Abstract
Cytochrome P450 46A1 (cholesterol 24-hydroxylase) is an important brain enzyme that may be inhibited by structurally distinct pharmaceutical agents both in vitro and in vivo. To identify additional inhibitors of CYP46A1 among U.S. Food and Drug Administration-approved therapeutic agents, we used in silico and intuitive predictions and evaluated some of the predicted binders in the enzyme and spectral binding assays. We tested a total of 298 marketed drugs for the inhibition of CYP46A1-mediated cholesterol hydroxylation in vitro and found that 13 of them reduce CYP46A1 activity by >50%. Of these 13 inhibitors, 7 elicited a spectral response in CYP46A1 with apparent spectral Kd values in a low micromolar range. One of the identified tight binders, the widely used antidepressant fluvoxamine, was cocrystallized with CYP46A1. The structure of this complex was determined at a 2.5 Å resolution and revealed the details of drug binding to the CYP46A1 active site. The NH2-containing arm of the Y-shaped fluvoxamine coordinates the CYP46A1 heme iron, whereas the methoxy-containing arm points away from the heme group and has multiple hydrophobic interactions with aliphatic amino acid residues. The CF3-phenyl ring faces the entrance to the substrate access channel and has contacts with the aromatic side chains. The crystal structure suggests that only certain drug conformers can enter the P450 substrate access channel and reach the active site. Once inside the active site, the conformer probably further adjusts its configuration and elicits the movement of the protein side chains.
Introduction
Cytochrome P450 46A1 converts cholesterol to 24S-hydroxycholesterol, thereby initiating the major pathway of cholesterol removal from the brain (Björkhem et al., 1997; Lund et al., 2003). Abrogation of CYP46A1 activity, as exemplified by Cyp46a1 knockout mice, leads to a compensatory reduction of cholesterol biosynthesis in the brain and the rate at which cholesterol is turned over in this organ (Lund et al., 2003; Kotti et al., 2006). Likewise, enhanced production of 24S-hydroxycholesterol in transgenic animals overexpressing Cyp46a1 increases the rate of cerebral cholesterol synthesis and enhances cholesterol turnover (Shafaati et al., 2011). Unexpectedly, cerebral cholesterol turnover was discovered to be linked to memory and cognition because severe deficiencies in spatial, associative, and motor learning were observed in Cyp46a1 knockout mice (Kotti et al., 2006; Russell et al., 2009). It was established that reduction of cerebral cholesterol biosynthesis also reduced the levels of geranylgeraniol, an intermediate in cholesterol biosynthesis, important for normal learning and hippocampal plasticity (Kotti et al., 2006, 2008). A positive effect of increased cholesterol 24-hydroxylation and cholesterol biosynthesis on cognitive function was demonstrated as well in gene therapy experiments on two mouse models of Alzheimer's disease (Hudry et al., 2010). Cerebral injections with Cyp46a1-containing adenovirus led to some cognitive improvements in these animals and also reduced amyloid deposits (Hudry et al., 2010). Although animal studies clearly demonstrate that CYP46A1 is important for higher-order brain function, numerous investigations in humans assessing the link between intronic polymorphisms in CYP46A1 and Alzheimer's disease are still conflicting (for a review, see Russell et al., 2009), probably because they were conducted on small patient populations. Studies using larger cohorts may be required to unambiguously establish the presence or lack of genetic linkage between CYP46A1 and Alzheimer's disease.
The results of biochemical and structural characterizations of CYP46A1 were surprising as well. While evaluating the properties of purified recombinant CYP46A1, we found that this endobiotic-metabolizing P450 interacts with compounds other than its endogenous substrate cholesterol. Some of these compounds, a number of sterols and marketed drugs, were even metabolized by CYP461 in vitro (Mast et al., 2003). Substrate-free and substrate-bound CYP46A1 crystal structures were then determined and revealed that the enzyme active site is plastic and could undergo a ligand-induced conformational fit (Mast et al., 2008). This finding explained the ability of CYP46A1 to bind structurally unrelated compounds and prompted evaluation of 52 chemicals, both drugs and nonpharmaceutical agents, for the inhibition of CYP46A1 activity in vitro. Nine strong inhibitors were identified (Mast et al., 2008; Shafaati et al., 2010). Of them, four [the antidepressant tranylcypromine (TCP), the anticonvulsant thioperamide, and two antifungals, voriconazole and clotrimazole] were cocrystallized with CYP46A1 to elucidate how compounds of different size, shape, hydrophobicity, and type of nitrogen-containing moiety interact with this key enzyme of cholesterol elimination from the brain. The data obtained confirmed the plasticity of CYP46A1 and demonstrated that induced conformational changes are specific to each inhibitor. Changes ranged from significant rearrangements of the loops and helical regions at the entrance of the active site to localized shifts in the protein backbone and side chains (Mast et al., 2010). In parallel, one of the in vitro inhibitors of CYP46A1, voriconazole, was investigated for the effect on CYP46A1 in vivo (Shafaati et al., 2010). Mice treated with intraperitoneal injections of voriconazole had a high concentration of the drug in the brain and a decrease in cerebral levels of the CYP46A1 product 24S-hydroxycholesterol. Cerebral concentrations of squalene, lathosterol (a marker for cholesterol biosynthesis), and HMG-CoA reductase mRNA were reduced as well, indicating that the decrease in 24S-hydroxycholesterol is probably due to the inhibition of CYP46A1 (Shafaati et al., 2010). Thus, voriconazole seems to inhibit CYP46A1 not only in vitro but also in vivo in mice, suggesting that it has the potential to inhibit CYP46A1 in humans because it is prescribed for prolonged periods of time to treat invasive fungal infection. In the present work, we continued our search of CYP46A1 inhibitors among therapeutic agents and identified additional strong inhibitors of CYP46A1. We used one of them, the antidepressant fluvoxamine (FLV), for cocrystallization with CYP46A1. Herein, we demonstrate that in silico predictions are difficult for CYP46A1 and describe the first structure of a FLV-protein cocomplex. The data obtained enhance our understanding of the structural basis of drug binding to CYP46A1 and provide new insight into interactions of drugs with a P450 enzyme.
Materials and Methods
Materials.
Pharmaceutical agents for screening were purchased from one of the following sources: Sigma-Aldrich (St. Louis, MO), Cayman Chemical Company (Ann Arbor, MI), Alfa Aesar (Ward Hill, MA), Waterstone Technology LLC (Carmel, IN), and Toronto Research Chemicals Inc. (North York, ON, Canada). Cholesterol and [3H]cholesterol were from Steraloids (Newport, RI) and American Radiolabeled Chemicals (St. Louis, MO), respectively. All other chemicals were from Sigma-Aldrich unless otherwise specified.
In Silico Screening.
Protein-drug interactions were evaluated by the Iron simulation software (Pharmatrope Ltd., http://pharmatrope.com/?q = node/17), which used the fragment-based approach (Clark et al., 2009a), calculating the binding free energies of the ligand from the binding free energies of its component fragments. The fragment-based approach is the emerging alternate strategy to high-throughput screening providing full sampling of possible ligand conformations with relatively little computation and therefore enabling screening of large virtual libraries at a high speed and low cost (Congreve et al., 2008; Clark et al., 2009a; Pellecchia, 2009). Two rounds of screening were performed. In the first round, the standard AMBER (Cornell et al., 1995) force field calculations were used, whereas in the second round, the calculations were revised as described below, and compounds were filtered for nitrogen-iron interactions. The crystal structure of CYP46A1 in complex with clotrimazole and all crystallographic water molecules [Protein Data Bank (PDB) code 3MDV] was used for modeling. Hydrogens were added to both protein and water, and their initial positions were adjusted by fixing the heavy atoms and allowing the hydrogens to move using the sculpting method in PyMOL. Because the protein structure adapts to the ligand during the simulation, it is not sensitive to the exact coordinates of the starting protein structure.
