Abstract
Songbirds provide rich natural models for studying the relationships between brain anatomy, behavior, environmental signals, and gene expression. Under the Songbird Neurogenomics Initiative, investigators from 11 laboratories collected brain samples from six species of songbird under a range of experimental conditions, and 488 of these samples were analyzed systematically for gene expression by microarray. ANOVA was used to test 32 planned contrasts in the data, revealing the relative impact of different factors. The brain region from which tissue was taken had the greatest influence on gene expression profile, affecting the majority of signals measured by 18,848 cDNA spots on the microarray. Social and environmental manipulations had a highly variable impact, interpreted here as a manifestation of paradoxical “constitutive plasticity” (fewer inducible genes) during periods of enhanced behavioral responsiveness. Several specific genes were identified that may be important in the evolution of linkages between environmental signals and behavior. The data were also analyzed using weighted gene coexpression network analysis, followed by gene ontology analysis. This revealed modules of coexpressed genes that are also enriched for specific functional annotations, such as “ribosome” (expressed more highly in juvenile brain) and “dopamine metabolic process” (expressed more highly in striatal song control nucleus area X). These results underscore the complexity of influences on neural gene expression and provide a resource for studying how these influences are integrated during natural experience.
Keywords: mRNA, avian, neuroanatomy, systems biology, neuroplasticity
The genome plays an active role in the biological embedding of social experience. Social interactions and variations in abiotic and biotic environmental conditions can trigger both transient and lasting changes in gene expression in specific parts of the brain (1–5). Conversely, preexisting variations in brain gene expression or genotype may influence how an individual reacts to particular social and environmental challenges (6, 7). These different forces—gene, environment, and life history—intersect in the biological tissue of the brain, itself an exceedingly complex physical entity. Studies of brain gene expression have typically found that most genes are expressed in the brain but in many different patterns according to brain cell subtype and subregion, developmental age, and physiological state of the organism (8–11). A coherent view has yet to emerge for how these many axes of variation are produced from the common genome present in all cells of each individual and how they are influenced by natural experience.
Songbirds (oscines of the order Passeriformes) are uniquely attractive organisms for studying how genes, environment, and life history interact in the complex anatomy of the brain (12). Like humans, songbirds mediate social interactions through learned vocalizations (13–15). Also like humans, most songbirds are altricial and their young require a prolonged period of parental care during which vocal learning is most active. Brain circuits involved in songbird vocal learning have been defined with unparalleled precision and detail (e.g., 16–19). This circuitry is organized into a set of interconnected nuclei (the song control system) that can be readily identified and even dissected for analyses of local gene expression. There are ∼4,000 songbird species, with significant differences in social organization, behavioral ontogeny, and sensitivity to environmental cues. Because these are all members of a single monophyletic lineage, they have a shared genetic background, affording rich opportunities for interpreting variations in gene expression in a functional context (12, 20).
To advance the development of genome-informed research using songbirds, a consortium of investigators organized the Songbird Neurogenomics (SoNG) Initiative (21), and an initial draft sequence of the zebra finch genome was produced (22). Through the SoNG Initiative, the “SoNG 20K microarray” was generated (21) with cDNA probes measuring gene expression at more than 15,000 chromosomal loci (22), and this array was validated for use in studies of other songbird species as well (21). Through a set of structured collaborations, this array was used to assess variations in gene expression in brain tissue samples contributed by multiple laboratories and representing a diversity of songbird species, brain subregions, developmental stages, and physiological or behavioral states. These collaborations were organized so that individual datasets could be analyzed independently, as shown in several reports to date (23–27). However, a unique opportunity was anticipated for an integrated global analysis to evaluate the relative impact on gene expression of the multiple biological and experiential dimensions represented in the combined dataset (21). Here, we develop such an analysis by combining two complementary approaches. First, we used a standardized data preprocessing and analysis method for each experiment so that the numbers of genes found “significant” can be compared between experiments. Second, we conducted a cross-experiment analysis to cluster genes with similar expression patterns across all treatment manipulations considered at once.
Our results answer fundamental questions about gene expression, brain organization, and experience. Do different regions of the brain differ significantly in their patterns of gene expression, or is all neural tissue mostly similar? Do the distinctive, interconnected nuclei of the song control circuit share a gene expression pattern that links them together and distinguishes them from the rest of the brain? Do different signals trigger similar changes in gene expression across the different song nuclei (e.g., a common “growth and regression” program), or is there significant heterogeneity in the way different brain regions respond at the genomic level to different experiential cues? Can species and age differences in learning ability and responsiveness to social cues be linked to specific variations in gene expression? Most generally, how much variation in brain gene expression can be attributed to differences in anatomy, sex, age, species, environment, and experience?
Results
Statistical Analyses of Variations in Gene Expression Across all Experiments.
RNA samples were collected from six different songbird species (phylogeny in SI Appendix, Fig. S1) and represent 80 different “treatments” [i.e., combinations of species, brain region, sex, age, behavioral and physiological (or reproductive) state]. The tissue samples were organized around 15 stand-alone experiments (Table 1 and SI Appendix, Table S1) contributed by investigators in 11 different laboratories but analyzed under uniform conditions in a single laboratory as originally planned under the SoNG Initiative (21). Each sample was hybridized to a zebra finch cDNA array along with a universal reference (a pool of zebra finch telencephalic RNA). Independent analyses of seven of the individual experiments have already been published (23–27), but all data were reanalyzed here under uniform procedures. The data from all 488 arrays, each representing one contributed sample (SI Appendix, Table S2), were first standardized and then assessed for relationships between the samples using principal components analysis (PCA) (SI Appendix, Fig. S2). We then conducted two types of analyses to assess the relative impact of different factors on gene regulation across the entire dataset: one using ANOVA to evaluate each experiment independently, followed by tabulation of results across experiments, and the other using weighted gene coexpression network analysis (WGCNA) (28, 29) to identify genes that shared similar expression patterns across all treatments and all experiments considered at once.
Table 1.
