFE concentrations (50–100%), both substrates exhibited spectra typical for an α-helix with two minima at λ = 208 and 222 nm. The plot of molar ellipticity at λ = 222 nm as a function of TFE concentration (Fig. 1D) revealed that both substrates had similarly low α-helical content at 0–25% TFE, whereas the non-glycosylated substrate had a higher α-helical content than the glycosylated substrate at 50–100% TFE.
The lower α-helical content of glycosylated substrate compared with a non-glycosylated substrate at higher TFE concentrations is consistent with previous observations of the effects of O-glycosylation on peptide conformation (54, 55). This suggests that TF-antigen interferes with formation of the hydrogen bond pattern of an α-helix, which is most likely attributable to steric hindrance. The calculated van der Waals volume of the Ser side chain is increased from 26.1 to 168.7 Å due to the presence of a single carbohydrate moiety (56, 57).
Substrate Cleavage Sites
ADAM and MMP cleavage sites in glycosylated and non-glycosylated substrates were identified. ADAM10 and -17 cleaved only at the Ala↓Val bond in both substrates. ADAM8 also cleaved only the Ala↓Val bond of the non-glycosylated substrate but was able to cleave three different bonds (Ala↓Val, Gln↓Ala, and Val↓Arg) in the glycosylated substrate. ADAM12 cleaved at two positions of the non-glycosylated substrate (Ala↓Val and Gln↓Ala) and three positions in the case of the glycosylated substrate (Ala↓Gln, Gln↓Ala, and Val↓Arg). ADAM9 cleaved only at the Ala↓Val bond of the non-glycosylated substrate, but because there was very little hydrolysis of the glycosylated substrate, the exact cleavage site was not determined.
In the case of MMPs and the non-glycosylated substrate, the number and position of cleavage sites varied widely. MMP-1 and MMP-9 cleaved mainly at the Ala↓Val bond with secondary cleavage sites at Ala↓Gln (MMP-1 and MMP-9) and Arg↓Ser (MMP-1 only). In contrast, MMP-2 and MMP-13 cleaved at three different positions. MMP-2 had two major (Ala↓Val and Ser↓Ser) and one minor (Arg↓Ser) cleavage site, whereas MMP-13 appeared to cleave three positions (Ala↓Val, Ala↓Gln, and Val↓Arg) at the same rate. MMP-8 was able to hydrolyze the Ala↓Val bond only, thus exhibiting the most similarity with ADAM proteases.
Interestingly, in the presence of TF-antigen, all tested MMPs cleaved equally at just two positions (Ala↓Val and Gln↓Ala). Overall, only the ADAM10 and -17 cleavage site specificity was not affected by the presence of a carbohydrate moiety at the Ser80 residue.
Enzyme Kinetics
In order to quantify the effect of glycosylation on ADAM proteolysis, kinetic parameters for hydrolysis of glycosylated and non-glycosylated substrates were determined. The Km value for ADAM17 hydrolysis of the non-glycosylated substrate (Table 2) was in line with the published Km value of 19 ± 5 μm (58). ADAM8 and -17 exhibited higher activity toward the glycosylated substrate as evidenced by 16- and 6-fold higher kcat/Km values, respectively (Table 2). The ADAM12 kcat/Km value was slightly increased for the glycosylated substrate (Table 2). In the case of ADAM17, the increase in activity was due to the cumulative effects of changes in both Km and kcat, whereas ADAM8 improvement of activity was almost entirely due to an increase in kcat. ADAM10 activity toward the glycosylated substrate was ∼3-fold lower than for the non-glycosylated substrate due to a change in kcat rather than Km, which suggests that ADAM10 can accommodate the carbohydrate moiety without the loss of affinity, albeit not as well as ADAM17 (Table 2). Because the carbohydrate moiety is accommodated by ADAM10, it is reasonable to hypothesize that ADAM10 contains a site that can be explored for exosite inhibitor discovery. Kinetic parameters of ADAM9-catalyzed hydrolysis of the glycosylated substrate were not determined due to very low activity. Approximately 10% of substrate was converted after 24 h of incubation with 25 nm ADAM9. For comparison, 25 nm ADAM8 converted 10% of glycosylated substrate within 30 min.
TABLE 2.
Kinetic parameters for ADAM hydrolysis of glycosylated and non-glycosylated substrates
| Enzyme |
kcat/Km |
kcat |
Km |
|||
|---|---|---|---|---|---|---|
| Glycosylated | Non-glycosylated | Glycosylated | Non-glycosylated | Glycosylated | Non-glycosylated | |
| m−1 s−1 | s−1 | μm | ||||
| ADAM8 | 1.4 ± 0.1 × 103 | 2.2 ± 0.0 × 102 | 0.04 ± 0.01 | 0.01 ± 0.03 | 31 ± 11 | 46 ± 15 |
| ADAM9 | NDa | 1.2 ± 0.1 × 103 | ND | 0.06 ± 0.01 | ND | 49 ± 6 |
| ADAM10 | 0.8 ± 0.3 × 104 | 2.5 ± 0.7 × 104 | 0.06 ± 0.01 | 0.28 ± 0.03 | 8.5 ± 1.3 | 12 ± 3 |
| ADAM12 | 1.3 ± 0.4 × 103 | 8.5 ± 0.5 × 102 | 0.02 ± 0.01 | 0.01 ± 0.00 | 20 ± 11 | 12 ± 1 |
| ADAM17 | 7.6 ± 1.7 × 104 | 1.2 ± 0.1 × 104 | 0.25 ± 0.03 | 0.14 ± 0.01 | 3.0 ± 0.5 | 12 ± 3 |
a ND, not determined due to low reaction velocity.
In order to determine whether substrate glycosylation has an effect on MMP catalysis, we determined kinetic parameters for MMP hydrolysis of glycosylated and non-glycosylated substrates. Interestingly, all tested MMPs, with the exception of MMP-14, exhibited very similar kinetic parameters for hydrolysis of either substrate (Table 3). In the case of MMP-14, the rate of hydrolysis of both substrates was significantly lower than for the rest of the tested MMPs, which made kinetic evaluation unfeasible.
TABLE 3.