Binding simulations were performed on a set of 1147 structures of marketed drugs in the U.S. Food and Drug Administration Adverse Event Reporting System (http://www.fda.gov/cder/aers), prepared by downloading three-dimensional structures from PubChem (http://pubchem.ncbi.nlm.nih.gov/) and minimizing them where necessary with obminimize from the OpenBabel toolset (Guha et al., 2006). The Pharmatrope Iron software sampled randomly in a selected region, using a Metropolis Monte Carlo acceptance criteria based on energy and counting the ratio of rejected and accepted poses. This random sampling consisted of random selection of a Cartesian point in the sampling region for insertion, random selection of an allowed conformation, and random selection of a rotation of the ligand. A new conformation was generated for each insertion attempt by randomizing the torsion angles of all single bonds. Conformations were allowed when the internal molecular mechanics nonbonded interaction energy was less than a preset threshold of 50 kcal/mol. Otherwise, the torsions were randomized again until an allowed conformation was generated. After at least one insertion into the binding site was accepted, the protein structure was periodically relaxed by randomly perturbing the position of the protein atoms in the binding site by small movements, followed by energy minimization of the protein-ligand complex to optimize the protein-ligand interaction (Ferguson and Raber, 1989). If the resulting energy was lower than the initial energy, the new, slightly modified, protein structure was used for future insertion attempts.
The entropy of binding with respect to the free state was computed using the following formulas: Entropyrotation−confrontation = kT · ln (accepted insertions/attempted insertions) (Guha et al., 2006) and Entropytranslation = kT · ln (sampling volume/1660.533) (Clark et al., 2009b). The overall free energy was computed using the relation: ΔGbinding = Einteraction − Entropyrotation−confrontation − Entropytranslation (Brooks et al., 1983). The Pharmatrope Iron free energy simulation was run at 300K using the AMBER (Cornell et al., 1995) force field, augmented by a CHARMM explicit hydrogen bond function (Brooks et al., 1983). Instead of the normal mixing rules that use the geometric average of atomic sigma parameters and the sum of the atomic radii, the Lennard-Jones interaction term between the heme iron and any nitrogen was assigned a σ of 10 kcal/mol and an equilibrium distance of 2.1 Å. That is, all nitrogen-iron interactions used this nonbonded parameter, whereas the same atoms involved in any other interaction used the normally computed Lennard-Jones parameters. A dielectric of 4.0 was used for the electrostatic function, with a nonbonded cutoff of 9 Å. The Iron free energy simulation was run until 25 random insertions were accepted, which required approximately 20 million steps at a binding entropy of approximately 10 kcal/mol. A maximum of 5000 poses were stored for each simulation. After the first run, 468 molecules had accepted poses within the binding site. These were further filtered for those that had one or more poses with a nitrogen-iron contact closer than 2.3 Å. This filter provided 107 compounds as candidates for experimental validation.
Heterologous Expression and Purification of Full-Length Human CYP46A1.
The expression construct was as described previously (Mast et al., 2003), except that a 4xHis tag was added at the C terminus of CYP46A1 to facilitate purification. The construct was transformed into GC5 cells (PGC Scientifics, Palm Desert, CA) in the presence of the chaperones GroEL and GroES (Thermo Fisher Scientific, Waltham, MA) and expressed in Escherichia coli (White et al., 2008). E. coli cells were harvested, and spheroplasts were isolated (Mast et al., 2003) and suspended in 100 mM potassium phosphate buffer (KPi), pH 7.2, containing 20% glycerol, a cocktail of EDTA-free protease inhibitors (Roche Applied Science, Indianapolis, IN), 1% sodium cholate, 5 mM β-mercaptoethanol (BME), and 0.5 mg/ml DNase. The spheroplast suspension was sonicated on ice using six 20-s pulses at 1-min intervals and subjected to ultracentrifugation at 106,000g for 60 min at 4°C. The resulting supernatant was diluted 2-fold with 10 mM KPi, pH 7.2, containing 5 mM BME and 20% glycerol and mixed with DE-cellulose equilibrated with 50 mM KPi, pH 7.2, 0.5% sodium cholate, 10 mM BME, and 20% glycerol. The P450-resin suspension was rotated for 30 min at 4°C, followed by sedimentation of DE-cellulose at 4000g for 5 min at 4°C and collection of the supernatant. The DE-cellulose was then washed with 5 resin volumes of the equilibration buffer and sedimented as before. The supernatants were combined and supplemented with 1 mM imidazole, pH 7.0, 0.2 M KCl, and 100 mM KPi, pH 7.2, and then were mixed with Ni-NTA agarose (QIAGEN, Valencia, CA) equilibrated with 100 mM KPi, pH 7.2, containing 0.2 M KCl, 20% glycerol, 1 mM imidazole, 10 mM BME, and 0.5% sodium cholate. After overnight incubation, the Ni-NTA agarose was sedimented at 4000g for 5 min at 4°C and sequentially washed with 5 resin volumes of equilibration buffer; 5 resin volumes of 50 mM KPi, pH 7.2, containing 20% glycerol, 1 mM imidazole, 10 mM BME, and 0.5% sodium cholate; and 5 resin volumes of 50 mM KPi, pH 7.2, containing 20% glycerol, 1 mM imidazole, 10 mM BME, and 0.5% CYMAL-6 (Affymetrix, Maumee, OH). The Ni-NTA agarose was then placed in a column, and CYP46A1 was eluted with the last wash buffer except that the imidazole concentration was increased to 300 mM. After elution, P450 was diluted 10-fold with 50 mM KPi, pH 7.2, containing 20% glycerol, 0.2 mM DTT, 1 mM EDTA, and 0.5% CYMAL-6 and mixed with SP-Sepharose, equilibrated with the dilution buffer. After a 3-h rotation at 4°C, the SP-Sepharose was washed with 5 resin volumes of equilibration buffer and 10 volumes of 50 mM KPi, pH 7.2, containing 20% glycerol, 0.2 mM DTT, 1 mM EDTA, and 0.2% CYMAL-6. CYP46A1 was eluted with the same buffer containing 0.4 M NaCl. The purified protein was diluted with 50 mM KPi, pH 7.2, containing 20% glycerol, 0.2 mM DTT, 1 mM EDTA, and 0.2% CYMAL-6, to decrease the NaCl concentration to 100 mM, and concentrated using an Amicon Ultra centrifugal filter. CYP46A1 was ∼80% pure as assessed by SDS-polyacrylamide gel electrophoresis and had a ratio of absorbances at 418 to 280 nm equal to ∼1.
Screening of Drugs for Their Effect on CYP46A1 Activity.
The conditions of the in vitro assay were as described previously (Mast et al., 2008). In brief, the reconstituted system contained 0.5 μM full-length recombinant CYP46A1, 1.0 μM NADPH cytochrome P450 oxidoreductase, 2.7 μM cholesterol as a substrate (a concentration equal to 0.5 Km of CYP46A1 for cholesterol), trace amounts of [3H]cholesterol (250,000 cpm), and 43 μM test drug (8 Km for cholesterol). Cholesterol hydroxylation was initiated by the addition of 1 mM NADPH and proceeded for 20 min at 37°C in 1 ml of 50 mM KPi, pH 7.2, containing 100 mM NaCl, 0.02% CYMAL-6, 40 μg of dilauroylglycerol-3-phosphatidylcholine, and 2 U of catalase. The enzyme reaction was terminated by the addition of 5 ml of CH2Cl2. The organic phase was isolated and evaporated. The residue was dissolved in CH3CN and analyzed by high-performance liquid chromatography (Pikuleva et al., 1997). The cholesterol in this assay was added from a 10 mM stock in 45% aqueous 2-hydroxypropyl-β-cyclodextrin. The drugs were dissolved according to the manufacturer's recommendations either in water, methanol, or 45% 2-hydroxypropyl-β-cyclodextrin.
Determination of the In Vitro IC50 and Ki.
The incubation conditions were the same as in the screening assay except that the drug concentrations varied from 1 pM to 100 μM, and the cholesterol concentration was 5.4 μM. The data were fit to the following equation by Graph-Pad Prism software:
in which [I] is the inhibitor concentration, IC50 is the inflection point, A is the CYP46A1 activity (100%) in the absence of inhibitor, and B is the residual CYP46A1 activity (percentage) at maximum inhibitor concentrations. The IC50 was determined at a cholesterol concentration equal to the Km; therefore, Ki = IC50/2, assuming a model of competitive inhibition.