Number of “significant” cDNAs for each of the 32 planned contrasts in the 15 experiments
| Exp. no. | Species | n (g) | Age | Sex | Region | Dataset S1 column | Contrast (no. of levels) | P < 0.001 | P < 2.6 E-06 |
| e01 | Gambel’s white-crowned sparrow | 71 (12) | Adult | M | HVC, RA | e01.A | Time after T (6) | 689 | 75 |
| e01.B | Region (2) | 7,112 | 4,413 | ||||||
| e01.C | Interaction | 57 | 2 | ||||||
| e02 | Zebra finch | 16 (3) | p35 | M | Area X | e02.A | Tutor experience (3) | 21 | 0 |
| e03 | House finch, Red crossbill | 24 (5) | Adult | M | Diencephalon | e03.A | Species (2) | 4,322 | 1,448 |
| e03.B | Food (2) | 53 | 1 | ||||||
| e03.C | Interaction | 55 | 2 | ||||||
| e03.D | Day length (2) | 66 | 1 | ||||||
| e04 | Zebra finch | 48 (8) | p1, p25, p45, Adult | M, F | Telencephalon | e04.A | Age (4) | 8,234 | 4,675 |
| e04.B | Sex (2) | 1,192 | 652 | ||||||
| e04.C | Interaction | 266 | 55 | ||||||
| e05 | Zebra finch | 24 (4) | p1, p7 | F | Telencephalon | e05.A | E-silastic (2) | 15 | 0 |
| e05.B | Age (4) | 2,715 | 653 | ||||||
| e05.C | Interaction | 20 | 0 | ||||||
| e06 | House finch | 24 (3) | Adult | M | HVC | e06.A | Time after T (3) | 15 | 0 |
| e07 | Zebra finch | 24 (4) | p55, Adult | M | HVC | e07.A | Age (2) | 682 | 43 |
| e07.B | Food access (2) | 47 | 0 | ||||||
| e07.C | Interaction | 15 | 0 | ||||||
| e08 | Starling | 120 (20) | Adult | M | Area X, HVC, POA, RA | e08.A | Region (2) | 13,245 | 10,511 |
| e08.B | Time (5) | 871 | 36 | ||||||
| e08.C | Interaction | 201 | 8 | ||||||
| e09 | Zebra finch | 12 (2) | Adult | M | HVC, shelf | e09.A | Region (2) | 1,038 | 139 |
| e10 | Zebra finch | 36 (6) | Adult | F | NCM, L2a | e10.A | Song playback (3) | 356 | 23 |
| e10.B | Region (2) | 8,963 | 5,874 | ||||||
| e10.C | Interaction | 48 | 4 | ||||||
| e11 | Zebra finch | 18 (3) | Adult | M | AL | e11.A | Song playback (3) | 2,585 | 417 |
| e12 | Zebra finch | 12 (2) | p20 | M | AL | e12.A | Song playback (2) | 19 | 1 |
| e13 | Zebra finch | 16 (2) | p25, p45 | M | LMAN | e13.A | Age (2) | 156 | 3 |
| e14 | Song sparrow | 31 (4) | Adult | M | Hypothalamus | e14.A | Season (2) | 123 | 6 |
| e14.B | STI (2) | 11 | 0 | ||||||
| e14.C | Interaction | 6 | 0 | ||||||
| e15 | Starling | 12 (2) | Adult | M | AL | e15.A | Song playback (2) | 37 | 1 |
Exp. no. gives the designated number of each experiment, and the other columns show the species, number of birds (n), number of treatment groups (g), age, sex, brain regions, and contrasts evaluated by each experiment. We provide both rigorous Bonferroni-corrected P values and a more liberal empirical standard (raw P < 0.001) derived from previous analyses of individual experiments in this dataset (23–27). We compared the numbers of significant probes based on raw P values instead of FDR-corrected P values because a cDNA could have the same raw P value in two experiments yet have vastly different FDR P values depending on how many other cDNAs have expression differences in each experiment. Full results are given in Dataset S1. AL, auditory lobule; F, female; LMAN, lateral magnocellular nucleus of the anterior nidopallium; M, male; p, posthatch day; POA, preoptic area; T, testosterone.
For the ANOVA, Table 1 summarizes the numbers of probes significant for each planned contrast in each experiment (full results for each probe and contrast are given in Dataset S1). The number of genes affected by a given type of factor can be taken as a measure of the relative impact of specific factors on specific genes (Table 2). With certain caveats (Discussion), we believe this numerical comparison of gene lists across experiments is valid due to the use of a common microarray, hybridization, and analysis pipeline.
Table 2.
Number of “significant” cDNAs for each main factor, averaged over the number of treatment levels in each contrast
| Factor (no. of contrasts) | No. of levels per contrast | Avg. no. sig. cDNAs per level per contrast P < 0.001 | Avg. no. sig. cDNAs per level per contrast P < 2.6 E-06 |
| Region (4) | 2, 2, 4, 2 | 2,967 | 1,960 |
| Species (1) | 2 | 2,161 | 724 |
| Age (4) | 4, 2, 2, 2 | 959 | 380 |
| Sex (1) | 2 | 596 | 326 |
| Social (6) | 3, 3, 3, 2, 2, 2 | 170 | 25 |
| Environment (7) | 6, 2, 2, 3, 2, 5, 2 | 63 | 3 |
Avg. no. sig., average number of significant.
For the WGCNA analysis, we began with the mean expression values for each treatment group derived from the ANOVA analysis and normalized them as described in Materials and Methods and SI Appendix, Fig. S3. WGCNA algorithms were then used to calculate the similarity of expression patterns for each pair of cDNAs considered across all treatment conditions (28, 29). This resulted in the identification of 95 “modules,” with each cDNA assigned to one primary module, such that all cDNAs in a given module showed more similar expression patterns to each other than to the patterns of other modules (details are provided in SI Appendix, SI Materials and Methods). For each module, a single eigenvalue describes the relative expression of all cDNAs in that module under a given treatment condition and the set of eigenvalues for all treatment conditions describes the overall expression pattern of that module. Large positive eigenvalues mean relatively higher expression values, and large negative eigenvalues mean relatively lower expression values (see SI Appendix, Fig. S6 for an example comparing expression profiles with eigenvalues for module 1; all eigenvalues for all cDNAs are in Dataset S2, and expression profiles for all modules are given in Dataset S3]. To assess major factors that might be responsible for each module’s expression pattern, we then performed ANOVA on the eigengene values for all 95 modules × 80 treatment groups for effects of species (5 levels), region (12 levels), age (8 levels), sex (2 levels), photoperiod at sacrifice (6 levels), or song exposure (2 levels) (Fig. 1). To gain insight into the possible functional significance of the various modules, we performed gene ontology (GO) enrichment analyses on the genes in each module (Dataset S4). Ten modules were enriched for specific GO terms at a false discovery rate (FDR) <0.02. These modules are described in Table 3, where they have been further organized into four groups based on their expression patterns as assessed from their eigenvalue bar plots (Dataset S3) and ANOVA factor analysis (SI Appendix, Table S4).