Kinetic parameters for MMP hydrolysis of glycosylated and non-glycosylated substrates
| Enzyme |
kcat/Km |
kcat |
Km |
|||
|---|---|---|---|---|---|---|
| Glycosylated | Non-glycosylated | Glycosylated | Non-glycosylated | Glycosylated | Non-glycosylated | |
| m−1 s−1 | s−1 | μm | ||||
| MMP-1 | 1.1 ± 0.1 × 103 | 1.6 ± 0.0 × 103 | 0.03 ± 0.02 | 0.02 ± 0.00 | 12 ± 0.2 | 12 ± 0.0 |
| MMP-2 | 0.4 ± 0.7 × 104 | 0.7 ± 0.0 × 104 | 0.02 ± 0.01 | 0.02 ± 0.01 | 5.9 ± 0.6 | 2.8 ± 0.6 |
| MMP-8 | 1.1 ± 0.1 × 104 | 1.3 ± 0.2 × 104 | 0.2 ± 0.03 | 0.3 ± 0.2 | 17 ± 2.5 | 13 ± 1.3 |
| MMP-9 | 1.9 ± 0.2 × 103 | 1.2 ± 0.1 × 103 | 0.01 ± 0.00 | 0.01 ± 0.00 | 7.4 ± 2.1 | 7.3 ± 2.7 |
| MMP-13 | 1.7 ± 0.2 × 104 | 1.7 ± 0.0 × 104 | 0.1 ± 0.04 | 0.07 ± 0.02 | 6.8 ± 3 | 4.4 ± 0.9 |
| MMP-14 | NDa | ND | ND | ND | ND | ND |
ND, not determined.
ADAM10 and -17 comprise a separate branch of the ADAM phylogenetic tree (9). They share 39% sequence similarity, but despite this low homology, it has proved difficult to design an isoform-selective inhibitor of ADAM17 (8). This situation is partially complicated by the absence of a crystal structure of the ADAM10 catalytic domain, which would allow the evaluation of structural differences between ADAM10 and -17.
Our kinetic data suggest differences in ADAM10 and -17 substrate binding site(s) corresponding to the Ser residue in position P4′ of the TNFα-based substrate. Existing structural and modeling studies reveal the presence of a large S3′ cleft in the ADAM17 structure that can potentially accommodate a bulky disaccharide (βGal-1,3-αGalNAc) (59–61). Alignment of ADAM10 and -17 sequences shows that residues forming the ADAM17 S3′ pocket are not conserved between ADAM10 and -17 (189EADLV versus 189VSHIT for ADAM17 and -10, respectively) (60). Interestingly, the ADAM10 I192L/T193V double mutant cleavage specificity profile was indistinguishable from that of ADAM17 WT (38).
Summary of Screening of Torrey Pines Institute for Molecular Studies (TPIMS) Library (Fig. 2)
FIGURE 2.
Summary of screening of TPIMS library for selective inhibitors of ADAM17.
Primary screen (Scaffold Ranking) was conducted using 27 representative mixture libraries. Library 1344 was chosen for further studies (deconvolution by positional scanning). 120 mixture samples representative of library 1344 were screened first in single dose, and the 18 most active and selective mixture samples were screened in dose-response experiments against ADAM10 and -17 and MMP-8 and -14. Based on the most selective and active moieties in each defined R position, 36 individual compounds were synthesized and screened in dose-response experiments against ADAM10 and -17 and MMP-8 and -14.
Screening for Inhibitors of ADAM10 and -17 Using Glycosylated and Non-glycosylated Substrates
Differences in kinetic parameters for ADAM17 hydrolysis of glycosylated and non-glycosylated substrates suggested that the carbohydrate moiety potentially interacts with hypothetical exosite and, therefore, could be exploited for inhibitor discovery. In order to test this hypothesis, high throughput screening assays were developed for ADAM10 and -17 using both glycosylated and non-glycosylated substrates. A pilot screen (termed the “scaffold ranking screen”) was conducted for 27 mixture libraries representative of the TPIMS proprietary drug-like collection (1, 2). Parallel screens of libraries against ADAM10 and -17 yielded three libraries (Fig. 3A, compounds 1172, 1174, and 1347) preferentially inhibiting ADAM10 in the glycosylated substrate assay and one library (Fig. 3B, compound 1344) preferentially inhibiting ADAM17 in the glycosylated substrate assay, suggesting the possibility of the presence of exosite inhibitors in the above mentioned library mixtures. Most interestingly, the same libraries were not active against ADAM10 and -17 in the non-glycosylated substrate assays and, therefore, would have been discarded from further studies if just the conventional active site-only substrate was utilized. Although not highly active against either target (maximum inhibition ≤50%), these mixtures exhibited good selectivity, which, in combination with preferential inhibition of glycosylated substrate hydrolysis, suggests that these mixtures potentially inhibit ADAM10 and -17 via novel mechanisms. In order to confirm the selective profile of library 1344 (Fig. 3C), a structure-activity relationship study was conducted using a positional scan approach. A positional scan is a screen of a systematically formatted collection of compounds that allows for the rapid identification of the active functionalities around a core scaffold (1, 62).
FIGURE 3.
Results of the pilot “scaffold ranking” screen of TPIMS drug-like library against ADAM10 and 17. Shown is an ADAM10 (A) and ADAM17 (B) screen using glycosylated (checked bars) and non-glycosylated substrate (small checked bars). The arrows indicate libraries containing potential exosite inhibitors of ADAM10 and 17. All assays were performed in triplicate. Activity and selectivity of all libraries were confirmed in reversed-phase HPLC-based assays. (C), basic scaffold of library 1344.
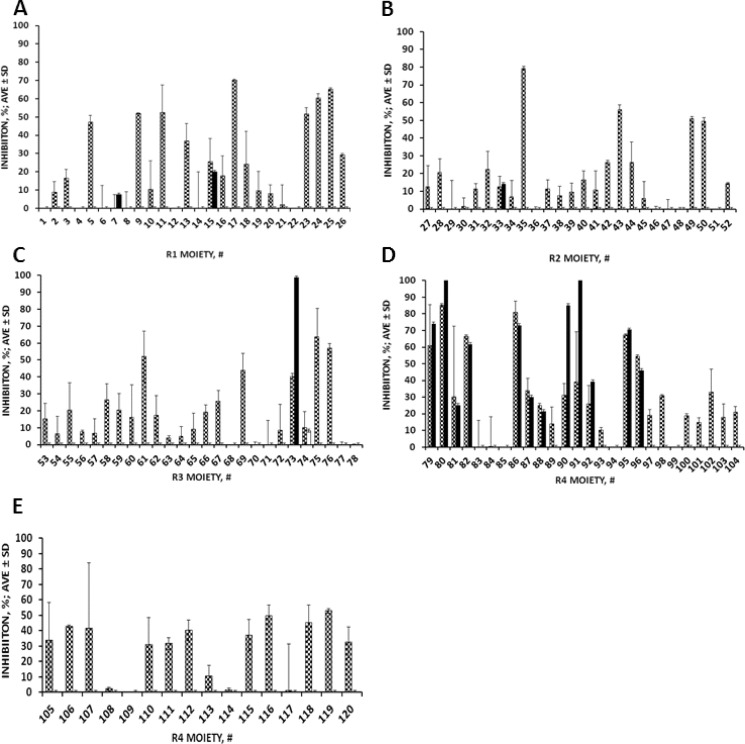
The basic scaffold of library 1344 (Fig. 3C), composed of 738,192 (26 × 26 × 26 × 42) members, has four sites of diversity (R1, R2, R3, and R4) and, therefore, is made up of four separate sublibraries, each having a single defined position (R) and three mixture positions (X). Screening the four sets of mixtures, totaling 120 mixtures (26 + 26 + 26 + 42) against ADAM10 and -17 can provide information about the groups most important for activity and selectivity in each R position in library 1344 (1). Most of functional groups in positions R1, R2, and R3 yielded libraries that either selectively inhibited ADAM17 or were inactive (Fig. 4, A–C). Functionalities in position R4 appear to influence the selectivity for ADAM17 to the greatest degree (Fig. 4, D and E). Approximately 50% of all substitutions tested in this position were inhibiting both enzymes to an equal extent.
