Abstract
Single-stranded DNA (ssDNA) recombineering is a technology which is used to make subtle changes in the chromosome of several bacterial genera. Cells which express a single-stranded DNA binding protein (RecT or Bet) are transformed with an oligonucleotide which is incorporated via an annealing and replication-dependent mechanism. By in silico analysis we identified ssDNA binding protein homologs in the genus Lactobacillus and Lactococcus lactis. To assess whether we could further improve the recombineering efficiency in Lactobacillus reuteri ATCC PTA 6475 we expressed several RecT homologs in this strain. RecT derived from Enterococcus faecalis CRMEN 19 yielded comparable efficiencies compared with a native RecT protein, but none of the other proteins further increased the recombineering efficiency. We successfully improved recombineering efficiency 10-fold in L. lactis by increasing oligonucleotide concentration combined with the use of oligonucleotides containing phosphorothioate-linkages (PTOs). Surprisingly, neither increased oligonucleotide concentration nor PTO linkages enhanced recombineering in L. reuteri 6475. To emphasize the utility of this technology in improving probiotic features we modified six bases in a transcriptional regulatory element region of the pdu-operon of L. reuteri 6475, yielding a 3-fold increase in the production of the antimicrobial compound reuterin. Directed genetic modification of lactic acid bacteria through ssDNA recombineering will simplify strain improvement in a way that, when mutating a single base, is genetically indistinguishable from strains obtained through directed evolution.
Keywords: Lactobacillus reuteri, Lactococcus lactis, RecT, genetic engineering, phosphorothioate, recombineering, reuterin
Introduction
Lactic acid bacteria (LAB) are Gram-positive bacteria and members of this group have been used for fermentation of foods for thousands of years. It is because of this long history of human consumption that many strains are now generally recognized as safe (GRAS).1,2 In addition, several strains of LAB are being investigated for the potential health-promoting properties they elicit.3-5 The combination of safe use and potential health-promoting abilities have led to exploration and application of these organisms as a platform for the delivery of therapeutics.6-9 In order to gain a better understanding of how these bacteria interact with their environment, and the ability to modify strains for subsequent use as therapeutic delivery vehicles, development of powerful genetic tools is of pivotal importance.
ssDNA recombineering has been applied in E. coli for over a decade, and we recently introduced this technology to a range of lactobacilli and Lactococcus lactis.10,11 The methodology relies on the expression of a homolog of the lambda phage-derived ssDNA binding protein (Bet) or a homolog derived from the Rac prophage (RecT). These proteins have collectively been referred to as recombinases.10,12 In cells expressing a recombinase one can introduce an oligonucleotide that subsequently is incorporated in the chromosome. The role of the recombinase is proposed to bind to the oligonucleotide to protect it from nuclease degradation and to promote annealing between the oligonucleotide and the template strand.13 Rather than a homologous recombination event, incorporation of the oligonucleotide occurs via an annealing and replication-dependent mechanism.10,14-17 Thus, the oligonucleotide may serve as a primer for lagging strand replication or possibly as a small Okazaki fragment.
Several strategies have been described which improve the ssDNA recombineering efficiency in E. coli, including the use of oligonucleotides with modified bases.18 The most dramatic increase in recombineering efficiency is obtained with an oligonucleotide which avoids the mismatch repair system.19,20 In L. reuteri we were able to show that the mismatch repair system is completely avoided with an oligonucleotide that yields five adjacent mismatches. Avoiding the mismatch repair system is critical to generate mutants in L. reuteri without the need for antibiotic selection. Moreover, whole genome sequence analysis of L. reuteri and L. lactis strains that had undergone a recombineering event showed that ssDNA recombineering is specific and not hypermutagenic.11 ssDNA recombineering is thus an efficient tool to make chromosomal mutations in lactic acid bacteria, and other Gram-positive organisms, to study gene function and to improve (probiotic) strain characteristics.
Herein, we discuss optimization parameters to increase recombineering efficiencies in L. lactis and L. reuteri and show how this technology may improve existing properties of a strain by introducing base changes in the chromosome.
Results and Discussion
RecT and Bet recombinases are not widely distributed in lactobacilli.
We recently established ssDNA recombineering in three Lactobacillus species and Lactococcus lactis by expressing a recombinase, RecT1, that was derived from L. reuteri ATCC PTA 6475.11 Previous studies have shown that different recombinase homologs have the ability to function in a wide range of hosts and thus screening multiple recombinases would be a useful approach when establishing or optimizing ssDNA recombineering in any organism.12,21 We focused on lactobacilli and L. lactis for the identification of recombinases which may serve as a useful database to improve or establish recombineering. As recombinases are highly diverse in their sequence content we identified homologs by utilizing the Position Specific Iteration Basic Local Alignment Search Tool (PSI-BLAST).22 To this end we used RecT1 derived L. reuteri ATCC PTA 6475, and the Bet homolog ORF245 derived from L. lactis SMQ-86 as initial query sequences.11,12
The genus Lactobacillus represents the largest genus within the phylum Firmicutes, and the NCBI currently holds 192 genomes encompassing 46 species in their databases. Our search within this genus resulted in the identification of 19 recombinase homologs that are present in 16 Lactobacillus strains encompassing 10 species (Table 1). In three strains two RecT homologs were identified: L. reuteri DSM 20016, L. casei BL23 and L. paracasei 8700:2 (Table 1). When we used the Bet homolog ORF245 as a query sequence no additional recombinases were identified. Thus, RecT and Bet recombinases are not widely distributed in the genus Lactobacillus.
