Abstract
The ability to generate tailor-made, functionalized polyester (polyhydroxyalkanoate, PHA) beads in bacteria by harnessing their natural carbon-storage granule production system is an exciting recent development. Proteins that naturally attach to the polyester granule core were rationally engineered to enable in vivo production of PHA beads which are applicable in bioseparation, protein purification, enzyme immobilization and diagnostics and which show advantageous properties toward the development of safe and efficient particulate vaccines. These beads are recombinantly produced as fully functional, insoluble polyester inclusions that can be easily separated from the cell. This simple one-step production of functionalized beads provides a tantalizing alternative to current commercial functional beads, for which proteins must be expressed, purified and then chemically attached to solid supports. The recent success in generating antigen-displaying PHA granules in the food-grade bacterium Lactococcus lactis capable of mediating protective immunity against Mycobacterium tuberculosis infection highlights the promise and flexibility of this new technology.
Keywords: PHA synthase, PHA, PHB, bacterial inclusions, beads, biopolymer, bioseparation, polyester, polyhydroxyalkanoate, vaccines
Introduction
Bacterial polymer inclusions
Many bacteria and archaea are capable of producing polymer inclusions, which serve as stockpiled carbon storage material during periods of nutrient imbalance.1 These can be made of glycogen (polysaccharide), polyphosphate (polyanhydride), cyanophycin (polyamide), or polyhydroxyalkanoate (PHA, polyester).2 Bacterial polyester granules have generated considerable industrial interest; PHA produced by bacterial fermentation using renewable and waste materials are a desirable alternative to petroleum-based plastics. Additionally, the biodegradability and biocompatibility of PHA make them attractive for medical applications.2-4 Recently, the discovery that PHA inclusions can be propagated as stable beads and can be engineered to show a variety of functionalities has created significant commercial interest.2-6
PHA synthesis
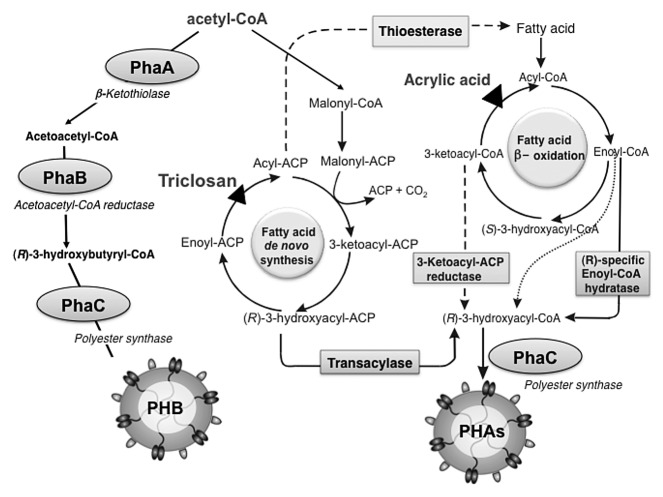
The key enzyme for PHA biosynthesis is the PHA synthase, which polymerizes (R)-3-hydroxyacyl-CoA thioesters into polyester with the concomitant release of coenzyme A. There are several classes of PHA synthases, which preferentially utilize different carbon chain length (R)-3-hydroxyacyl-CoA precursors, thus generating a variety of polyesters with differing side chain lengths and consequently different material properties.7
The Ralstonia eutropha enzyme PhaC is the most characterized PHA synthase. It catalyzes the formation of the polyester poly(3-hydroxybutyrate) (PHB) from (R)-3-hydroxybutyryl-CoA monomers. The (R)-3-hydroxybutyryl-CoA molecule is produced by two other enzymes encoded in the R. eutropha PHB biosynthesis operon: PhaA, a β-kethothiolase which condenses two molecules of acetyl-CoA into acetoacetyl-CoA, and PhaB, an NADPH-dependent acetoacetyl-CoA reductase which reduces acetoacetyl-CoA into the (R)-3-hydroxybutyryl-CoA utilized by the synthase PhaC. Together, these three proteins, PhaA, PhaB and PhaC, are sufficient for production of PHB granules in the presence of acetyl-CoA.