FIGURE 4.
Results of the positional scan analysis of library 1344 against ADAM10 and -17. Positional scan of R1 (A), R2 (B), R3 (C), and R4 (D and E) defined moieties against ADAM10 (black bars) and ADAM17 (checked bars) using glycosylated substrate.
We chose the library samples that were the most selective for and active against ADAM17 for each R position (total of 18) to be studied in dose-response assays against ADAM10, ADAM17, MMP-8, and MMP-14. Additionally, we also chose R4-80, which inhibited both ADAM10 and -17 equipotently (Fig. 4D), to act as a broad spectrum in-screen control. These samples exhibited 100–200 nm IC50 values against ADAM17 while sparing the three other metalloproteases (Fig. 5). To our knowledge, there are no publicly available selective ADAM17 inhibitors that spare ADAM10. In addition, these samples had little or no activity against MMP-8 and MMP-14, which are important anti-targets in skin and breast cancer (63–65). In contrast, sample R4-80 exhibited a broad spectrum inhibition profile with IC50 values in the 100 nm range for all four metalloproteases tested (Fig. 5E), suggesting the importance of position R4 for selectivity among the metalloproteases. We used actinonin and MMP-9/-13 inhibitor as pharmacological assay controls for MMP-8 and MMP-14 and ADAM10 and -17, respectively (Fig. 5F). Both compounds are well known broad spectrum metalloprotease inhibitors (39, 40, 66, 67) that act via binding of the active site zinc. Based on the dose-response experiments with mixture libraries, we synthesized 36 individual compounds (2 R1 × 3 R2 × 2 R3 × 3 R4) containing functional groups for each defined R position that exhibited the most selectivity and potency toward ADAM17 (Table 4) and tested them against the target enzyme (ADAM17) and counter targets (ADAM10, MMP-8, and MMP-14) in a dose-response experiment. Only one of the 36 tested individual compounds (Tables 5 and 6) reached 25% inhibition at the highest tested concentration (40 μm) in the counter target assays (Table 5, sample 15), suggesting a highly selective nature of these compounds. The most active of the tested compounds had low micromolar potency against ADAM17, whereas the mixture libraries had IC50 values in 100 nm range.
FIGURE 5.
Results of dose-response study of most ADAM17-selective and potent mixture samples for each defined R position against metalloprotease panel. Shown are test libraries (A–E) and pharmacological assay controls (F) against ADAM10 (□), ADAM17 (♦), MMP-8 (♢), and MMP-14 (○). Structures of functional groups present in the defined R position for each sublibrary are shown as insets. Structures of MMP-9/-13 inhibitor (G) and actinonin (H) are shown. Note that sample R2-35 (see Fig. 5B) exhibits inhibition of MMP-14 only at the highest dose tested (1 μm).
TABLE 4.
Most selective and potent functionalities against ADAM17 derived from positional scan of library 1344
| Sample no. | Moiety in defined position |
|||
|---|---|---|---|---|
| R1 | R2 | R3 | R4 | |
| 25 | (S)-4-Hydroxybenzyl | Mix | Mix | Mix |
| 24 | (S)-Propyl | Mix | Mix | Mix |
| 35 | Mix | (S)-4-Hydroxybenzyl | Mix | Mix |
| 43 | Mix | (R)-4-Hydroxybenzyl | Mix | Mix |
| 49 | Mix | (S)-2-Naphthylmethyl | Mix | Mix |
| 61 | Mix | Mix | (S)-4-Hydroxybenzyl | Mix |
| 69 | Mix | Mix | (R)-4-Hydroxybenzyl | Mix |
| 80 | Mix | Mix | Mix | 2-Phenylbutyl |
| 119 | Mix | Mix | Mix | 2-Adamantan-1-yl-ethyl |
| 106 | Mix | Mix | Mix | Cyclopentyl-methyl |
TABLE 5.
SAR study results of individual compounds synthesized based on positional scan of 1344
Percentage inhibition data reported as a mean of three experiments ± S.D. μm IC50 values for the inhibition of ADAM17 are in parenthesis.
TABLE 6.
SAR study results of individual compounds synthesized based on positional scan of 1344
Percentage inhibition data reported as a mean of three experiments ± S.D. μm IC50 values for the inhibition of ADAM17 are in parenthesis.
According to the harmonic mean model (68), although mixture library 1344 has ∼1000 individual constituents, the activity of the mixture can be attributed to the presence of one or few very potent compounds. Although we chose the functionalities for each R position that exhibited the most potency against ADAM17, it is entirely possible that the combination of these functionalities did not recapture the structure of the most potent individual constituent from each mixture library. Our future studies will therefore focus on the systematic iterative deconvolution of each position at a time.
SAR Analysis
Based on the positional scan of library 1344, position R1 appears to be the most tolerant to the type of functionality present (Fig. 4A). The most active moieties are both aromatic (compound 5, (S)-phenyl; compound 9, (R)-2-naphtylmethyl; compound 25, (S)-4-hydroxybenzyl) and aliphatic (compound 13, (S)-isopropyl; compound 17, (R)-methyl; compound 23, (R)-cyclohexyl). Position R2 has a strong preference for aromatic groups (compounds 35 and 43, (S)- and (R)-4-hydroxybenzyl; compounds 49 and 50, (S)- and (R)-naphthylmethyl) (Fig. 4B). Interestingly, (S)-4-hydroxybenzyl (compound 35) was preferred over (R)-4-hydroxybenzyl (compound 43), suggesting the importance of proper orientation of ligand in this binding site. Position R3 exhibited preferences similar to R2 (compounds 61 and 69, (S)- and (R)-4-hydroxybenzyl; compounds 75 and 76, (S)- and (R)- naphthylmethyl), suggesting the similarities between R2 and R3 binding sites within ADAM17 (Fig. 4C). The most potent substitutions in position R4 were aromatic moieties (compounds 79, 80, 82, 86, 90, 91, 95, and 96; Fig. 4D), which, however, inhibited ADAM10 and -17 almost equipotently. In striking contrast, the most selective and potent ADAM17 substitutions were bulky cyclic aliphatic residues (compound 119, 2-adamantan-1-yl-ethyl; compound 116, 4-tert-butyl-cyclohexyl-methyl; compound 106, cyclopentyl-methyl).