Enzyme Assay Using Isolated Brain Microsomes.
Bovine brain was obtained from a local slaughterhouse 3 h after the animals were sacrificed. The temporal lobe cortex (∼100 g) was isolated, rinsed in ice-cold 0.9% NaCl, blotted, and homogenized with a Teflon grinder in 200 ml of 50 mM Tris-HCl (pH 7.4), containing 250 mM sucrose, 5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 100 μg/ml butylated hydroxytoluene, and a cocktail of protease inhibitors. This tissue homogenate was subjected to centrifugation at 9000g for 20 min to remove unbroken cells, nuclei, cell debris, and mitochondria. The resultant supernatant was centrifuged at 106,000g for 60 min at 4°C to pellet the microsomes. The pelleted microsomes were resuspended in 3 ml of 50 mM KPi, pH 7.2, containing 1 mM EDTA and used (1 mg of total protein) for reconstitution with 1 μM cytochrome P450 oxidoreductase and various concentrations of FLV (10–100 μM, added from 5 mM stock in water) in a total volume of 1 ml of 50 mM KPi, pH 7.2, containing 100 mM NaCl. Metabolism of endogenous cholesterol present in the microsomes was initiated by the addition of a NADPH-regenerating system (5 mM NADPH, 50 mM glucose 6-phosphate, and 10 U of glucose-6-phosphate dehydrogenase) and proceeded for 30 min at 37°C. The enzyme reaction was terminated by addition of 6 ml of Folch reagent (chloroform-methanol, 2:1, v/v) followed by determination of the content of 24-hydroxycholesterol by gas chromatography-mass spectrometry as described previously (Mast et al., 2011) using deuterated 24S-hydroxycholesterol as an internal standard.
Spectral Binding Assay.
Binding affinities of different drugs for full-length CYP46A1 were estimated as described previously (Mast et al., 2003) except that the concentration of CYP46A1 was 0.25 μM. The buffer (50 mM KPi, pH 7.2) contained 100 mM NaCl and 0.02% CYMAL-6, and the temperature was 18°C. When the effect of dilauroylglycerol-3-phosphatidylcholine was tested, phospholipid vesicles were prepared as described previously (Heo et al., 2011) and added to the buffer at a final concentration of 40 μg/ml. The conditions for titration of truncated Δ(2–50)CYP46A1 varied but always used 50 mM KPi containing 100 mM NaCl. Apparent binding constants (Kd) were calculated using either the ΔA = (ΔAmax[L])/(Kd + [L]) or ΔA = 0.5ΔAmax (Kd + [E] + [L] − (Kd + [E] + [L]2 − + [E][L]) equation, in which ΔA is the spectral response at different ligand (drug) concentrations [L], and ΔAmax is the maximal amplitude of the spectral response.
Crystallization of CYP46A1 in Complex with FLV.
The truncated Δ(2–50)CYP46A1 variant was used, which was expressed and purified as described previously (White et al., 2008). The CYP46A1-FLV complex was obtained by saturating the enzyme with 50 μM FLV during the last purification step, chromatography on CM-cellulose. Crystals were grown at 18°C in sitting drops after mixing 1 μl of an ∼40 mg/ml P450 solution with 1 μl of precipitant solution (15% polyethylene glycol 8000, 50 μM FLV, 50 mM KPi, pH 5.8, and 20% glycerol) and microseeding with a cat whisker.
X-Ray Data Collection, Processing, and Structure Determination.
X-ray diffraction data were collected from a single crystal at Stanford Synchrotron Radiation Lightsource (SSRL) beam line 7-1 at 100 K (Soltis et al., 2008). The diffraction limit was 2.5 Å. The data set was processed with Mosflm (Leslie, 1999) and Collaborative Computational Project, Number 4 (1994) programs. The structure of FLV-bound CYP46A1 was determined by molecular replacement using cholesterol 3-sulfate-bound structure as the search model with cholesterol 3-sulfate omitted (Protein Data Bank code 2Q9F) and Phaser (McCoy et al., 2007). The model was refined using Refmac (Murshudov et al., 1997) and Coot for electron density fitting (Emsley and Cowtan, 2004). The FLV complex crystallized in space group I4122 with one molecule per asymmetric unit. Data collection and refinement statistics are given in Supplemental Table 1; coordinates and structure factors have been deposited in the PDB (code 4ENH).
Modeling of FLV Conformers in Water.
The calculations were performed using the MacroModel package and the OPLS-2005 (Optimized Potential for Liquid Simulations) force field within Maestro 9.3 (Schrödinger LLC, Portland, OR; http://www.schrodinger.com). Each run contained a conformational search of 1000 steps by the PRCG (Polak-Ribiere conjugate gradient) minimization algorithm. Extended cutoff distances (equivalent to a van der Waals cutoff of 8.0 Å, an electrostatic cutoff of 20.0 Å, and a hydrogen bond cutoff of 4.0 Å) were used. Each minimized conformer was filtered through a relative energy window of 21 kJ/mol (5.02 kcal/mol), and redundant conformers were eliminated using a maximum atom deviation cutoff of 0.5 Å.
Results
Testing of Compounds Generated by the First Round of In Silico Predictions.
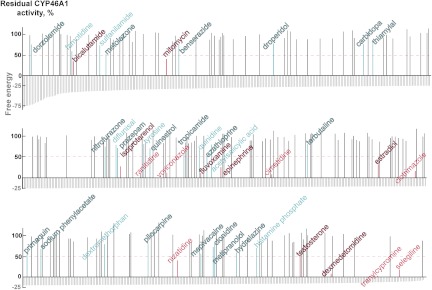
The computational analysis of 1147 drugs for binding to CYP46A1 identified 784 compounds with docking scores within 1 log scale: from the lowest (−80.3) to −6.9 (Fig. 1 gray lines, the lower docking score indicates higher binding affinity). From these 784 drugs, we then tested experimentally only 202 (Supplemental Table 2) because our enzyme assay is labor-intensive and low-throughput. The drugs tested met at least two of the following six criteria: 1) had a high negative docking score; 2) had a distant similarity to the cholesterol molecule; 3) had a nitrogen-containing functionality that can potentially coordinate the P450 heme iron; 4) were known to cross the blood-brain barrier; 5) were used on a chronic basis; and 6) were known to inhibit one or several P450 enzymes. These selection criteria enabled identification of 27 pharmaceutical agents that inhibited CYP46A1-mediated cholesterol hydroxylation by more than 20% (Fig. 1, cyan and magenta lines, drug names are in the stroked font), reflective of a change of enzyme activity corresponding to 2 times the S.D. value of our enzyme assay and indicative of compounds having some affinity for CYP46A1 (Mast et al., 2008, 2010). Of these 27 drugs, which we define as actives, 8 inhibited CYP46A1 activity by more than 50%, an effect usually observed with nanomolar to low micromolar inhibitors of CYP46A1 (Mast et al., 2008, 2010) (Fig. 1, magenta lines, drug names are in the stroked font). We used 50% inhibition of CYP46A1 activity as an arbitrary cutoff limit for identification of hits and calculating the hit rate of our computational predictions. The computed binders also included 15 drugs (8 actives and 7 hits) that were tested by us previously (Fig. 1, cyan and magenta lines, drug names are in the regular font) (Mast et al., 2008, 2010), increasing the number of experimentally evaluated compounds to 217 (202 + 15). Of these 217 drugs, 42 (15 + 27) were actives and 15 (7 + 8) were hits. Therefore, the hit rate in this round of in silico screening was equal to 6.9% (15 of 217). To find additional hits for CYP46A1, we decided to revise the force field calculations and conduct the second round of computational dockings.
Fig. 1.