Table 1. Identification of recombinases in lactobacillus and Lactococcus lactis.
| strain | Accession nr. | AAæ | ID¶ | |
|---|---|---|---|---|
| Lactobacillus |
|
|
|
|
| reuteri |
DSM 20016 |
YP_001271411.1 |
309 |
309/309 (100%) |
| plantarum |
WCFS |
YP_004888633 |
295 |
136/298 (46%) |
| pentosus |
IG1 |
CCC18574 |
295 |
131/297 (44%) |
| reuteri |
DSM 20016 |
YP_001271733.1 |
329 |
183/297 (62%) |
| plantarum |
ATCC 14917 |
ZP_07078351 |
299 |
134/302 (44%) |
| plantarum |
ST-III |
YP_003925340 |
299 |
133/302 (44%) |
| buchneri |
NRLL B-30956 |
YP_004398761 |
315 |
142/315 (45%) |
| fermentum |
IFO 3956 |
YP_001843483 |
342 |
163/320 (51%) |
| buchneri |
ATCC 11577 |
ZP_03943265 |
333 |
141/287 (49%) |
| paracasei |
8700:2 |
ZP_04674145 |
284 |
47/303 (16%) |
| paracasei |
8700:2 |
ZP_04674324 |
279 |
45/273 (16%) |
| paracasei |
ATCC 25302 |
ZP_03964192 |
287 |
54/288 (19%) |
| mali |
KCTC 3596 |
ZP_09447952 |
274 |
57/295 (19%) |
| rhamnosus |
Lc 705 |
YP_003173539 |
291 |
137/285 (48%) |
| rhamnosus |
R0011 |
EHJ20790 |
291 |
136/285 (48%) |
| casei |
BL23 |
YP_001987247 |
287 |
54/288 (19%) |
| rhamnosus |
GG |
YP_003170850 |
291 |
136/285 (48%) |
| casei |
BL23 |
YP_001986912 |
295 |
53/314 (17%) |
| ruminis |
SPM0211 |
ZP_08564335 |
254 |
38/263 (14%) |
| Lactococcus |
|
|
|
|
| lactis subsp lactis |
KF147 |
YP_003353956 |
308 |
71/320 (22%) |
| lactis subsp lactis |
CV56 |
ADZ64766 |
308 |
70/320 (22%) |
| lactis subsp cremoris |
CNCM I-1631 |
EHE93774 |
308 |
68/311 (22%) |
| lactis subsp lactis |
Il1403 |
NP_266602 |
268 |
54/236 (23%) |
| lactis subsp cremoris |
A76 |
AEU39829 |
115 |
26/117 (22%) |
| lactis subsp cremoris | A76 | AEU41341 | 245 | 242/245 (99%) |
To identify recombinases in the the genus Lactobacillus, Lactococcus lactis we searched the NCBI protein databases of the respective genera using the tool PSI-BLAST with standard algorithm settings. A total of five iterations were performed. As a query sequence we used RecT1 derived from L. reuteri ATCC PTA 6475 (accession number ZP_03961322, 329 amino acids). Results highlighted in gray are the PSI-BLAST results using ORF245 (Lactococcus lactis strain SMQ-85, ϕUL36; accession number AAF74061, 245 amino acids) as a query. ælength of the recombinase homolog in amino acids; ¶number of identical residues compared with the (partial) amino acid sequence of L. reuteri ATCC PTA 6475 RecT (or L. lactis ORF245 in the gray rows), followed by the percentage of identical residues within the alignment in brackets.
L. lactis is represented by 16 subspecies and a total of 34 genomes in the NCBI databases. Using L. reuteri ATCC PTA 6475 RecT1 as a query we identified recombinase homologs in five different strains (Table 1). In three strains (KF147, CV56 and CNCM I-1631) the identified proteins are closely related as their amino acid sequences share > 95% identity. In L. lactis subsp cremoris A76 a putative recombinase was identified; however, an in-frame stop codon (TAA) yielded an open reading frame that is considerably shorter (115 amino acids) than the recombinases which have been experimentally validated. Downstream of the stop codon an open reading frame of 98 amino acids was identified. Using a pairwise BLASTP comparison between the deduced amino acid sequence of the 3'-end of the recombinase and L. reuteri RecT revealed 24% identity between the sequences. Using L. lactis ORF245 SMQ-86 (Bet) as a query we identified an additional recombinase in L. lactis subsp cremoris A76 which differs by only three amino acids from ORF245 (Table 1, highlighted in gray). When we searched the protein databases of the genus Lactobacillus and strains of L. lactis using the amino acid sequences of the second RecT homolog of L. reuteri or RecT derived from E. faecalis CRMEN 19 as initial queries we did not identify any additional homologs.
Expression of RecT derived from E. faecalis yields comparable recombineering efficiencies in L. reuteri compared with native RecT protein.
Previously we have shown that expression of a native recombinase (RecT1) of L. reuteri yields recombineering efficiencies ranging from 0.3–19%, which allows identification of mutants without the need for antibiotic selection.11 To investigate whether we could further increase the recombineering efficiency in L. reuteri by expressing different RecT homologs, we cloned genes putatively encoding RecT from a range of sources in the backbone of pSIP411. These are two RecT homologs derived from L. casei BL23, a single homolog derived from L. plantarum BAA-793 (pseudonymous to L. plantarum WCFS1), and a homolog derived from E. faecalis CRMEN 19 (a kind gift from Donald Court, NCI-Frederick, MD). The recombineering efficiency in L. reuteri expressing the different homologs was assessed as previously described.11 Briefly, competent cells were prepared in which expression of RecT was induced 20 min prior to harvesting. One hundred micrograms of oligonucleotide was transformed in cells which upon incorporation in the chromosome, yields an amino acid change in the RNA polymerase protein (RpoB) that confers resistance to the drug rifampicin. Thus, the level of rifampicin-resistant colonies is a measure for the efficiency of recombineering.
Expression of a RecT homolog derived from L. casei BL23 (accession number YP_001987247) from pJP046 was detrimental to L. reuteri cells as cells aggregated and could not be resuspended, and was therefore not included for further analysis. The highest recombineering efficiency was obtained in cells expressing L. reuteri RecT1 or RecT derived from E. faecalis (Table 2). E. faecalis RecT also proved efficient in E. coli, yielding recombinants at levels comparable to E. coli Bet, and higher than E. coli RecT.12 The observation that RecT from E. faecalis CRMEN 19 is efficient in phylogenetically distant bacteria suggests that this is a highly efficient recombinase. Direct comparison of the amino acid sequences of E. faecalis RecT with L. reuteri RecT1 revealed 52% identity. Although it is tempting to suggest that recombinases with highly similar sequences promote ssDNA recombineering at comparable levels in the same genetic background, careful selection of a recombinase is required. For example, we noted a 250-fold lower recombineering efficiency in L. reuteri expressing the L. plantarum RecT homolog from pJP052 compared with L. reuteri RecT expressed from pJP042, yet both proteins share 54% amino acid identity. Preliminary experiments in L. plantarum BAA-793 harboring either pJP052 or pJP042 also yielded higher recombineering levels with pJP042 (unpublished results). Thus, when establishing this technology in a new host it is therefore useful to screen a large panel of recombinases derived from different hosts.
