Abstract
The carbonate radical anion CO3•− is a decomposition product of nitrosoperoxycarbonate derived from the combination of carbon dioxide and peroxynitrite, an important biological byproduct of the inflammatory response. The selective oxidation of guanine in DNA by CO3•− radicals is known to yield spiroiminodihydantoin (Sp), guanidinohydantoin (Gh), and a novel intrastrand cross-linked product, 5’-d(CCATCG*CT*ACC) between guanine C8 (G*) and thymine N3 (T*) atoms in the oligonucleotide (Crean et al., Nucleic Acids Res., 2008, 36, 742–755). Involvement of the T-N3 (pKa of N3-H is 9.67) suggests that the formation of 5’-d(CCATCG*CT*ACC) might be pH – dependent. This hypothesis was tested generating CO3•− radicals by the photodissociation of carbonatotetramminecobalt(III) complexes by steady-state UV irradiation that allowed for studies of product yields in the pH 5.0 – 10.0 range. The yields of 5’-d(CCATCG*CT*ACC) is ~ 45 times greater at pH 10.0 than at pH 5.0, which is consistent with the proposed mechanism that requires N3(H) thymine proton dissociation followed by nucleophilic addition to the C8 guanine radical.
Keywords: DNA, oxidative damage, carbonate radical, hole transfer, kinetics
Introduction
The carbonate radical anion, CO3•−, is a biologically important oxidant that can initiate some of the oxidative reactions that have been commonly assigned to hydroxyl radicals,[1] and its role in vivo may have been underestimated.[2] In biological systems, the CO3•− radical can arise as a consequence of a cascade of events that occurs during chronic infection and inflammation. Because these response mechanisms have been implicated in the etiology of some cancers,[3, 4] the pathways of oxidative reactions that arise as a result of inflammation are of considerable interest.
The inflammatory response is correlated with a persistent oxidative and nitrosative stress that is associated with the overproduction of nitric oxide and superoxide radical anions followed by their rapid combination with formation of the highly toxic peroxynitrite.[5, 6] The major mode of peroxynitrite reactivity in vivo[2, 7, 8] is a fast reaction with carbon dioxide[9] to form a highly unstable intermediate nitrosoperoxycarbonate that rapidly decomposes homolytically to nitrogen dioxide and the carbonate radical anion.[10]