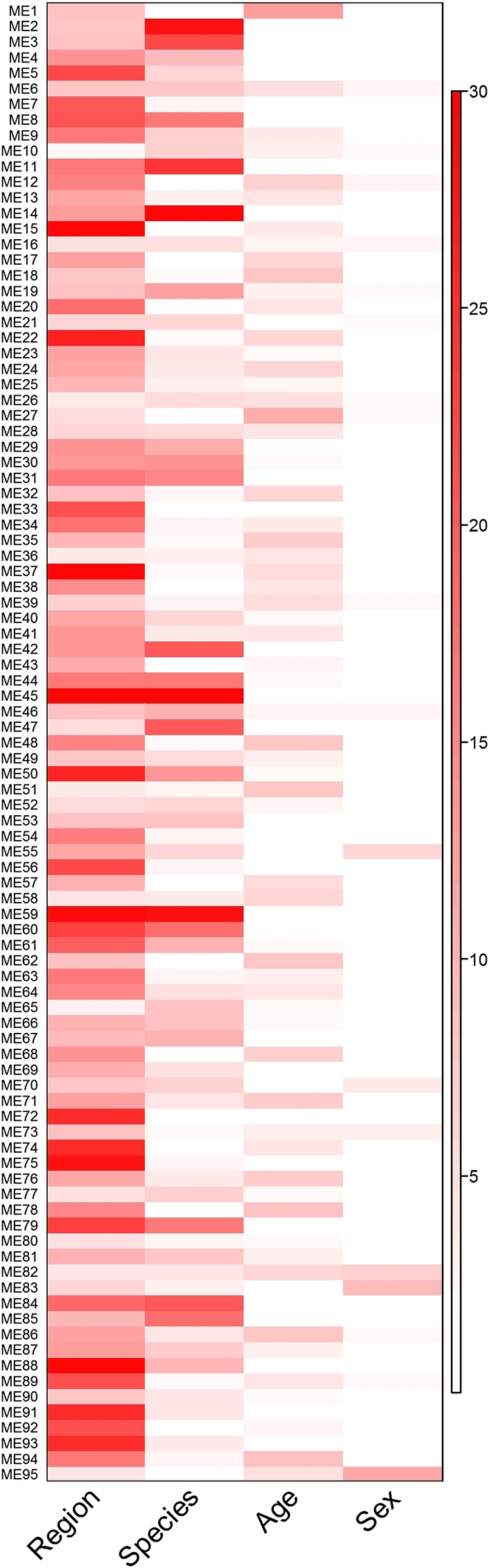
Fig. 1.
Heat map of the P values from separate one-way ANOVAs for region, species, age, and sex on the eigengene values for each module (ME1–ME95) from the WGCNA analysis. The color scale is −log10 (P value). Exact P values are provided in SI Appendix, Table S4.
Table 3.
Ten modules with significantly enriched GO terms
| Theme | Module | No. of cDNAs | Factor | Visual pattern | GO FDR | Expect | Observe | GO description |
| “Ribosome” | 1 | 829 | Age × region | p1 and p7 telencephalon | 2E-09 | 8 | 32 | Structural constituent of ribosome |
| 13 | 408 | Region × age | LMAN and juvenile telencephalon | 4E-10 | 4 | 25 | Ribosome | |
| “Area X” | 22 | 196 | Region | Area X low | 0.016 | 0 | 3 | Stereocilium |
| 89 | 47 | Region | Area X high, pallium low | 0.0001 | 0 | 6 | Ion transport | |
| 91 | 46 | Region | Area X high | 0.019 | 0 | 2 | Dopamine metabolic process | |
| “Sex” | 82 | 51 | Sex | M > F | 0.019 | 0 | 2 | Glutamate secretion |
| 95 | 43 | Sex | F > M | 0.016 | 0 | 2 | ssDNA binding | |
| “Contrast telencephalon and diencephalon” | 30 | 152 | Species × region | Fring. or Ember. X hypothalamus or diencephalon | 0.018 | 0 | 3 | Secretory granule |
| 71 | 62 | Region | Telencephalon high; diencephalon, HVC and X low | 0.0044 | 0 | 2 | Axis specification | |
| 61 | 75 | Region | Hypothalamus low | 0.0053 | 0 | 2 | Transmembrane receptor protein tyrosine kinase signaling protein activity |
Ember., Emberizidae; F, female; Fring., Fringillidae; LMAN, lateral magnocellular nucleus of the anterior nidopallium; M, male; p, posthatch day.
Comparisons of the individual ANOVA results and the WGCNA analyses lead us to the following overall conclusions.
Almost all Genes Vary in Expression Across Brain Regions, Ages, or States.
Of the 18,848 cDNA spots analyzed, only 1,660 reported no significant differences in expression by ANOVA within any of the individual experiments using the criterion of P < 0.001 (SI Appendix, Fig. S4). Similarly, by WGCNA, all 95 modules had highly significant P values (P < 1e-5) for at least one factor and only 316 cDNAs could not be assigned to any module.
WGCNA analysis was especially useful in revealing how multiple factors may interact to generate different patterns of gene expression, because 88 of the 95 modules showed significant P values for two or more factors (Fig. 1 and SI Appendix, Table S4). Module 1 (the single largest module) and module 13 are illustrative of this, because both “age” and “region” are highly significant interacting factors in their expression patterns. This is further illustrated in SI Appendix, Fig. S7, where eigenvalue plots for modules 1 and 13 have been colored to show how these two factors map onto the 80 different experimental treatment groups (compare this with SI Appendix, Fig. S6, where the same plot for module 1 is colored to show “experiment”). In both of these modules, samples from the youngest animals (posthatch days 1–7) have the highest eigenvalues, and samples from auditory lobule and its subregions caudomedial nidopallium (NCM) and L2a have low eigenvalues. Both of these modules are highly enriched for functions associated with ribosomes (Table 3).