PHA granule self-assembly and structure
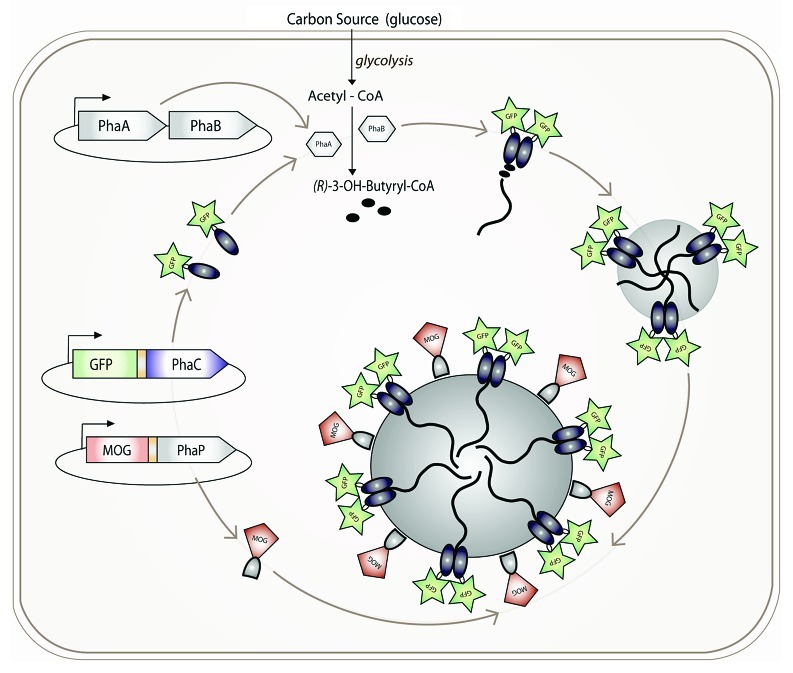
The PHA granule consists primarily of an amorphous, hydrophobic polyester core surrounded by an outer layer of proteins. It has been suggested that phospholipids might contribute to this outer layer; however, no discrimination between isolation dependent contamination and inherent presence has been undertaken. Several models for particle formation have been described.8,9 The clearest evidence supports the micelle model, by which the amphipathic nature of the PHA synthase, once it has begun synthesizing its attached hydrophobic polyester chain, is sufficient for granule formation (Fig. 1). Granules readily form in vitro10 in the absence of any cellular elements involved in other models. In vivo, granule formation is localized, occurring at cell poles11,12 or at “central mediation elements.”13
Figure 1.
Formation of functionalized PHB granules.
In natural PHA granules, PHA synthases, phasins, depolymerases and various regulator proteins coat the bead surface. In R. eutropha, these include the genetic regulator PhaR and the granule-coating structural phasin PhaP, which together affect the number and size of granules.14 In contrast to the other proteins which are attached to the bead surface via hydrophobic interactions, the PHA synthase PhaC remains covalently attached to the granule, anchored by its bound polyester chain. This presents a unique natural cross-link of a protein to a polymeric support structure.
The stability of the PHA beads outside of the cell, combined with the covalent attachment of the PHA synthase protein and/or the tight binding of the phasin proteins to the bead surface, has recently been exploited for generating functionalized nano-/micro-beads. Functional proteins of interest are produced as genetic fusions to the PhaC synthase and/or PhaP phasin proteins. When these proteins are expressed in the cell, granules form with the fusion proteins stably displayed on the granule surface, resulting in one-step production of functionalized beads. This process generates engineered biopolyester beads that are capable of binding IgG,15-17 inorganic substrates18 and biotin,19 displaying target antigens,20-23 catalyzing enzymatic reactions,24-26 acting as vaccines,27-29 enabling production of purified proteins30-32 and diagnostic imaging.11,12,18,21,23 The biobased, biodegradable and biocompatible properties of these granules make them an attractive alternative to chemically manufactured beads.