Table 2. Relative activity of RecT homologs derived from different lactic acid bacteria in L. reuteri ATCC PTA 6475.
| plasmid | source RecT | accession number‡ | relative activity (%) | SD |
|---|---|---|---|---|
| pJP042 |
L. reuteri ATCC PTA 6475 |
ZP_03961195 |
100 |
NA |
| pJP043 |
E. faecalis CRMEN 19 |
NA |
76 |
38 |
| pJP045 |
L. casei BL23 |
YP_001986912 |
15 |
8 |
| pJP046 |
L. casei BL23 |
YP_001987247 |
ND |
ND |
| pJP052 | L. plantarum BAA-793 | YP_004888633 | 0.4 | 0.3 |
The recombineering efficiencies in L. reuteri expressing the different RecT homologs. Data are expressed as percentage activity relative to cells harboring pJP042. Data presented are the averages of five independent experiments. ‡NA, not applicable; SD, standard deviation; ND, not determined. The E. faecalis CRMEN19 RecT only differs by two amino acids compared with RecT from E. faecalis V583 (accession number AE016830), as determined by Datta et al.12
The lagging strand bias of recombineering efficiency is approximately 4,000-fold in L. reuteri but 25-fold in L. lactis.
The highest efficiency of ssDNA recombineering is obtained with an oligonucleotide that is—apart from the mutations to be incorporated—identical to the lagging strand of replication. The lagging strand bias in E. coli, Pseudomonas syringae and in Mycobacterium tuberculosis is approximately 3 to 10-fold, 10-fold and 1,000 to 10,000-fold, respectively.12,21,23 One hypothesis for this lagging strand bias is that more single-stranded DNA is exposed during replication on the lagging strand because of the formation of the different Okazaki fragments. At present it is unknown why the magnitude of strand bias in recombineering in Mycobacterium tuberculosis is different from E. coli.
To determine to what extent there is a strand bias in recombineering in L. reuteri and L. lactis we transformed cells expressing RecT with 100 µg oligonucleotide that is incorporated in either the leading strand [oJP610 (L. reuteri) or oJP1157 (L. lactis)] or the lagging strand [oJP577 (L. reuteri) or oJP563 (L. lactis)] of replication. The leading and lagging strand oligonucleotides yield the same amino acid changes, namely T487S and H488R for L. reuteri and H486N for L. lactis. The level of rifampicin-resistant colonies per 109 cells was determined (Fig. 1). In L. reuteri approximately 4,000-fold more rifampicin-resistant colonies were obtained with the lagging strand oligonucleotide (oJP577) compared with the leading strand oligonucleotide (oJP610). In L. lactis the bias is not as great since the difference is 25-fold higher for the lagging strand compared with the leading strand. It is unclear why L. lactis shows a relatively small lagging strand bias (similar to E. coli) whereas L. reuteri displays a large bias, similar to the lagging strand bias observed in Mycobacterium tuberculosis. One possibility is the replication forks of L. reuteri and Mycobacterium may function in a way such that larger regions of single-stranded DNA are exposed compared with E. coli or L. lactis. Although the rpoB genes are located in similar regions of the chromosome, located at 559 Kb (L. lactis) and 455 Kb (L. reuteri) from the ori, the strand bias might be affected by the specific position of the target site that is being recombineered in a chromosome. Regardless of the size of the lagging strand bias, these results show that also in lactic acid bacteria the highest efficiency of recombineering is achieved with an oligonucleotide that is incorporated in the lagging strand of replication.
Figure 1.
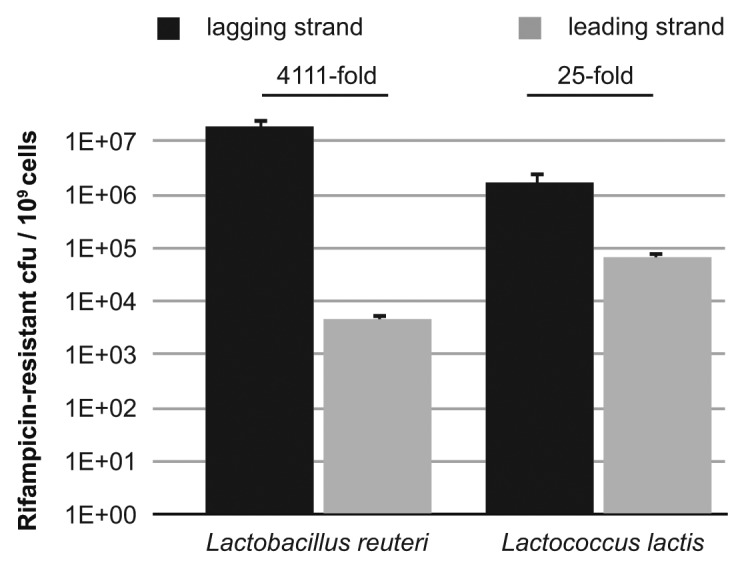
Lagging strand bias of ssDNA recombineering in L. reuteri and L. lactis. One hundred micrograms of oligonucleotide, incorporated in either the leading or lagging strand of DNA replication, was transformed in L. reuteri and L. lactis to quantify the level of recombineering. Incorporation of the oligonucleotide yields a rifampicin-resistant phenotype, and the level of rifampicin-resistant colonies was expressed per 109 cells. The black bars represent the number of rifampicin-resistant colonies obtained from transforming an oligonucleotide that is incorporated in the lagging strand of DNA replication (oJP577 and oJP563 for L. reuteri and L. lactis, respectively) and the gray bars represent the number of rifampicin-resistant colonies obtained from transforming a leading strand oligonucleotide (oJP610 and oJP1157 for L. reuteri and L. lactis, respectively). The fold difference in the lagging strand bias is indicated above the bars. Data presented are the averages of three independent experiments and the error bars indicate standard deviation.
Increased levels of oligonucleotide with phosphorothioate linkages increase the recombineering efficiency in L. lactis but not in L. reuteri.