Brain Region Is a Dominating Factor in Differential Gene Expression.
Whether summing the numbers of genes significant for all contrasts (by ANOVA; Table 2), or the numbers of gene expression modules significant for each major treatment factor (by WGCNA; Fig. 1), we find that the major determinant of gene expression pattern is the region from which the brain sample was drawn. The effect of brain region is enormous, with more than 70% of all probes producing different signals among just the four brain regions compared in experiment e08. Although within-subject designs may have enhanced the sensitivity of individual experiments focused on regional comparisons, region also emerged as the dominant factor in the WGCNA analysis, which compared group mean expression values across all experiments at once: Region had the lowest (most significant) P value for the most modules (n = 68), followed by species (n = 19), age (n = 6), and then sex (n = 3).
The comparatively limited effect of sex is interesting, given the chromosomal differences between the sexes and the absence of robust dosage compensation in birds (30, 31). In the one experiment where sex was an explicit contrast (e04), we detected 652 significant differences after Bonferroni correction, or 1,192 using the criterion of raw P < 0.001. This estimate is somewhat more conservative than a previous analysis of the same microarray data (27), although our analyses detected a greater number of cDNAs showing interaction between sex and age (266 at P < 0.001). Two WGCNA modules, module 82 and module 95, showed strong effects of sex and also evidence of functional enrichment by GO analysis. Module 82 genes are expressed higher in males (SI Appendix, Fig. S8), and the two genes that contribute to its annotation for glutamate secretion (Table 3) are both present on the Z chromosome (consistent with overall higher expression in males). They are APBA1, a neuronal adaptor protein that interacts (in humans) with the Alzheimer’s disease amyloid precursor protein, and NTRK2, a BDNF/NT-3 growth factor receptor. Module 95 genes are more highly expressed in females, and include genes for ssDNA binding proteins ERCC5 (DNA excision repair) and SUB1 (an RNA polymerase II cofactor).
Great Variation Across the Song Control System.
The song control nuclei have been described only in oscine songbirds, raising questions about their ontological and evolutionary origins. Here, we find that the song nuclei differ from each other as much as they do from the rest of the brain. Thousands of genes differentiate nucleus HVC (letters used as proper name) and its immediately adjacent and functionally related surrounding shelf region (experiment e09), and even more differentiate HVC from the motor output nucleus to which it projects, the robust nucleus of the arcopallium (RA) (experiment e01). In the experiment using starlings (e08), comparing three major song nuclei (HVC, RA, and X) and the preoptic area of the hypothalamus, roughly 55% of all spots on the microarray showed a significant effect of brain region. These results do not disprove the hypothetical existence of one or a few general markers common to all song nuclei (e.g., 32), but they do suggest that each song nucleus is highly differentiated not only from the surrounding tissue but from every other nucleus in the circuit.
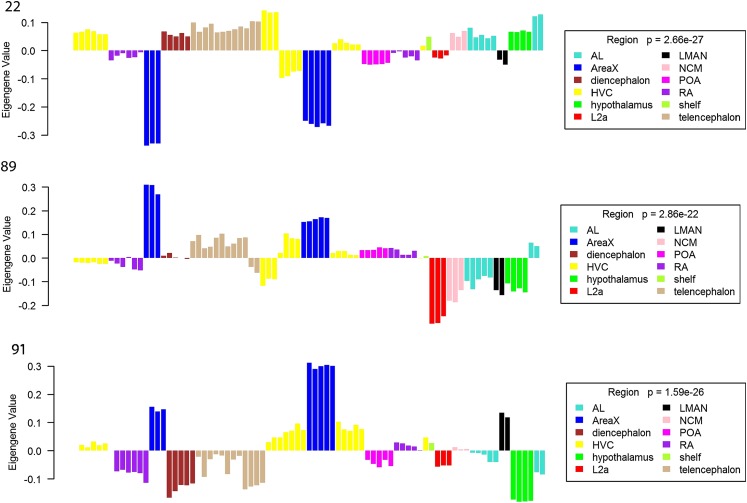
Study of gene expression patterns that distinguish individual song nuclei may be uniquely informative about the functional and biological properties of each nucleus. A detailed analysis of HVC gene expression from experiment e09 has been presented previously (23), and we also note a very strong signature in the combined data for another song nucleus, area X. In the WGCNA analysis, module 22 genes are expressed at low levels in this nucleus, whereas module 89 and 91 genes are expressed at high levels (Fig. 2 and Table 3). In contrast to the pallial (or “cortical”) nuclei HVC and RA, area X is a striatal nucleus that is densely populated with small neurons and receives rich dopaminergic innervation (33). Consistent with this and with prior evidence for enrichment of various neurotransmitter receptors in area X (23, 33), module 89 contains 6 ion channel-associated genes, including two glutamate receptors (GRIA1 and GRIK3), a GABA receptor (GABRB3), a cyclic nucleotide gated channel (CNGA3), a monoamine transporter (SLC22A3), and a transient receptor potential cation channel (TRPM1). Module 91 includes another glutamate receptor (GRINA) and is enriched for the term “dopamine metabolic process.”
Fig. 2.
Plots of the representative expression patterns for modules 22 (Top), 89 (Middle), and 91 (Bottom), colored by brain region. Each bar represents one of the 80 treatment groups (treatment names are provided in SI Appendix, Fig. S6), and the height of the bar indicates the relative expression level for each group as estimated by the eigengene value. All three module types had significantly overrepresented GO terms and highly significant P values for brain region ANOVA. The expression patterns show that all three distinguish area X from the other regions. AL, auditory lobule; LMAN, lateral magnocellular nucleus of the anterior nidopallium; POA, preoptic area.
Large Differences in Gene Expression Even Among Closely Related Songbird Species.