Individual compounds synthesized on the basis of this positional scan analysis exhibited good selectivity for ADAM17. Both aromatic ((S)-4-hydroxybenzyl) and aliphatic ((S)-isopropyl) groups preserved ADAM17 selectivity when present in R1 (Fig. 6, A and B, compounds 9 and 27). Substitution of the hydroxybenzyl of compound 9 for the bulkier naphthylmethyl in R2 yielded more potent compound 15 without the loss of selectivity. Interestingly, a similar substitution in compound 27 did not lead to the significant changes in activity or selectivity (Fig. 6, B and D, compounds 27 and 34). Similarly, substitution of the R4 2-adamantan-1-yl-ethyl group found in compounds 15 and 34 for cyclopentyl-methyl (compounds 17 and 36) resulted in unchanged potency and selectivity (Fig. 6, C–F).
FIGURE 6.
Results of dose response study of most ADAM17 selective and potent individual compounds. ADAM10 (□), ADAM17 (♦), MMP-8 (♢), and MMP-14 (○). Structures of individual compounds (compounds 9 (A), 27 (B), 15 (C), 34 (D), 17 (E), and 36 (F)) are shown as insets.
Overall, individual compounds derived from the positional scan exhibited micromolar potency and good selectivity toward ADAM17. To our knowledge, there are no reports of ADAM17 inhibitors that spare ADAM10, MMP-8, and MMP-14. One of the most promising ADAM inhibitors that is currently in clinical trial for breast cancer, INCB7839 (Incyte Corp., Wilmington, DE), inhibits multiple metalloproteases (69), including equipotent low nanomolar range activity against both ADAM10 and -17.
Inhibition Kinetics
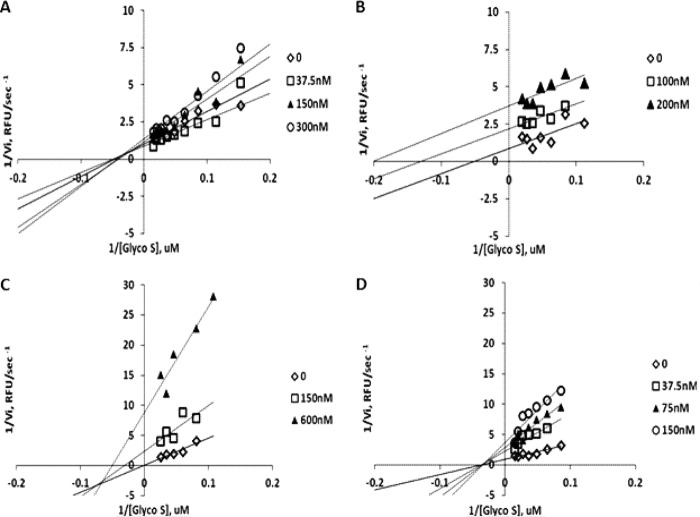
To interrogate the mechanism of inhibition for the most selective and potent libraries, inhibition kinetic studies were performed with samples R1-24, R2-35, R3-61, and R4-119. Non-linear regression analysis suggested pure non-competitive inhibition mechanisms for samples R1-24, R3-61, and R4-119 (Ki/Ki′ = 1.1 ± 1.2, 0.91 ± 2.5, and 0.79 ± 0.52, respectively), and an uncompetitive mechanism for R2-35 (Ki/Ki′ = 0.1 ± 0.3). Additional evaluation of inhibition mechanisms by the model comparison routine of GraphPad Prism software indicated that these are indeed preferred inhibition models. Examination of the kinetic data by a linearized Lineweaver-Burk graphic approach confirmed the pure non-competitive inhibition modality for R1-24 and R4-119 (Fig. 7, A and D) and uncompetitive modality for R2-35 (Fig. 7B). In the case of R3-61, the Lineweaver-Burk plot is suggestive of mixed inhibition (Fig. 7C) with elements of uncompetitive inhibition.
FIGURE 7.
Lineweaver-Burk plots of glycosylated substrate hydrolysis by ADAM17 in the presence of representative members of library 1344. A, compound 24; B, compound 35; C, compound 61; D, compound 119.