Recombinant Production of PHB Inclusions
E. coli
Most of the research on recombinantly produced PHB inclusions as functional beads uses E. coli to express the phaCAB (PHB biosynthesis) operon from R. eutropha and, if required, the phaP gene from R. eutropha. For production of most of the polyester beads described below, the PHB synthase gene (phaC) from R. eutropha was cloned into the pET14b expression vector under the control of the T7 promoter,28 and the phaA and phaB genes mediating synthesis of (R)-3-hydroxybutyryl-CoA were cloned into a separate vector under the control of the lac promoter.33 Upon IPTG induction and provision with an excess carbon source (e.g., glucose), recombinant E. coli BL21(DE3) harboring these plasmids readily formed PHB inclusions, which were separated from the cell by lysis and gradient centrifugation.18 Granule formation can be conveniently monitored in vivo by staining cells with the fluorescent lipophilic dye Nile red.34
The extensive understanding of E. coli genetics and metabolism, its long-established use for large-scale recombinant protein production, and the many strains available for specific recombinant protein features make E. coli a very adaptable host for PHA bead bioengineering, and it has been the primary host for functionalized recombinant PHB bead production to date.
Lactococcus lactis
However, generating beads for biomedical applications in E. coli or other Gram-negative bacteria is problematic, because of the presence of lipopolysaccharide (LPS) endotoxins, which require extensive purification processes to remove. To get around this limitation, the generally-regarded-as-safe, food-grade bacterium Lactococcus lactis has recently been engineered to produce PHA beads.35 The phaCAB genes from R. eutropha were synthesized to adapt the codon bias to L. lactis, cloned into the pNZ8148 vector as an operon under the control of the nisA promoter and transformed into the L. lactis strain NZ9000. Induction with nisin in glucose–supplemented M17 media resulted in PHB granule formation. However, the beads were both smaller in size (100–200 nm) and contributed less PHB per dried biomass (6% of cellular dry weight by gas chromatography/mass spectrometry (GC-MS) analysis) than reported for recombinant E. coli.
The feasibility of commercial-scale production of functionalized PHA beads in L. lactis remains hampered by the low PHB yield obtained in this organism and hence is currently limited to the production of high-value medical products, such as vaccines. Initial attempts to adjust media, carbon source and aeration, as well as supplementation with growth enhancers l-arginine or hemin, did not improve PHB yield beyond the 6% mark.35 Improvement of yield will likely require more fundamental measures than modified growth conditions, possibly including re-engineering metabolic flux to push carbon utilization away from lactate production and toward the PHB biosynthesis pathway.35
Other bugs and systems
The broad range of bacteria capable of producing PHA naturally or recombinantly, as well as the success of producing recombinant PHA granules in a range of hosts as far-reaching as yeasts36 and plants,37 suggests that many organisms with desirable host properties could be exploited for functionalized PHA bead production. Although E. coli remains the most commercially viable host for large-scale production, functionalized PHB beads have been also been generated in R. eutropha31 and Pseudomonas aeruginosa,25 as well as L. lactis.
Additionally, recombinant PHA synthase proteins from other organisms with preferences for different (R)-3-hydroxyacyl-CoA precursors can generate polyester granules with varied material properties.7 Many PHA synthases are also fairly promiscuous in their use of 3-hydroxyacyl-CoA thioester precursors, and metabolic engineering to influence precursor composition can result in tailor-made PHAs (Fig. 2).2,8,13
Figure 2.
Metabolic pathways of PHA formation.