Besides the use of an oligonucleotide that is incorporated in the lagging strand of replication, there are a range of options that have been utilized in E. coli to increase the recombineering efficiency. Among these are co-transformation of carrier DNA or the use of phosphorothiotate oligonucleotides (PTOs) to avoid host nucleases, oligonucleotides with other chemical modifications, or design of oligonucleotides that avoid the mismatch repair system (MMR).18-20 We have previously shown how the MMR in L. reuteri may be avoided using oligonucleotides that have multiple adjacent mismatches, however, we had not investigated whether we can increase the recombineering efficiency in either L. reuteri or L. lactis by means of avoiding the host nucleases.11
PTOs are oligonucleotides that contain a phosphorothioate (PT) bond by replacing an oxygen atom with a sulfur atom in the phosphate backbone of the oligonucleotide, which yields a nuclease-resistant oligonucleotide.24 We designed three oligonucleotides which have five PT-linkages located either at the 5'-end, the 3'-end or at both termini (Fig. 2A). When incorporated in the chromosome all PT-containing oligonucleotides will yield the same base changes compared with the respective control oligonucleotide that lacks PT-linkages, subsequently resulting in a rifampicin-resistant phenotype. The resulting amino acid changes for L. lactis and L. reuteri are identical as described above. Recombineering efficiency is determined by plating cells on antibiotic-free plates and on plates containing rifampicin, followed by expressing the number of rifampicin-resistant colonies per 109 viable cells.
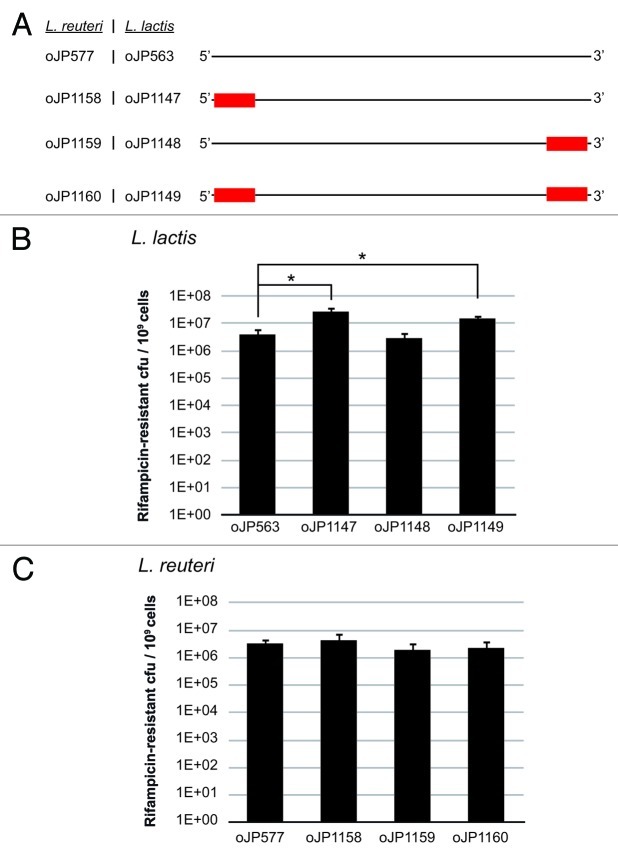
Figure 2.
Increased ssDNA recombineering efficiency in L. lactis with oligonucleotides harboring phosphorothioate-linkages. (A) Schematic overview of the different phosphorothioate oligonucleotides (PTOs). The red rectangle indicates the presence of 5 PT linkages in the oligonucleotide at either the 5’-end, the 3’-end or at both proximal ends. The control oligonucleotide (listed on top) does not have any PT linkages. The two columns left of each oligonucleotide list the corresponding names for L. reuteri and L. lactis. The panel of oligonucleotides for L. reuteri or for L. lactis have the same nucleotide sequence and yield a rifampicin-resistant phenotype once incorporated in the chromosome. (B) Recombineering efficiencies in L. lactis transformed with 200 μg control oligonucleotide (oJP563) or 200 μg PTOs. Statistical significance (p < 0.05) is indicated with an asterisk between oJP563 and oJP1147, and oJP563 and oJP1149. All data presented are the averages of three independent experiments, and error bars represent standard deviation. (C) Recombineering efficiencies in L. reuteri transformed with 100 μg control oligonucleotide (oJP577) or 100 μg PTOs. Recombinants were expressed as the number of rifampicin-resistant colonies per 109 cells. All data presented are the averages of three independent experiments, and error bars represent standard deviation.
Increasing the oligonucleotide concentration to 200 ∝g in L. reuteri resulted in cell lysis and negatively impacted recombineering efficiency (unpublished results). In L. lactis increasing the levels of oJP563 from 100 ∝g to 200 ∝g yielded on average (n = 3) an increase from 2E + 06 ± 0.7 to 4E + 06 ± 2.8 rifampicin-resistant colonies per 109 cells, respectively. We were surprised to note that further increasing the oligonucleotide concentration to 500 ∝g yielded on average 1.6E + 07 ± 0.3 rifampicin-resistant colonies per 109 cells (~10-fold increase over 100 ∝g). For construction of mutants in L. lactis NZ9000 it would be useful to use 500 ∝g oligonucleotide so that fewer colonies need to be screened; however, for the purpose of identifying the effect of PTOs (Fig. 2A) on the recombineering efficiency in L. lactis NZ9000, we initially used 200 ∝g to minimize cost for oligonucleotide synthesis.
The presence of 5 PT-linkages on the 5'-end of the oligonucleotide (oJP1147) improves recombineering efficiency 6.8-fold (p = 0.03) compared with the control oligonucleotide lacking the PT-linkages (oJP563) (Fig. 2B). Thus, oJP1147 yielded recombinants in 2.7% of the total population of cells that survived the electroporation. Compared with the control no increase in the number of recombinants was observed with an oligonucleotide harboring PT-linkages on the 3'-end (oJP1148) but placing five PT-linkages on either end (oJP1149) yielded a 3.8-fold increase (p = 0.01) in the number of recombinants (Fig. 2B). Having established that oJP1147 increases the recombineering efficiency we transformed 500 ∝g oJP563 and oJP1147 accordingly which yielded recombinants in 1.6% and 5.6%, respectively, of the population of cells that survived the transformation.