Most ADAM17 inhibitors described in the literature act via zinc binding due to the presence of zinc-binding moieties (e.g. hydroxamates, phosphinates) in their structures (5, 8, 61, 70). The prominent feature of the basic scaffold of library 1344 (Fig. 3C) is two piperazine-2,3-dione moieties. Piperazine- and diketo-piperazine-based MMP and ADAM inhibitors have been described in the literature; however, they also have other zinc-binding groups present and as a consequence have broad inhibition profiles (71, 72). The piperazine group was shown to interact with catalytic zinc of farnesyl protein transferase via the nitrogen atom (73). Piperazine derivatives have not been co-crystallized with ADAM17; therefore, it is unknown whether piperazine moieties can contribute to the interactions with zinc of an ADAM17 active site. However, the prevalence of non- and uncompetitive inhibition modalities among selective ADAM17 inhibitors suggests that this chemotype acts via a non-zinc-binding mechanism, possibly by binding outside of the ADAM17 active site. In order to investigate this possibility, we performed dual inhibition kinetics using N-hydroxyacetamide (AHA) (Fig. 8A), a known zinc binder and competitive millimolar range inhibitor of many metalloproteases, in combination with inhibitor compound 15, following a methodology described previously (74). AHA was used in the concentration range of 0–2.5 mm, in combination with compound 15 at concentrations between 0 and 7.5 μm. When initial velocities from this experiment were organized in a Yonetani-Theorell plot, they formed a series of intersecting lines of best fit (Fig. 8B). In Yonetani-Theorell plots, the intersecting lines suggest simultaneous (i.e. mutually non-exclusive) binding of both inhibitors to the enzyme (49). Because AHA is known to bind to zinc, compound 15 most likely acts via a non-zinc-binding mechanism and, potentially, outside of the active site and beyond the catalytic domain, as suggested by a recent report (33). Single inhibition kinetics of compound 15 suggest non-competitive inhibition (Ki/Ki′ = 0.5 ± 0.6; Fig. 8C) consistent with mutually non-exclusive AHA binding. We investigated the possibility of compound 15 binding outside of the catalytic domain by performing dose-response experiments with compound 15 against ADAM17 ectodomain (ECD) and catalytic domain-only constructs. AHA and MMP-9/-13 inhibitor were utilized as controls. Both controls exhibited nearly identical IC50 values for inhibition of either construct (AHA IC50 = 1.5 ± 0.2 mm versus 1.6 ± 0.2 mm for ADAM17 ECD and catalytic domain-only, respectively; MMP-9/-13 inhibitor IC50 = 0.3 ± 0.03 nm versus 0.5 ± 0.04 nm for ADAM17 ECD and catalytic domain-only (CD), respectively; Fig. 8, D and E), whereas compound 15 inhibited ADAM17 ECD 10-fold more potently (compound 15 IC50 = 4.2 ± 0.4 μm versus 47 ± 4 μm for ADAM17 ECD and catalytic domain-only (CD), respectively; Fig. 8F). This suggests certain cooperativity between catalytic and non-catalytic domains of ADAM17 in the binding of compound 15. One possibility is that compound 15 can bind across domains due to the spatial proximity of the non-catalytic and catalytic domains, as described in the case of the inhibitory antibody reported by Tape et al. (33). The existence of a binding pocket within the catalytic domain capable of accommodating compound 15, whose size is affected by the presence of a non-catalytic domain, is yet another possibility. The small size of compound 15, as compared with an antibody, makes the latter model more likely. Further structural studies are needed to ascertain the exact ADAM17 binding site of these novel, selective ADAM17 inhibitors.
FIGURE 8.
Characterization of mechanism of inhibition of ADAM17 catalytic domain and ectodomain by compound 15. A, structure of AHA; B, Yonetani-Theorell plot of glycosylated substrate hydrolysis by ADAM17 in the presence of AHA and compound 15. Note the non-parallel lines of best fit, indicating mutually non-exclusive binding by two inhibitors. C, Lineweaver-Burk plot of glycosylated substrate hydrolysis by ADAM17 in the presence of compound 15. Shown is a dose-response study of inhibition of ADAM17 catalytic domain and ectodomain by AHA (D), MMP-9/-13 inhibitor (E), and compound 15 (F). CD, catalytic domain-only.
Significance
Selective inhibitors of ADAM proteases can be useful research and therapeutic tools to study the roles of individual ADAM isoforms in diseases and normalcy. ADAM inhibitors reported to date act via active site zinc chelation and as a result inhibit multiple related zinc proteases (MMPs, ADAMs, and ADAMTSs). Here we report the discovery of a novel class of ADAM17-selective inhibitors that act via a non-zinc-binding mechanism.
This discovery was enabled by the development of a glycosylated, potentially exosite-binding substrate. Differences in the abilities of ADAM proteases to accommodate substrate carbohydrates point to the existence of unique exosites within ADAM structures that can be explored for discovery of isoform-selective non-zinc-binding inhibitors. These inhibitors were also able to inhibit ADAM17 hydrolysis of various non-glycosylated substrates (data not shown), suggesting their potential usefulness for a variety of applications that require ADAM17 inhibition.
Most cell surface proteins are glycosylated in one or more positions. Types of carbohydrate moieties present on the same cell surface protein can vary in disease and normalcy and thus can potentially be used as disease biomarkers. Results presented here suggest that differential glycosylation of substrates affects not only the rate of their hydrolysis but also the cleavage sites by different enzymes, which may lead to a release of different neoepitopes in disease and normalcy.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R03DA033985-01 (to D. M.), CA098799 (to G. B. F.), and 1R01DA031370 (to R. A. H.). This work was also supported by James and Esther King Biomedical Research Program Grant 2KN05 (to D. M.), by the Multiple Sclerosis National Research Institute (to G. B. F.), and by the State of Florida, Executive Office of the Governor's Office of Tourism, Trade, and Economic Development.

This article contains supplemental Materials and Methods.
- ADAM