Applications
Protein purification
Early examples of functionalized PHB beads exploited the phasin protein PhaP’s affinity for the hydrophobic bead surface as an affinity tag for protein purification. The enhanced green fluorescent protein (EGFP) gene was fused to phaP, separated by an intein encoding sequence. Beads expressing the PhaP-intein-EGFP fusion protein were produced in R. eutropha using the native phaCAB PHB synthesis operon. After bead isolation and induction of intein self-cleavage, pure, functional EGFP protein was released from the PHB granules. This was repeated successfully with maltose-binding protein (MBP) and β-galactosidase (lacZ) as the fusion partners (Fig. 3).30,31
Figure 3.
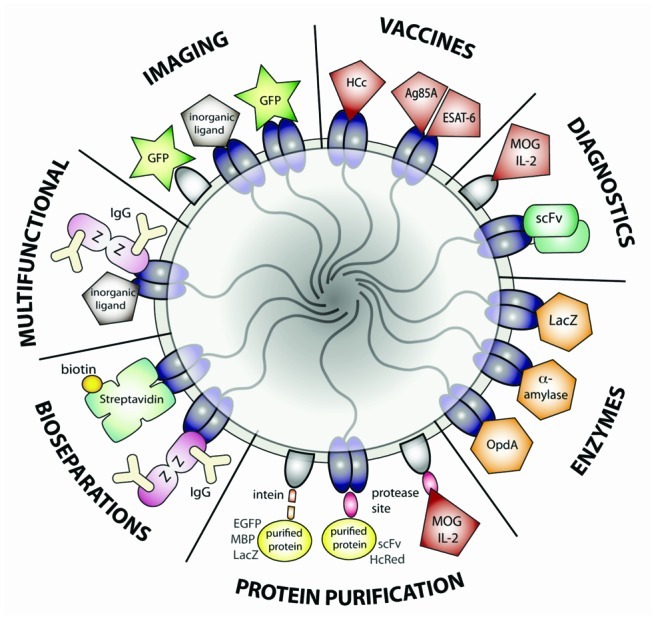
Current applications of functionalized PHA granules.
Likewise, fusion proteins containing proteolytic cleavage sites between PhaC/PhaP and a protein of interest have been used to isolate recombinant proteins. The PhaP-MOG/IL2 diagnostic beads discussed below (see Diagnostics) contained an enterokinase cleavage site between the fusion partners; incubation with enterokinase resulted in complete cleavage of the functional fusion partner (MOG/IL2).21 However, PhaP is only attached to the beads by hydrophobic interactions and thus may potentially contaminate the soluble fraction. Therefore, this method was recently extended to fusions with the covalently-attached, PhaC synthase, using HcRed (a fluorescent protein) or scFv (an anti-β-galactosidase antibody fragment). Incubation of these beads with enterokinase resulted in release of pure functional HcRed or scFv proteins.38
Enzymes
Several PHA beads with active immobilized enzymes have been developed so far. The first proof-of-concept experiment demonstrated that an N-terminal fusion of lacZ to PhaC expressed in Pseudomonas aeruginosa produced granules with functional β-galactosidase activity, which were stable in storage for several months.25
Next, an optimized α-amylase variant (TermamylTM) was fused to PhaC. The resulting beads exhibited α-amylase activity consistent with the enzyme kinetics of free α-amylase.26 These beads were reusable and stable up to 85°C, demonstrating their suitability for industrial use.
Recently, enzyme-displaying polyester beads have been used for bioremediation. The Agrobacterium radiobacter organophosphohydrolase enzyme OpdA, which hydrolyzes neurotoxic organophosphorous pesticides, was immobilized on PHA beads by recombinant expression as a PhaC fusion protein.39 The OpdA-expressing beads exhibited similar enzymatic properties to free OpdA protein, were functional up to 65°C, and retained functionality after storage in tap water for up to 11 d. The OpdA beads successfully detoxified the pesticide coumaphos in wool scour effluent, a toxic byproduct of the wool industry, demonstrating their industrial utility.