It needs to be emphasized that these numbers are obtained by a direct comparison of the number of colonies that are formed on non-selective and rifampicin-containing plates. Thus, cells are only resistant to rifampicin which have already incorporated the oligonucleotide during the recovery period after the transformation. When we screen 200 colonies from the non-selective plates for rifampicin-resistance we note that the level of rifampicin-resistant colonies is about 2-fold higher, yielding ~3% and ~12% recombinants obtained with 500 ∝g oJP563 and oJP1147, respectively. This is indicative of the fact that during growth on non-selective plates the oligonucleotides may still be incorporated into the chromosome. However, prolonging the outgrowth of transformed L. lactis cells did not further increase the efficiency when cells are plated on rifampicin-containing plates (unpublished results). This may suggest that either the growth rate of rifampicin-resistant mutants is different to that of wild type cells, or that binding of the oligonucleotide to the template strand hampers the replication. Further experiments are required to evaluate whether fewer PT-linkages may yield a similar result or whether increasing the number of PT-linkages may further increase the level of recombinants. Based on three independent experiments we also noted that the fold difference between oJP563 and oJP1147 is 6.7- and 3.5-fold when transforming 200 ∝g and 500 ∝g oligonucleotide, respectively. It is plausible that exonucleases in the cell are being saturated by the higher level of oligonucleotides that are transformed. In E. coli, for example, co-transformation of carrier-DNA also increases the recombineering efficiency and also here the rationale is saturating the host nucleases.20
In L. reuteri no increase in the recombineering efficiency was obtained when transforming PTOs. One hundred micrograms of a PTO harboring five PT-linkages on the 5'-end (oJP1158), the 3'-end (oJP1159) or on both termini (oJP1160) yielded comparable recombineering efficiencies compared with the control oligonucleotide oJP577 (Fig. 2C). In three independent experiments we noted that the recombineering efficiency with the control oligonucleotide oJP577 is slightly lower (3.3E + 06 ± 1.3 recombinants per 109 cells) compared with efficiencies reported in Figure 1 which exceed 1E + 07 recombinants per 109 cells, and these differences may be attributed to different batches of oligonucleotides.
As the presence of five PT linkages did not increase level of recombinants compared with the control oligonucleotide oJP577, we subsequently assessed recombineering efficiencies with 100 ∝g oligonucleotide harboring 10 PT linkages. However, no increase in recombineering efficiency was obtained (unpublished results). An oligonucleotide which only contained PT linkages completely abolished the recombineering activity in L. reuteri yielding 100,000-fold fewer rifampicin-resistant colonies compared with oJP577, corresponding to levels which are comparable to the negative control (no DNA) (unpublished results). Some bacteria have the ability to naturally incorporate sulfur-linkages in the DNA backbone, a characteristic which is encoded by a cluster of dnd genes which is hypothesized to serve as a restriction modification system.25,26 More recently an endonuclease has been identified in Streptomyces coelicor A(3)2 (Sco4631) that cleaves DNA near a PT-linkage thereby excluding the possibility for uptake of the dnd gene cluster.27 Based on BLAST analyses and searching the database for identification of dnd genes we did not identify a dnd gene cluster in L. reuteri.28 Although we could not identify a homolog of the S. coelicor endonuclease Sco4631 by BLAST analysis it is plausible that L. reuteri encodes other endonucleases which confer a similar function. If an oligonucleotide that only has PT-linkages anneals to the template strand, cleavage by an endonuclease would subsequently result in ablation of ssDNA recombineering. Thus, a panel of different optimizations may be required for different strains of lactic acid bacteria to improve/establish ssDNA recombineering and the rules established in E. coli will not always apply.
Subtle base changes in the chromosome of L. reuteri increase the reuterin production levels.
Single-stranded recombineering presents the ability to optimize probiotic properties in a subtle and time-efficient manner without the need for antibiotic selection. We previously showed that mutations in the D-Ala-D-Ala ligase gene, yielding a single amino acid change, converted L. reuteri to a vancomycin-sensitive phenotype.11 Here we provide an example of how ssDNA recombineering may be used to modulate a putative probiotic factor: the production of an antimicrobial compound.
Reuterin (3-hydroxypropionaldehyde) is a broad-spectrum antimicrobial compound that is produced by L. reuteri.29 It has proven effective in the inhibition of a range of intestinal pathogens, including Vibrio cholerae, Salmonella enterciae, enterohemorrhagic E. coli (EHEC) and enteropathogenic E. coli (EPEC).30 Reuterin is produced as an intermediate in the production of 1,3-propanediol from glycerol, which is converted to reuterin by the enzyme glycerol dehydratase. In cells derived from the mid-logarithmic growth phase, lower levels of reuterin are produced whereas levels are increased when cells reach the stationary phase. Sriramulu et al. reported the presence of CRE (catabolite repression element) sequence located downstream of the predicted -10 promoter region (Fig. 3A) of the glycerol dehydratase locus. CRE loci are binding sites for carbon catabolite control protein A (CcpA), which is an important regulator in the carbon catabolite repression in Gram-positive bacteria.32 In L. lactis a correlation was found between the location of the CRE sequence relative to the promoter sequence and subsequent repression or activation.33 Activation was predicted when the CRE sequence was located upstream of the -35 region, whereas repression was predicted when the CRE sequence was located in or downstream of the -35 and -10 region. We hypothesized that mutating the CRE locus would increase the reuterin levels in L. reuteri cells grown in MRS medium, which contains a high concentration of glucose.