We also detected a very large effect of species in our data, although this effect was numerically smaller than the effect of region (i.e., there are more differences between 2 regions of the same species than between the same region of 2 different species). One might question whether the lower effect of species could reflect reduced sensitivity of cross-species hybridizations; however, the ANOVA contrast that detected the greatest number of differences across all the experiments in the entire dataset (contrast e08.A) was a comparison within starlings, supporting prior conclusions that the SoNG Initiative 20K cDNA array is useful for all passerines (21). Any comparison of expression differences between species runs the risk of confounding expression changes with sequence divergence, but one experiment in our dataset compared two closely related species from the same family (house finches and red crossbills, family Fringillidae). These two species are equally distant from the zebra finch (SI Appendix, Fig. S1); thus, one would expect that they should share similar patterns of sequence divergence relative to the probes on the zebra finch microarray. The 1,448 (4,322) observed differences in hybridization signal seem higher than expected for sequence divergence between two members of the same family (21). This suggests there may be true species differences in their brain gene expression patterns, although this needs to be confirmed experimentally [e.g., using methods like RNA sequencing, which avoid the problem of species cross-hybridization].
Effects of Experience Are Variable and Often Subtle.
Against these large effects of brain region and species on gene expression, we observed fewer and more variable effects of experience, even though most experiments involved some manipulation of experience-dependent factors. In Table 2, we classified experience-dependent factors as either social (i.e., exposure to another bird through sound or sight) or environmental (i.e., alteration of light cycle or food availability). Unlike the experience-independent factors above, however, we could link none of these factors to specific modules in WGCNA (SI Appendix, SI Materials and Methods).
Nevertheless, by ANOVA, we did detect strong effects in some but not all experiments. Prior analysis of experiment e11 (comparing males hearing novel song, familiar song or silence) had reported very large effects, with thousands of cDNAs varying under these three conditions (24), and our analysis of the same data are in agreement (contrast e11.A). Experiment e10 (contrast e10.A) is an independent replication of the primary effect of social isolation followed by song exposure, using females hearing only novel song or silence and separately analyzing two subregions (NCM and field L2a) that are combined in the “auditory lobule” dissection used in experiment e11. In marked contrast to these two experiments, four others also used song playback to assess effects of social interactions (e02, e12, e14, and e15) but found only modest or even negligible changes in gene expression (Discussion).
Six experiments involved natural or artificial changes in photoperiod (some with additional testosterone manipulation) to stimulate changes in behavior (contrasts e03.D and e14.A) or growth and regression in the song control nuclei (contrasts e01.A, e06.A, and e08.B). Effects on expression profile ranged from minimal (0 or 1 gene expression differences after Bonferroni correction in e03.D and e06.A) to moderate (dozens to hundreds of changes in e01.A and e08.B). The primary difference in results here may be time: Experiments e01 and e08 both assayed multiple time points ranging from 3 to 56 d after a shift in photoperiod and found differences emerging at multiple points all across this period, whereas experiment e03 only assessed 7 d after a shift to long days and experiment e06 only assessed 24 h and 48 h. These results indicate that photoperiod shifts can indeed cause large changes in brain gene expression but may do so on a prolonged time course.
Together, these experiments indicate that gene responsiveness to acute experience does not necessarily correlate with our perceptions of behavioral responsiveness but may vary in ways more broadly linked to the developmental, environmental, and perhaps social contexts in which the organism is embedded.
Frequent Regulation of Cell-Matrix and Peptidergic Signaling Genes.
The 12 most commonly regulated genes in our dataset (SI Appendix, Fig. S5) all show strong effects of both development (contrast e04.A) and brain region (contrasts e08.A and e10.B). Additionally, 4 of these genes map to the Z sex chromosome and all these show main effects of sex in their expression (CRHBP, GPR98, PCSK1, and JUND; contrast P). Three genes encode transcription factors often described as immediate early genes (EGR1, NR4A3, and JUND). Interestingly, however, the largest numerical subcategory of the most highly regulated genes comprises 7 genes that encode proteins involved in cell-matrix interactions and cell-cell (mainly peptidergic) signaling. Three of these (ADAMTS1, APOH, and PCSK1) are found in the same module in the WGCNA analysis (module 26), indicating they have very similar overall expression patterns in this dataset. ADAMTS1, an ECM protein, is a target of regulation by mineralocorticoid; APOH is a phospholipid binding protein typically described in serum; and PCSK1 is a proprotein convertase that activates proopiomelanocortin, proenkephalin, and other neuropeptide precursors. The others in this functionally related set, found in different expression modules, include cathepsin B (CTSB), an extracellular protease; corticotropin-releasing hormone binding protein (CRHBP), whose regulation in HVC (contrast e09.A) was independently confirmed by in situ hybridization (23); G protein-coupled receptor (GPR98); and calcipressin, regulator of calcineurin (RCAN2).
Brain Genes Responding to Food Manipulations.
Experiment e03 (contrast e03.B and interaction e03.C) tested the effects of an enriched diet on the red crossbill (Loxia curvirostra), an opportunistic breeder that responds to food with increased reproductive behavior even on the shortest and longest days of the year (34). Fifty-three cDNAs were significant in the diencephalon for the effect of food at P < 0.001, but only 1 cDNA remained significant after Bonferroni correction. It maps to ENSTGUG00000011402, which is described as a novel gene in Ensembl and may be unique to birds based on lack of alignments to other well-curated genomes. A different gene, ENSTGUG00000007155 [macrophage-expressed gene 1 protein (MPEG1)] remained significant after Bonferroni correction for the interaction of species and food (there are two separate cDNAs on the array, and both were significant in this test). These are some interesting leads to pursue in regard to the evolution of linkages between environmental signals and reproductive behavior.
