Abstract
While nitrogen availability is known to limit primary production in large parts of the ocean, vitamin starvation amongst eukaryotic phytoplankton is becoming increasingly recognized as an oceanographically relevant phenomenon. Cobalamin (B12) and thiamine (B1) auxotrophy are widespread throughout eukaryotic phytoplankton, with over 50% of cultured isolates requiring B12 and 20% requiring B1. The frequency of vitamin auxotrophy in harmful algal bloom species is even higher. Instances of colimitation between nitrogen and B vitamins have been observed in marine environments, and interactions between these nutrients have been shown to impact phytoplankton species composition. This review surveys available data, including relevant gene expression patterns, to evaluate the potential for interactive effects of nitrogen and vitamin B12 and B1 starvation in eukaryotic phytoplankton. B12 plays essential roles in amino acid and one-carbon metabolism, while B1 is important for primary carbohydrate and amino acid metabolism and likely useful as an anti-oxidant. Here we will focus on three potential metabolic interconnections between vitamin, nitrogen, and sulfur metabolism that may have ramifications for the role of vitamin and nitrogen scarcities in driving ocean productivity and species composition. These include: (1) B12, B1, and N starvation impacts on osmolyte and antioxidant production, (2) B12 and B1 starvation impacts on polyamine biosynthesis, and (3) influence of B12 and B1 starvation on the diatom urea cycle and amino acid recycling through impacts on the citric acid cycle. We evaluate evidence for these interconnections and identify oceanographic contexts in which each may impact rates of primary production and phytoplankton community composition. Major implications include that B12 and B1 deprivation may impair the ability of phytoplankton to recover from nitrogen starvation and that changes in vitamin and nitrogen availability may synergistically impact harmful algal bloom formation.
Keywords: cobalamin, thiamine, S-adenosylmethionine, nitrogen, sulfur, urea cycle, microbial interactions, harmful algal blooms
Introduction
The rate, magnitude, and species composition of marine primary production has a profound influence of global carbon cycling and therefore climate. As a result, factors controlling the growth of marine primary producers are of considerable interest. While nitrogen and iron availability are often considered the primary bottom-up controls on short-term marine primary productivity, the importance of organic growth factors received considerable early attention (Cowey, 1956; Droop, 1957, 1962; Menzel and Spaeth, 1962; Provasoli, 1963; Gold, 1968; Carlucci and Silbernagel, 1969; Carlucci and Bowes, 1970; Swift and Taylor, 1972; Swift, 1981) and is the subject of renewed interest (e.g., Sañudo-Wilhelmy et al., 2012).
Recent developments in analytical techniques (Okbamichael and Sañudo-Wilhelmy, 2004; Sañudo-Wilhelmy et al., 2012), application of trace metal clean bioassay experiments (Panzeca et al., 2006; Sañudo-Wilhelmy et al., 2006; Bertrand et al., 2007; Gobler et al., 2007; Koch et al., 2011), and culture-based surveys of vitamin requirements (Croft et al., 2005; Tang et al., 2010) have identified B12 (cobalamin) and B1 (thiamine) as highly important growth factors for eukaryotic phytoplankton and suggest that these micronutrients have the potential to broadly influence marine productivity and species composition. Due to the fact that B12 and B1 both play numerous essential roles in cellular biochemistry, starvation for these nutrients has the potential to impact phytoplankton cellular metabolism through a range of mechanisms. The increase in available genome and transcriptome data for relevant organisms has opened doors for new modes of inquiry into the role of these micronutrients in phytoplankton metabolism as well as their potential for interaction with additional states of nutrient deprivation (Croft et al., 2006; Helliwell et al., 2011; Bertrand et al., 2012). Here we review available data to examine potential interactions between B12, B1, and nitrogen deprivation and sulfur metabolism in eukaryotic phytoplankton communities and provide insight into the potential implications of these interactions for phytoplankton evolutionary trajectories and biogeochemical cycling.
Cobalamin and thiamine
Production, demand, and biochemical function
Cobalamin, B12, is a cobalt-containing organometallic micronutrient that conducts elegant chemistry facilitated by the controlled reactivity of the axial Co-C bond in methyl and adenosylcobalamin (Schrauzer and Deutsch, 1969; Lexa and Savant, 1983; Drennan et al., 1994). The resulting reactivity provides the biochemical capacity for methylation and rearrangement reactions, where a hydrogen atom on one carbon constituent is exchanged for another functional group, typically a methyl, amine, or alcohol group. Cobalamin is believed to be produced only by select bacteria and archaea (Roth et al., 1996; Martens et al., 2002) and is required by humans and other metazoans, by an estimated half of all eukaryotic phytoplankton (Tang et al., 2010) and by some bacteria that are not able to synthesize it (Rodionov et al., 2003; Zhang et al., 2009). Vitamin B12 biosynthesis requires over 30 enzymatic steps and significant consumption of cellular energy, carbon, nitrogen, cobalt, zinc, and in some cases iron (Roth et al., 1996; Raux et al., 2000).
Vitamin B12 demand by eukaryotic phytoplankton is thought to arise from its role as a cofactor in the enzyme methionine synthase, which catalyzes the conversion of homocysteine and methyl-tetrahydrofolate to tetrahydrofolate and methionine (Table 1). The active form of B12 in methionine synthase is methylcobalamin. Algae that require B12 absolutely posses only the B12-dependant version of this enzyme (MetH), while those that do not have an absolute requirement have the ability to use an alternative B12 independent version (MetE) (Croft et al., 2005). Phylogenetic analysis of metE and metH coding sequences support a complex evolutionary history of metE gene gain and loss within eukaryotic organisms. In contrast, the phylogeny of metH is well resolved and apparently monophyletic in eukaryotes (Helliwell et al., 2011). These analyses suggest that absolute B12 requirements in eukaryotic algae have likely arisen as a result of multiple independent loses and acquisitions of metE from eukaryotic genomes. Indeed, under high B12 concentrations, metH is continually expressed by algal strains, whereas metE, if present, is repressed until B12 is depleted (Croft et al., 2005; Helliwell et al., 2011; Bertrand et al., 2012). These results suggest that B12 auxotrophy in eukaryotic algae arose as a function of variable B12 availability in the environment. This is supported by observations that the distribution of metE in eukaryotic phytoplankton does not follow phylogenetic lines. Importantly, there is strain level variability in whether or not phytoplankton exhibit an absolute requirement for B12 (Tang et al., 2010). In addition, B12 is a cofactor in the enzyme methylmalonyl coA mutase (mmcM), which is encoded in some but not all B12-requiring phytoplankton genomes (Table 1). mmcM's function in eukaryotic phytoplankton remains somewhat unclear, though it likely plays a role in the citric acid cycle as well as fatty acid and propionate metabolism. However, the presence of mmcM genes in phytoplankton genomes does not confer a B12 requirement under typical laboratory growth conditions (Croft et al., 2006).
Table 1.
Vitamin B12-related genes in sequenced marine eukaryotic phytoplankton genomes and select marine prokaryotic genomes.
| Auran | Phatr | Thaps | Psemu | Fracy | Emihu | Ostta | Ostlu | MicPu | Chlre | ChlNC | Syn 8102 | Pro 9313 | P. ubique | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MetH | 34875 | 23399 | 693 | 213031 | 207237 | 423073 | 16287 | 45056 | 148156 | 76715 | 36916 | SYNW 1238 | PMT0729 | x |
| MetE | x | 28056 | x | x | 228154 | x | x | x | x | 154307 | 141995 | x | x | x |
| MmcM | 26280 | 51830 | 33685 | 261420 | 273786 | 120906 417351 | x | x | x | x | 18280 | x | x | x |
| CBA1 | 63075 | 48322 | 11697 | 235642 | 241429, 246327, 273295, 269995 | x | x | x | x | x | x | x | x | x |
| RNR Class 2 B12 | x | x | x | x | x | x | x | x | x | x# | x | SYNW 1147 | PMT 0793 | x |
| RNR Class 1 Fe; small | 65685 59025 | 39306 17523 | 32555 8522 3367 | 67342 252139 | 268008 206256 | 470988 469622 200748 | 22908 8886 | 32923 39468 | 155636 174818 | 188785 144621 | 34102 10712 57791 | x | x | PB7211_302 |
| RNR Class 1 Fe; Large | 30730, 37557, 24558 | 42726, 45529 | 370, 268807 | 223844 319245 | 260490, 262570, 205957 | 449248, 212824 | 22667 | 48569 | 167892 | 185583 | 32953 | x | x | PU1002_00625 |
| B12 biosynthesis | No | No | No | No | No | No | No | No | No | No | No | Yes | Yes | No |
| B12 Aux. by genome | Yes | No | Yes | Yes | No | Yes | Yes | Yes | Yes | No | No | No | No | No |
| B12 Aux. by culture | Yes (Tang et al., 2010) | No (Droop, 1958) | Yes (Guillard and Ryther, 1962) | Yes (Tang et al., 2010)* | No (Helliwell et al., 2011) | See (Helliwell et al., 2011) | Yes (Helliwell et al., 2011) | Yes (Helliwell et al., 2011) | No (Provasoli and Carlucci, 1974) | No (Shihira and Krauss, 1965) |
The hypothesized vitamin requirements of each strain are also given, along with whether culture-based confirmation of auxotophic status is available. This table represents an expansion of information given in Croft et al., 2006 and Helliwell et al., 2011. No eukaryote is known to make vitamin B12; B12 auxotrophy in eukaryotic algae appears to depend on the presence or absence of B12-independent methionine synthase (Croft et al., 2005; Helliwell et al., 2011). Auran, Aureococcus anophagefferans; Phatr, Phaeodactylum tricornutum; Thaps, Thalassiosira pseudonanna; Psemu, Pseudo-nitzschia multiseries CLN-47; Fracyl, Fragilariopsis cylindrus; Chlre, Chlamydomonas reinhardtii (v4; filtered or best proteins); Emihu, Emiliania huxleyi; Ostta, Ostreococcus taurii; Ostlu, Ostreococcus lucimarinus V2 filtered model proteins; MicPu, Micromonas pusilla CCMP1545 c3.0, filtered model proteins; ChlNC, Chlorella sp. NC64A filtered proteins; Syn8102, Synechococcus sp. WH8102; Pro9313, Prochlorococcus marinus MIT 9313; P. ubique, Candidatus Pelagibacter ubique SAR11 HTCC1002.
Auxotrophy tested in culture of a different strain.
Has a protein with substantial sequence similarity but missing active site: (154521).
Thiamine, B1, is a cofactor required by all organisms and produced by many prokaryotes as well as by fungi, plants, and some eukaryotic phytoplankton (Webb et al., 2007). It is a sulfur-containing compound, produced though joining of a pyrimidine and a thiazole moiety, and is phosphorylated in its coenzyme form (thiamine diphosphate). In bacterial biosynthetic pathways, thiazole biosynthesis requires six distinct enzymatic steps and pyrimidine synthesis requires two (Rodionov et al., 2002; Jurgenson et al., 2009). While the bacterial thiamine biosynthesis pathway is well characterized, eukaryotic biosynthesis pathways remain poorly understood and appear to be distinct in plants and fungi (Jurgenson et al., 2009). Algal thiamine biosynthesis is even less well-characterized but likely conducted by some enzymes similar to bacterial thiamine biosynthesis genes and some enzymes similar to the yeast and plant pathways (Croft et al., 2006), though this remains to be conclusively demonstrated.