A summary of the first round of in silico predictions. The docking scores of the 784 computed binders are shown as gray vertical lines. The effects of drugs on CYP46A1 activity are also shown: black lines correspond to the drugs that altered CYP46A1 activity by ≤20%, and cyan and magenta lines are the pharmaceutical agents that inhibited CYP46A1 by >20% (actives) and 50% (hits), respectively. The magenta dashed line indicates the inhibition cutoff limit for the hits. Drugs tested in our previous studies and the present work are shown in the regular and stroked fonts, respectively. Data on all tested drugs are given in Supplemental Table 2.
Testing of Compounds Generated by the Second Round of In Silico Predictions.
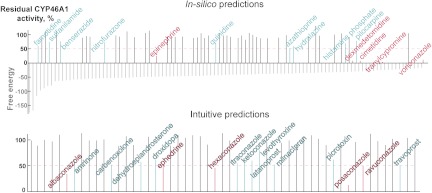
Enhancing the nitrogen-iron interactions in the force field calculations and filtering compounds based on the nitrogen-iron distance yielded 107 binders, 14 of which (9 actives and 5 hits) we already tested either in the first round of screening or in our previous studies (Fig. 2, cyan and magenta lines, drug names are in the regular font). Of the remaining 93, 43 were then selected using the same criteria as those for the first round of predictions and evaluated in the enzyme assay (Supplemental Table 2). However, no additional actives or hits were found, although the overall hit rate increased to 8.8%, (4 + 1)/(14 + 43), for all drugs tested.
Fig. 2.
A summary of the second round of in silico predictions and intuitive predictions. The docking scores are shown as gray vertical lines and effects on enzyme activity as lines in black (no CYP46A1 inhibition), cyan (>20% inhibition), and magenta (>50% of inhibition). The magenta dashed line indicates the inhibition cutoff limit for the hits. Drugs tested in our previous studies and during the first round are shown in the regular font and drugs tested in this round are in the stroked font. Data on all tested drugs are given in Supplemental Table 2.
Testing of Compounds Based on Intuitive Predictions.
In parallel with testing the in silico predictions, we conducted a search of CYP46A1 inhibitors based on intuitive predictions, a strategy used in our previous studies (Mast et al., 2008; Shafaati et al., 2010). Our intuitive predictions were applied to drugs that were not among the candidates in the in silico screenings, yet were worth testing because they were either known P450 inhibitors (http://medicine.iupui.edu/clinpharm/ddis/table.aspx) or were pharmaceutical agents that are used on a long-term or chronic basis such as steroids, antifungals, chemotherapeutic agents, antidepressants, sedatives, and antihypertensives. We visually inspected the chemical structures of these therapeutic agents, looking for “fragments” of drugs known to bind to CYP46A1 from our previous structural studies (Mast et al., 2008, 2010). Particular attention was given to flexible linear structures that can fit the banana-shaped active site of CYP46A1. The emphasis was on compounds that can interact with the protein side chains and/or coordinate the P450 heme iron via their nitrogen- or oxygen-containing functionalities. This is usually possible when there are no bulky substitutions in the vicinity of these functionalities that obstruct interactions with CYP46A1. A total of 53 compounds were selected and tested in the enzyme assay (Supplemental Table 2). Of these, 16 were actives and 5 were hits (Fig. 2), which corresponds to the hit rate of 9.4%, slightly higher than that in the second round of the in silico predictions.
Binding Properties of the Newly Identified Hits.
These were assessed by a spectral assay, in which ligand binding is detected by a shift in the P450 spectrum due to the displacement of the water molecule coordinating the heme iron in ligand-free P450. However, some ligands bind to the P450 active site, but do not elicit a spectral response. Of the 13 hits identified in the present study, seven induced a spectral response, providing insight into the type of binding and enabling determination of their apparent Kd values (Table 1). The spectral response for all of the spectrally detectable drugs, except bicalutamide, indicated that the P450 heme iron was coordinated with a nitrogen atom from the drug (the so-called type II spectral response). However, the Kd values varied and were from an intermediate nanomolar to a low micromolar range (0.33–2.5 μM). Only one drug, dexmedetomidine, had a low nanomolar affinity (0.02 μM). We then determined the Ki of dexmedetomidine and also of FLV, which we had decided to cocrystallize with CYP46A1. The Ki values were 0.13 and 1.7 μM, respectively, reflective of the Kd values of the drugs (Table 1), yet the difference in the Ki (∼13-fold) was not identical to the difference in the Kd (35-fold).
TABLE 1.
Inhibitory and binding properties of the hits identified in the present study
Assay conditions are described under Materials and Methods. The results represent the average of triplicate measurements ± S.D.
| Hit | Residual CYP46A1 Activitya | Kdb | Ki | Indication or Use |
|---|---|---|---|---|
| % | μM | |||
| In silico prediction | ||||
| Bicalutamide | 35 ± 2.0 | 1.2 ± 0.05 | NM | Antineoplastic |
| Dexmedetomidine | 8.0 ± 0.3 | 0.02 ± 0.005 | 0.13 ± 0.01 | Analgesic, sedative |
| Epinephrine | 25 ± 2.0 | NSP | NM | Adrenergic stimulant |
| Estradiol | 32 ± 6.0 | NSP | NM | Steroid hormone |
| FLV (E-isomer) | 11 ± 0.2 | 0.7 ± 0.1 | 1.7 ± 0.3 | Antidepressant |
| Isoproterenol | 25 ± 2.0 | NSP | NM | Bronchodilator |
| Mitomycin | 41 ± 1.0 | NSP | NM | Antineoplastic |
| Testosterone | 49 ± 4.0 | NSP | NM | Steroid hormone |
| Intuitive predictions | ||||
| Albaconazole | 35 ± 3.0 | 2.5 ± 0.1 | NM | Antifungal |
| Ephedrine | 49 ± 2.0 | NSP | NM | Bronchodilator |
| Hexaconazole | 46 ± 3.0 | 0.4 ± 0.02 | NM | Antifungal |
| Posaconazole | 13 ± 1.2 | 0.3 ± 0.03 | NM | Antifungal |
| Ravuconazole | 50 ± 3.0 | 0.6 ± 0.02 | NM | Antifungal |
NM, not measured; NSP, weak or no spectral response.
Activity in the screening enzyme assay.
Calculated on the basis of spectral binding.
FLV Effects on Truncated Δ(2–50)CYP46A1, Full-Length CYP46A1, and Cholesterol 24-Hydroxylation in Isolated Brain Microsomes.
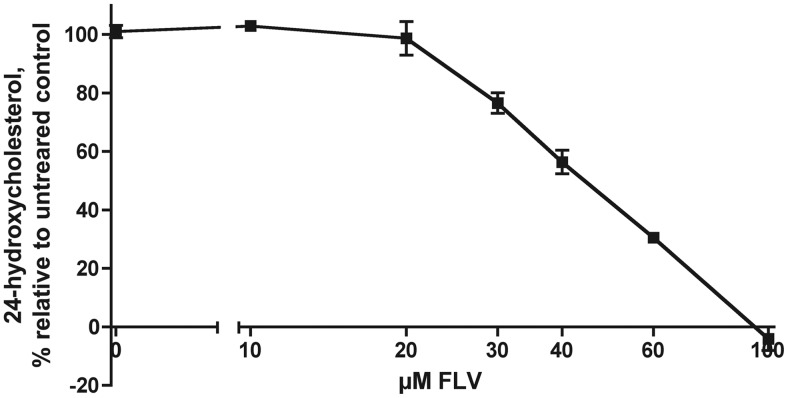
All studies described in the previous sections were conducted on full-length CYP46A1. However, for crystallization we used truncated Δ(2–50)CYP46A1. Therefore, we repeated some experiments on truncated CYP46A1 using exactly the same assay conditions as in studies of full-length CYP46A1. Similar to full-length CYP46A1, FLV inhibited the truncated enzyme, but the extent of this inhibition was greater with no enzyme activity detected. However, the spectral parameters of FLV binding were very similar to those of full-length CYP46A1, with the Kd being 0.8 μM and ΔAmax being 0.11. Furthermore, to gain insight into whether FLV reaches the active site of membrane-bound CYP46A1, we also assessed FLV binding by full-length CYP46A1 in the presence of phospholipid vesicles. The Kd increased to 1.2 μM and ΔAmax decreased to 0.08. We then used a more physiologically relevant system (isolated bovine microsomes) and found that FLV inhibited CYP46A1-mediated 24-hydroxylation of endogenous cholesterol in a concentration-dependent manner (Fig. 3), indicating the potential of inhibiting CYP46A1 in vivo.