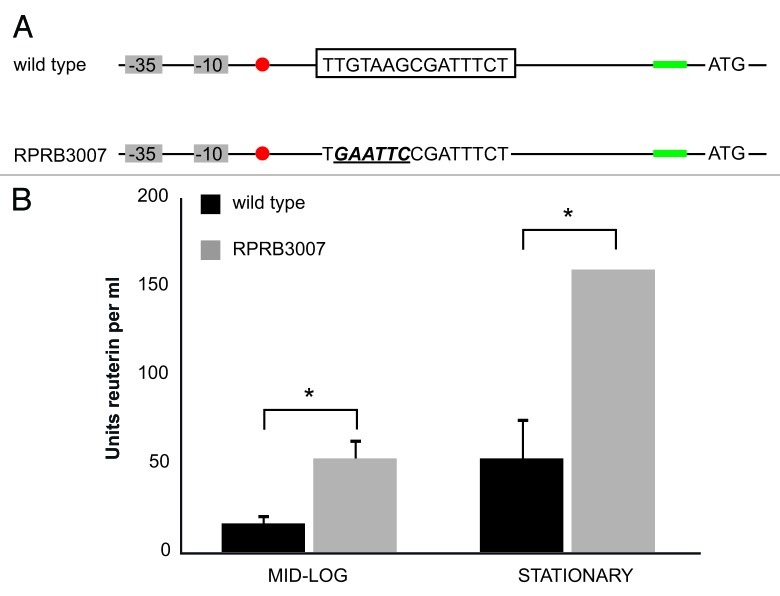
Figure 3.
Increased reuterin production followed by modification of the CRE region upstream of the pdu operon. (A) Schematic overview of the transcriptional regulatory elements located upstream of the pdu operon as determined before in L. reuteri wild type (top).31 Grey rectangles indicate the predicted -35 and -10 regions, the red dot indicated the transcriptional start, boxed sequence represents the catabolite response element (CRE), the green rectangle represents the Shine-Dalgarno sequence, and ATG is the translational start site. Bottom: upon incorporation of oJP675 six adjacent bases are modified in the CRE region. The newly incorporated sequence is italicized and underlined. (B) Reuterin minimum inhibitory concentration assay with L. reuteri wild type and RPRB3007. Cells were harvested at growth phases representative for the mid-logarithmic phase (OD600 = 0.6) or stationary phase (OD600 = 2) followed by a reuterin assay, a minimum inhibitory concentration assay, and conversion to Units reuterin per ml as described in Materials and Methods. The Units reuterin/ml for the wild type strain (black bars) and strain RPRB3007 (gray bars) were determined for both growth phases. Asterisks indicates statistical significance (p < 0.05). Data presented are averages of three independent experiments, and error bars indicate standard deviation.
We designed an oligonucleotide (oJP675) that, when incorporated in the chromosome, changes six adjacent bases in the CRE locus (Fig. 3A). We identified a mutant genotype by MAMA-PCR, and following isolation of a pure genotype the strain was designated RPRB3007. To assess reuterin production, cells were harvested during mid-logarithmic growth phase (OD600 = 0.6) and at the onset of stationary phase (OD600 = 2), and reuterin production was monitored. Using a minimum inhibitory concentration (MIC) assay we found that approximately 3-fold more reuterin is produced by RPRB3007 compared with the wild type with cells derived from both growth phases (p < 0.05) (Fig. 3B). The fact that modification of the CRE region does not yield comparable reuterin levels in both growth phases suggests that other regulatory elements play a role.
In summary, we identified recombinase homologs in Lactobacillus and Lactococcus lactis. Expression of RecT homologs derived from L. casei and L. plantarum did not improve the recombineering efficiency in L. reuteri ATCC PTA 6475; however, RecT derived from E. faecalis CREM 19 was equally efficient compared with L. reuteri RecT1. Increasing the oligonucleotide concentration to 500 ∝g combined with the use of PTOs only proved useful in L. lactis, yielding recombinants in 12% of the population. This improvement will make the identification of mutations without the need for any type of selection much easier in L. lactis. To demonstrate the utility of ssDNA recombineering we made six base changes in the promoter region upstream of the pdu-operon, yielding a 3-fold increase in the production of the antimicrobial reuterin. In both L. reuteri ATCC PTA 6475 and L. lactis NZ9000 ssDNA recombineering is currently the most time-efficient methodology to make targeted subtle genome changes without the need for antibiotic selection, and we envisage the methodology will be a useful approach for several lactic acid bacteria.
Materials and Methods
Bacterial strains, plasmids and media.
All strains and plasmids used in this study are listed in Table S1. All lactobacilli used in this study were cultured under anaerobic conditions at 37°C in deMan Rogosa Sharpe (MRS) medium (Difco, BD BioSciences). Lactococcus lactis NZ9000 was cultured statically at 30°C in M17 broth (Difco, BD BioSciences) supplemented with 0.5% (w/v) glucose. E. coli was cultured in Luria-Bertani (LB) broth (Difco, BD BioSciences) at 37°C with vigorous shaking. Electrocompetent cells of L. reuteri and L. lactis were prepared as described before.34,35 For selection of L. reuteri harboring derivatives of pSIP411, 5 ∝g/ml erythromycin was used, and rifampicin-resistant colonies were selected on plates harboring 25 ∝g/ml rifampicin. Selection for L. lactis NZ9000 harboring pJP005 was achieved using 5 ∝g/ml chloramphenicol, and rifampicin-resistant resistant colonies were selected on plates supplemented with 50 ∝g/ml rifampicin.
Reagents and enzymes.
All restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (NEB). PCR amplification for cloning purposes were performed with KOD hot start DNA polymerase (Novagen). Prior to restriction digestion, ligation or transformation DNA was precipitated with Pellet Paint Co-precipitant (Novagen).
Identification of RecT homologs by PSI-BLAST.
The level of homology between RecT homologs is low, and we therefore opted for PSI-BLAST to identify homologs in the genera Lactobacillus, and in Lactococcus lactis strains. As queries, the amino acid sequences of the RecT homologs derived from Lactobacilllus reuteri ATCC PTA 6475 (accession number ZP_03961195 [RecT1]; ZP_03961322 [RecT2]), Enterococcus faecalis V583 (accession number AE016830 [EF2132]) and a Bet homolog of Lactococcus lactis SMQ-85, ϕUL36 (accession number AAF74061 [ORF245]) were used. For each search a total of five iterations using a standard e-value cut-off of 0.005 was performed. Results were further filtered by setting a cut-off to 10% amino acid identity. In the Lactobacillus database there are several L. reuteri genomes that are highly similar (strains DSM20016, JCM 1112, ATCC PTA 6475, ATCC PTA 4659). We chose to only present the RecT homologs identified in L. reuteri DSM 20016 but similar results are obtained for the other strains mentioned.
Construction of RecT expression vectors.