While thiamine was one of the first organic cofactors identified as important for algal growth, early work showed that there are some phytoplankton strains that produce thiamine de novo, and some that can scavenge and salvage either the thiazole or pyrimidine moieties from the environment in order to construct a functional cofactor (Droop, 1958; Provasoli and Carlucci, 1974). Preliminary inquiry into eukaryotic phytoplankton genomes conducted via identification of coding sequences similar to those encoding known bacterial, fungi, and plant thiamine biosynthesis enzymes suggests that there are potentially different pathways for thiamine production in stramenopiles versus the green algal lineage (Table 2, McRose et al., 2012). The absence of a gene encoding ThiC, a protein involved in pyrimidine biosynthesis, appears to correlate with B1 auxotrophy in algae with sequenced genomes, regardless of lineage (Table 2). This intriguing observation warrants further exploration. Since ThiC is involved in pyrimidine biosynthesis, the relationship between ThiC gene presence and thiamine auxotrophy is likely to hold only for auxotrophs with the ability to synthesize thiamine diphosphate when provided the pyrimidine moiety, not those that can synthesize the vitamin when provided with the thiazole moiety, such as some dinoflagellates and cryptophytes (Droop, 1958). ThiC is an interesting protein; it requires S-adenosyl methionine (SAM) for activity (Chatterjee et al., 2008), is an iron-sulfur cluster protein, and is present in both the plant and bacterial thiamine biosynthesis pathways (Goyer, 2010).
Table 2.
Vitamin B1 (Thiamine)-related genes in sequenced eukaryotic phytoplankton genomes.
| Auran | Phatr | Thaps | Psemu | Fracy | Emihu | Ostta | Ostlu | MicPu 1545 | MicPu 299 | Chlre | ChlNC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ThiC | x | 38085 | 41733 | 255053 | 225659 | x | x | x | x | x | 192720 | 136333 |
| thiD+thiE/Thi6/TenI | x | 47293 | 262964-3 | 320126 | 153126, 161112 | 102278 | 20618, 6224 | 17535 | 52893 | x | 390684 | 58425 |
| ThiF* | 31873, 32858 | 34373, 20318 | 261602, 35049 | 207357, 293997 | 194811, 275015 | 68584 | 19906 | 38170 | 51160 | 113992 | 138485 | 22673 |
| dsx | 59650 | bd1689 | 574 | 65889 | 206898 | 440786 | 15650 | 48774 | 121145 | 107366 | 196568 | 59788 |
| ThiG% | x | PhtrCp129 | ThpsCp126, bd1620 | – | Scaffold 95, 27066–27869 | Emhu Cp072 | x | – | x | – | x | x |
| ThiS% | AuanCp078 | PhtrCp091 | ThpsCp091 | – | Scaffold 95, 5640–5849 | x | x | – | x | – | x | x |
| ThiO/H** | 72208 | 31544 | 263655 | 230060 | 241529 | 53832 | x | x | x | x | 196226 | 30311 |
| Thi4 | x | x | x | x | x | x | 20276∧ | x | 52894∧ | x | 185190 | 22703 |
| TPK | 20636 | 5423 | 262503@ | 264355 | 86232 | 56054 | 10431 | 12109 | 163134 | 109022 | 72868 | 11702 |
| ThiM/10 | x | x | x | x | x | x | x | x | x | x | 126905$ | 53510 |
| B1 Aux. by culture | Yes (Tang et al., 2010) | No (Droop, 1958) | No (Guillard and Ryther, 1962) | No (Tang et al., 2010)*** | No (Bertrand, unpublished) | Yes (Carlucci and Bowes, 1970)*** | Yes (McRose et al., 2012) | Yes (McRose et al., 2012) | No (Provasoli and Carlucci, 1974) | No (Shihira and Krauss, 1965) |
The hypothesized vitamin requirements of each strain are also given, along with whether culture-based confirmation of auxotophic status is available. This table expands information given in Croft et al., (2006). Auran = Aureococcus anophagefferans; Phatr = Phaeodactylum tricornutum; Thaps = Thalassiosira pseudonanna; Psemu = Pseudo-nitzschia multiseries CLN-47; Fracyl = Fragilariopsis cylindrus; Chlre = Chlamydomonas reinhardtii (v14; filtered or best proteins); Emihu = Emiliania huxleyi; Ostta = Ostreococcus taurii; Ostlu = Ostreococcus lucimarinus V2 filtered model proteins; MicPu = Micromonas pusilla CCMP1545 c3.0, filtered model proteins; ChlNC = Chlorella sp. NC64A filtered proteins.
X = not found, “–” = search not possible (chloroplast genome not available)
ThiF is not easily assigned because of similarities with MoeB/Z
ThiG and ThiS are often chloroplast encoded
unclear, potential Thi4 (similarity to tenA proteins too)
uncertain assignment
mutants of this are thiamine auxotrophs
The diatoms appear to have ThiO, Chlre, and ChlNC have thiH
Auxotrophy tested in culture of a different strain.
Thiamine catalyzes a number of transformations that are important in carbohydrate and branched amino acid metabolism including those involved in glycolysis, the pentose phosphate pathway, and the tricarboxylic acid pathway. These notably include 2-oxoglutarate dehydrogenase (ODG), pyruvate dehydrogenase/decarboxylase, branched-chain α-ketoacid dehydrogenase, as well as transketolases, acetolactate synthase, and alpha-keto acid dehydrogenase. The chemistry involved in these reactions often includes two carbon group transfers or dehydrogenation reactions (Frank et al., 2007). There is mounting evidence that thiamine may play additional, non-cofactor roles as well. In plants, thiamine has been tied to cellular responses to oxidative stress and disease. Plants subjected to hydrogen peroxide, salt stress, and high light stress, for example, all showed enhanced thiamine production and increased thiamine biosynthesis protein transcripts, such as ThiC (reviewed in Goyer, 2010; Rapala-Kozik et al., 2012). It is possible that this increase in thiamine under stress results from demand for transketolase activity in the pentose phosphate pathway which regenerates NADPH required for activity of some antioxidants (Goyer, 2010). However it is also possible that thiamine itself functions as an antioxidant in these cells, as thiamine compounds have antioxidant capacities, likely through the transfer of H+ from amino groups on the thiazole and pyrimidine rings to reactive species (Hu et al., 1995; Lukienko et al., 2000; Bettendorff and Wins, 2009). While there has been comparatively little study of these potential roles of thiamine in algae, available evidence suggests that thiamine biosynthesis per cell in diatoms increases as a function of increasing cell density and nutrient depletion, which may be caused by the increase in oxidative stress (Pinto et al., 2003).
Thiamine auxotrophy is strikingly different from B12 auxotrophy in algae; while B12 requirements are determined by the ability of a phytoplankton strain to replace B12-requiring metabolisms (Table 1), B1 auxotrophy is defined by whether or not an algal strain is able to synthesize the vitamin de novo (Table 2). Considering that the enzymes for B1 biosynthesis are not yet completely elucidated in algae, it is difficult to discern, through analysis of protein coding sequences, the evolutionary origin of B1 auxotrophy. However, observations concerning the phylogenetic distribution of thiamine auxotrophy support the notion that biosynthesis potential may have also been lost and acquired multiple times. For instance, in the case of two strains of the same species of dinoflagellate, isolated from the same site, one is a B1 auxotroph and one is not (Tang et al., 2010). Among Micromonas spp. strains with thiamine requirements, one is missing more of the biosynthetic pathway than the other (McRose et al., 2012; Table 2). These data suggests that like B12, B1 auxotrophy in algae has likely arisen numerous times though gene loss events. Such loss events could be driven by chronically high thiamine availability coupled to transcriptional repression and associated loss of purifying selection and gene erosion. While this repression is yet to be documented, eukaryotic phytoplankton genomes encode thiamine riboswitches (Croft et al., 2007; Worden et al., 2009); which offer a mechanism by which high thiamine bioavailability can regulate gene transcription.
Oceanographic distributions and cycling
In the ocean, dissolved (0.2 μm) vitamin B12 and B1 show variable but often nutrient-like depth profiles and are thought to be present in sub-picomolar quantities to up to 30 pM for B12 and 500 pM for B1 (Sañudo-Wilhelmy et al., 2012). Concentrations of these vitamins in coastal waters are generally higher than in open ocean regions (Panzeca et al., 2009). Measurement techniques for B vitamins in seawater remained restricted to bioassays (Menzel and Spaeth, 1962; Carlucci, 1966) until solid phase extraction, high pressure liquid chromatography methods were developed (Okbamichael and Sañudo-Wilhelmy, 2004; Okbamichael and Sanudo-Wilhelmy, 2005). Development of these techniques, coupled with mass spectrometry, has fostered more efficient and accurate methods for vitamin detection and quantitation in seawater. Such methods however still require inconveniently large volumes and are not currently optimized to detect B12 with different α or β axial groups or differentially phosphorylated forms of thiamine, which may be present in seawater and could be important for bioactivity as well as biogeochemical cycling. In addition, concentration measurements alone may not be an informative measure of the impact of vitamins on marine biogeochemical processes since their concentrations are low and they may be cycled and regenerated rapidly in the euphotic zone as a function of biological production and consumption as well as abiotic processing. The halflife of B12 in the surface ocean with respect to photodegradation alone is approximately 4 days, while B1 is more resistant to abiotic transformations in seawater (Gold et al., 1966; Carlucci et al., 1969). This, along with differences in production and consumption of vitamins by different components of marine microbial communities, may explain the observation that B12 and B1 concentrations and cycling may be decoupled in the water column (Panzeca et al., 2008; Sañudo-Wilhelmy et al., 2012). It remains a challenge to reconcile the interesting observation that B vitamin abundance patterns are associated with basin-scale water mass origin (Sañudo-Wilhelmy et al., 2012) with the likely rapid changes in production and consumption of these vitamins. To address this question, continued efforts to measure these vitamins, along with assessments of microbial community composition and vitamin acquisition rates, should include assessments of variability on short (hours to days) as well as seasonal timescales.
Either through dissolved organic matter exudation and cell lysis via the cycling of the microbial loop, (Azam, 1998; Karl, 2002; Droop, 2007) or through direct symbiotic interaction (Croft et al., 2005), some portion of the bacterial and archaeal community must be the ultimate source of vitamin B12 to eukaryotic phytoplankton. The genetic potential for vitamin B12 production remains largely uncharacterized in any marine environment (Bertrand et al., 2011a). This is in part because the occurrence of the biosynthesis pathway among bacterial and archaeal lineages is extremely variable and is not easily queried using typical phylogenetic profiling techniques. An exception to this is the marine cyanobacteria, where all sequenced genomes appear to contain the B12 biosynthetic pathway (Rodionov et al., 2003), and numerous strains have been shown to produce significant amounts of B12 (Bonnet et al., 2010). The identity of other groups that contribute significantly to oceanic B12 production remains unclear, however, and is of particular importance in regions with scarce cyanobacterial populations such as the polar oceans (Caron et al., 2000; Marchant, 2005). The extremely abundant SAR11 group appears to neither synthesize nor require the vitamin (Table 1). In addition, there are examples from many sequenced marine bacterioplankton genomes of strains that either cannot produce the vitamin themselves but require it for various metabolisms, or those that can salvage degraded B12 for repair and reuse (Bertrand et al., 2011a). In sum, B12 uptake by marine bacteria and archaea can be as significant as uptake by eukaryotic phytoplankton (Bertrand et al., 2007; Koch et al., 2011). This results in a scenario in which eukaryotic phytoplankton likely compete for B12 resources with some components of the prokaryotic community (Bertrand et al., 2011b; Sañudo-Wilhelmy et al., 2012).