Fig. 3.
FLV inhibition of CYP46A1-mediated cholesterol hydroxylation in isolated bovine brain microsomes. Assay conditions are described under Materials and Methods. The results represent the average of triplicate measurements ± S.D. Some of the error bars are not seen because they are smaller than the symbol size.
Crystal Structure of CYP46A1 in Complex with FLV.
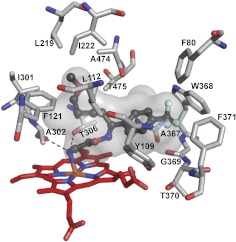
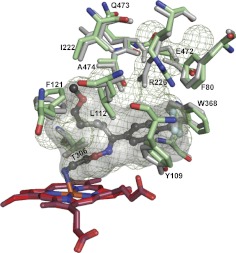
FLV was chosen for cocrystallization with CYP46A1 because it has an interesting Y-shape structure with the CF3-phenyl ring linked to the two alkyl chains, the NH2- and methoxy-containing (Supplemental Fig. 1). Perhaps more important was the fact that this antidepressant is widely used and known to inhibit several drug-metabolizing P450s (CYP1A2, -2C19, -2C9, and -3A4), causing drug-drug interactions (Muscatello et al., 2012). However, structural information about FLV binding to a P450 or any other enzyme or protein is currently lacking. Hence, we crystallized and determined the crystal structure of the FLV-CYP46A1 complex. In this complex, FLV is bound to the CYP46A1 active site and has a well defined electron density, allowing unambiguous fitting of the drug (Supplemental Fig. 1). The NH2-containing chain of FLV is directed toward the heme and coordinates the heme Fe with the nitrogen-iron distance being 2.29 Å (Fig. 4). The primary amine has two additional interactions: hydrogen bonds with Ala302 carbonyl and Thr306 hydroxyl located at 2.82 and 3.02 Å, respectively. The methoxy-containing chain of FLV is positioned at almost a 90° angle relative to the NH2-containing chain and points away from the heme iron. This chain is surrounded by a cluster of mainly aliphatic amino acid residues (Leu112, Phe121, Leu219, Ile222, Ile301, Ala474, and Thr475), which form a pocket and restrain this chain on all sides. The CF3-phenyl group of FLV is also located in a pocket formed by a cluster of aromatic residues (Phe80, Tyr109, Trp368, and Phe371), as well as the side chain of Ala367 and backbone atoms of Gly369 and Thr370. Overall, there is good fit between the shape of the active site cavity and FLV. A comparison with the ligand-free CYP46A1 (Fig. 5) indicates that this fit is achieved by readjustments of the side chains of Arg226, Glu472, Leu112, Phe 80, Phe121, Ile222, Gln473, Ala474, Thr306, Trp368, and Tyr109, of which the shifts of Arg226 and Glu472 lead to major changes in the shape and volume of the active site (470 Å3 in ligand-free CYP46A1 versus 302 Å3 in FLV-bound enzyme). Of significance is a 0.5 Å shift of the Thr306 hydroxyl, enabling a hydrogen bond interaction with the FLV primary amine.
Fig. 4.
View of the CYP46A1 active site illustrating interactions with FLV. FLV is in dark gray, and amino acid residues in contact with FLV are in light gray. Dashed black lines indicate hydrogen bonds. The heme group is in red. The nitrogen, oxygen, fluorine, and iron atoms are in blue, red, pale cyan, and orange, respectively. The enclosed volume of the active site is shown as a semitransparent surface.
Fig. 5.
Superimposed views of the active sites in FLV-bound (light gray) and ligand-free (light green) CYP46A1 showing the amino acid residues undergoing conformational changes upon FLV binding. The active site volumes (semitransparent surface in FLV-bound and mesh in ligand-free CYP46A1) are also shown. FLV is in dark gray. The heme group in FLV and ligand-free CYP46A1 are in red and raspberry, respectively. Coloring of atoms is the same as in Fig. 4.
Structural Differences in Binding of FLV and TCP to CYP46A1.
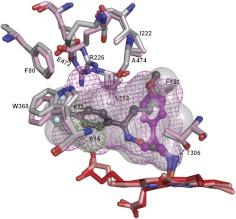
To better understand FLV binding to CYP46A1, we compared the crystal structure of the FLV-CYP46A1 complex with that of TCP-bound CYP46A1 determined by us previously (Mast et al., 2010). Like FLV, TCP is an antidepressant drug containing an NH2 group. However, TCP has a different mechanism of action and inhibits the monoamine oxidase enzyme, not the transporters, as does FLV. In addition, TCP is smaller and more compact than FLV and has ∼80-fold lower spectral Kd for CYP46A1 than FLV (Mast et al., 2010). Despite these differences, the shape and volume of the active site are very similar in FLV- and TCP-bound complexes (Fig. 6). This is probably because in both, drug binding induces conformational changes that involve the same set of amino acid residues shifting either significantly (Arg226, Glu472, Leu112, and Phe121) or only a little (Ile222, Phe 80, Trp368, Ala474, and Thr306) compared with ligand-free CYP46A1. Although the shape of the active site is similar in the two crystal structures, the position of the phenyl rings of the drugs is very different. In the CYP46A1-FLV complex, the phenyl ring is away from the heme because of the long NH2-containing chain, whereas in the CYP46A1-TCP complex, the ring is closer and positioned orthogonally. Therefore, the occupancy of the active site volume is >90% in FLV-bound enzyme and <50% in TCP-bound CYP46A1. Furthermore, upon binding, FLV may displace more water molecules from the active site than TCP because no water molecules are present in the active site of FLV-bound CYP46A1 determined at a 2.5 Å resolution versus two water molecules in the TCP-bound CYP46A1 at a 2.0 Å resolution. Another important difference is the position of the primary amine. In FLV, it is closer to the I helix by 0.78 Å and forms a hydrogen bond with the Thr306 hydroxyl, an interaction absent in the TCP-bound structure. However, both drugs have a hydrogen bond to the Ala302 carbonyl. Thus, FLV has three NH2 group interactions (with the heme Fe, Thr306 hydroxyl, and Ala302 carbonyl), whereas TCP has only two (with the heme Fe and Ala302 carbonyl).
Fig. 6.
Superimposed views of FLV-bound (light gray) and TCP-bound (pink) CYP46A1 showing that the same amino acid residues undergo conformational changes upon drug binding. The active site volumes (semitransparent surface in FLV-bound and mesh in TCP-bound CYP46A1) are also shown. Two active site water molecules, 613 and 614, in TCP-bound CYP46A1 are shown as green dotted spheres. The dashed black line indicates an additional hydrogen bond between FLV-NH2 and CYP46A1. FLV is in dark gray, and TCP is in magenta. The heme groups in FLV- and TCP-bound CYP46A1 are in red and salmon, respectively. Coloring of atoms is the same as in Fig. 4.
Determinants of FLV Binding to CYP46A1 Compared with Those of TCP Binding.