We have devised a photochemical method for generating CO3•− radicals in aqueous solution at pH 7.5 and showed that only guanine in DNA can be oxidized via a one-electron abstraction mechanism by this radical.[11–14] In these experiments, irradiation with either steady-state[15, 16] or pulsed UV light (e.g., 308 nm excimer laser pulses[11–14]) of a buffered solution of bicarbonate HCO3− and persulfate anions, causes the dissociation of the S2O82− ions to sulfate radical anions, SO4•−. In turn, the sulfate radical anions oxidize the HCO3− ions by a one-electron transfer mechanism to generate the CO3•− radicals. The latter induce the site-selective oxidation of guanine bases in 2’-deoxyoligonucleotides in either the single- or double-stranded forms. The formation and decay of the neutral guanine radicals, G(-H)• thus formed, can be conveniently monitored via their UV absorption maximum at 315 nm.[11, 12] The chemical end-products of these latter radicals are mostly the diastereomeric pair of spiroiminodihydantoin (Sp) and the guanidinohydantoin (Gh) lesions, the latter normally formed in smaller yields.[12]
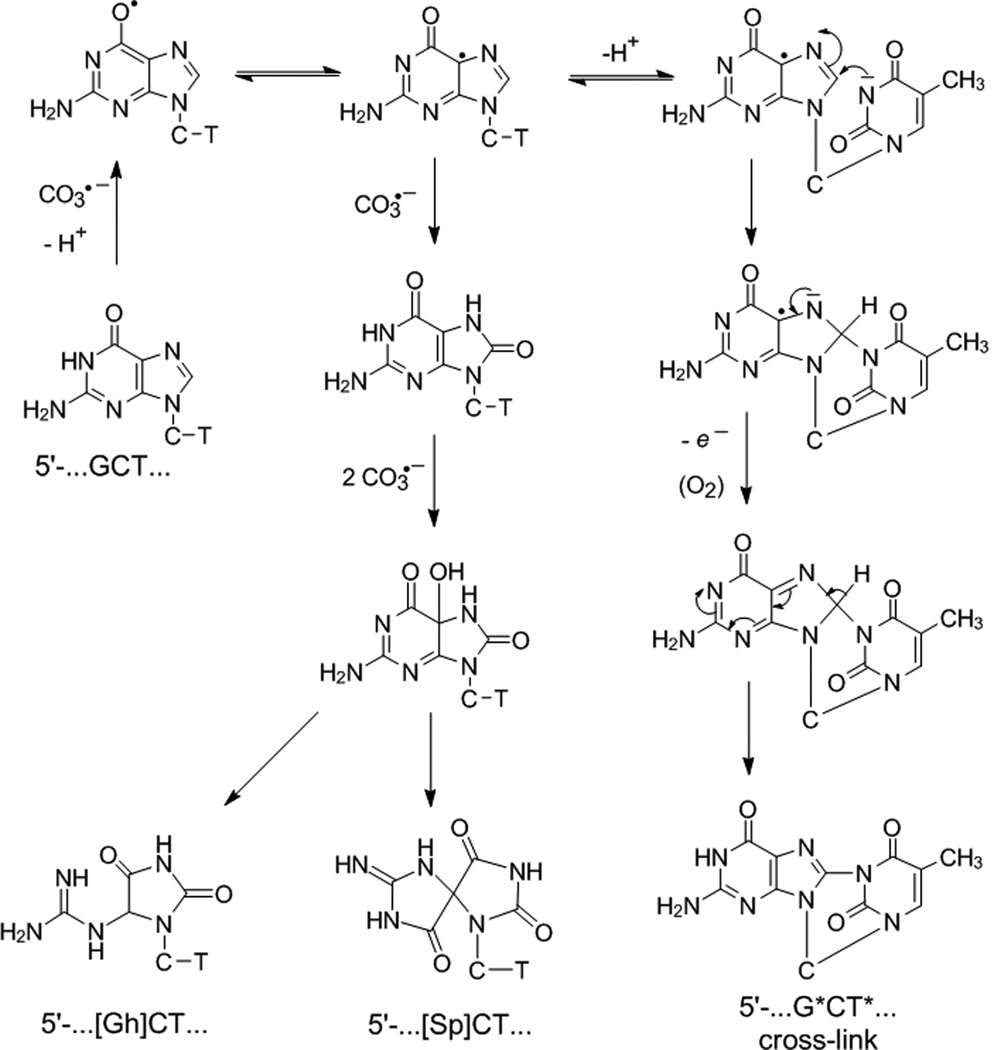
More recently, using continuous illumination methods for generating CO3•− radicals, we found that, in addition to Sp and Gh, the oxidation of the single guanine in the sequence 5’-CCATCGCTACC by CO3•− radicals yields the intrastrand cross-linked 5’-CCAT*CG*CTACC (minor) and 5’-CCATCG*CT*ACC (major) products in yields comparable to those of the diastereomeric Sp products.[16] Analysis of the intrastrand cross-linked products by NMR and mass spectrometry methods indicate that the G* and T* bases are covalently linked via the C8 atom of guanine (G*) and the N3-atom of thymine (T*) in the 5’-d(CCATCG*CT*ACC) sequence (G*CT*). A mechanism of formation was previously proposed by us,[16] and a modified version of the reactions that lead to the G*CT* cross-linked product is shown in Scheme 1 (see below for further details). The proposed mechanisms require the consecutive abstraction of 2 – 4 electrons associated with proton transfer reactions and therefore the distribution of the G*CT*, Sp and Gh products might be pH – dependent. However, using our previously published method of generating CO3•− radicals,[11–14] lowering the pH of the solution is not feasible because of the unfavorable acid-base equilibrium of the bicarbonate anion. At pH < 7.5 the HCO3− anions are transformed to CO2 and thus the yield of CO3•− radicals decreases as the pH is decreased. Although SO4•− radicals can oxidize both the HCO3− and CO32− anions, CO2 is unreactive.[17]
Scheme 1.