Construction of an entire enzymatic pathway on PHB beads is also possible. Recently, a single genetic fusion of the PhaA-PhaB-PhaC PHB synthesis pathway was demonstrated to produce PHB granules in recombinant E. coli.24
Bioseparations
The immunoglobulin G (IgG) binding ZZ domain of Protein A from Staphylococcus aureus has been successfully fused to PhaC. The resulting ZZ-PhaC beads, recombinantly expressed in E. coli, had similar IgG binding capacity and purification power to commercial protein-A sepharose.15-17 Recently, functional ZZ-PhaC beads have been expressed recombinantly in Lactococcus lactis as well.35 Interestingly, the L. lactis beads displayed improved IgG-binding capacity, which may be due to the reduced size (increased protein-domain displaying surface area per bead mass) and/or higher density of the ZZ-PhaC fusion protein at the bead surface.
Additionally, a fusion of PhaC and streptavidin has produced PHB beads capable of binding biotin. These beads were successfully used to bind biotinylated antibodies and to purify biotinylated DNA.19
Diagnostics
Antigen-displaying PHB beads can be used in Fluorescence-Activated Cell Sorting (FACS) to detect the presence of specific antibodies. Fusions of mouse interleukin-2 (IL2) or myelin oligodendrocyte glycoprotein (MOG) to PhaP produced beads recognized by monoclonal anti-IL2 or anti-MOG antibodies conjugated to a fluorescent dye. FACS analysis showed clear separation of the beads according to expressed antigen; furthermore, the beads retained this capability even after one year in storage.21 Additionally, FACS analysis of MOG-PhaP showed specific and sensitive detection of anti-MOG serum antibodies from MOG-immunized mice.
Approaching diagnostics from the other angle, an anti-β-galactosidase single-chain antibody variable fragment (scFv) has been successfully expressed on PHB beads as a PhaC fusion. The PhaC-scFv beads bound β-galactosidase both in vitro and in vivo.22 This technique has clear implications for diagnostics; beads containing immobilized diagnostic antibody fragments to detect specific molecular antigens could be recombinantly produced in one-step, eliminating the need for separate antibody purification and crosslinking to supports.
Imaging
Several groups have created fluorescent PHB granules exhibiting functional fluorescent proteins such as GFP and YFP, through both PhaC and PhaP fusions.11,12,30,32
Additionally, many of the beads with bioseparation functionality can be used for imaging. For example, Protein A beads can be used in combination with labeled antibodies for ELISA, FACS or in vivo visualization. PHB beads displaying both the Protein A antibody-binding ZZ domain and functional binding peptides for inorganic gold and silica particles have been developed;18 such granules may prove useful for targeted deposition of inorganic compounds or medical imaging contrast agents.
Multifunctional beads expressing both specific binding functionality and self-labeling such as GFP expression could also be used for imaging and diagnostics. One example has already been produced: co-expressing PhaC-GFP and MOG-PhaP fusion proteins in recombinant E. coli resulted in fluorescent beads recognized by anti-MOG and anti-GFP antibodies.23
Vaccines
The suitability of antigen-displaying PHB beads as particulate vaccines has been demonstrated. PHB beads carrying the Ag85A-ESAT-6 antigens from Mycobacterium tuberculosis fused to PhaC induced a mixed Th1/Th2 immune response and showed no adverse effects in mice.35 However, the potential presence of LPS endotoxins in the E. coli-produced beads limits their applicability for medical applications.