Experiment e07 (contrast e07.B and interaction e07.C) also used food manipulation to manipulate behavior, not by varying nutritional content but only by varying the daily rhythm of food availability in this case. Restriction of access to the last 6 h of a 14-h light phase does not affect daily food intake or body mass but reduces daily song production by more than two-thirds (35, 36). Here, the primary goal was to test the interaction of the amount of singing with juvenile development of song nucleus HVC. Forty-seven and 15 genes were significant (P < 0.001) for the main effect of food manipulation and interaction with age, respectively, although none was significant after Bonferroni correction. Nevertheless, the 47 apparent food-sensitive (singing-sensitive) genes do show an intriguing and highly significant enrichment (FDR of P < 0.05) for 2 voltage-gated potassium channel genes (KCNS1, KCNC2), one calcium-gated potassium channel (KCNN2), two serotonin receptors (HTR1D, HTR2A), and 3 other G protein-coupled receptor signaling pathway genes (INSR, ZMPSTE24, and C5orf32). There is no overlap in the cDNAs significant in these two experiments (e03 and e07) using food manipulation.
Discussion
Here, we performed a synthetic statistical analysis of a unique dataset collected under the SoNG Initiative (21). The SoNG Initiative was designed with two goals: (i) to engage the participation of a wide range of songbird biologists, allowing individuals to choose the experimental questions of greatest interest within their own subfields, and (ii) to minimize the sources of technical variation between the various experiments so that they could be analyzed as a group to gain broad insight into the relative influence of diverse factors on brain gene expression, as we have done here. The rationale for this metaanalysis was the potential for extracting information from the whole that could not be obtained within any individual planned experiment by itself. Our results affirm this expectation and also provide a platform for bringing forward observations within individual experiments that were beyond or tangential to the experimenter’s initial objectives. Although we did not conduct new independent experiments to validate specific microarray results presented here, extensive validations have been performed previously on subsets of these data (23–27, 30), and these consistently found very high agreement with measurements based on in situ hybridization and PCR.
By design, the SoNG Initiative aimed for breadth over depth, and breadth was indeed achieved by drawing from samples contributed by 11 different laboratories. However, this also resulted in a sparse dataset with frequent confounding of factors (e.g., all hypothalamic samples were from non-zebra finches, thus confounding the effects of species and brain region when comparing these experiments with the rest of the amassed data). Moreover, even though we minimized sources of technical variation by using a single analysis pipeline, the effect of experiment (i.e., the laboratory that provided the brain samples) is still primary in the initial clustering of data (SI Appendix, SI Materials and Methods and Table S4) echoing a prior study comparing data from different laboratories using the same mouse strains and experimental protocols (37). Future studies will benefit from both deeper sampling and more systematic control of experimental variables to describe gene expression configurations uniquely associated with specific developmental or functional states in the brain accurately. Future studies are also likely to benefit from the rapidly developing techniques for direct sequencing of RNA (38) as a potential improvement over the microarray technology used in the SoNG Initiative.
As a complement to ANOVA, we used a relatively recently developed statistical method, WGCNA, because it extracts patterns from all the combined data without requiring a formal specification of the expected factors and interactions. Thus, it may be especially useful as a discovery tool in data mining. Here, for example, we found three modules that were dominated by higher or lower expression in area X, suggesting that this song nucleus may be unusually distinctive, even though no experiment was explicitly designed to test this idea. WGCNA also identifies gene sets that are consistently coexpressed, which may define common functional processes, states, or cell types that vary systematically across the different treatment groups. The module 1 genes, for example, tend to share roles in ribosomal function and protein synthesis, processes that are most active in juvenile brain development. Interestingly, only 10 of the 95 modules have significant functional internal relationships as assessed by GO. We can only speculate whether the lack of significant annotations for the remaining modules is a limitation of the annotations currently available in GO or evidence for coexpression of highly cellular disparate processes in most of the modules. Another use of WGCNA data is to identify central or “hub” genes within the gene expression network, which may serve as network control points and are operationally defined by each gene’s module membership or connectivity value (the correlation between a gene’s actual expression pattern and the module’s eigengene value). For example, transcription factor genes that closely match a module’s eigengene pattern might be responsible for regulating the other genes in that module. We explored that possibility for the modules in Table 3; transcription factor genes with high connectivity values (among the top 5 within each module) are found in area X modules 22 (NRF2) and 91 (HLF), as well as in the male-increased module 82 (SIX6). Functional manipulations of these genes would be informative as to their potential role in area X differentiation and function. A fourth demonstrated use of WGCNA data is to compare modules in different networks built from different datasets to look for evidence of deep conservation. For example, Oldham et al. (39) used this approach to find evidence of conserved modules that correspond to particular brain cell types. Here, we just created a single network to describe the expression pattern across all our data at once. A productive future analysis might be to construct separate networks for the same brain region in different species to define conserved functional modules more sharply.
Both ANOVA and WGCNA approaches gave similar results with respect to the relative impact of the major factors represented in the 488 brain tissue samples analyzed, with both finding brain region to be the major determinant. A dominant effect of brain region has also been described in the mammalian brain (40–42). Regional variations in gene expression probably reflect both developmental differences (e.g., the relative abundance of different cell types in the various brain regions) and physiological differences (i.e., local microenvironments, modulatory inputs, hormonal signals). In any case, the strong effect of brain region is a major factor to consider in the search for core regulatory mechanisms common in the nervous system, as well as in considering how environmental factors reach the genome. Genes are expressed at different levels in different brain circuits and systems, and even a broad organismal factor like “stress” will thus engage with a different genomic substrate in different parts of the brain.
Songbirds have attracted much attention in scientific research as models for studying the interplay between development, social experience, neural circuitry, and behavior. Our results indicate that experience-dependent factors interact powerfully with the dominating experience-independent factors, such that the effects vary greatly with brain region, species, age, and sex. For example, the largest social effect (most regulated genes) occurred in the adult auditory forebrain of zebra finches in response to song playbacks (experiment e11). However, no significant differences were detected in response to song or tutor exposure in the juvenile zebra finch (experiments e02 and e12), and effects were much reduced in the starling auditory forebrain (experiment e15) and in the song sparrow hypothalamus (experiment e14). A reduced genomic response to song playbacks was the a priori prediction in the experiment with juvenile zebra finches (experiment e12), based on the hypothesis of age-dependent “constitutive plasticity” during the critical period for juvenile song learning (25). According to this hypothesis, gene expression patterns associated with enhanced information storage are sustained (constitutive) throughout the developmental period when the bird is most sensitive to song tutoring, whereas they become suppressed in the adult and are induced only when the bird experiences a strong, salient social stimulus (e.g., isolation followed by sudden exposure to a new conspecific).