Thiamine sources to eukaryotic phytoplankton include de novo production, uptake or salvage from bacterial production, or uptake and salvage of thiamine produced by other algae (Carlucci and Bowes, 1970; Provasoli and Carlucci, 1974). Similar to B12, competition likely occurs for B1 amongst microalgae as well as between algal and bacterial groups since not all prokaryotes have the ability to produce thiamine (Rodionov et al., 2002). It remains unclear, however, what the relative importance of these uptake vectors are and how this varies across oceanic regions. A striking difference between B12 and B1 auxotropy is that B1 requirements in algae could potentially be supplied by growth with B1 producing algal strains as well as with some bacteria (Table 2). This is in contrast to B12 where the only potential source of B12 to auxotrophic algae is bacterial and archaeal production. This opens interesting avenues for exploration of species succession and potential commensalism between not only algae and bacteria but also between different algal strains.
Bottle incubation bioassay experiments have suggested that availability of B12 and to some degree B1 influence overall rates of primary production as well as phytoplankton community composition in regions ranging from the Southern Ocean to temperate coastal environments (Panzeca et al., 2006; Sañudo-Wilhelmy et al., 2006; Bertrand et al., 2007; Gobler et al., 2007; Koch et al., 2011). In many cases, addition of B vitamins to communities resulted in the proliferation of diatoms or dinoflagellates and larger groups of eukaryotic phytoplankton (Table 3). This may have important implications for carbon and nitrogen export as well as silica cycling, since larger phytoplankton tend to support a higher percentage of organic matter export. In addition, coastal and open ocean North Atlantic studies revealed that regions with higher B12 concentrations correlated with regions with high bacterioplankton productivity or density (Gobler et al., 2007; Panzeca et al., 2008), though it remains unclear whether these correlations are due to bacterial production of the vitamin or enhanced bacterial abundance as a result of higher B12 availability. In the Ross Sea of the Southern Ocean, bacterial abundance was shown to be low where primary production was stimulated by B12, meaning that where bacterioplankton communities were more numerous, B12 was less likely to limit primary production (Bertrand et al., 2011b). This suggests that bacterioplankton have an important impact on B12 supply to eukaryotic phytoplankton, at least in polar regions. However, intimate associations between bacteria and eukaryotic phytoplankton are known to occur (Figure 1; Cole, 1982; Grossart et al., 2005); the importance of these associations to B vitamin cycling and availability to phytoplankton in the marine environment are just beginning to be explored and offer numerous exciting avenues for continued research.
Table 3.
Results of B-vitamin supplementation in published marine bottle incubation bioassays.
| Location | Experiments with stimulation of Chl a production by a B vitamin | B vitamin changed community composition? | Size class or functional group with biggest response | Notes | Interactions with N | References |
|---|---|---|---|---|---|---|
| Long Island embayments | 1/1 | 1/1 | >5 μm | Observed correlation between dissolved B12, B12 drawdown and growth of large phytoplankton | Yes | Sañudo-Wilhelmy et al., 2006 |
| Antarctic Peninsula | 1/1 | 1/1 | nd | Primary and secondary limitation by B1 + B12 | nd | Panzeca et al., 2006 |
| Ross Sea | 2/3 | 3/3 | Diatoms | – | nd | Bertrand et al., 2007 |
| Long Island embayments | 4/14 | – | >5 μm | Fall experiments: large size fraction B vitamin limited | Yes | Gobler et al., 2007 |
| Ross Sea | 2/5 | 5/5 | Diatoms | B12 uptake rates Fe limited | nd | Bertrand et al., 2011a,b |
| Gulf of Alaska | 1/2 | 2/2 | Dinoglagellates in coastal, diatoms in upwelling | N and Fe co-limitation with B12 | Yes | Koch et al., 2011 |
nd = no data.
Figure 1.
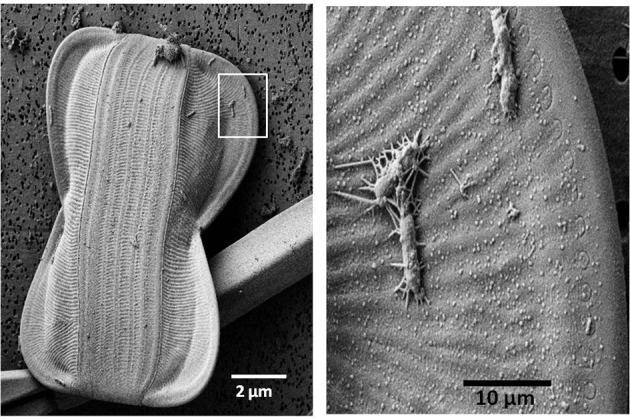
Bacteria can be intimately associated with diatoms. This sea ice Amphiprora diatom cell has bacterial cells attached through an apparently tight association likely via the use extracellular polymeric substances (EPS). SEM micrographs were collected at the UC Riverside Center for Nanoscale Science and Engineering. Samples were filtered, critical point dried to preserve cellular structures, coated with Pt:Pd to prevent charging, and imaged at 2 kv on a Zeiss 1540 FE-SEM.
The importance of nitrogen to eukaryotic phytoplankton
Nitrogen is an essential component of all life. The availability of nitrogen is thought to limit the productivity of marine microbial communities in large portions of the ocean (McCarthy and Carpenter, 1983; Hecky and Killam, 1988; Moore et al., 2004). Oceanic dissolved nitrogen distributions are driven in large part by coupled biological processing and large scale patterns in ocean circulation. Dissolved inorganic nitrogen (DIN) is generally considered to be the major source of nitrogen to marine microbial communities; the availability of these compounds (nitrate, nitrite, and ammonia), particularly in the oligotrophic ocean, can be depleted below 0.03 μM (Capone, 2000). Phytoplankton growth limitation by inorganic nitrogen availability has also been observed in coastal and upwelling environments (Kudela and Dugdale, 2000). The availability of this inorganic nitrogen has long been used, via nitrogen balancing calculations, to estimate organic matter export from the surface ocean (Eppley and Peterson, 1979), a concept which has profoundly influenced the field of biogeochemical oceanography. Models of the role of different nitrogen sources to phytoplankton and their microbial transformations have evolved to include additional processes, yet this conceptualization of balance between dissolve nitrate upwelled into the euphotic zone and export of biogenic and dissolved organic nitrogen (Bronk et al., 1994) continues to shape our understanding of controls on marine primary production and carbon cycling.
The relative availability of different N sources is now known to play a role in structuring phytoplankton species composition. Though reduced N compounds require less energy to assimilate, there are differences between taxonomic groups in terms of the impact of these differences on growth rate and the impact of ammonia availability on oxidized N acquisition (Dortch, 1990). In addition, differences in the ability of varying phytoplankton functional groups to respond to variable nitrogen concentrations and sources can create important niche dimensions. For instance, diatoms are a particularly successful group of eukaryotic phytoplankton that tend to dominate in coastal and upwelling regions. These locations are often characterized by highly variable nitrogen sources and concentrations. The ability of diatoms to respond quickly to pulsed nitrogen additions can, in part, explain a portion of their success in such environments. Their successful responses to these pulsed additions are partially explained by their ability to tightly couple anabolic and catabolic nitrogen transformations through incorporation of a complete urea cycle into central metabolism (Allen et al., 2011). Diatoms also tend to exhibit their maximal growth rates when grown on reduced nitrogen sources such as ammonia and urea (Dortch, 1990; Bender et al., 2012), but also in some cases dominate environments when nitrate is the dominant source of DIN. Their ability to take up and flexibly utilize a range of nitrogen sources also likely contributes to their role as a dominant phytoplankton group. We suggest that B vitamin deprivation may impair the ability of diatoms to effectively respond to and recover from nitrogen deprivation and that this may have important implications for interactions between marine microbial groups. This results from the fact that metabolisms impacted by B1 and B12 have important roles in pathways and mechanisms for allocation of cellular N recovery from N starvation.
B vitamin and N interactions in oceanic environments
It is clear that marine bacterial communities, in some cases, compete with eukaryotic phytoplankton for inorganic nitrogen sources, including nitrate (Kirchman and Wheeler, 1998; Kirchman, 2000; Allen et al., 2001, 2005). These heterotrophic bacterial communities also conduct the canonical transformation of organic N sources to ammonia and dissolved organic nitrogen via cycling within the microbial loop. As a result, bacterial communities can be either net sources or net sinks of available N to phytoplankton communities (Kirchman, 2000; Zehr and Ward, 2002). This may vary as a function of the C:N ratio of available organic matter as well as the community composition of microbial assemblages (Kirchman, 2000). There are clear parallels between N and B vitamin availability in the ocean; the interaction between marine microbial groups plays a key role in shaping the influence these chemicals have on productivity. As a result, the implications of combined nitrogen starvation and B vitamin deprivation for eukaryotic phytoplankton will clearly be interactively impacted by bacterial communities. Intimately associated bacterial communities, such as those shown in Figure 1, have the potential to impact vitamin availability as well as nitrogen resources to phytoplankton; interactions between B vitamin and N dynamics in algal bacterial associations have yet to be explored, but are intriguing areas for research.
There have been two studies examining interactive impact of DIN and B vitamin addition on phytoplankton communities. In Long Island embayments, shifts from dinoflagellate dominated, primarily N limited communities in summer to diatom dominated blooms in fall coincided with decreases in B12 and B1 availability and increases in chlorophyll production upon B vitamin additions, suggesting that N and B vitamin availability both influence coastal phytoplankton species succession and biomass. Interestingly, in several instances, B12 or B1 and nitrate, when added together, stimulated chlorophyll production to a greater degree than adding either nutrient alone (Gobler et al., 2007). This interactive effect has yet to be mechanistically explored, but could be a function of vitamins being independently secondarily limiting, or could be explained by biochemical interactions between nitrogen and B vitamin production or demand (Saito et al., 2008). In a series of bottle incubation studies in the coastal, nitrogen limited region of the Gulf of Alaska, the addition of nitrate alone yielded enhanced productivity, and a shift from a dinoflagellate to diatom dominated community. In contrast, the addition of B12 and nitrate together yielded a community dominated by dinoflagellates (Koch et al., 2011). This striking result suggests that B vitamin availability severely impacted the response of the coastal phytoplankton community to nitrogen availability. This response suggests that the dinoflagellate community could not respond to nitrogen addition under B12 starvation conditions, either due to secondary, independent limitation of dinoflagellate growth by B12 availability or due to biochemical interactions between nitrogen and B12 metabolism leading to colimitation. Since a higher proportion of dinoflagellates are B12 auxtrophs (90%) than are diatoms (60%) (Tang et al., 2010), this response may be expected. However, diatom ability to respond to nitrogen additions over dinoflagellates under low B12 availability may not be entirely explained by differences in auxotrophy and warrants further exploration. Here we examine potential biochemical mechanisms for interaction between B vitamin and nitrogen metabolism.