Proposed mechanism of guanine oxidation in the 5’-d(CCATCGCTACC) sequence context by carbonate radical anions. Although only the formation of the 5’-d(CCATCG*CT*ACC) product is shown, minor amounts of the isomeric 5’-d(CCAT*CG*CTACC) are also formed (Supporting Information. This Scheme is a modified version of the one published originally.[16] The main modification in Scheme involves the mechanism of deprotonation of the N3-thymine, and the resonance forms of the G(-H)• radical, which are now represented by the two conjugated resonance forms shown.[35]. The O6 and C5 radical forms of G(-H)• are expected to have lower energies than the C8-centered σ-radical protonated at N1 that we proposed earlier.[16]
In order to overcome this unfavorable pH dependence, we have employed an alternative photochemical method for generating CO3•− radicals by photolysis of metal complexes.[17] As a source of carbonate radical anions, we selected carbonatotetrammineCo(III) complexes that upon photolysis readily yield CO3•− radicals.[18–26] The latter radicals generated by this method have been successfully used in studies of oxidation reactions of diverse organic molecules that include amino acids[21–23] and short peptides.[26] Recently, the oxidation of guanine by CO3•− radicals derived from the photolysis of [Co(NH3)4CO3]+ complexes has been monitored by ultrafast infrared laser spectroscopy.[27] However, the kinetics of guanine oxidation and the nature of the guanine oxidation end-products were not studied.
In this work, we show that the photodissociation of [Co(NH3)4CO3]+ complexes is indeed suitable for studying the reaction kinetics of CO3•− radicals with the 5’-d(CCATCGCTACC) oligonucleotide sequence. Furthermore, the distribution of Sp, Gh, and G*CT* oxidation products at pH 7.5 is similar and independent of the method of generating CO3•− radicals by the photodissociation of [Co(NH3)4CO3]+ complexes or the sulfate radical-mediated oxidation of HCO3− ions. However, the distributions of the end products, Sp, Gh and the cyclic intramolecular G*CT* cross-links, are markedly different at pH 5.0 and 10.0. The mechanistic implications of these observations for the mechanisms of guanine radical-mediated reactions that lead to intrastrand cross-link formation are discussed.
Results and Discussion
Monitoring of radical intermediates by transient absorption spectroscopy
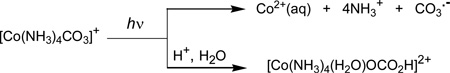
The CO3•− radical with a reduction potential[28] of Eo(CO3•−/CO32−) = 1.59 V vs NHE can efficiently oxidize appropriate electron donors.[17] In contrast, hydrogen atom abstraction by the CO3•− radicals is generally slow.[17] The conjugate acid (HCO3•) is a strong acid (pKa < 0) and, at pH > 0, these radicals exist in the anion form, CO3•−.[29] The UV irradiation of [Co(NH3)4CO3]+ complexes has been shown to generate different products according to two pathways :[19, 24, 25]
In one of these pathways, CO3•− radicals together with Co2+(aq) ions are produced, while in the other the hydrated complex [Co(NH3)4(H2O)OCO2H]2+ is generated. Time-resolved, transient absorption experiments have shown that the photolysis of [Co(NH3)4CO3]+ complexes induced by 248 nm KrF excimer, or 266 nm Nd: Yag laser pulses, yields CO3•− radicals.[19, 24, 25] Here we show that CO3•− radicals can also be produced easily by 308 nm XeCl excimer laser pulse excitation because the UV absorption spectrum of [Co(NH3)4CO3]+ extends beyond 300 nm (i.e., beyond the absorption threshold of the normal DNA bases).