To improve the potential for PHB biobeads as viable human vaccines, the Ag85A-ESAT-6 genes were synthesized to adapt the codon bias to L. lactis and fused to the phaC gene in the pNZ8148 vector previously shown to generate PHB beads in L. lactis.35 Functional beads displaying the mycobacterial antigens were isolated from both L. lactis and the previously reported E. coli and used to immunize mice. Bead-based vaccines from both organisms generated an immune response and protected against Mycobacterium bovis challenge to a similar degree as the current human TB vaccine, M. bovis Bacille Calmette-Guérin (BCG).29 However, the L. lactis—produced beads had a reduced effect on IL-10 production and less protection against spleen infection than E. coli—produced beads. Both beads were significantly more effective than recombinant protein alone, confirming previous findings that particulate vaccines are more effective than soluble antigens.40
Recently, PHB beads in E. coli and L. lactis were engineered to display the Hepatitis C virus core antigen (HCc).28 As with the M. tuberculosis Ag85A-ESAT-6-PhaC bead vaccines, the HCc-PhaC bead vaccines produced significant and specific immune responses in vaccinated mice. Interestingly, while the L. lactis beads generated a Th1 specific immune response, the E. coli beads triggered a more general response; this may be due to an adjuvant-like effect of contaminating LPS or E. coli host proteins, or to the larger size of the E. coli-produced beads.
Conclusion and Future Directions
Functionalized PHA biobeads are becoming increasingly recognized for their potential in biotechnological and biomedical applications. The many successes highlighted above indicate the adaptability of this new technology, by which a variety of functional particles can be designed “to order” by rational engineering of functional proteins into the bead-forming PHA synthase or other granule-attached proteins. However, much of this potential remains untapped. E. coli, while a well-established system for recombinant protein production, is far from the only suitable source of tailor-made PHB granules and is especially problematic for medical applications due to the potential for endotoxin contamination. The recent production of PHB bead vaccines in the generally-regarded-as-safe bacterium Lactococcus lactis28,29 exemplifies the potential benefits of further bioengineering and ongoing efforts to push the boundaries of this new technology.
Acknowledgments
This work was supported by research grants from New Zealand Ministry of Science and Innovation, Massey University and Polybatics Ltd.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/19567
References
- 1.Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–72. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rehm BHA. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol. 2010;8:578–92. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- 3.Chen GQ. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev. 2009;38:2434–46. doi: 10.1039/b812677c. [DOI] [PubMed] [Google Scholar]
- 4.Rodríguez-Carmona E, Villaverde A. Nanostructured bacterial materials for innovative medicines. Trends Microbiol. 2010;18:423–30. doi: 10.1016/j.tim.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Grage K, Jahns AC, Parlane N, Palanisamy R, Rasiah IA, Atwood JA, et al. Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules. 2009;10:660–9. doi: 10.1021/bm801394s. [DOI] [PubMed] [Google Scholar]
- 6.Jahns AC, Rehm BHA. Relevant uses of surface proteins—display on self-organized biological structures. Microb Biotechnol. 2011 doi: 10.1111/j.1751-7915.2011.00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rehm BH. Polyester synthases: natural catalysts for plastics. Biochem J. 2003;376:15–33. doi: 10.1042/BJ20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehm BHA. Biogenesis of microbial polyhydroxyalkanoate granules: a platform technology for the production of tailor-made bioparticles. Curr Issues Mol Biol. 2007;9:41–62. [PubMed] [Google Scholar]