All oligonucleotides were ordered from IDTDNA Technologies and are listed in Supplemental Table 2. We amplified and cloned recT genes from different sources in the backbone of the sakacin-based expression vector pSIP411.36 Using conventional colony PCR the recT genes were amplified with oligonucleotide pairs oJP413-oJP414 (accession number YP_004888633, L. plantarum BAA-793), oJP409-oJP410 (accession number YP_001986912, L. casei BL23), oJP411-oJP412 (accession number YP_001987247, L. casei BL23) and oJP373-oJP161.1 (recT E. faecalis CRMEN 19 from E. coli SIMD44).12 The backbone of pSIP411 was amplified with oligonucleotide pair oJP367-oJP371, followed by DpnI treatment to digest template DNA. The different amplicons were purified by Pellet Paint precipitation (Novagen) and subsequently blunt-end ligations were performed using molar ratio’s of 1:1 of vector:insert. The orientation of each of the inserts was verified by PCR using oligonucleotide pairs oJP414-oJP415 (L. plantarum), oJP409-oJP416 and oJP411-oJP416 (L. casei) to yield constructs pJP052, pJP045 and pJP046, respectively. The amplicon corresponding to the recT gene of E. faecalis was digested with NcoI-XbaI followed by ligation into the backbone of pSIP411 that was generated with oligonucleotide pair oJP367-oJP368 and treated with the same enzymes. Ligation was performed using a molar ratio of 1:1 vector:insert. Insertion was verified by PCR analysis (oJP373-oJP161.1) and restriction digest analysis, and the resulting construct was named oJP043.
Recombineering experiments.
Cells for recombineering purposes were prepared as described before.11 All oligonucleotides were ordered from IDTDNA Technologies at a 100 nmol scale and desalted.
To test the recombineering efficiency in L. reuteri ATCC PTA 6475 expressing the different RecT homologs we first established pJP043, pJP045, pJP046 and pJP052 in this strain. As previously described, the recombineering efficiency was determined by transforming 100 ∝g oJP577 in the different strains.11 This oligonucleotide, when incorporated, yields an amino acid change yielding a rifampicin-resistant phenotype. The level of rifampicin-resistant colonies was adjusted per 109 cells, and the efficiency of the different proteins was expressed relative to the levels obtained with pJP042.
To determine the lagging strand bias of recombineering in L. reuteri and L. lactis, we used the same approach as described above. Oligonucleotides which are incorporated in the lagging strand of replication (oJP577 and oJP563 for L. reuteri and L. lactis, respectively) or the leading strand of replication (oJP610 and oJP1157 for L. reuteri and L. lactis, respectively) were used. One hundred micrograms oligonucleotide was transformed in cells, and the number of rifampicin-resistant colonies per 109 cells were determined.
Oligonucleotides with phosphorothioate (PT) linkages were designed such that 5 consecutive PT linkages were present either on the 5'-end, the 3'-end or on both termini of the oligonucleotide (see Fig. 2A). In L. reuteri expressing RecT from pJP042 100 ∝g of each oligonucleotide was transformed and levels were expressed relative to the control oligonucleotide oJP577. L. lactis NZ9000 expressing RecT from pJP005 was transformed with 200 ∝g or 500 ∝g oligonucleotide and the experiment was performed in an identical manner as described before.
Mutating CRE locus upstream of pdu operon.
The Catabolite Response Element (CRE) located upstream of the pdu operon was previously described.31 We designed an oligonucleotide (oJP675) that, when incorporated, makes six adjacent base changes within the CRE locus thereby creating an EcoRI restriction endonuclease site that is unique in the 500 bp flanks. L. reuteri ATCC PTA 6475 competent cells harboring pJP042 were prepared upon induction for expression of RecT and subsequently transformed with 100 ∝g oJP675. From the pool of viable cells we screened 300 colonies by PCR (oligonucleotide pair oJP673-oJP674) to yield a 1 kb amplicon. The amplicons were subsequently subjected to restriction digest analysis with EcoRI, and a digested amplicon (a 500 bp fragment) was indicative for incorporation of oJP675. The corresponding culture was re-streaked in order to separate the mixed genotypes and a pure genotype was identified by subsequent PCR amplification and restriction digest analysis as described above. The newly constructed strain was denoted RPRB3007.
Reuterin production assay.
Reuterin production was measured as previously described with minor modifications.30,37,38 Briefly, L. reuteri wild type and strain RPRB3007 were grown overnight, and subsequently the strains were diluted to OD600 = 0.05. At OD600 = 0.6 and OD600 = 2, representative for the mid-logarithmic and the stationary growth phase, respectively, cells were harvested by centrifugation (10 min 4,000 × g). The pellets were washed twice with 50 mM sodium phosphate buffer (pH 7.4), and wet weight of the pellet was determined. The pellets were resuspended in 250 mM glycerol to a concentration of 3.3 mg/ml, and 15 ml was transferred to a 15 ml conical screw cap tube (corresponding to 50 mg cell pellet). The cell suspension was incubated for 1.5 h at 37°C, followed by centrifugation (10 min 4,000 × g). The reuterin-containing supernatant was filter-sterilized by passing it through a 0.22 ∝m filter (Millipore), and the filtered supernatant was stored at 4°C for up to a week prior to use.
Minimum inhibitory concentration (MIC) assay to assess antimicrobial reuterin levels.
This assay was performed as described before with minor modifications.30,37,38 Briefly, 2-fold dilutions were prepared of reuterin in water in a total volume of 100 ∝l, and added in triplicate to a 96-well plate (VWR Scientific). A fresh overnight of E. coli DH5〈 cells were diluted to ~104 cells per ml in 2× Luria-Bertani broth, and 100 ∝l was transferred to all wells. As a control, 100 ∝l of bacterial culture was added to 100 ∝l water. The relative reuterin concentration levels were determined as the reciprocal of the reuterin dilution which did not yield E. coli growth.
Supplementary Material
Acknowledgments
This work was supported in part by a subcontract to R.A.B. by National Institutes of Health [R01AT004326] to James Versalovic; and by a Strategic Partnership Grant by Michigan State University. The authors thank Donald Court (Center for Cancer Research, National Cancer Institute, Frederick, MD) for providing E. coli SIMD44, David Mills (Department of Viticulture and Enology, University of California at Davis, CA) for providing L. casei BL23, and Eamonn Connolly (Biogaia AB) for providing L. reuteri ATCC PTA 6475.