Molecular responses of eukaryotic phytoplankton to B vitamin starvation
There is some information available concerning the molecular responses of eukaryotic phytoplankton to B-vitamin starvation. While studies that examine the response of phytoplankton to B1 deprivation have not been described in detail (McRose et al., 2012), there has been comparatively extensive inquiry into the molecular response of phytoplankton to B12 deprivation. An important consequence of B12 deprivation in eukaryotic phytoplankton appears to be impaired methionine synthase activity and the use of B12-independant MetE over dependent MetH (Croft et al., 2005; Helliwell et al., 2011; Bertrand et al., 2012). Methionine serves not only as a protein-building amino acid but as the precursor to S-adenosylmethionine (AdoMet or SAM), an important methylating agent, propylamine donor, and radical source. Indeed, there is evidence that SAM deprivation is an important consequence of low B12 availability in diatoms (Bertrand et al., 2012; Figure 2). Notably, ThiC, an important algal thiamine biosynthesis protein, is SAM-dependant and responds to B12 deprivation in diatom cultures, suggesting that there may be consequences of B12 deprivation for thiamine production. In addition, dinoflagellate transcriptome and metatranscriptome sequencing studies reveal that SAM cycling genes are among the most highly expressed transcripts in multiple dinoflagellate species (Lidie et al., 2005; Moustafa et al., 2010; Toulza et al., 2010). These data, along with the high percentage of surveyed dinoflagellates that exhibit an obligate B12 requirement (>90%; Tang et al., 2010) suggest that B12 availability may have important implications for dinoflagellate SAM metabolism; perhaps a disproportionately important process relative to diatoms. This could potentially result from extensive dinoflagellate DNA methylation or increased demand due to toxin production, which has a high SAM requirement (Lin, 2011). In addition, impaired methionine synthase activity prevents efficient folate recycling, which has important implications for nucleic acid biosynthesis (Scott and Weir, 1981; Croft et al., 2005). Molecular evidence for altered folate metabolism has also been documented as a significant component of the diatom response to B12 deprivation (Bertrand et al., 2012). This likely holds true for other algal groups as well since the diatom response is similar to distantly related organisms such as humans and other metazoans (e.g., Scott and Weir, 1981).
Figure 2.
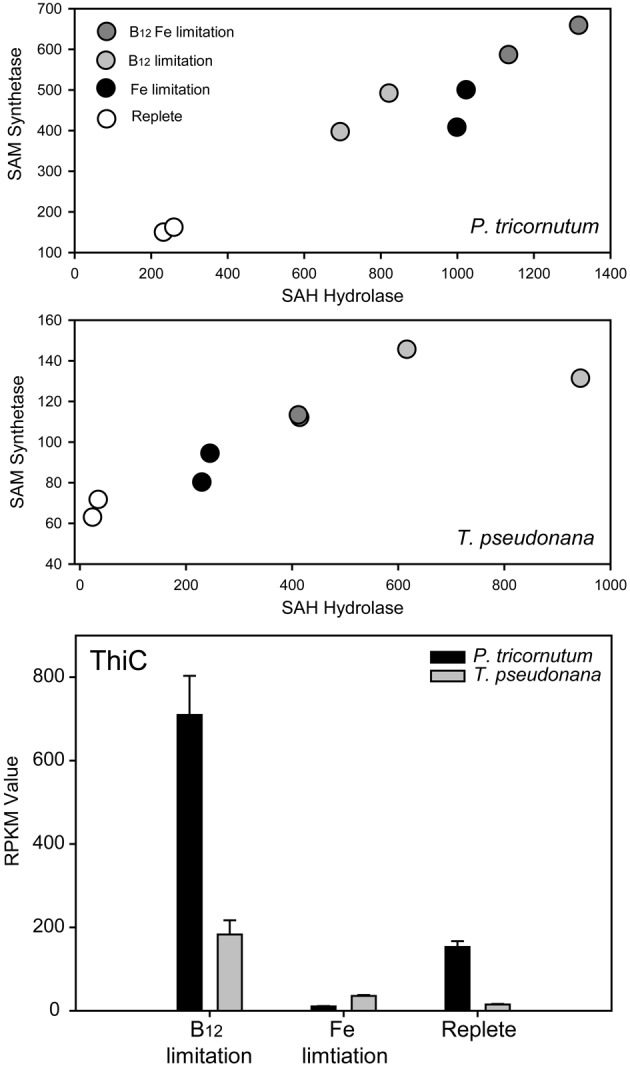
Evidence from Bertrand et al., 2012 that AdoMet SAM starvation is an important consequence of B12 deprivation, with implications for thiamine biosynthesis. SAM synthetase (Tp 39946, Pt 18319) converts methionine and ATP to SAM. SAM, after use for methylation reactions, is converted to S-adenosylhomocysteine (SAH). SAH can act as an inhibitor to methylation reactions because of its high affinity for most methyltranserfases. SAH hydrolase (Tp 28496; Pt bd 913) catalyzes the reversible interconversion of SAH to homocysteine and adenosine. The expression of the genes encoding these proteins in two diatoms appears to correlate. RPKM (Reads Per Kilobase of exon model per Million mapped reads) gene expression values are plotted against each other for each of eight samples in two diatoms, duplicates of replete, low B12, low B12 with low iron, and low iron alone. Expression under iron limited conditions was examined along with B12 to verify whether changes induced were likely a general stress response or more specific to the vitamin. In both diatoms, cells grown under nutrient replete conditions express these genes at the lowest level. Iron and B12 availability both influence the expression of these genes, with B12 having a greater impact of gene expression the B12 requiring diatom T. pseudonana. ThiC is a SAM-dependent protein required for pyrimidine moiety synthesis in thiamine biosynthesis. The expression of genes encoding ThiC in both these diatoms is elevated under low B12 availability and not under low iron availability, suggesting that thiamine biosynthesis, and B12 availability may be linked in these diatoms, potentially through B12 impacts on SAM availability.
Molecular aspects of acquisition of these vitamins in eukaryotic algae remains poorly understood. An important result of these inquiries into the molecular response of algae to vitamin deprivation has been the identification of proteins that are potentially involved in B12 or B1 acquisition. Bertrand et al. (2012) identified a previously uncharacterized protein, deemed CBA1, that is directly involved in B12 acquisition by diatoms and that is much more abundant in diatoms when they are experiencing B12 deprivation. This protein, however, appears to be restricted to the stramenopile lineage, suggesting that other eukaryotic algal groups utilize different, as of yet unidentified, pathways for B12 uptake (Bertrand et al., 2012). Several candidate proteins involved in thiamine trafficking have been identified in whole genome sequencing projects (Worden et al., 2009) and transcriptomic analyses of Micromonas cultures under thiamine deprivation have also resulted in identification of additional putative thiamine transporters in this B1 auxotrophic group (McRose et al., 2012).
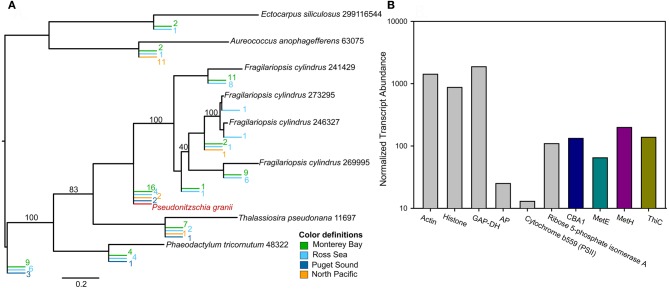
Examination of transcripts encoding CBA1, ThiC, MetE, and MetH in natural Antarctic diatom communities revealed that all these transcripts are relatively abundant and therefore that B vitamin metabolism is likely an important component of the molecular physiology of field communities (Figure 3). These expression patterns suggest that Antarctic diatom communities are experiencing B12 deprivation (expressing MetE and CBA1) and that some subset of the community is able to potentially utilize B12 for methionine regeneration (MetH expression). This suggests that there are potentially different B12 quotas for different diatom species, with species experiencing starvation at varying intersections of cellular demand relative to ambient B12 availability. Additionally, there may be a subset of diatoms with localized B12 sources, such as intimately attached bacteria (Figure 1). These results suggest, however, that examining the distribution of B12—responsive transcripts in field populations will yield important insights into the impact of vitamin availability on community structure and primary productivity. For these analyses, it would be useful to know what percentage of the eukaryotic phytoplankton community possesses the ability to produce MetE; this would allow for more extensive interpretation of MetE and MetH transcript expression patterns. Similar analyses may be possible with thiamine—responsive genes in the future. Potential candidates for this include the recently identified putative transporters as well as ThiC, which appears to be present in genomes that are not auxotrophs and absent from genomes of organisms that require exogenous thiamine (Table 2).
Figure 3.
The abundance and diversity of CBA1, B12 starvation indicator, and the abundance of ThiC, MetH, and MetE transcripts attributable to diatoms in Antarctic transcriptomic datasets, compared to the expression of transcripts encoding other well-characterized proteins (modified from Bertrand et al., 2012). (A) Phylogenetic tree containing CBA1 sequences from 454 metatranscriptomic (cDNA) libraries from the Ross Sea of the Southern Ocean, Monterey Bay, Puget Sound, and the North Pacific. Reference sequences from Phaeodactylum tricornutum, Fragilariopsis cylindrus, Thalassiosira pseudonana, Aureococcus anophagefferenas, and Ectocarpus siliculosus genomes were used to construct these trees and are shown in black. CBA1-like sequences from environmental samples are shown in color, as described in the key. CBA1 transcripts were detectable in diverse marine environments, suggesting that cobalamin acquisition is an important component of diatom molecular physiology. (B) The normalized abundance of Open Reading Frames (ORFs) assigned to CBA1 from within the Ross Sea is shown in blue, MetE: PF01717 is shown in green, MetH:PF02965 is shown in purple, ThiC:PF01964 in yellow, while the abundance of read counts assigned to diatom ORFs containing well-characterized pfam domains for comparison [Actin: PF00022, Histone:PF00125, GAP-DH: PF02800, Alkaline Phosphatase: PF00245, Flavodoxin: PF00258, Cytochrome b559 (PSII): PF00283, Ribose 5-phosphate isomerase A: PF06026] are shown in gray. Read counts for each ORF where summed across six libraries from Ross Sea samples and RPKM values were calculated. RPKMs were then summed across all diatom ORFs that contained that a domain of interest. CBA1, MetE, MetH, and ThiC are not among the extremely abundant transcripts (e.g., those encoding Actin, GAP-DH, Histone) but are comparable to those encoding Calvin Cycle protein Ribose 5-phosphate isomerase A, and are more abundant than the transcripts encoding a cytochrome required for photosystem II activity (b559) as well as alkaline phosphatase (AP), suggesting that they are of importance to the molecular physiology of natural diatom communities.