- 9.Tian J, Sinskey AJ, Stubbe J. Kinetic studies of polyhydroxybutyrate granule formation in Wautersia eutropha H16 by transmission electron microscopy. J Bacteriol. 2005;187:3814–24. doi: 10.1128/JB.187.11.3814-3824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerngross TU, Martin DP. Enzyme-catalyzed synthesis of poly[(R)-(-)-3-hydroxybutyrate]: formation of macroscopic granules in vitro. Proc Natl Acad Sci U S A. 1995;92:6279–83. doi: 10.1073/pnas.92.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters V, Rehm BHA. In vivo monitoring of PHA granule formation using GFP-labeled PHA synthases. FEMS Microbiol Lett. 2005;248:93–100. doi: 10.1016/j.femsle.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 12.Peters V, Becher D, Rehm BHA. The inherent property of polyhydroxyalkanoate synthase to form spherical PHA granules at the cell poles: the core region is required for polar localization. J Biotechnol. 2007;132:238–45. doi: 10.1016/j.jbiotec.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Aldor IS, Keasling JD. Process design for microbial plastic factories: metabolic engineering of polyhydroxyalkanoates. Curr Opin Biotechnol. 2003;14:475–83. doi: 10.1016/j.copbio.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Pötter M, Madkour MH, Mayer F, Steinbüchel A. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology (Reading, Engl.) 2002; 148:2413-2426. [DOI] [PubMed]
- 15.Jahns AC, Rehm BHA. Tolerance of the Ralstonia eutropha class I polyhydroxyalkanoate synthase for translational fusions to its C terminus reveals a new mode of functional display. Appl Environ Microbiol. 2009;75:5461–6. doi: 10.1128/AEM.01072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brockelbank JA, Peters V, Rehm BHA. Recombinant Escherichia coli strain produces a ZZ domain displaying biopolyester granules suitable for immunoglobulin G purification. Appl Environ Microbiol. 2006;72:7394–7. doi: 10.1128/AEM.01014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis JG, Rehm BHA. ZZ polyester beads: an efficient and simple method for purifying IgG from mouse hybridoma supernatants. J Immunol Methods. 2009;346:71–4. doi: 10.1016/j.jim.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Jahns AC, Haverkamp RG, Rehm BHA. Multifunctional inorganic-binding beads self-assembled inside engineered bacteria. Bioconjug Chem. 2008;19:2072–80. doi: 10.1021/bc8001979. [DOI] [PubMed] [Google Scholar]
- 19.Peters V, Rehm BHA. Protein engineering of streptavidin for in vivo assembly of streptavidin beads. J Biotechnol. 2008;134:266–74. doi: 10.1016/j.jbiotec.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Shang G, You M, Peng S, Wang Z, Wu H, et al. Endotoxin removing method based on lipopolysaccharide binding protein and polyhydroxyalkanoate binding protein PhaP. Biomacromolecules. 2011;12:602–8. doi: 10.1021/bm101230n. [DOI] [PubMed] [Google Scholar]
- 21.Bäckström BT, Brockelbank JA, Rehm BHA. Recombinant Escherichia coli produces tailor-made biopolyester granules for applications in fluorescence activated cell sorting: functional display of the mouse interleukin-2 and myelin oligodendrocyte glycoprotein. BMC Biotechnol. 2007;7:3. doi: 10.1186/1472-6750-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grage K, Rehm BHA. In vivo production of scFv-displaying biopolymer beads using a self-assembly-promoting fusion partner. Bioconjug Chem. 2008;19:254–62. doi: 10.1021/bc7003473. [DOI] [PubMed] [Google Scholar]
- 23.Atwood JA, Rehm BHA. Protein engineering towards biotechnological production of bifunctional polyester beads. Biotechnol Lett. 2009;31:131–7. doi: 10.1007/s10529-008-9836-9. [DOI] [PubMed] [Google Scholar]