Higher occupancy of the active site volume, displacement of more water molecules, and an additional hydrogen bond suggest that FLV should bind tighter to CYP46A1 than TCP to CYP46A1. In fact, it binds more weakly, indicating that other factors affect FLV binding. One of them could be the protonation of the NH2 group because only unprotonated primary amines can ligate the heme Fe. In aqueous solutions, the concentration of unprotonated amine is determined by its pKa and the pH of the solution. The pKa of 8.7 of FLV (Foda et al., 1996) is higher than that of TCP (8.4). Hence, at physiological pH, the effective concentration of an unprotonated amine will be lower in an FLV solution compared with that in a TCP solution, thus increasing the apparent spectral Kd of FLV. To assess the effect of pKa, we performed titrations with FLV at pH 6.5 and 8.0, values that do not denature CYP46A1, and compared the Kd values obtained with that at pH 7.2 (Table 2). Titration with cholesterol served as a control for possible changes in the protein elicited by the altered pH. The Kd values of FLV for CYP46A1 were very similar at different pHs, as were the Kd values of cholesterol, suggesting that coordinate bonding to iron is not a major factor determining the difference in affinities of FLV and TCP. Therefore, we next evaluated the contribution of the additional hydrogen bond to the Thr306 hydroxyl relative to that of TCP. The spectral Kd of FLV for the T306A mutant increased only 3.6-fold (Table 2), indicating a relatively weak contribution of this bond to the strength of FLV binding to CYP46A1, consistent with the increased length of this bond, 3.02 Å versus 2.8 Å of a regular hydrogen bond.
TABLE 2.
Effect of different factors on FLV binding to truncated CYP46A1 as assessed by spectral assay
The results represent the average of triplicate measurements ± S.D.
| Ligand, P450 | pH | Temperature | Kd | ΔAmax/nmol P450 |
|---|---|---|---|---|
| °C | μM | |||
| Effect of pH | ||||
| FLV (E-isomer), ΔWT | 6.5 | 18 | 0.6 ± 0.08 | 0.09 ± 0.01 |
| CHO, ΔWT | 6.5 | 18 | 0.1 ± 0.05 | 0.11 ± 0.007 |
| FLV (E-isomer), ΔWT | 7.2 | 18 | 0.8 ± 0.05 | 0.12 ± 0.004 |
| CHO, ΔWT | 7.2 | 18 | 0.1 ± 0.03 | 0.12 ± 0.02 |
| FLV (E-isomer), ΔWT | 8.0 | 18 | 0.7 ± 0.01 | 0.06 ± 0.005 |
| CHO, ΔWT | 8.0 | 18 | 0.1 ± 0.01 | 0.05 ± 0.001 |
| Effect of hydrogen bonding to T306A | ||||
| FLV (E-isomer), ΔT306A | 7.2 | 18 | 2.9 ± 0.03 | 0.12 ± 0.006 |
| Effect of temperature | ||||
| FLV (E-isomer), ΔWT | 7.2 | 4 | 2.3 ± 0.2 | 0.10 ± 0.002 |
| CHO, ΔWT | 7.2 | 4 | 0.1 ± 0.01 | 0.10 ± 0.001 |
| FLV (E-isomer), ΔWT | 7.2 | 18 | 0.8 ± 0.05 | 0.12 ± 0.004 |
| CHO, ΔWT | 7.2 | 18 | 0.1 ± 0.02 | 0.11 ± 0.001 |
| FLV (E-isomer), ΔWT | 7.2 | 37 | 0.4 ± 0.03 | 0.11 ± 0.01 |
| CHO, ΔWT | 7.2 | 37 | 0.1 ± 0.01 | 0.10 ± 0.005 |
| Effect of FLV isomerization | ||||
| FLV (E-isomer), ΔWT | 7.2 | 18 | 0.8 ± 0.05 | 0.12 ± 0.004 |
| FLV (Z-isomer), ΔWT | 7.2 | 18 | 2.1 ± 0.03 | 0.08 ± 0.003 |
CHO, cholesterol; ΔWT, truncated CYP46A1 wild type; ΔT306A, truncated CYP46A1 mutant.
We also assessed the contribution of entropy, because FLV is expected to be floppy with multiple rotatable bonds. Therefore, when restrained in the CYP46A1 active site, FLV could become more ordered and lose greater entropy than TCP, thus explaining its diminished binding. The effect of entropy was evaluated by titrating CYP46A1 with FLV and cholesterol (as a control) at different temperatures (Table 2). The Kd of FLV decreased almost 8-fold as the titration temperature increased from 4 to 37°C, whereas the Kd of cholesterol remained unchanged. These data suggest that entropy does contribute to FLV binding to CYP46A1, raising a question about the conformation of FLV in solution.
FLV has a C=N bond and can exist as two geometric isomers, E (trans) or Z (cis). The former is a much more effective inhibitor of the serotonin transporter than the latter (Miolo et al., 2002) and is the major component of FLV. Contamination with the Z-isomer is usually minor (≤0.5–0.7%). Thus, if the Z-isomer preferentially binds to CYP46A1, the apparent Kd of FLV will be high because of the low effective concentration of the binding isomer. To test this possibility, we irradiated a FLV solution with ultraviolet light to convert the E-isomer into the Z-isomer (Supplemental Fig. 2) (Deubner and Holzgrabe, 2002) and used this solution for titration of CYP46A1. Binding of the Z-isomer was a little weaker than that of the E-isomer, as indicated by the 2.6-fold increase in Kd (Table 2). The major difference was in the time to obtain a CYP46A1 spectral response, which developed over a much longer period in the case of the Z-isomer (2–3 min) compared with the instant response of the E-isomer. Thus, the Kd of the FLV preparation could not be explained by the preferential binding of the low-abundance Z-isomer. However, the slower spectral response to the Z-isomer is interesting, reflecting either slower isomer binding or, if the rates of the isomer binding are the same, the time required for positioning of the Z isomer inside the active site to coordinate the heme iron. These experiments indicate that not only isomerization but also conformation of the isomers could play a role in the binding of FLV to CYP46A1.
Conformations of FLV in Water.
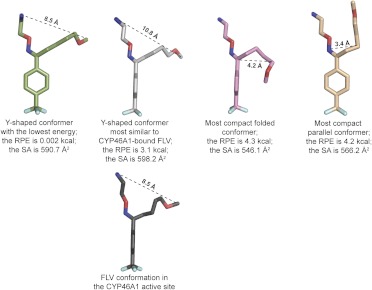
The computational search of FLV conformations in solution yielded 584 conformers with relative potential energy from 0.002 to 5.0 kcal and surface area from 541 to 609 Å2. On the basis of shape, these conformers could be broadly divided into three groups: 1) those of a Y-shape similar to the structure of FLV in the FLV-CYP46A1 complex; 2) conformers with the two alkyl chains in parallel orientation; and 3) conformers with the methoxy-containing chain folded over the phenol ring (Fig. 7). In general, conformers of the first group had lower relative potential energy than conformers of the second and third groups, suggesting that they would be the most abundant conformers in solution. The data obtained indicate conformational heterogeneity of FLV in solution and support the notion that this conformational heterogeneity could affect the Kd of FLV for CYP46A1.
Fig. 7.
Possible conformations of the FLV E-isomer in solution as suggested by computational modeling. Dashed black lines indicate the maximal width of the conformers. RPE, the relative potential energy; SA, surface area.
Discussion
In the present work, we capitalized on our prior crystallographic studies of CYP46A1 (Mast et al., 2008, 2010) and used fragment-based virtual ligand screening to identify additional marketed drugs that bind tightly to CYP46A1, a key enzyme of cholesterol elimination from the brain. In the first round of in silico predictions, the standard AMBER (Cornell et al., 1995) force field, which does not consider the nitrogen-iron interaction to be a significant contributor to the overall computed energy, was used. The hit rate was only 6.9%. Nevertheless, we were able to identify eight new CYP46A1 inhibitors including FLV. In the second round of screening, the force field was revised to enhance nitrogen-iron interactions, and compounds that did not have their nitrogen within the bonding distance from the iron of ≤2.3 Å were discarded. This bonding distance filter eliminated from consideration ∼80% of the initially computed binders and significantly reduced the number of compounds for subsequent experimental validations. The hit rate was increased to 8.8% but did not lead to identification of additional binders compared with those in the first round of screening. In part, this result could be due to a lesser number of compounds tested in the enzyme assay (43 versus 202 tested in the first round) or to the bias of the investigators (N.M. and I.A.P.) who selected the drugs for experimental validation. This bias or investigator intuition is characterized by the hit rate for our intuitive predictions, which was 9.4%, comparable to that in the second round of screenings (8.8%). Because our intuitive predictions were performed on a set of drugs that were not predicted to bind to CYP46A1 computationally, there is no overlap between the molecules selected by the two approaches; in silico and intuitive predictions are thus complementary rather than redundant.