Potential B12 and B1 metabolic interactions with nitrogen in eukaryotic phytoplankton
B vitamin and N starvation impacts on osmolyte production and utilization
Osmolytes are molecules that serve roles in osmoregulation. In eukaryotic phytoplankton, these include proline, glycine betaine (GBT), dimethylsulfonium propionate (DMSP), homarine, and isethionic acid (Boroujerdi et al., 2012). There are potentially important roles for B12, B1, methionine, SAM, and nitrogen metabolism in osmolyte production that likely result in interactive biochemical effects. One example is that methionine and SAM are both required for DMSP production, which is used by a subset of diatoms possibly as a cryoprotectant, osmolyte (Stefels, 2000), or antioxidant (Sunda et al., 2002), and is the precursor to the climatically important gas dimethylsulfide (Charlson et al., 1987). SAM recycling genes appear to play a role in the response of diatoms to low nitrogen, suggesting that there may be synergistic impacts of nitrogen and B12 depletion on SAM availability (Table 4). This observation is intriguing and warrants further exploration via SAM metabolite analysis under conditions of varying B12 and nitrogen availability.
Table 4.
B12 and B1 related genes from published P. tricornutum EST libraries.
| Original | Si− | Si+ | Low Fe | n replete | N-starved | Urea, low N | Ammonia, | Description | |
|---|---|---|---|---|---|---|---|---|---|
| standard | chemostat | low N | |||||||
| os | sm | sp | fl | nr | ns | ua | aa | ||
| B12-RELATED | |||||||||
| G18319 | 0 | 0 | 1 | 9 | 4 | 15 | 20 | 33 | s-adenosyl homocysteine hydrolase |
| G48322 | 3 | 2 | 0 | 0 | 1 | 0 | 2 | 3 | CBA1 |
| G18665 | 1 | 0 | 1 | 1 | 1 | 3 | 10 | 11 | Glycine hydroxymethyltransferase |
| G28056 | 0 | 7 | 11 | 0 | 0 | 2 | 0 | 1 | MetE |
| G913.1 | 1 | 0 | 0 | 5 | 2 | 3 | 0 | 1 | S-adenosylmethionine synthetase |
| G54015 | 0 | 0 | 1 | 4 | 1 | 6 | 0 | 0 | Glycine hydroxymethyltransferase |
| G23399 | 1 | 0 | 0 | 9 | 0 | 1 | 5 | 5 | MetH |
| G51830 | 4 | 0 | 3 | 0 | 0 | 3 | 7 | 4 | Methylmalonyl co a mutase |
| G30471 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 2 | Methylenetetrahydrofolate reductase |
| B1 USE | |||||||||
| G20183 | 2 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | Transketolase |
| G20360 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | Pyruvate dehydrogenase e1 component beta subunit |
| G12375 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | Pyruvate dehydrogenase e1 component alpha subunit |
| G29016 | 2 | 0 | 0 | 0 | 0 | 4 | 0 | 2 | 2-oxoglutarate dehydrogenase e1 oxoglutarate alpha-ketoglutaric |
| G37341 | 2 | 1 | 3 | 0 | 0 | 0 | 2 | 7 | Acetolactate synthase |
| G48444 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 2-oxoglutarate dehydrogenase e1 component |
| G46387 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | Dehydrogenase, E1 component |
| G36257 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | Fructose-6-phosphate phosphoketolase |
| G9476 | 1 | 2 | 4 | 2 | 0 | 1 | 0 | 0 | 2-oxoisovalerate dehydrogenase alpha, mitochondrial expressed |
| G41856 | 14 | 0 | 2 | 1 | 2 | 3 | 12 | 3 | Plastid transketolase |
| G29260 | 5 | 0 | 0 | 2 | 1 | 2 | 11 | 6 | Probable transketolase |
| G11021 | 0 | 3 | 2 | 3 | 1 | 0 | 0 | 0 | Branched-chain alpha-keto acid decarboxylase e1 beta subunit |
| B1 SYNTHESIS | |||||||||
| G34373 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | Possible ThiF |
| G1689.1 | 3 | 0 | 0 | 2 | 2 | 5 | 1 | 0 | Possible Dsx |
| G31544 | 4 | 2 | 0 | 1 | 0 | 4 | 3 | 2 | Possible ThiO |
| G38085 | 9 | 1 | 0 | 0 | 1 | 1 | 3 | 4 | ThiC |
| G47293 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Possible ThiD/E |
| G5423 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | TPK |
| LCPA BIOSYNTHESIS | |||||||||
| G7617 | 0 | 0 | 1 | 0 | 0 | 3 | 4 | 0 | s-adenosylmethionine decarboxylase proenzyme |
| G7910 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | Spermine synthase |
| G3362 | 0 | 2 | 0 | 0 | 0 | 4 | 9 | 0 | S-adenosylmethionine decarboxylase |
| G7621 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | s-adenosylmethionine decarboxylase proenzyme |
Treatment descriptions and labels can be found in Maheswari et al. (2010).
In addition, nitrogen limitation has been previously identified as an important factor driving DMSP and DMS produced by phytoplankton populations. Nitrogen deprivation, more than any other nutrient starvation scenario tested, led to enhanced DMSP production per cell in an important oceanic diatom (Bucciarelli and Sunda, 2003; Sunda et al., 2007). A possible explanation for this trend is that under nitrogen starvation, N-containing osmolytes such as proline, homarine, and GBT are replaced by DMSP, which does not contain nitrogen (Bucciarelli and Sunda, 2003). There is some evidence that under N-replete conditions, GBT and homarine replace DMSP in T. pseudonana and that GBT concentrations increase upon addition of N to N-starved cultures of diatoms and coccolithophores (Keller et al., 1999). If DMSP is in fact used to replace N-containing osmolytes, B12 starvation coupled with N-limitation has the potential to negatively impact that substitution in at least two ways. The first is by potentially limiting the amount of DMSP produced due to restricted methionine availability. The second is again through SAM deprivation, which has been hypothesized to play an important role in diatom metabolism under low B12 conditions (Figure 3; Bertrand et al., 2012). These metabolic connections suggest that there may be synergistic impacts of B12 and N starvation on DMSP-producing algal strains. Alternatively, if the primary function for DMSP is as an antioxidant, increases in DMSP as a function of nitrogen starvation could be due to elevated demand for DMSP under the oxidative stress induced by nitrogen deprivation (Sunda et al., 2002, 2007). If DMSP in fact serves an important antioxidant role and if thiamine is shown to be an important algal antioxidant as well, this suggests that there could be potentially important interactions between B1, B12, and N availability in algal cells in response to oxidative stress.
Synthesis of GBT, in many organisms, also requires SAM as a methyl group donor. Also like DMSP, there is evidence that GBT production is tied to nitrogen metabolic status of individual cultures, (Keller et al., 1999, 2004). Since GBT synthesis requires nitrogen and is likely SAM dependent, B12 starvation may prompt substitution of other osmolytes, such as proline, for GBT as well as DMSP. This may have important implications for cellular nitrogen cycling. Notably, proline is generated from ornithine via activity of ornithine cyclodeaminase. Ornithine is an important metabolite in the urea cycle, which is a major pathway for nitrogen recycling in diatoms and potentially other algae (Fernie et al., 2012). If the proline balance were significantly impacted as a result of a metabolic cascade resulting from changes in the osmolyte balance, this could have significant impacts on overall cellular nitrogen metabolism.
B12 and B1 starvation impacts on amino acid and polyamine biosynthesis
The major organic constituent of diatom silica frustules are a series of long chain polyamines (LCPAs). Different diatoms synthesize different suites of LCPAs (Kroger et al., 2000). These molecules, along with silica deposition proteins called silafins and silafin-like girdle band and nanopattern-associated proteins call cingulins (Scheffel et al., 2011), induce biomineralization, and are responsible for differences in frustule morphology between diatom groups. LCPAs vary in chain length and degree of methylation, but appear to all be synthesized from putricine, spermidine, or spermine precursors. These precursors are synthesized sequentially from ornithine, with spermidine, and spermine production both requiring SAM as a propylamine donor. Subsequent steps in LCPA formation likely require SAM as well (Kroger and Poulsen, 2008). LCPAs recovered in net tows are mostly putracine-based, with varying degrees of methylation, suggesting that SAM is an important component of LCPA production for field diatom populations as well (Bridoux et al., 2012). Conceivably, reduced SAM production through B12 starvation could induce changes in silica frustule formation by decreasing the pool of available LCPAs. Indeed, reduction of LCPA production as a result of the addition of an inhibitor for ornithine decarboxylase, which is known to be involved in polyamine biosynthesis, dramatically reduced biogenic silica formation in T. pseudonana (Frigeri et al., 2006). In diatoms, possible LCPA biosynthesis genes have been identified. These are potentially gene fusions of bacterially derived polyamine biosynthetic enzymes S-adenosylmethionine decarboxylase (SAM DC) and an aminopropyltransferase (Michael, 2011), which require input of SAM. The Met salvage pathway would need to be efficient, and if SAM starvation does result from B12 deprivation, there could be substantial implications of low B12 for LCPA biosynthesis. Ornithine represents a significant component of the carbon and nitrogen pool within phytoplankton cells and is a centrally important metabolite in the ornithine urea cycle (OUC), which is the major distribution hub for nitrogen in diatom cells (Allen et al., 2011; Bender et al., 2012). If SAM starvation results in major changes in ornithine balance through alterations in polyamine biosynthesis, this would hold substantial ramifications for the impact of B12 deprivation on nitrogen cycling.
Overall, it seems that N starvation could induce up-regulation in pathways that demand B12, such as methionine and SAM synthesis. This would potentially be reflected in elevated expression of B12 acquisition proteins under nitrogen limitation, and elevation of proteins required to generate methionine.
Links between N, B1 and B12 through sulfur metabolism in eukaryotic phytoplankton
Connections between nitrate reduction and sulfur assimilation are well known. Sulfate reduction is thought to be regulated by nitrogen nutrition in plants (Koprivova et al., 2000; Takahashi et al., 2011); this may also be true for phytoplankton, as sulfur uptake and assimilation genes in diatoms appear to be responsive to nitrogen availability. Both B1 and B12 have important ties to sulfur metabolism, since B12 is important for sulfur amino acid cycling and DMSP synthesis and B1 is produced from thiazole, a sulfur containing moiety. Indeed, in plants, methionine synthesis and other aspects of sulfur metabolism are very tightly regulated by SAM availability. If the B12 dependence of SAM availability hypothesized for phytoplankton is verified (Bertrand et al., 2012), this suggests that B12 availability may influence additional aspects of sulfur and nitrogen metabolism.
Influence of B12 and B1 starvation on the diatom urea cycle through impacts on the citric acid cycle and amino acid cycling
Important impacts of vitamins on amino acid and amine cycling include the previously discussed impact of B12 on cysteine and methionine cycling and the impact of B1 on branched amino acid synthesis. B1 contributes to the first step in valine synthesis as well as important steps in amino acid degradation and recycling via keto acid dehydrogenase activity (Binder et al., 2007). In addition, B1 appears to impact nitrogen assimilation and amino acid recycling though the dependence of 2-oxoglutarate dehydrogenase (OGDHC) on the cofactor (Bunik and Fernie, 2009). For instance, potato OGDHC inhibition causes reductions in nitrate assimilation as well as increases in glutamate and GABA accumulation (Araujo et al., 2008). This suggests that disturbances in B1 metabolism may have profound affects for nitrogen assimilation and amino acid recycling, though this has yet to be confirmed for phytoplankton.