- 24.Mullaney JA, Rehm BHA. Design of a single-chain multi-enzyme fusion protein establishing the polyhydroxybutyrate biosynthesis pathway. J Biotechnol. 2010;147:31–6. doi: 10.1016/j.jbiotec.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Peters V, Rehm BHA. In vivo enzyme immobilization by use of engineered polyhydroxyalkanoate synthase. Appl Environ Microbiol. 2006;72:1777–83. doi: 10.1128/AEM.72.3.1777-1783.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasiah IA, Rehm BHA. One-step production of immobilized alpha-amylase in recombinant Escherichia coli. Appl Environ Microbiol. 2009;75:2012–6. doi: 10.1128/AEM.02782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parlane NA, Wedlock DN, Buddle BM, Rehm BHA. Bacterial polyester inclusions engineered to display vaccine candidate antigens for use as a novel class of safe and efficient vaccine delivery agents. Appl Environ Microbiol. 2009;75:7739–44. doi: 10.1128/AEM.01965-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parlane NA, Grage K, Lee JW, Buddle BM, Denis M, Rehm BHA. Production of a particulate hepatitis C vaccine candidate by an engineered Lactococcus lactis strain. Appl Environ Microbiol. 2011;77:8516–22. doi: 10.1128/AEM.06420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parlane NA, Grage K, Mifune J, Basaraba RJ, Wedlock DN, Rehm BHA, et al. Vaccines displaying mycobacterial proteins on biopolyester beads stimulate cellular immunity and induce protection against tuberculosis. Clin Vaccine Immunol. 2012;19:37–44. doi: 10.1128/CVI.05505-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banki MR, Gerngross TU, Wood DW. Novel and economical purification of recombinant proteins: intein-mediated protein purification using in vivo polyhydroxybutyrate (PHB) matrix association. Protein Sci. 2005;14:1387–95. doi: 10.1110/ps.041296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnard GC, McCool JD, Wood DW, Gerngross TU. Integrated recombinant protein expression and purification platform based on Ralstonia eutropha. Appl Environ Microbiol. 2005;71:5735–42. doi: 10.1128/AEM.71.10.5735-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Wu H, Chen J, Zhang J, Yao Y, Chen GQ. A novel self-cleaving phasin tag for purification of recombinant proteins based on hydrophobic polyhydroxyalkanoate nanoparticles. Lab Chip. 2008;8:1957–62. doi: 10.1039/b807762b. [DOI] [PubMed] [Google Scholar]
- 33.Amara AA, Rehm BHA. Replacement of the catalytic nucleophile cysteine-296 by serine in class II polyhydroxyalkanoate synthase from Pseudomonas aeruginosa-mediated synthesis of a new polyester: identification of catalytic residues. Biochem J. 2003;374:413–21. doi: 10.1042/BJ20030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiekermann P, Rehm BHA, Kalscheuer R, Baumeister D, Steinbüchel A. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol. 1999;171:73–80. doi: 10.1007/s002030050681. [DOI] [PubMed] [Google Scholar]
- 35.Mifune J, Grage K, Rehm BHA. Production of functionalized biopolyester granules by recombinant Lactococcus lactis. Appl Environ Microbiol. 2009;75:4668–75. doi: 10.1128/AEM.00487-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leaf TA, Peterson MS, Stoup SK, Somers D, Srienc F. Saccharomyces cerevisiae expressing bacterial polyhydroxybutyrate synthase produces poly-3-hydroxybutyrate. Microbiology (Reading, Engl.) 1996; 142 (Pt 5):1169-1180. [DOI] [PubMed]
- 37.John ME, Keller G. Metabolic pathway engineering in cotton: biosynthesis of polyhydroxybutyrate in fiber cells. Proc Natl Acad Sci U S A. 1996;93:12768–73. doi: 10.1073/pnas.93.23.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grage K, Peters V, Rehm BHA. Recombinant protein production by in vivo polymer inclusion display. Appl Environ Microbiol. 2011;77:6706–9. doi: 10.1128/AEM.05953-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blatchford PA, Scott C, French N, Rehm B. Immobilization of organophosphohydrolase OpdA from Agrobacterium radiobacter by overproduction at the surface of polyester inclusions inside engineered Escherichia coli. Biotechnol Bioeng. 2011 doi: 10.1002/bit.24402. In press. [DOI] [PubMed] [Google Scholar]
- 40.Rice-Ficht AC, Arenas-Gamboa AM, Kahl-McDonagh MM, Ficht TA. Polymeric particles in vaccine delivery. Curr Opin Microbiol. 2010;13:106–12. doi: 10.1016/j.mib.2009.12.001. [DOI] [PubMed] [Google Scholar]