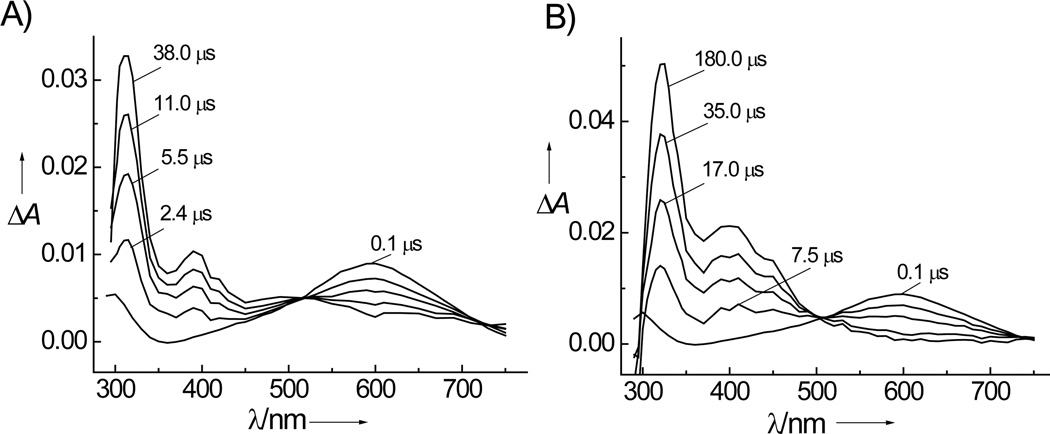
The irradiation of air-equlibrated buffer solutions (pH 7.5) containing [Co(NH3)4CO3]+ and dGuo or 8-oxodGuo solutions by 308 nm excimer laser pulses generates the time-dependent transient absorption spectra shown in Figure 1. The decay of the CO3•− transient absorbance at 600 nm is correlated with the growth of a narrow absorption band at 315 nm (Figure 1A) due to guanine radicals that are formed by the rapid deprotonation of the guanine radical cations (pKa = 3.9) in neutral solutions.[11, 12, 30] In the case of solutions with 8-oxodGuo, the decay of CO3•− radicals is associated with the rise of a narrow absorption band due to 8-oxodGuo radicals at 325 nm (Figure 1B) that, at pH 7.5, exist mostly in the neutral form since the pKa of the radical cation is 6.6.[31] The spectral characteristics of the G(-H)• and 8-oxoGua(-H)• radicals produced by the oxidation of the dGuo or 8-oxodGuo nucleosides by CO3•− radicals generated by the photolysis of [Co(NH3)4CO3]+ are identical to those obtained in our previous experiments in which CO3•− radicals were derived from the oxidation of HCO3− anions by SO4•− radicals.[11, 12] Analogous results are obtained with the single-stranded oligonucleotide sequences 5’-d(CCATCGCTACC) and 5’-d(CCATC[8-oxoGua]CTACC) (data not shown).
Figure 1.
Kinetics of dGuo (A) and 8-oxodGuo (B) oxidation by CO3•− radicals generated by the photolysis of [Co(NH3)4CO3]+. Transient absorption spectra were recorded at fixed time intervals after the 308 nm laser pulse excitation (60 mJ pulse−1cm−2) of air-equilibrated phosphate buffer solution (pH 7.5) containing 2 mM dGuo or 0.1 mM 8-oxodGuo, and 2 mM [Co(NH3)4CO3]+ and 300 mM NaCl.
The rate constants of oxidation of free nucleosides and oligonucleotides by CO3•− radicals were extracted from the decay curves of CO3•− radicals by methods that have been previously described in detail,[11, 12] and the rate constants thus obtained are summarized in Table 1. Briefly, the decay of CO3•− radicals occurs via two competitive reactions: (i) oxidation of DNA (reactions 1–4) and (ii) bimolecular recombination of CO3•− radicals (reaction 5), as shown in Table 1. We found that the rate constants of G or 8-oxoGua oxidation by CO3•− radicals in the form of nucleosides, or embedded in the oligonucleotides (Table 1), are identical within experimental error with the values obtained in experiments where the CO3•− radicals are generated by the photolysis of [Co(NH3)4CO3]+ or by oxidation of HCO3− with photochemically generated SO4•− radicals.[11, 12]
TABLE 1.
Rate constants of the one-electron oxidation of free nucleosides and single-stranded oligonucleotides by CO3•− radicals in air-equilibrated buffer solutions (pH 7.5) generated either by photolysis of [Co(NH3)4CO3]+ or S2O82−/HCO3−.
| N | Reaction | k, M−1s−1[a] | |
|---|---|---|---|
| [Co(NH3)4CO3]+ | S2O82−/HCO3− | ||
| 1 | CO3•− + dGuo → CO32− + dGuo(-H)• | (6.8±0.7)×107 | (6.7±0.7)×1;107[b] |
| 2 | CO3•− + 8-oxodGuo → CO32− + 8-oxodGuo(-H)• | (7.6±0.8)×108 | (7.9±0.8)×108[b] |
| 3 | CO3•− + 5’-CCATCGCTACC → CO32− + 5’-CCATC[G(-H)•]CTACC | (2.3±0.3)×107 | (2.4±0.3)×107[c] |
| 4 | CO3•− + 5’-CCATC[8-oxoGua]CTACC → CO32− + 5’-CCATC[8-oxoGua(-H)•]CTACC | (3.3±0.4)×108 | (3.2±0.4)×108[c] |
| 5 | CO3•− + CO3•− → C2O62− → CO42− + CO2 | (1.3±0.1)×107 | (1.3±0.1)×107[b] |
The rate constants were measured in air-equilibrated buffer solutions (pH 7.5) containing either 2 mM [Co(NH3)4CO3]+ and 300 mM NaCl or 10 mM Na2S2O8 and 300 mM NaHCO3−. The uncertainties represent standard errors for the best least-squares fits of the appropriate kinetic equations (Supporting Information) to the experimental decay profiles of CO3•− radicals monitored at 600 nm.