On the basis of combined predictions from both approaches, a total of 13 new pharmaceutical agents were found to bind to CYP46A1 with low micromolar affinities, and many of them represent interesting targets for cocrystallization with CYP46A1. Herein we describe the complex with FLV. This is the first report on crystal structure of FLV bound to a biomolecule and a second CYP46A1 structure in complex with the primary amine, thus enabling comparative analysis of amine binding to CYP46A1. Although the architecture of the active site is very similar in FLV- or TCP-bound CYP46A1, drug positioning is different, as is the strength of interaction with CYP46A1. We examined possible factors that underlie this difference in affinities and obtained evidence pointing to substrate entropy and conformational heterogeneity in solution.
The FLV-bound CYP46A1 structure raises an interesting question about the role of substrate conformation in solution and conformer selection at the entrance to the substrate access channel. Indeed, inside the CYP46A1 active site, FLV has its aliphatic chains spread apart by ∼8.5 Å. However, in such a conformation the drug cannot fit into the substrate access channel whose narrowest portion is only 6.9 Å as indicated by the crystal structure of ligand-free CYP46A1 (Mast et al., 2008). This suggests that in solution FLV exits as a mixture of conformers including those with a more compact positioning of the two alkyl chains (Fig. 7), which is supported by our computational analysis. We envision that in both folded and parallel conformations FLV can enter the substrate access channel. In the folded conformation, the amine-containing chain will enter the access channel first and coordinate the iron after reaching the active site. Once FLV is in the active site and anchored to the iron, the methoxy-containing chain unfolds and is stabilized by hydrophobic interactions with protein amino acid residues. In contrast, the conformer in the parallel configuration would have to enter the substrate access channel with the CF3-phenyl group first and then, once inside the active site, flip over to position the NH2 group for coordination with the iron. Binding of this isomer would require energy to widen the entrance to the substrate access channel and flip inside the active site. Experiments are in progress to investigate whether FLV shows conformational preferences in solution and whether only some of the possible FLV conformers are selected by CYP46A1 because of interactions at the entrance to the access channel. The existence of conformers and conformer selection by CYP46A1 would also account for the increased Kd of FLV relative to that of TCP.
In vivo, FLV is known to undergo intensive biotransformation, leading to at least 11 different metabolites, probably involving two P450s, CYP2D6 and CYP1A2 (Overmars et al., 1983; Carrillo et al., 1996; Spigset et al., 2001). The main route of FLV metabolism is oxidative elimination of the methoxyl group yielding the carboxylic acid. The primary amine could also be the target, either for acetylation or oxidative removal, but these products are minor. Despite FLV preparations containing only very little of the Z-isomer, some FLV metabolites occur in both E and Z forms. Crystal structures of CYP2D6 and CYP1A2 are available (Rowland et al., 2006; Sansen et al., 2007; Wang et al., 2012). CYP2D6, crystallized in substrate-free (PDB code 2F9Q) (Rowland et al., 2006) and substrate (prinomastat)-bound forms (PDB code 3QM4) (Wang et al., 2012), has a large active site (582 and 712 Å3, respectively), yet the shape of the cavity is significantly different in the two forms because of conformational changes induced by substrate binding (Wang et al., 2012). The substrate access channel is open in the substrate-free form but closed in substrate-bound CYP2D6. Binding of the parallel conformer of FLV (Fig. 7) would be consistent with the specificity of CYP2D6 for the methoxyl group. Although the entrance to this channel is relatively wide in substrate-free CYP2D6 (>8 Å), the enzyme, nevertheless, preferentially binds pharmaceutical agents positively charged at neutral pH, like FLV. This selectivity is probably due to the interaction with a negatively charged amino acid residue (Glu222) in the substrate access channel, which serves as a “bait” for the initial binding of positively charged compounds (Wang et al., 2012). Thus, in contrast to CYP46A1 with its relatively narrow substrate access channel and no negatively charged residues at the channel entrance or anywhere inside the active site, CYP2D6 has a means of selection for the positively charged compounds. The other FLV-metabolizing P450 CYP1A2 has only been crystallized in complex with one compound, α-naphthoflavone (Sansen et al., 2007). The active site cavity in this cocomplex is compact and uniformly narrow, consistent with the shape of CYP1A2 substrates, which are relatively large planar compounds. There are two negatively charged residues inside the active site, Asp313 and Asp320. However, their role in the interaction with the substrate primary amine is unclear because the route of substrate entry is not evident from the CYP1A2 structure. In any case, the presence of the negatively charged residues inside the active site raises the possibility that CYP1A2 also has a mechanism for recognition of the NH3+ group. CYP46A1 does not have any negatively charged amino acid residues inside the active site. It would therefore be an example of a P450 with a different mechanism for selection of the positively charged amines.
Relative to P450s 2D6 and 1A2, CYP46A1 may also have a different route of FLV entry. Cholesterol, the endogenous substrate for CYP46A1, probably enters the P450 through the membrane, where it is in abundance, in agreement with our studies of cholesterol metabolism by isolated brain microsomes. However, it not clear how FLV reaches the CYP46A1 active site. This drug is amphipathic in both folded and parallel conformations and thus may partition to the membrane and compete with cholesterol in the membrane. It is also possible that FLV binds in the vicinity of the cytosolic channel (Pikuleva, 2008). If so, FLV should induce conformational changes to widen this narrow channel. Separate studies are required to investigate how FLV might inhibit CYP46A1 in vivo.
In summary, the present work demonstrates that computational predictions of drug binding to CYP46A1 are not straightforward, and intuitive predictions could be of value as well. Taken together, both types of predictions resulted in the identification of new binders to CYP46A1 among marketed drugs, providing targets for subsequent cocrystallization. New structures of CYP46A1-drug complexes continue to lead to a better understanding of drug inhibition of an important brain enzyme and identify areas that are not yet sufficiently studied in the P450 field: the role of a drug's conformation and its route of entry into the protein and the mechanism of ligand selection at the entrance to the substrate access channel. CYP46A1 seems to be a good model for investigating these topics because it acquires cholesterol from the membrane (endoplasmic reticulum) and, nevertheless, is inhibited by different pharmaceutical agents.
Supplementary Material
Acknowledgments
We thank Casey D. Charvet for calculation of the active site volumes, Dr. M. Shimoji for generating the T306A CYP46A1 mutant, and Dr. W. Zheng for purification of the T306A CYP46A1 mutant. We thank the staff at the SSRL for their assistance in data collection.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM62882]. I.A.P. is a recipient of the Jules and Doris Stein Professorship from Research to Prevent Blindness. Portions of this research were carried out at SSRL, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research and by the National Institutes of Health National Institute of General Medical Sciences [Grant P41-GM103393] and National Institutes of Health National Center for Research Resources [Grant P41-RR001209].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- P450
- cytochrome P450
- TCP
- tranylcypromine
- FLV
- fluvoxamine
- KPi
- potassium phosphate
- NTA
- nitrilotriacetic
- BME
- β-mercaptoethanol
- DTT
- dithiothreitol
- SSRL
- the Stanford Synchrotron Radiation Lightsource
- PDB
- Protein Data Bank.
Authorship Contributions
Participated in research design: Mast, Stout, and Pikuleva.
Conducted experiments: Mast, Linger, Clark, and Stout.
Performed data analysis: Mast, Clark, Wiseman, Stout, and Pikuleva.
Wrote or contributed to the writing of the manuscript: Mast, Linger, Clark, Wiseman, Stout, and Pikuleva.