- a disintegrin and metalloprotease
- ADAMTS
- ADAM with thrombospondin motifs
- MMP
- metalloprotease
- TFE
- trifluoroethanol
- TPIMS
- Torrey Pines Institute for Molecular Studies
- AHA
- N-hydroxyacetamide
- ECD
- ectodomain
- SAR
- structure-activity relationship.
REFERENCES
- 1. Houghten R. A., Pinilla C., Giulianotti M. A., Appel J. R., Dooley C. T., Nefzi A., Ostresh J. M., Yu Y., Maggiora G. M., Medina-Franco J. L., Brunner D., Schneider J. (2008) Strategies for the use of mixture-based synthetic combinatorial libraries. Scaffold ranking, direct testing in vivo, and enhanced deconvolution by computational methods. J. Comb. Chem. 10, 3–19 [DOI] [PubMed] [Google Scholar]
- 2. Pinilla C., Appel J. R., Borràs E., Houghten R. A. (2003) Advances in the use of synthetic combinatorial chemistry. Mixture-based libraries. Nat. Med. 9, 118–122 [DOI] [PubMed] [Google Scholar]
- 3. Moss M. L., Stoeck A., Yan W., Dempsey P. J. (2008) ADAM10 as a target for anti-cancer therapy. Curr. Pharm. Biotechnol. 9, 2–8 [DOI] [PubMed] [Google Scholar]
- 4. Kataoka H. (2009) EGFR ligands and their signaling scissors, ADAMs, as new molecular targets for anticancer treatments. J. Dermatol. Sci. 56, 148–153 [DOI] [PubMed] [Google Scholar]
- 5. Moss M. L., Sklair-Tavron L., Nudelman R. (2008) Drug insight. Tumor necrosis factor-converting enzyme as a pharmaceutical target for rheumatoid arthritis. Nat. Clin. Pract. Rheumatol. 4, 300–309 [DOI] [PubMed] [Google Scholar]
- 6. Asai M., Hattori C., Szabó B., Sasagawa N., Maruyama K., Tanuma S., Ishiura S. (2003) Putative function of ADAM9, ADAM10, and ADAM17 as APP α-secretase. Biochem. Biophys. Res. Commun. 301, 231–235 [DOI] [PubMed] [Google Scholar]
- 7. Kenny P. A., Bissell M. J. (2007) Targeting TACE-dependent EGFR ligand shedding in breast cancer. J. Clin. Invest. 117, 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Georgiadis D., Yiotakis A. (2008) Specific targeting of metzincin family members with small-molecule inhibitors. Progress toward a multifarious challenge. Bioorg. Med. Chem. 16, 8781–8794 [DOI] [PubMed] [Google Scholar]
- 9. Edwards D. R., Handsley M. M., Pennington C. J. (2008) The ADAM metalloproteinases. Mol. Aspects Med. 29, 258–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fingleton B. (2007) Matrix metalloproteinases as valid clinical targets. Curr. Pharm. Des. 13, 333–346 [DOI] [PubMed] [Google Scholar]
- 11. Dennis M. S., Eigenbrot C., Skelton N. J., Ultsch M. H., Santell L., Dwyer M. A., O'Connell M. P., Lazarus R. A. (2000) Peptide exosite inhibitors of factor VIIa as anticoagulants. Nature 404, 465–470 [DOI] [PubMed] [Google Scholar]
- 12. Roberge M., Peek M., Kirchhofer D., Dennis M. S., Lazarus R. A. (2002) Fusion of two distinct peptide exosite inhibitors of Factor VIIa. Biochem. J. 363, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roberge M., Santell L., Dennis M. S., Eigenbrot C., Dwyer M. A., Lazarus R. A. (2001) A novel exosite on coagulation factor VIIa and its molecular interactions with a new class of peptide inhibitors. Biochemistry 40, 9522–9531 [DOI] [PubMed] [Google Scholar]
- 14. Izaguirre G., Rezaie A. R., Olson S. T. (2009) Engineering functional antithrombin exosites in α1-proteinase inhibitor that specifically promote the inhibition of factor Xa and factor IXa. J. Biol. Chem. 284, 1550–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scheer J. M., Romanowski M. J., Wells J. A. (2006) A common allosteric site and mechanism in caspases. Proc. Natl. Acad. Sci. U.S.A. 103, 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hosseini M., Jiang L., Sørensen H. P., Jensen J. K., Christensen A., Fogh S., Yuan C., Andersen L. M., Huang M., Andreasen P. A., Jensen K. J. (2011) Elucidation of the contribution of active site and exosite interactions to affinity and specificity of peptidylic serine protease inhibitors, using non-natural arginine analogs. Mol. Pharmacol. 80, 585–597 [DOI] [PubMed] [Google Scholar]
- 17. Johnson A. R., Pavlovsky A. G., Ortwine D. F., Prior F., Man C. F., Bornemeier D. A., Banotai C. A., Mueller W. T., McConnell P., Yan C., Baragi V., Lesch C., Roark W. H., Wilson M., Datta K., Guzman R., Han H. K., Dyer R. D. (2007) Discovery and characterization of a novel inhibitor of matrix metalloprotease-13 that reduces cartilage damage in vivo without joint fibroplasia side effects. J. Biol. Chem. 282, 27781–27791 [DOI] [PubMed] [Google Scholar]
- 18. Engel C. K., Pirard B., Schimanski S., Kirsch R., Habermann J., Klingler O., Schlotte V., Weithmann K. U., Wendt K. U. (2005) Structural basis for the highly selective inhibition of MMP-13. Chem. Biol. 12, 181–189 [DOI] [PubMed] [Google Scholar]
- 19. Baragi V. M., Becher G., Bendele A. M., Biesinger R., Bluhm H., Boer J., Deng H., Dodd R., Essers M., Feuerstein T., Gallagher B. M., Jr., Gege C., Hochgurtel M., Hofmann M., Jaworski A., Jin L., Kiely A., Korniski B., Kroth H., Nix D., Nolte B., Piecha D., Powers T. S., Richter F., Schneider M., Steeneck C., Sucholeiki I., Taveras A., Timmermann A., Van Veldhuizen J., Weik J., Wu X., Xia B. (2009) A new class of potent matrix metalloproteinase 13 inhibitors for potential treatment of osteoarthritis. Evidence of histologic and clinical efficacy without musculoskeletal toxicity in rat models. Arthritis Rheum. 60, 2008–2018 [DOI] [PubMed] [Google Scholar]
- 20. Lauer-Fields J. L., Minond D., Chase P. S., Baillargeon P. E., Saldanha S. A., Stawikowska R., Hodder P., Fields G. B. (2009) High throughput screening of potentially selective MMP-13 exosite inhibitors utilizing a triple-helical FRET substrate. Bioorg. Med. Chem. 17, 990–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roth J., Minond D., Darout E., Liu Q., Lauer J., Hodder P., Fields G. B., Roush W. R. (2011) Identification of novel, exosite-binding matrix metalloproteinase-13 inhibitor scaffolds. Bioorg Med. Chem. Lett. 21, 7180–7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wittwer A. J., Hills R. L., Keith R. H., Munie G. E., Arner E. C., Anglin C. P., Malfait A. M., Tortorella M. D. (2007) Substrate-dependent inhibition kinetics of an active site-directed inhibitor of ADAMTS-4 (aggrecanase 1). Biochemistry 46, 6393–6401 [DOI] [PubMed] [Google Scholar]