A variant of the constitutive plasticity hypothesis could also explain the lower numbers of song-responsive genes seen in experiment e15.A with starlings (not previously published) and experiment e14.B with song sparrows [independently described by Mukai et al. (26)]. Starlings, unlike the zebra finch, are lifelong “open learners” in which functional sensitivity to new song models is sustained into adulthood. The constitutive plasticity hypothesis thus predicts that juvenile patterns of basal gene expression should be sustained in the adult starling, whereas they become suppressed in the adult zebra finch; a direct comparison of gene expression in adult and juvenile starlings and zebra finches would be informative in this regard. The song sparrow experiment involved a different brain region (hypothalamus instead of auditory forebrain) and a different behavioral context [aggressive response to simulated territorial intrusion (STI)]. The initial hypothesis in that experiment was that gene responses to STI would be greatest in spring during the season of enhanced behavioral aggression, but the opposite was observed (26). Thus, gene expression patterns appear to become more “constitutive” and less immediately “experience-dependent” during precisely those ages or seasons when a focused response to immediate experience may be most critical to the organism.
The diversity of songbirds and their various adaptations to different ecological and behavioral niches have inspired many proposals of comparative experiments to correlate genetic or gene expression differences with specific functional or behavioral differences (e.g., 12, 43). Our data here suggest that differences in brain gene expression between even relatively closely related species may be quite large, which will challenge any simple species comparisons attempting to correlate gene expression variation with particular traits. This also raises the question of whether there are conserved transcriptional networks for embedding of social experience in the brain, and if so, what would be required to detect them? If such networks exist, we suggest that future research may find them by a process of triangulation, combining both comparative studies of species that differ in responsiveness to social experience (e.g., open vs. closed learners in songbirds) with careful developmental analyses of changing social sensitivities within a laboratory-reared species like the zebra finch and looking for correlated patterns of regulation that extend across both evolutionary and developmental time scales.
Our analysis shows that experience-dependent changes in gene expression occur against a background of enormous structured variation in the brain. Although all nervous tissue obviously shares some intrinsic commonalities, our results highlight how much variation there is at the molecular level, even among regions that are functionally related. Against this baseline of intrinsic variation, some experiential treatments result in large changes in gene expression in a particular brain region, whereas other similar manipulations may result in much smaller effects. These variations may be especially important for understanding the constraints and mechanisms that underlie the biological embedding of social and environmental conditions.
Material and Methods
Tissue Samples and Microarray Hybridizations.
Tissues were provided and analyzed under the SoNG Initiative program as described (21). SI Appendix, Table S1 describes the general design of each of the completed experiments that contributed to the final results of this analysis. The provider of each tissue is indicated in SI Appendix, Table S1, and in each case the provider obtained approval for the animal experiments from the Institutional Animal Care and Use Committee of the provider's home institution (see author list for affiliations). SI Appendix, Table S2 presents technical details for all 488 samples representing the set of 80 treatment groups and 32 planned contrasts. See SI Appendix, SI Materials and Methods for details on sample preparation, hybridization, and scanning.
Data Preprocessing.
All 488 arrays were initially preprocessed together in R (44) with the limma package (45), using a standardized method, including background correction and within-array normalization (details are provided in SI Appendix, SI Materials and Methods).
ANOVA.
Further data preprocessing and statistical analysis were done separately for each experiment. The sample/reference log2 ratios were between-array normalized using the scale method; the log2 transformation of the ratio values is the common practice for microarray data before use of parametric statistics (45). Next, a statistical model was fit, either a one-way ANOVA F test for those experiments with one factor or with the main effects and interaction F tests for those experiments with two factors, plus the reference X dye interaction and any additional factors as necessary (SI Appendix, Table S3).
The statistical models were fit using all 20,160 addresses on the array, but the comparisons of significant probes were limited to the 18,848 addresses spotted with cDNA clones from the nonredundant “songbird” (SB) series of cDNAs. In total, 13,859 of these SB sequences were mapped to Ensembl genes in the zebra finch genome assembly, 8,521 of which were unique, as previously described (22). Mapping of multiple SB cDNAs to the same Ensembl gene may indicate alternative transcripts (e.g., splicing) or representation of different portions of the same transcript. Thus, in the ANOVA analysis, we treated each SB spot independently.
WGCNA.
WGCNA applies a variety of algorithms to calculate the similarity of expression patterns among all pairs of genes across all treatment conditions, assigning each gene to a “module” based on shared expression patterns. To reduce the significant computational burden of WGCNA, we used the mean values for each treatment group as derived from the ANOVA models, which collapsed the 488 samples down to 80 treatment groups. The 80 group mean values were further normalized to remove the batch effects of species and cDNA amplification as described in SI Appendix, SI Materials and Methods. Of the 18,848 SB cDNAs on each array, 17,175 were used in the WGCNA analysis because they had at least 1 of the 32 within-experiment contrasts with a raw P value <0.001 and an estimated group-level value for all 80 treatment groups. By WGCNA, these 17,175 individual expression patterns were collapsed down to 95 modules ranging from 829 to 43 cDNAs each, plus the “0” module with 316 cDNAs that did not fit any of the 95 patterns well enough. Each cDNA’s module assignment is indicated in Dataset S1, and a complete list of kME values (the module eigengene-based connectivity measure) for all genes in all modules is given in Dataset S2.
GO Enrichment Tests.
GO analyses of specific gene sets were performed using the Web-based GOfinch tool (http://bioinformatics.iah.ac.uk/tools/GOfinch). This analysis relies on the gene-level annotation supplied by Ensembl, and performs both Fisher and Hypergeometric tests of enrichment for terms in the input list against the terms in the reference (the full list of Ensembl gene identifiers represented on the 20K microarray). Because ANOVA and WGCNA both generated lists of individual SB cDNAs, these identifiers were subsequently converted to Ensembl gene names. Thus, SB cDNAs that mapped to redundant Ensembl genes were collapsed into a single gene entry for GO analysis.
Supplementary Material
Acknowledgments
Michaela Hau contributed to the design of experiment e03. The SoNG Initiative was supported by Public Health Service Grant R01 NS 045264.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Biological Embedding of Early Social Adversity: From Fruit Flies to Kindergartners,” held December 9–10, 2011, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/biological-embedding.