The OUC is of central importance to diatoms and potentially other phytoplankton as a nitrogen assimilation and repackaging hub. The OUC and the citric acid (TCA) cycles are linked (Allen et al., 2011). Mitochondrial amino acid catabolism yield carbon skeletons for the TCA cycle as well as ammonia and bicarbonate that is shunted into the OCU. This connection is supported by metabolic data suggesting that fumarate and malate, important TCA cycle intermediates, display similar patterns as OUC metabolites in diatom cell lines with altered urea cycle pathways (CPS knockdowns; Allen et al., 2011). Both B12 and B1 play important roles in the citric acid (TCA) cycle. For example, B12 is a cofactor for mmcM which generates succinyl coA from methylmalonyl coA, an amino acid degradation product. Expression of the gene encoding mmcM is upregulated under—N conditions in P. tricornutum EST libraries (Table 4), suggesting that there could be consequences of reduced mmcM activity for cells experiencing nitrogen deprivation. B12 availability does not appear to influence mmcM expression (Bertrand et al., 2012). It is notable that mmcM expression levels are not insignificant in diatom transcriptome studies, suggesting that this gene product may be of utility to phytoplankton despite the fact that the presence of this gene in phytoplankton genomes does not confer an absolute B12 demand (Table 1). There are numerous connections between B1 and the citric acid cycle. B1 is required for the generation of acetyl CoA from pyruvate via the pyruvate dehydrogenase complex. The enzyme ODG is also thiamine-dependent and plays a important role in the citric acid cycle. This enzyme is also thought to be a important player in plant nitrogen assimilation though its impact on glutamine stores (Bunik and Fernie, 2009). Interestingly, the reactant consumed by this protein, 2-oxoglutarate, accumulates strongly in diatom cell lines with impacted urea cycle (Allen et al., 2011). These data suggest that B1- and B12-dependent metabolisms play key roles in steps that maintain cellular carbon and nitrogen recycling; synergistic impacts of B vitamin deprivation and N starvation are therefore likely.
Synthesis and implications for eukaryotic phytoplankton ecology
Many of the interactions between B vitamins and N metabolism described above have the potential to profoundly influence eukaryotic phytoplankton ecology and are summarized in Figure 4. From these interactions, we can hypothesize that nitrogen limitation, experienced by phytoplankton in much of the ocean, may induce enhanced demand for B12 and B1 via a variety of mechanisms. These include substitution of N-containing osmolytes with DMSP, substituting N-containing antioxidants and DMSP with thiamine, and effectively recycling amino acids and glutamine stores utilizing high amounts of B1. There are also interactions discussed above that would result in B vitamin deprivation leading to impaired nitrogen recycling which could conceivably increase nitrogen demand in phytoplankton cells. These include impaired glutamine recycling due to reduced 2-oxogultarate dehydrogenase and pyruvate dehydrogenase activity and impaired ornithine and proline cycling due to B1 and B12 impacts on the TCA cycle as well as through potential imbalances in the methyl cycle due to B12 deprivation. These mechanisms all suggest that biochemical interactions between B vitamin and N limitation have the potential to lead to interactive colimitation and thus that the B vitamin and N colimitations observed in field studies (Koch et al., 2011; Gobler et al., 2007) may be due to both independent secondary limitation or dependent colimitation scenarios (Saito et al., 2008).
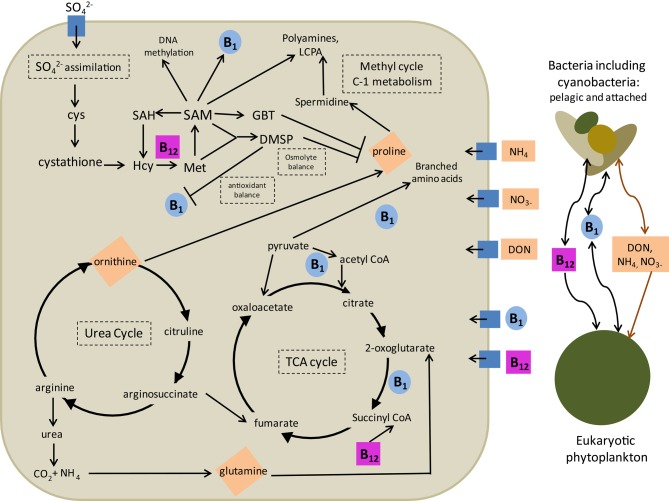
Figure 4.
An overview of B12 and B1 interactions with nitrogen metabolism in eukaryotic phytoplankton. Four major intracellular mechanisms are outlined: (1) impacts on osmolyte and antioxidant production and utilization and (2) impacts on polyamine biosynthesis via the methyl cycle, (3) impacts on the urea cycle and amino acid recycling through impacts on the citric acid cycle, and (4) impacts of nitrogen balance on sulfur assimilation. Major cellular nitrogen stores impacted by B1 and B12 availability are shown in orange diamonds. Arrows denote direction of reaction, production, or consumption. Bars denote potential negative feedbacks, where increases in originating compound may decrease abundance or importance of the connected compound. Also described are major interactions with other microbial groups outside the cell in terms of production and consumption of B1, B12, and nitrogen sources. Groups considered include pelagic and attached bacteria, including cyanobacteria (brown) and other eukaryotic algae (green). Relevant acquisition pathways are denoted by blue boxes. Cys, cysteine; Hcy, homocysteine; SAH, S-adenosyl homocysteine; SAM, S-adenosyl methionine; GBT, glycine betaine; DMSP, dimethylsulfonium propionate.
These biochemical dependencies have the potential to impart changes in phytoplankton species composition based on differences in B1 and B12 demand between phytoplankton groups. Diatoms are thought to rely on an efficient urea cycle for distributing and recycling nitrogen (Allen et al., 2011; Bender et al., 2012). The impacts of B12 and B1 deprivation on the efficiency of the urea cycle, therefore, may disproportionately impact diatoms. In addition, if dinoflagellate SAM demand is indeed elevated over other phytoplankton as hypothesized, it is possible that B12 deprivation could disproportionately impact dinoflagllate strategies for coping with nitrogen deprivation, such as the use of DMSP to replace N-containing osmolytes. These impacts may be of particular importance to harmful algal bloom species, which are known to have disproportionately high instances of B1 and B12 auxotrophy (Tang et al., 2010). Additionally, toxin production by some dinoflagellate species has been shown to increase under N-limitation (Ransom Hardison et al., 2012); synthesis pathways of many dinoflagellate toxins such as saxitoxin and brevetoxin are thought to be SAM-dependent (Lin, 2011). Together, these data suggests that HAB species may be more susceptible than others to impacts of these dependent colimitations between N and B vitamins and that these colimitations may additionaly impact toxin production rates. This is further evidence that B vitamin dynamics should be considered when predicting and evaluating potential for harmful algal bloom scenarios.
Given that B12, B1, and nitrogen availability to eukaryotic phytoplankton all have potential to be impacted by bacterial community composition and activity, the bacterial community is likely an important driver of when and where instances of these dependent colimitations may be important. This may be especially true when considering timing and species composition in spring bloom scenarios, which is an active area of continued research today (Mahadevan et al., 2012). Swift and Guillard (1978) determined that spring bloom diatom species, though not B12 auxotrophs, grew faster and experienced shorter lag phases in the presence of the vitamin, suggesting that possible interactions between N and S metabolism, and B12 utilization could be important for bloom timing and species composition. Recent work also suggests that bacterioplankton respond to various phases in spring blooms by changing both metabolic potential and species composition over time (Teeling et al., 2012). This could have important impacts for B12 and B1 production and consumption as well as for nitrogen availability and recycling. Mounting evidence suggests that there could be synergistic interactions of these impacts on eukaryotic phytoplankton that could influence not only species composition but also bloom timing and overall productivity. This suggests that time series measurements, over both day to week and seasonal timescales, which include B12, B1, and nitrogen species concentration measurements and uptake rates as well as protein or transcript-based indicators of nitrogen and vitamin deprivation, would be useful, particularly in conjunction with bacterioplankton community composition assessments and implementation of B vitamin biosynthesis indicators. Locations where this would be of considerable interest include high latitude ecosystems, which largely lack B12 producing cyanobacteria, coastal locations with HAB blooming dinoflagellates, and diatoms as well as the North Atlantic, before during and after bloom scenarios.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Greg Wanger for the use of his electron microscopy images. We are indebted to Mak Saito, Peter Lee, Ruben Valas, Stephane Lefebvre, James McCarthy, Jeff McQuaid, and Karen Beeri for helpful discussions. This work was funded by an NSF Office of Polar Programs Postdoctoral Fellowship to Erin M. Bertrand (ANT-1103503) and NSF-MCB-1024913, NSF-ANT-1043671, and DOE-DE-SC0006719 (Andrew E. Allen).