- 23. Yegneswaran S., Tiefenbrunn T. K., Fernandez J. A., Dawson P. E. (2007) Manipulation of thrombin exosite I, by ligand-directed covalent modification. J. Thromb. Haemost. 5, 2062–2069 [DOI] [PubMed] [Google Scholar]
- 24. Sela-Passwell N., Rosenblum G., Shoham T., Sagi I. (2010) Structural and functional bases for allosteric control of MMP activities. Can it pave the path for selective inhibition? Biochim. Biophys. Acta 1803, 29–38 [DOI] [PubMed] [Google Scholar]
- 25. Rzychon M., Chmiel D., Stec-Niemczyk J. (2004) Modes of inhibition of cysteine proteases. Acta Biochim. Pol. 51, 861–873 [PubMed] [Google Scholar]
- 26. Ishii R., Minagawa A., Takaku H., Takagi M., Nashimoto M., Yokoyama S. (2007) The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prudent R., Sautel C. F., Cochet C. (2010) Structure-based discovery of small molecules targeting different surfaces of protein-kinase CK2. Biochim. Biophys. Acta 1804, 493–498 [DOI] [PubMed] [Google Scholar]
- 28. Pantke M. M., Reif A., Valtschanoff J. G., Shutenko Z., Frey A., Weinberg R. J., Pfleiderer W., Schmidt H. H. (2001) Pterin interactions with distinct reductase activities of NO synthase. Biochem. J. 356, 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vogel A., Schilling O., Späth B., Marchfelder A. (2005) The tRNase Z family of proteins. Physiological functions, substrate specificity, and structural properties. Biol. Chem. 386, 1253–1264 [DOI] [PubMed] [Google Scholar]
- 30. Gurard-Levin Z. A., Mrksich M. (2008) The activity of HDAC8 depends on local and distal sequences of its peptide substrates. Biochemistry 47, 6242–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takeda S., Igarashi T., Mori H. (2007) Crystal structure of RVV-X. An example of evolutionary gain of specificity by ADAM proteinases. FEBS Lett. 581, 5859–5864 [DOI] [PubMed] [Google Scholar]
- 32. Hall T., Pegg L. E., Pauley A. M., Fischer H. D., Tomasselli A. G., Zack M. D. (2009) ADAM8 substrate specificity. Influence of pH on preprocessing and proteoglycan degradation. Arch. Biochem. Biophys. 491, 106–111 [DOI] [PubMed] [Google Scholar]
- 33. Tape C. J., Willems S. H., Dombernowsky S. L., Stanley P. L., Fogarasi M., Ouwehand W., McCafferty J., Murphy G. (2011) Cross-domain inhibition of TACE ectodomain. Proc. Natl. Acad. Sci. U.S.A. 108, 5578–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lauer-Fields J. L., Minond D., Sritharan T., Kashiwagi M., Nagase H., Fields G. B. (2007) Substrate conformation modulates aggrecanase (ADAMTS-4) affinity and sequence specificity. Suggestion of a common topological specificity for functionally diverse proteases. J. Biol. Chem. 282, 142–150 [DOI] [PubMed] [Google Scholar]
- 35. Minond D., Lauer-Fields J. L., Cudic M., Overall C. M., Pei D., Brew K., Moss M. L., Fields G. B. (2007) Differentiation of secreted and membrane-type matrix metalloproteinase activities based on substitutions and interruptions of triple-helical sequences. Biochemistry 46, 3724–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Minond D., Lauer-Fields J. L., Cudic M., Overall C. M., Pei D., Brew K., Visse R., Nagase H., Fields G. B. (2006) The roles of substrate thermal stability and P2 and P1′ subsite identity on matrix metalloproteinase triple-helical peptidase activity and collagen specificity. J. Biol. Chem. 281, 38302–38313 [DOI] [PubMed] [Google Scholar]
- 37. Minond D., Lauer-Fields J. L., Nagase H., Fields G. B. (2004) Matrix metalloproteinase triple-helical peptidase activities are differentially regulated by substrate stability. Biochemistry 43, 11474–11481 [DOI] [PubMed] [Google Scholar]
- 38. Caescu C. I., Jeschke G. R., Turk B. E. (2009) Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem. J. 424, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moss M. L., Rasmussen F. H. (2007) Fluorescent substrates for the proteinases ADAM17, ADAM10, ADAM8, and ADAM12 useful for high-throughput inhibitor screening. Anal. Biochem. 366, 144–148 [DOI] [PubMed] [Google Scholar]
- 40. Moss M. L., Rasmussen F. H., Nudelman R., Dempsey P. J., Williams J. (2010) Fluorescent substrates useful as high-throughput screening tools for ADAM9. Comb. Chem. High Throughput Screen. 13, 358–365 [DOI] [PubMed] [Google Scholar]
- 41. Imperiali B., Rickert K. W. (1995) Conformational implications of asparagine-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 92, 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Otvos L., Jr., Cudic M. (2003) Conformation of glycopeptides. Mini Rev. Med. Chem. 3, 703–711 [DOI] [PubMed] [Google Scholar]
- 43. Bann J. G., Peyton D. H., Bächinger H. P. (2000) Sweet is stable: glycosylation stabilizes collagen. FEBS Lett. 473, 237–240 [DOI] [PubMed] [Google Scholar]
- 44. Tagashira M., Iijima H., Isogai Y., Hori M., Takamatsu S., Fujibayashi Y., Yoshizawa-Kumagaye K., Isaka S., Nakajima K., Yamamoto T., Teshima T., Toma K. (2001) Site-dependent effect of O-glycosylation on the conformation and biological activity of calcitonin. Biochemistry 40, 11090–11095 [DOI] [PubMed] [Google Scholar]
- 45. Meldal M., Bock K. (1994) A general approach to the synthesis of O- and N-linked glycopeptides. Glycoconj. J. 11, 59–63 [DOI] [PubMed] [Google Scholar]
- 46. Takakura-Yamamoto R., Yamamoto S., Fukuda S., Kurimoto M. (1996) O-Glycosylated species of natural human tumor necrosis factor-α. Eur. J. Biochem. 235, 431–437 [DOI] [PubMed] [Google Scholar]
- 47. Bringman T. S., Lindquist P. B., Derynck R. (1987) Different transforming growth factor-α species are derived from a glycosylated and palmitoylated transmembrane precursor. Cell 48, 429–440 [DOI] [PubMed] [Google Scholar]
- 48. Wollscheid B., Bausch-Fluck D., Henderson C., O'Brien R., Bibel M., Schiess R., Aebersold R., Watts J. D. (2009) Mass spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 27, 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martinez-Irujo J. J., Villahermosa M. L., Mercapide J., Cabodevilla J. F., Santiago E. (1998) Analysis of the combined effect of two linear inhibitors on a single enzyme. Biochem. J. 329, 689–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Copeland R. (2000) Enzymes, 2nd Ed., pp. 289–290, Wiley-VCH, New York [Google Scholar]
- 51. Chalaris A., Garbers C., Rabe B., Rose-John S., Scheller J. (2011) The soluble interleukin 6 receptor. Generation and role in inflammation and cancer. Eur. J. Cell Biol. 90, 484–494 [DOI] [PubMed] [Google Scholar]