This article is a PNAS Direct Submission.
Data deposition: The microarray hybridization data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE36748).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200655109/-/DCSupplemental.
References
- 1.Mello CV, Vicario DS, Clayton DF. Song presentation induces gene expression in the songbird forebrain. Proc Natl Acad Sci USA. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burmeister SS, Jarvis ED, Fernald RD. Rapid behavioral and genomic responses to social opportunity. PLoS Biol. 2005;3:e363. doi: 10.1371/journal.pbio.0030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitfield CW, et al. Genomic dissection of behavioral maturation in the honey bee. Proc Natl Acad Sci USA. 2006;103:16068–16075. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cummings ME, et al. Sexual and social stimuli elicit rapid and contrasting genomic responses. Proc Biol Sci. 2008;275:393–402. doi: 10.1098/rspb.2007.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322:896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champoux M, et al. Serotonin transporter gene polymorphism, differential early rearing, and behavior in rhesus monkey neonates. Mol Psychiatry. 2002;7:1058–1063. doi: 10.1038/sj.mp.4001157. [DOI] [PubMed] [Google Scholar]
- 7.Belsky J, et al. Vulnerability genes or plasticity genes? Mol Psychiatry. 2009;14:746–754. doi: 10.1038/mp.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jongeneel CV, et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome Res. 2005;15:1007–1014. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 10.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W, et al. The functional landscape of mouse gene expression. J Biol. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clayton DF, Balakrishnan CN, London SE. Integrating genomes, brain and behavior in the study of songbirds. Curr Biol. 2009;19:R865–R873. doi: 10.1016/j.cub.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marler P. Birdsong and speech development: Could there be parallels? Am Sci. 1970;58:669–673. [PubMed] [Google Scholar]
- 14.Doupe AJ, Kuhl PK. Birdsong and human speech: Common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- 15.Jarvis ED. Learned birdsong and the neurobiology of human language. Ann N Y Acad Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- 17.Brenowitz EA, Margoliash D, Nordeen KW. An introduction to birdsong and the avian song system. J Neurobiol. 1997;33:495–500. [PubMed] [Google Scholar]
- 18.Aronov D, Andalman AS, Fee MS. A specialized forebrain circuit for vocal babbling in the juvenile songbird. Science. 2008;320:630–634. doi: 10.1126/science.1155140. [DOI] [PubMed] [Google Scholar]
- 19.Bottjer SW, Altenau B. Parallel pathways for vocal learning in basal ganglia of songbirds. Nat Neurosci. 2010;13:153–155. doi: 10.1038/nn.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brenowitz EA, Beecher MD. Song learning in birds: Diversity and plasticity, opportunities and challenges. Trends Neurosci. 2005;28:127–132. doi: 10.1016/j.tins.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Replogle K, et al. The Songbird Neurogenomics (SoNG) Initiative: Community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovell PV, Clayton DF, Replogle KL, Mello CV. Birdsong “transcriptomics”: Neurochemical specializations of the oscine song system. PLoS ONE. 2008;3:e3440. doi: 10.1371/journal.pone.0003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong S, et al. Discrete molecular states in the brain accompany changing responses to a vocal signal. Proc Natl Acad Sci USA. 2009;106:11364–11369. doi: 10.1073/pnas.0812998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.London SE, Dong S, Replogle K, Clayton DF. Developmental shifts in gene expression in the auditory forebrain during the sensitive period for song learning. Dev Neurobiol. 2009;69:437–450. doi: 10.1002/dneu.20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukai M, et al. Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. PLoS ONE. 2009;4:e8182. doi: 10.1371/journal.pone.0008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomaszycki ML, et al. Sexual differentiation of the zebra finch song system: Potential roles for sex chromosome genes. BMC Neurosci. 2009;10:24. doi: 10.1186/1471-2202-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 30.Itoh Y, et al. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh Y, et al. Sex bias and dosage compensation in the zebra finch versus chicken genomes: General and specialized patterns among birds. Genome Res. 2010;20:512–518. doi: 10.1101/gr.102343.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akutagawa E, Konishi M. A monoclonal antibody specific to a song system nuclear antigen in estrildine finches. Neuron. 2001;31:545–556. doi: 10.1016/s0896-6273(01)00388-9. [DOI] [PubMed] [Google Scholar]
- 33.Wada K, Sakaguchi H, Jarvis ED, Hagiwara M. Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J Comp Neurol. 2004;476:44–64. doi: 10.1002/cne.20201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn TP. Integration of photoperiodic and food cues to time changes in reproductive physiology by an opportunistic breeder, the Red Crossbill, Loxia-Curvirostra (Aves, Carduelinae) J Exp Zool. 1995;272:213–226. [Google Scholar]
- 35.Johnson F, Rashotte M. Food availability but not cold ambient temperature affects undirected singing in adult male zebra finches. Physiol Behav. 2002;76:9–20. doi: 10.1016/s0031-9384(02)00685-6. [DOI] [PubMed] [Google Scholar]
- 36.Rashotte ME, Sedunova EV, Johnson F, Pastukhov IF. Influence of food and water availability on undirected singing and energetic status in adult male zebra finches (Taeniopygia guttata) Physiol Behav. 2001;74:533–541. doi: 10.1016/s0031-9384(01)00600-x. [DOI] [PubMed] [Google Scholar]
- 37.Crabbe JC, Wahlsten D, Dudek BC. Genetics of mouse behavior: Interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- 38.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 39.Oldham MC, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nadler JJ, et al. Large-scale gene expression differences across brain regions and inbred strains correlate with a behavioral phenotype. Genetics. 2006;174:1229–1236. doi: 10.1534/genetics.106.061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zapala MA, et al. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci USA. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brenowitz EA. Comparative approaches to the avian song system. J Neurobiol. 1997;33:517–531. doi: 10.1002/(sici)1097-4695(19971105)33:5<517::aid-neu3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 44.R_Development_Core_Team . Vienna, Austria: R Foundation for Statistical Computing; 2008. R: A language and environment for statistical computing. [Google Scholar]
- 45.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.