References
- Allen A., Dupont C. L., Obornik M., Horak A., Nunes-Nesi A., McCrow J. P., et al. (2011). Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473, 203–206 10.1038/nature10074 [DOI] [PubMed] [Google Scholar]
- Allen A. E., Booth M. G., Frischer M. E., Verity P. G., Zehr J. P., Zani S. (2001). Diversity and detection of nitrate assimilation genes in marine bacteria. Appl. Environ. Microbiol. 67, 5343–5348 10.1128/AEM.67.11.5343-5348.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen A. E., Booth M. G., Verity P. G., Frischer M. E. (2005). Influence of nitrate availability on the distribution and abundance of heterotrophic bacterial nitrate assimilation genes in the Barents Sea during summer. Aquat. Microb. Ecol. 39, 247–255 [Google Scholar]
- Araujo W. L., Nunes-Nesi A., Trenkamp S., Bunik V. I., Fernie A. R. (2008). Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation. Plant Physiol. 148, 1782–1796 10.1104/pp.108.126219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696 [Google Scholar]
- Bender S. J., Parker M. S., Armbrust E. (2012). Coupled effects of light and nitrogen source on the urea cycle and nitrogen metabolism over a diel cycle in the marine diatom Thalassiosira pseudonana. Protist 163, 232–251 10.1016/j.protis.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Bertrand E. M., Allen A. E., Dupont C. L., Norden-Krichmar T. M., Bai J., Valas R. E., et al. (2012). Influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein. Proc. Natl. Acad. Sci. U.S.A. 109, E1762–E1771 10.1073/pnas.1201731109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E. M., Saito M. A., Jeon Y. J., Neilan B. A. (2011a). Vitamin B12 biosynthesis gene diversity in the Ross Sea: the identification of a new group of putative polar B12-biosynthesizers Environ. Microbiol. 13, 1285–1298 10.1111/j.1462-2920.2011.02428.x [DOI] [PubMed] [Google Scholar]
- Bertrand E. M., Saito M. A., Lee P. A., Dunbar R. B., Sedwick P. N., DiTullio G. R. (2011b). Iron limitation of a springtime bacterial and phytoplankton community in the Ross Sea: implications for vitamin B12 nutrition. Front. Aquat. Microbiol. 2:160 10.3389/fmicb.2011.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E. M., Saito M. A., Rose J. M., Riesselman C. R., Lohan M. C., Noble A. E., et al. (2007). Vitamin B12 and iron co-limitation of phytoplankton growth in the Ross Sea. Limnol. Oceanogr. 52, 1079–1093 [Google Scholar]
- Bettendorff L., Wins P. (2009). Thiamin diphosphate in biological chemistry: new aspects of thiamin metabolism, especially triphosphate derivatives acting other than as cofactors. FEBS J. 76, 2917–2925 10.1111/j.1742-4658.2009.07019.x [DOI] [PubMed] [Google Scholar]
- Binder S., Knill T., Schuster J. (2007). Branched-chain amino acid metabolism in higher plants. Physiol. Plant. 129, 68–78 [Google Scholar]
- Bonnet S., Webb E. A., Panzeca C., Karl D. M., Capone D. G., Sanudo-Wilhelmy S. A. (2010). Vitamin B12 excretion by cultures of the marine cyanobacteria Crocosphaera and Synechococcus. Limnol. Oceanogr. 55, 1959–1964 [Google Scholar]
- Boroujerdi A. F. E., Lee P. A., DiTullio G. R., Janech M. G., Vied S. B., Bearden D. W. (2012). Identification of isethionic acid and other small molecule metabolites of Fragilariopsis cylindrus with nuclear magnetic resonance Anal. Bioanal. Chem. 404, 777–784 10.1007/s00216-012-6169-2 [DOI] [PubMed] [Google Scholar]
- Bridoux M. C., Keil R., Ingalls A. E. (2012). Analysis of natural diatom communities reveals novel insights into diversity of long chain polyamine structures involved in silica precipitation. Org. Geochem. 47, 9–21 [Google Scholar]
- Bronk D. A., Glibert P. M., Ward B. B. (1994). Nitrogen uptake, dissolved organic nitrogen release, and new production. Science 265, 1843–1846 10.1126/science.265.5180.1843 [DOI] [PubMed] [Google Scholar]
- Bucciarelli E., Sunda W. G. (2003). Influence of CO2, nitrate, phosphate, and silicate limitation on intracellular DMSP in batch cultures of the coastal diatom Thalassiosira pseudonana. Limnol. Oceanogr. 48, 2256–2265 [Google Scholar]
- Bunik V. I., Fernie A. R. (2009). Metabolic control exerted by the 2-oxoglutarate dehydrogenase reaction: a cross-kingdom comparison of the crossroads between energy production and nitrogen assimilation. Biochem. J. 422, 405–421 10.1042/BJ20090722 [DOI] [PubMed] [Google Scholar]
- Capone D. G. (2000). “The marine microbial nitrogen cycle,” in Microbial Ecology of the Oceans, ed Kirchman D. (New York, NY: Wiley Liss; ), 455–494 [Google Scholar]
- Carlucci A. F. (1966). Bioassay of seawater. II. Methods for the determination of vitamin B1 in seawater. Can. J. Microbiol. 12, 1079–1089 [DOI] [PubMed] [Google Scholar]
- Carlucci A. F., Bowes P. M. (1970). Production of vitamin B12, thiamine, and biotin by phytoplankton. J. Phycol. 6, 351–357 [Google Scholar]
- Carlucci A. F., Silbernagel S. B. (1969). The effect of viamin concentrations on growth and development of vitamin-requiring algae. J. Phycol. 5, 64–67 [DOI] [PubMed] [Google Scholar]
- Carlucci A. F., Silbernagel S. B., McNally P. M. (1969). The influence of temperature and solar radiation on persistence of vitamin B12, thiamine, and biotin in seawater. J. Phycol. 5, 302–305 [DOI] [PubMed] [Google Scholar]
- Caron D. A., Dennett M. A., Lonsdale D. J., Moran D. M., Shalapyonok L. (2000). Microzooplankton herbivory in the Ross Sea, Antarctica. Deep Sea Res. II 47, 3249–3272 [Google Scholar]
- Charlson R. J., Lovelock J. E., Andreae M. O., Warren S. G. (1987). Oceanic phytoplankton, atmospheric sulphur, cloud albedo, and climate. Nature 326, 655–661 [Google Scholar]
- Chatterjee A., Li S., Zhang Y., Grove T. L., Lee M., Krebs C., et al. (2008). Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat. Chem. Biol. 4, 758–765 10.1038/nchembio.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. J. (1982). Interactions between bacteria and algae in aquatic systems. Annu. Rev. Ecol. Sys. 13, 291–314 [Google Scholar]
- Cowey C. B. (1956). A preliminary investigation of the variaton of vitamin B-12 in oceanic and coastal waters. J. Mar. Biol. Assoc. U.K. 35, 609–620 [Google Scholar]
- Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93 10.1038/nature04056 [DOI] [PubMed] [Google Scholar]
- Croft M. T., Moulin M., Webb M. E., Smith A. G. (2007). Thiamine biosynthesis in algae is regulated by riboswitches. Proc. Natl. Acad. Sci. U.S.A. 104, 20770–20775 10.1073/pnas.0705786105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft M. T., Warren M. J., Smith A. G. (2006). Algae need their vitamins. Eukaryot. Cell 5, 1175–1184 10.1128/EC.00097-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortch Q. (1990). The interaction between ammonium and nitrate uptake in phytoplankton. MEPS 61, 183–201 [Google Scholar]
- Drennan C. L., Matthews R. G., Ludwig M. L. (1994). Cobalamin-dependent methoionine synthase: the structure of a methylcobalamin-binding fragment and implicatiosn for other B12-dependent enzymes. Curr. Opin. Struct. Biol. 4, 919–929 [DOI] [PubMed] [Google Scholar]
- Droop M. R. (1957). Vitamin B12 in marine ecology. Nature 180, 1041–1042 [Google Scholar]
- Droop M. R. (1958). Requirement for thiamine among some marine and supralittoral protists. J. Mar. Biol. Assoc. U.K. 37, 323–329 [Google Scholar]
- Droop M. R. (1962). “Organic micronutrients,” in Physiology and Biochemistry of Algae, ed Lewin R. A. (New York, NY: Academic Press; ), 141–159 [Google Scholar]
- Droop M. R. (2007). Vitamins, phytoplankton and bacteria: symbiosis or scavenging? J. Plankton Res. 29, 107–113 [Google Scholar]
- Eppley R. W., Peterson B. J. (1979). Particulate organic matter flux and planktonic new production in the deep ocean. Nature 282, 677–680 [Google Scholar]
- Fernie A. R., Obata T., Allen A. E., Araújo W. L., Bowler C. (2012). Leveraging metabolomics for functional investigations in sequenced marine diatoms. Trends Plant Sci. 17, 395–403 10.1016/j.tplants.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Frank R., Leeper F., Luisi B. (2007). Structure, mechanism and catalytic duality of thiamine-dependent enzymes. Cell. Mol. Life Sci. 64, 892–905 10.1007/s00018-007-6423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigeri L. G., Radabaugh T. R., Haynes P. A., Hildebrand M. (2006). Identification of Proteins from a cell wall fraction of the diatom Thalassiosira pseudonana. Mol. Cell. Proteomics 5, 182–193 10.1074/mcp.M500174-MCP200 [DOI] [PubMed] [Google Scholar]
- Gobler C. J., Norman C., Panzeca C., Taylor G. T., Sanudo-Wilhelmy S. A. (2007). Effect of B-vitamins and inorganic nutrients on algal bloom dynamics in a coastal ecosystem. Aquat. Microb. Ecol. 49, 181–194 [Google Scholar]
- Gold K. (1968). Some factors affecting the stability of thiamine. Limnol. Oceanogr. 13, 185–188 [Google Scholar]
- Gold K., Roels O. A., Bank H. (1966). Temperature dependent destruction of thiamine in seawater. Limnol. Oceanogr. 12, 410–413 [Google Scholar]
- Goyer A. (2010). Thamine in plants: aspects of its metabolism and functions. Phytochemistry 71, 1615–1624 10.1016/j.phytochem.2010.06.022 [DOI] [PubMed] [Google Scholar]
- Grossart H., Levold F., Allgaier M., Brinkhoff S. M. (2005). Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7, 860–873 10.1111/j.1462-2920.2005.00759.x [DOI] [PubMed] [Google Scholar]
- Guillard R. R. L., Ryther J. H. (1962). Studies of marine planktonic diatoms 1. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can J. Microbiol. 8, 229–239 [DOI] [PubMed] [Google Scholar]
- Hecky R. E., Killam (1988). Nitrogen limitation of phytoplankton in freshwater and marine environments: a review of recent evidence on the effects of enrichment. Limnol. Oceanogr. 33, 796–822 [Google Scholar]
- Helliwell K. E., Wheeler G. L., Leptos K. C., Goldstein R. E., Smith A. G. (2011). Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol. Biol. Evol. 28, 2921–2933 10.1093/molbev/msr124 [DOI] [PubMed] [Google Scholar]
- Hu M., Chen Y., Lin Y. (1995). The antioxidant and prooxidant activity of some B vitamins and vitamin-like compounds. Chem. Biol. Interact. 97, 63–73 10.1016/0009-2797(95)03608-8 [DOI] [PubMed] [Google Scholar]
- Jurgenson C. T., Begley T. P., Ealick S. E. (2009). The structural and biochemical foundations of thiamine biosynthesis. Annu. Rev. Biochem. 78, 569–603 10.1146/annurev.biochem.78.072407.102340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D. M. (2002). Nutrient dynamics in the deep blue sea. Trends Microbiol. 10, 410–418 [DOI] [PubMed] [Google Scholar]
- Keller M. D., Keine R. P., Matrai P. A., Bellows W. K. (1999). Production of glycine betaine and DMSP in marine phytoplankton 1. Batch cultures. Mar. Biol. 135, 237–248 [Google Scholar]
- Keller M. D., Matrai P. A., Keine R. P., Bellows W. K. (2004). Responses of coastal phytoplankton populations to nitrogen addition: dynamics of cell-associated DMSP, GBT and homarine. Can. J. Fish. Aquat. Sci. 61, 685–699 [Google Scholar]
- Kirchman D. (2000). “Uptake and regeneration of inorganic nutrients by marine heterotrophic bacteria,” in Microbial Ecology of the Oceans, ed Kirchman D. (New York, NY: Wiley Liss; ), 261–288 [Google Scholar]
- Kirchman D. L., Wheeler P. A. (1998). Uptake of ammonium and nitrate by heterotrophic bacteria and phytoplankton in the sub-Arctic Pacific. Deep Sea Res. I 45, 347–365 [Google Scholar]