- 52. Müllberg J., Oberthür W., Lottspeich F., Mehl E., Dittrich E., Graeve L., Heinrich P. C., Rose-John S. (1994) The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. J. Immunol. 152, 4958–4968 [PubMed] [Google Scholar]
- 53. Cole A. R., Hall N. E., Treutlein H. R., Eddes J. S., Reid G. E., Moritz R. L., Simpson R. J. (1999) Disulfide bond structure and N-glycosylation sites of the extracellular domain of the human interleukin-6 receptor. J. Biol. Chem. 274, 7207–7215 [DOI] [PubMed] [Google Scholar]
- 54. Wormald M. R., Petrescu A. J., Pao Y. L., Glithero A., Elliott T., Dwek R. A. (2002) Conformational studies of oligosaccharides and glycopeptides. Complementarity of NMR, x-ray crystallography, and molecular modeling. Chem. Rev. 102, 371–386 [DOI] [PubMed] [Google Scholar]
- 55. McManus A. M., Otvos L., Jr., Hoffmann R., Craik D. J. (1999) Conformational studies by NMR of the antimicrobial peptide, drosocin, and its non-glycosylated derivative. Effects of glycosylation on solution conformation. Biochemistry 38, 705–714 [DOI] [PubMed] [Google Scholar]
- 56. Falenski J. A., Gerling U. I., Koksch B. (2010) Multiple glycosylation of de novo designed α-helical coiled coil peptides. Bioorg. Med. Chem. 18, 3703–3706 [DOI] [PubMed] [Google Scholar]
- 57. Zhao Y. H., Abraham M. H., Zissimos A. M. (2003) Fast calculation of van der Waals volume as a sum of atomic and bond contributions and its application to drug compounds. J. Org. Chem. 68, 7368–7373 [DOI] [PubMed] [Google Scholar]
- 58. Patel I. R., Attur M. G., Patel R. N., Stuchin S. A., Abagyan R. A., Abramson S. B., Amin A. R. (1998) TNF-α convertase enzyme from human arthritis-affected cartilage. Isolation of cDNA by differential display, expression of the active enzyme, and regulation of TNF-α. J. Immunol. 160, 4570–4579 [PubMed] [Google Scholar]
- 59. Huang A., Joseph-McCarthy D., Lovering F., Sun L., Wang W., Xu W., Zhu Y., Cui J., Zhang Y., Levin J. I. (2007) Structure-based design of TACE selective inhibitors. Manipulations in the S1′–S3′ pocket. Bioorg. Med. Chem. 15, 6170–6181 [DOI] [PubMed] [Google Scholar]
- 60. Healy E. F., Romano P., Mejia M., Lindfors G., 3rd. (2010) Acetylenic inhibitors of ADAM10 and ADAM17. In silico analysis of potency and selectivity. J. Mol. Graph. Model. 29, 436–442 [DOI] [PubMed] [Google Scholar]
- 61. Bahia M. S., Silakari O. (2010) Tumor necrosis factor α-converting enzyme. An encouraging target for various inflammatory disorders. Chem. Biol. Drug Des. 75, 415–443 [DOI] [PubMed] [Google Scholar]
- 62. Houghten R. A., Pinilla C., Appel J. R., Blondelle S. E., Dooley C. T., Eichler J., Nefzi A., Ostresh J. M. (1999) Mixture-based synthetic combinatorial libraries. J. Med. Chem. 42, 3743–3778 [DOI] [PubMed] [Google Scholar]
- 63. Balbín M., Fueyo A., Tester A. M., Pendás A. M., Pitiot A. S., Astudillo A., Overall C. M., Shapiro S. D., López-Otin C. (2003) Loss of collagenase-2 confers increased skin tumor susceptibility to male mice. Nat. Genet. 35, 252–257 [DOI] [PubMed] [Google Scholar]
- 64. Palavalli L. H., Prickett T. D., Wunderlich J. R., Wei X., Burrell A. S., Porter-Gill P., Davis S., Wang C., Cronin J. C., Agrawal N. S., Lin J. C., Westbroek W., Hoogstraten-Miller S., Molinolo A. A., Fetsch P., Filie A. C., O'Connell M. P., Banister C. E., Howard J. D., Buckhaults P., Weeraratna A. T., Brody L. C., Rosenberg S. A., Samuels Y. (2009) Analysis of the matrix metalloproteinase family reveals that MMP8 is often mutated in melanoma. Nat. Genet. 41, 518–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Szabova L., Chrysovergis K., Yamada S. S., Holmbeck K. (2008) MT1-MMP is required for efficient tumor dissemination in experimental metastatic disease. Oncogene 27, 3274–3281 [DOI] [PubMed] [Google Scholar]
- 66. Wang Z., Herzog C., Kaushal G. P., Gokden N., Mayeux P. R. (2011) Actinonin, a meprin A inhibitor, protects the renal microcirculation during sepsis. Shock 35, 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Becker-Pauly C., Bruns B. C., Damm O., Schütte A., Hammouti K., Burmester T., Stöcker W. (2009) News from an ancient world. Two novel astacin metalloproteases from the horseshoe crab. J. Mol. Biol. 385, 236–248 [DOI] [PubMed] [Google Scholar]
- 68. Santos R. G., Giulianotti M. A., Dooley C. T., Pinilla C., Appel J. R., Houghten R. A. (2011) Use and implications of the harmonic mean model on mixtures for basic research and drug discovery. ACS Comb. Sci. 13, 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Witters L., Scherle P., Friedman S., Fridman J., Caulder E., Newton R., Lipton A. (2008) Synergistic inhibition with a dual epidermal growth factor receptor/HER-2/neu tyrosine kinase inhibitor and a disintegrin and metalloprotease inhibitor. Cancer Res. 68, 7083–7089 [DOI] [PubMed] [Google Scholar]
- 70. Murumkar P. R., DasGupta S., Chandani S. R., Giridhar R., Yadav M. R. (2010) Novel TACE inhibitors in drug discovery. A review of patented compounds. Expert Opin. Ther. Pat. 20, 31–57 [DOI] [PubMed] [Google Scholar]
- 71. Letavic M. A., Barberia J. T., Carty T. J., Hardink J. R., Liras J., Lopresti-Morrow L. L., Mitchell P. G., Noe M. C., Reeves L. M., Snow S. L., Stam E. J., Sweeney F. J., Vaughn M. L., Yu C. H. (2003) Synthesis and biological activity of piperazine-based dual MMP-13 and TNF-α-converting enzyme inhibitors. Bioorg. Med. Chem. Lett. 13, 3243–3246 [DOI] [PubMed] [Google Scholar]
- 72. Szardenings A. K., Antonenko V., Campbell D. A., DeFrancisco N., Ida S., Shi L., Sharkov N., Tien D., Wang Y., Navre M. (1999) Identification of highly selective inhibitors of collagenase-1 from combinatorial libraries of diketopiperazines. J. Med. Chem. 42, 1348–1357 [DOI] [PubMed] [Google Scholar]
- 73. Njoroge F. G., Vibulbhan B., Pinto P., Strickland C., Bishop W. R., Nomeir A., Girijavallabhan V. (2006) Enhanced FTase activity achieved via piperazine interaction with catalytic zinc. Bioorg. Med. Chem. Lett. 16, 984–988 [DOI] [PubMed] [Google Scholar]
- 74. Gooljarsingh L. T., Lakdawala A., Coppo F., Luo L., Fields G. B., Tummino P. J., Gontarek R. R. (2008) Characterization of an exosite binding inhibitor of matrix metalloproteinase 13. Protein Sci. 17, 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]