References
- Björkhem I, Lütjohann D, Breuer O, Sakinis A, Wennmalm A. (1997) Importance of a novel oxidative mechanism for elimination of brain cholesterol. Turnover of cholesterol and 24(S)-hydroxycholesterol in rat brain as measured with 18O2 techniques in vivo and in vitro. J Biol Chem 272:30178–30184 [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri EE, Olafson BD, States DJ, Swaminathan S, Karplus M. (1983) Charm—a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217 [Google Scholar]
- Carrillo JA, Dahl ML, Svensson JO, Alm C, Rodríguez I, Bertilsson L. (1996) Disposition of fluvoxamine in humans is determined by the polymorphic CYP2D6 and also by the CYP1A2 activity. Clin Pharmacol Ther 60:183–190 [DOI] [PubMed] [Google Scholar]
- Clark M, Meshkat S, Talbot GT, Carnevali P, Wiseman JS. (2009a) Fragment-based computation of binding free energies by systematic sampling. J Chem Inf Model 49:1901–1913 [DOI] [PubMed] [Google Scholar]
- Clark M, Meshkat S, Wiseman JS. (2009b) Grand canonical free-energy calculations of protein-ligand binding. J Chem Inf Model 49:934–943 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography.Acta Crystallogr D Biol Crystallogr 50:760–763 [DOI] [PubMed] [Google Scholar]
- Congreve M, Chessari G, Tisi D, Woodhead AJ. (2008) Recent developments in fragment-based drug discovery. J Med Chem 51:3661–3680 [DOI] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. (1995) A second generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J Am Chem Soc 117:5179–5197 [Google Scholar]
- Deubner R, Holzgrabe U. (2002) Quantitative 1H NMR spectroscopic determination of the E/Z isomer ratio of the antidepressant drug fluvoxamine for use in pharmaceutical analysis. Magn Reson Chem 40:762–766 [Google Scholar]
- Emsley P, Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 (Pt 12 Pt1):2126–2132 [DOI] [PubMed] [Google Scholar]
- Ferguson DM, Raber DJ. (1989) A new approach to probing conformational space with molecular mechanics: random incremental pulse search. J Am Chem Soc 111:4371–4378 [Google Scholar]
- Foda NH, Radwan MA, Al Deeb OA. (1996) Fluvoxamine maleate, in Analytical Profiles of Drugs Substances and Excipients, vol 24 (Brittain HG. ed) pp 165–208, Academic Press, San Diego, CA [Google Scholar]
- Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. (2006) The Blue Obelisk-interoperability in chemical informatics. J Chem Inf Model 46:991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo GY, Bederman I, Mast N, Liao WL, Turko IV, Pikuleva IA. (2011) Conversion of 7-ketocholesterol to oxysterol metabolites by recombinant CYP27A1 and retinal pigment epithelial cells. J Lipid Res 52:1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudry E, Van Dam D, Kulik W, De Deyn PP, Stet FS, Ahouansou O, Benraiss A, Delacourte A, Bougnères P, Aubourg P, et al. (2010) Adeno-associated virus gene therapy with cholesterol 24-hydroxylase reduces the amyloid pathology before or after the onset of amyloid plaques in mouse models of Alzheimer's disease. Mol Ther 18:44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotti T, Head DD, McKenna CE, Russell DW. (2008) Biphasic requirement for geranylgeraniol in hippocampal long-term potentiation. Proc Natl Acad Sci USA 105:11394–11399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotti TJ, Ramirez DM, Pfeiffer BE, Huber KM, Russell DW. (2006) Brain cholesterol turnover required for geranylgeraniol production and learning in mice. Proc Natl Acad Sci USA 103:3869–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AG. (1999) Integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr 55 (Pt 10):1696–1702 [DOI] [PubMed] [Google Scholar]
- Lund EG, Xie C, Kotti T, Turley SD, Dietschy JM, Russell DW. (2003) Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover. J Biol Chem 278:22980–22988 [DOI] [PubMed] [Google Scholar]
- Mast N, Charvet C, Pikuleva IA, Stout CD. (2010) Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain. J Biol Chem 285:31783–31795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast N, Norcross R, Andersson U, Shou M, Nakayama K, Bjorkhem I, Pikuleva IA. (2003) Broad substrate specificity of human cytochrome P450 46A1 which initiates cholesterol degradation in the brain. Biochemistry 42:14284–14292 [DOI] [PubMed] [Google Scholar]
- Mast N, Reem R, Bederman I, Huang S, DiPatre PL, Bjorkhem I, Pikuleva IA. (2011) Cholestenoic acid is an important elimination product of cholesterol in the retina: comparison of retinal cholesterol metabolism with that in the brain. Invest Ophthalmol Vis Sci 52:594–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast N, White MA, Bjorkhem I, Johnson EF, Stout CD, Pikuleva IA. (2008) Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain. Proc Natl Acad Sci USA 105:9546–9551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. (2007) Phaser crystallographic software. J Appl Crystallogr 40 (Pt 4):658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miolo G, Caffieri S, Levorato L, Imbesi M, Giusti P, Uz T, Manev R, Manev H. (2002) Photoisomerization of fluvoxamine generates an isomer that has reduced activity on the 5-hydroxytryptamine transporter and does not affect cell proliferation. Eur J Pharmacol 450:223–229 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53 (Pt 3):240–255 [DOI] [PubMed] [Google Scholar]
- Muscatello MR, Spina E, Bandelow B, Baldwin DS. (2012) Clinically relevant drug interactions in anxiety disorders. Hum Psychopharmacol 27:239–253 [DOI] [PubMed] [Google Scholar]
- Overmars H, Scherpenisse PM, Post LC. (1983) Fluvoxamine maleate: metabolism in man. Eur J Drug Metab Pharmacokinet 8:269–280 [DOI] [PubMed] [Google Scholar]
- Pellecchia M. (2009) Fragment-based drug discovery takes a virtual turn. Nat Chem Biol 5:274–275 [DOI] [PubMed] [Google Scholar]
- Pikuleva IA. (2008) Cholesterol-metabolizing cytochromes P450: implications for cholesterol lowering. Expert Opin Drug Metab Toxicol 4:1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikuleva IA, Björkhem I, Waterman MR. (1997) Expression, purification, and enzymatic properties of recombinant human cytochrome P450c27 (CYP27). Arch Biochem Biophys 343:123–130 [DOI] [PubMed] [Google Scholar]
- Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, Lewis CJ, Tennant MG, Modi S, Eggleston DS, et al. (2006) Crystal structure of human cytochrome P450 2D6. J Biol Chem 281:7614–7622 [DOI] [PubMed] [Google Scholar]
- Russell DW, Halford RW, Ramirez DM, Shah R, Kotti T. (2009) Cholesterol 24-hydroxylase: an enzyme of cholesterol turnover in the brain. Annu Rev Biochem 78:1017–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2007) Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem 282:14348–14355 [DOI] [PubMed] [Google Scholar]
- Shafaati M, Mast N, Beck O, Nayef R, Heo GY, Björkhem-Bergman L, Lütjohann D, Björkhem I, Pikuleva IA. (2010) The antifungal drug voriconazole is an efficient inhibitor of brain cholesterol 24S-hydroxylase in vitro and in vivo. J Lipid Res 51:318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafaati M, Olin M, Båvner A, Pettersson H, Rozell B, Meaney S, Parini P, Björkhem I. (2011) Enhanced production of 24S-hydroxycholesterol is not sufficient to drive liver X receptor target genes in vivo. J Intern Med 270:377–387 [DOI] [PubMed] [Google Scholar]
- Soltis SM, Cohen AE, Deacon A, Eriksson T, González A, McPhillips S, Chui H, Dunten P, Hollenbeck M, Mathews I, et al. (2008) New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr D Biol Crystallogr 64 (Pt 12):1210–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spigset O, Axelsson S, Norström A, Hägg S, Dahlqvist R. (2001) The major fluvoxamine metabolite in urine is formed by CYP2D6. Eur J Clin Pharmacol 57:653–658 [DOI] [PubMed] [Google Scholar]
- Wang A, Savas U, Hsu MH, Stout CD, Johnson EF. (2012) Crystal structure of human cytochrome P450 2D6 with prinomastat bound. J Biol Chem 287:10834–10843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Mast N, Bjorkhem I, Johnson EF, Stout CD, Pikuleva IA. (2008) Use of complementary cation and anion heavy-atom salt derivatives to solve the structure of cytochrome P450 46A1. Acta Crystallogr D Biol Crystallogr 64 (Pt 5):487–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.