- Koch F., Marcoval M. A., Panzeca C., Bruland K. W., Sanudo-Wilhelmy S. A., Gobler C. J. (2011). The effect of vitamin B12 on phytoplankton growth and community structire in the Gulf of Alaska. Limnol. Oceanogr. 56, 1023–1034 [Google Scholar]
- Koprivova A., Suter M., Op den Camp R., Brunold C., Kopriva S. (2000). Regulation of sulfate assimilation by nitrogen in Arabadopsis. Plant Physiol. 122, 737–746 10.1104/pp.122.3.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger N., Deutzmann R., Bergsdorf C., Sumper M. (2000). Species-specific polyamines from diatoms control silica morphology. Proc. Natl. Acad. Sci. U.S.A. 97, 14133–14138 10.1073/pnas.260496497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger N., Poulsen N. (2008). Diatoms- from cell wall biogeneisis to nanotechnology. Annu. Rev. Genet. 42, 83–107 10.1146/annurev.genet.41.110306.130109 [DOI] [PubMed] [Google Scholar]
- Kudela R. M., Dugdale R. C. (2000). Nutrient regulation of phytoplankton productivity in Monterey Bay, CA. Deep Sea Res. II 47, 1023–1053 [Google Scholar]
- Lexa D., Savant J. M. (1983). The electrochemistry of vitamin B12. Acc. Chem. Res. 16, 235–243 [Google Scholar]
- Lidie K. B., Ryan J. C., Barbier M., Van Dolah F. M. (2005). Gene expression in Florida red tide dinoflagellate Karenia brevis. Mar. Biotechnol. 7, 481–493 10.1007/s10126-004-4110-1 [DOI] [PubMed] [Google Scholar]
- Lin S. (2011). Genomic understanding of dinoflagellates. Res. Microbiol. 162, 551–569 10.1016/j.resmic.2011.04.006 [DOI] [PubMed] [Google Scholar]
- Lukienko P. I., Melnichenko N. G., Zverinskii I. V., Zabrodskaya S. V. (2000). Antioxidant properties of thiamine. Bull. Exp. Biol. Med. 130, 874–876 [PubMed] [Google Scholar]
- Mahadevan A., D'Asaro E., Lee C., Perry M. J. (2012). Eddy-driven stratification initiates North Atlantic spring phytoplankton blooms. Science 337, 54–58 10.1126/science.1218740 [DOI] [PubMed] [Google Scholar]
- Maheswari U., Jabbari K., Petit J. L., Porcel B. M., Allen A. E., Cadoret J. P., et al. (2010). Digital gene expression profiling of novel diatom transcripts provides insight into their biological functions. Genome Biol. 11:R85 10.1186/gb-2010-11-8-r85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant H. J. (2005). “Cyanophytes,” in Antarctic Marine Protists, eds Scott F. J., Marchant H. J. (Hobart: Australian Biological Resources Study; ), 324–325 [Google Scholar]
- Martens J. H., Bargv H., Warren M. J., Jan D. (2002). Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 58, 275–285 10.1007/s00253-001-0902-7 [DOI] [PubMed] [Google Scholar]
- McCarthy J. J., Carpenter E. J. (1983). “Nitrogen cycling in the near surface waters of the open ocean,” in Nitrogen in the Marine Environment, eds Carpenter E. J., Capone D. G. (New York, NY: Academic Press; ), 487–572 [Google Scholar]
- McRose D., Yan S., Reistetter E., Worden A. Z. (2012). “Vitamin biosynthesis and regulation in marine algae,” in Genomics Science Contractors-Grantees Meeting, (Bethesda, MD: ). [Google Scholar]
- Menzel D. W., Spaeth J. P. (1962). Occurrence of vitamin B12 in the Sargasso Sea. Limnol. Oceanogr. 7, 151–154 [Google Scholar]
- Michael A. J. (2011). Molecular machines encoded by bacterially-derived multidomain gene fusions that potentially synthesize N- methylate and tranfer long chain polyamines in diatoms. FEBS Lett. 585, 2627–2634 10.1016/j.febslet.2011.07.038 [DOI] [PubMed] [Google Scholar]
- Moore J. K., Doney S. C., Lindsay K. (2004). Upper ocean ecosystem dynamics and iron cycling in a global three-dimensional model. Glob. Biogeochem. Cycles 18, 4028–4049 [Google Scholar]
- Moustafa A., Evans A. N., Kulis D. M., Hackett J. D., Erdner D. L., Anderson D. M., et al. (2010). Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS ONE 5:e9688 10.1371/journal.pone.0009688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbamichael M., Sañudo-Wilhelmy S. A. (2004). A new method for the determination of Vitamin B12 in seawater. Anal. Chim. Acta 517, 33–38 [Google Scholar]
- Okbamichael M., Sanudo-Wilhelmy S. A. (2005). Direct determination of vitamin B1 in seawater by solid phase extraction and high performance liquid chromatography quantification. Limnol. Oceanogr. Methods 3, 241–246 [Google Scholar]
- Panzeca C., Beck A., Leblanc K., Taylor G. T., Hutchins D. A., Sañudo-Wilhelmy S. A. (2008). Potential cobalt limitation of vitamin B12 synthesis in the North Atlantic Ocean. Glob. Biogeochem. Cycles 22, 2029–2036 [Google Scholar]
- Panzeca C., Beck A. J., Tovar-Sanchez A., Segovia-Zavala J., Taylor G. T., Gobler C. J., et al. (2009). Distributions of dissolved vitamin B12 and Co in coastal and open-ocean environments. Estuar. Coast. Shelf Sci. 85, 223–230 [Google Scholar]
- Panzeca C., Tovar-Sanchez A., Agusti S., Reche I., Duarte M., Taylor G. T., et al. (2006). B vitamins as regulators of phytoplankton dynamics. EOS 87, 593–596 [Google Scholar]
- Partensky F., Hess W. R., Vaulot D. (1999). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto E., Nieuwerburgh L., Barros M., Pedersen M., Colepicolo P., Snoeijs P. (2003). Density dependant patterns in thiamine and pigment production in the diatom Nitzschia microcephala. Phytochemistry 63, 155–163 10.1016/S0031-9422(03)00048-7 [DOI] [PubMed] [Google Scholar]
- Provasoli L. (1963). “Organic regulation of phytoplankton fertility,” in The Sea, ed Hill M. N. (New York, NY: Interscience; ), 165–219 [Google Scholar]
- Provasoli L., Carlucci A. F. (1974). “Vitamins and growth regulators,” in Algal Physiology and Biochemistry, ed Stewart W. P. D. (Oxford: Blackwell; ), 741–787 [Google Scholar]
- Ransom Hardison D., Sunda W. G., Litaker R. W., Shea D., Tester R. A. (2012). Nitrogen limitation increases brevetoxins in Karenia brevis: implications for bloom toxicity. J. Phycol. 48, 844–858 [DOI] [PubMed] [Google Scholar]
- Rapala-Kozik M., Wolak N., Kujda M., Banas A. K. (2012). Upregulation of thiamine biosynthesis in A. thaliana seedlings under salt and osmotic stress conditions is mediated by abscisic acid at the early stages of this stress response. BMC Plant Biol. 12:2 10.1186/1471-2229-12-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raux E., Schubert H. L., Warren M. J. (2000). Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell. Mol. Life Sci. 57, 1880–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. (2002). Comparative genomics of thiamine biosyntheisis in prokaryotes. J. Biol. Chem. 277, 48949–48959 10.1074/jbc.M208965200 [DOI] [PubMed] [Google Scholar]
- Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. (2003). Comparative genomics of the Vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278, 41148–41159 10.1074/jbc.M305837200 [DOI] [PubMed] [Google Scholar]
- Roth J. R., Lawrence J. G., Bobik T. A. (1996). Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50, 137–181 10.1146/annurev.micro.50.1.137 [DOI] [PubMed] [Google Scholar]
- Saito M. A., Goepfert T. J., Ritt J. T. (2008). Some thoughts on the concept of colimitation: three definitions and the importance of bioavailability. Limnol. Oceanogr. 53, 276–290 [Google Scholar]
- Sañudo-Wilhelmy S. A., Cutter L., Durazo R., Smail E., Gomez-Consarnau L., Webb E. A., et al. (2012). Multiple B-vitamin deficiency in large areas of the coastal ocean. Proc. Natl. Acad. Sci. U.S.A. 109, 14041–14045 10.1073/pnas.1208755109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sañudo-Wilhelmy S. A., Okbamichael M., Gobler C. J., Taylor G. T. (2006). Regulation of phytoplankton dynamics by vitamin B12. Geophys. Res. Lett. 33, LO4604. [Google Scholar]
- Scheffel A., Poulsen N., Shian S., Kroger N. (2011). Nanopatterned protein microrings from a diatom that direct silica morphognesis. Proc. Natl. Acad. Sci. U.S.A. 108, 3175–3180 10.1073/pnas.1012842108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauzer G. N., Deutsch E. (1969). Reactions of cobalt(I) supernucleophiles. the alkylation of vitamin B12s, cobaloximes (I), and related compounds. J. Am. Chem. Soc. 91, 3341–3350 [DOI] [PubMed] [Google Scholar]
- Scott J. M., Weir D. G. (1981). The methyl folate trap: a physiological response in man to prevent methyl group deficiency in kwashiorkor (methionine deficiency) and an explanation for folic-acid-induced exacerbation of subacute combined degeneration in pernicious anaemia. Lancet 318, 337–340 [DOI] [PubMed] [Google Scholar]
- Shihira I., Krauss R. W. (1965). Chlorella: Physiology and Taxonomy of 41 Isolates. College Park, PA: University of Maryland Press [Google Scholar]
- Stefels J. P. (2000). Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 43, 183–197 [Google Scholar]
- Sunda W., Kieber D. J., Kiene R. P., Huntsman S. (2002). An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320 10.1038/nature00851 [DOI] [PubMed] [Google Scholar]
- Sunda W. G., Hardison R., Kiene R., Bucciarelli E., Harada H. (2007). The effect of nitrogen limitation on cellular DMSP and DMS release in marine phytopankton: climate feedback implications. Aquat. Sci. 69, 341–351 [Google Scholar]
- Swift D. (1981). Vitamin levels in the Gulf of Maine and ecological significance of vitamin B12 there. J. Mar. Res. 39, 375–403 [Google Scholar]
- Swift D. G., Guillard R. R. (1978). Unexpected response to vitamin B12 of dominant centric diatoms from the spring bloom in the Gulf of Maine. J. Phycol. 14, 377–386 [Google Scholar]
- Swift D. G., Taylor W. R. (1972). Growth of vitamin B12 - limited cultures: Thalassiosira pseudonana, Monochyrsis lutheri, and Isochrysis galbana. J. Phycol. 10, 385–391 [Google Scholar]
- Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. (2011). Sulfur assimilarion in photosynthetic organisms: molecular function and regulation transporters and assimilatory proteins. Annu. Rev. Plant Biol. 62, 157–184 10.1146/annurev-arplant-042110-103921 [DOI] [PubMed] [Google Scholar]
- Tang Y. Z., Koch F., Gobler C. J. (2010). Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc. Natl. Acad. Sci. U.S.A. 107, 20756–20761 10.1073/pnas.1009566107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611 10.1126/science.1218344 [DOI] [PubMed] [Google Scholar]
- Toulza E., Shin M. S., Blanc G., Audic S., Laabir M., Collos Y., et al. (2010). Gene expression in proliferating cells of the dinoflagellate Alexandrium catenella. Appl. Environ. Microbiol. 76, 4521–4529 10.1128/AEM.02345-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb M., Marquet A., Mendel R., Rebeille F., Smith A. (2007). Elucidating biosynthetic pathways for vitamins and cofactors. Nat. Prod. Rep. 24, 988–1008 10.1039/b703105j [DOI] [PubMed] [Google Scholar]
- Worden A. Z., Lee J. H., Mock T., Rouzé P., Simmons M. P., Aerts A. L., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268–274 10.1126/science.1167222 [DOI] [PubMed] [Google Scholar]
- Zehr J. P., Ward B. B. (2002). Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl. Environ. Microbiol. 68, 1015–1024 10.1128/AEM.68.3.1015-1024.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Rodionov D. A., Gelfand M. S., Gladyshev V. N. (2009). Comparative genomics analysis of nickel, cobalt, and vitamin B12 utilization. BMC Genomics 10:78 10.1186/1471-2164-10-78 [DOI] [PMC free article] [PubMed] [Google Scholar]