Disclosure of Potential Conflicts of Interest
The authors declare that there is no conflict of interest.
Supplemental Material
Supplemental materials may be found here: http://www.landesbioscience.com/journals/bioe/article/21049/
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/21049
References
- 1.Mogensen G, Salminen S, O’Brien J, Ouwehand A, Holzapfel W, Shortt C, et al. Food microorganisms: Health benefits, safety evaluation and strains with documented history of use in foods. Bull Int Dairy Fed. 2002:4–9. [Google Scholar]
- 2.Mogensen G, Salminen S, O’Brien J, Ouwehand A, Holzapfel W, Shortt C, et al. Inventory of microorganisms with a documented history of use in food. Bull Int Dairy Fed. 2002:10–9. [Google Scholar]
- 3.Lebeer S, Vanderleyden J, De Keersmaecker SCJ. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8:171–84. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 4.Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol. 2012;10:66–78. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 5.Ventura M, O’Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 6.Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–5. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 7.Krüger C, Hu Y, Pan Q, Marcotte H, Hultberg A, Delwar D, et al. In situ delivery of passive immunity by lactobacilli producing single-chain antibodies. Nat Biotechnol. 2002;20:702–6. doi: 10.1038/nbt0702-702. [DOI] [PubMed] [Google Scholar]
- 8.Guimarães VD, Gabriel JE, Lefèvre F, Cabanes D, Gruss A, Cossart P, et al. Internalin-expressing Lactococcus lactis is able to invade small intestine of guinea pigs and deliver DNA into mammalian epithelial cells. Microbes Infect. 2005;7:836–44. doi: 10.1016/j.micinf.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto S, Wada J, Katayama T, Jikimoto T, Nakamura M, Kinoshita S, et al. Genetically modified Bifidobacterium displaying Salmonella-antigen protects mice from lethal challenge of Salmonella Typhimurium in a murine typhoid fever model. Vaccine. 2010;28:6684–91. doi: 10.1016/j.vaccine.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A. 2001;98:6742–6. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Pijkeren JP, Britton RA. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012;40:e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci U S A. 2008;105:1626–31. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–88. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Muyrers JPP, Rientjes J, Stewart AF. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol Biol. 2003;4:1. doi: 10.1186/1471-2199-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huen MSY, Li XT, Lu LY, Watt RM, Liu DP, Huang JD. The involvement of replication in single stranded oligonucleotide-mediated gene repair. Nucleic Acids Res. 2006;34:6183–94. doi: 10.1093/nar/gkl852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maresca M, Erler A, Fu J, Friedrich A, Zhang Y, Stewart AF. Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol Biol. 2010;11:54. doi: 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosberg JA, Lajoie MJ, Church GM. Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics. 2010;186:791–9. doi: 10.1534/genetics.110.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HH, Xu G, Vonner AJ, Church G. Modified bases enable high-efficiency oligonucleotide-mediated allelic replacement via mismatch repair evasion. Nucleic Acids Res. 2011;39:7336–47. doi: 10.1093/nar/gkr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci U S A. 2003;100:15748–53. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawitzke JA, Costantino N, Li XT, Thomason LC, Bubunenko M, Court C, et al. Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J Mol Biol. 2011;407:45–59. doi: 10.1016/j.jmb.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Kessel JC, Hatfull GF. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol Microbiol. 2008;67:1094–107. doi: 10.1111/j.1365-2958.2008.06109.x. [DOI] [PubMed] [Google Scholar]
- 22.Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 2010;38:3952–62. doi: 10.1093/nar/gkq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swingle B, Markel E, Cartinhour S. Oligonucleotide recombination: a hidden treasure. Bioeng Bugs. 2010;1:263–6. doi: 10.4161/bbug.1.4.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckstein F. Nucleoside phosphorothioates. Annu Rev Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 25.He X, Ou HY, Yu Q, Zhou X, Wu J, Liang J, et al. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol. 2007;65:1034–48. doi: 10.1111/j.1365-2958.2007.05846.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3:709–10. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- 27.Liu G, Ou HY, Wang T, Li L, Tan H, Zhou X, et al. Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA. PLoS Genet. 2010;6:e1001253. doi: 10.1371/journal.pgen.1001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou HY, He X, Shao Y, Tai C, Rajakumar K, Deng Z. dndDB: a database focused on phosphorothioation of the DNA backbone. PLoS One. 2009;4:e5132. doi: 10.1371/journal.pone.0005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talarico TL, Casas IA, Chung TC, Dobrogosz WJ. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob Agents Chemother. 1988;32:1854–8. doi: 10.1128/AAC.32.12.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spinler JK, Taweechotipatr M, Rognerud CL, Ou CN, Tumwasorn S, Versalovic J. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe. 2008;14:166–71. doi: 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sriramulu DD, Liang M, Hernandez-Romero D, Raux-Deery E, Lünsdorf H, Parsons JB, et al. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J Bacteriol. 2008;190:4559–67. doi: 10.1128/JB.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warner JB, Lolkema JS. CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol Rev. 2003;67:475–90. doi: 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J Bacteriol. 2007;189:1366–81. doi: 10.1128/JB.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holo H, Nes IF. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989;55:3119–23. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahrné S, Molin G, Axelsson L. Transformation of Lactobacillus reuteri with electroporation: Studies on the erythromycin resistance plasmid pLUL631. Curr Microbiol. 1992;24:199–205. doi: 10.1007/BF01579282. [DOI] [Google Scholar]
- 36.Sørvig E, Mathiesen G, Naterstad K, Eijsink VGH, Axelsson L. High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology. 2005;151:2439–49. doi: 10.1099/mic.0.28084-0. [DOI] [PubMed] [Google Scholar]
- 37.Chung TC, Axelsson L, Lindgren SE, Dobrogosz WJ. In Vitro Studies on Reuterin Synthesis by Lactobacillus reuteri. Microb Ecol Health Dis. 1989;2:137–44. doi: 10.3109/08910608909140211. [DOI] [Google Scholar]
- 38.Schaefer L, Auchtung TA, Hermans KE, Whitehead D, Borhan B, Britton RA. The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology. 2010;156:1589–99. doi: 10.1099/mic.0.035642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.