Data from ref 11.
Data from ref. 12.
Identification of guanine lesions produced by CO3•− radicals in single-stranded oligonucleotides
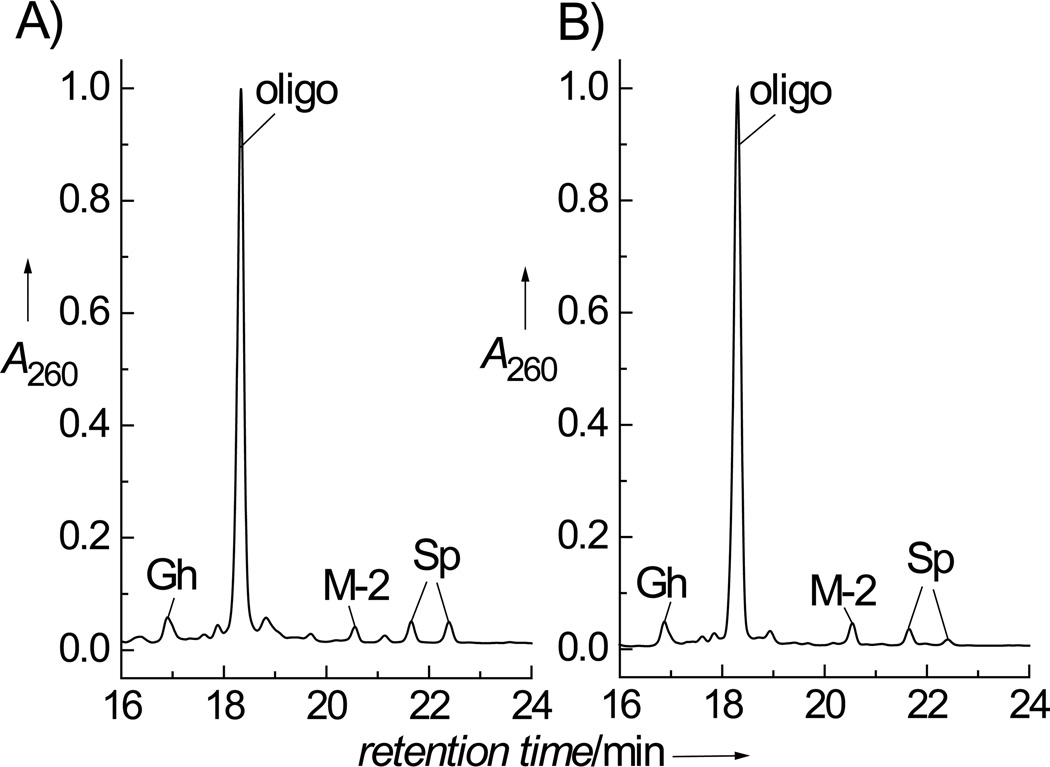
The end products of oxidation of guanine in the 5'-d(CCATCGCTACC) sequence generated by CO3•− radicals produced by the photolysis of either [Co(NH3)4CO3]+ or S2O82−/HCO3− were compared at pH 7.5 upon irradiation of the samples with continuous 300 – 340 nm light from a 100 W Xe arc lamp. In these experiments, the irradiation times were adjusted to limit the conversion of the original oligonucleotides to less than 10 – 20% of the starting material. Typical anion-exchange HPLC profiles of the irradiated solutions are shown in Figure 2. The distributions of oxidized end-products are the same and independent of the method of generating the CO3•− radicals either by photolysis of [Co(NH3)4CO3]+ (Figure 2A) or S2O82−/HCO3− (Figure 2B). Indeed, MS analysis showed that the products obtained in both experiments are identical. The unmodified 5’-d(CCATCGCTACC) sequence (mass, M: 3237.2) eluted at 18.3 min, the guanidinohydantoin adduct eluted at 17.0 min (M + 6: 3243.2), and the adducts containing the diastereomeric spiroiminodihydantoin adducts eluted at 21.7 and 22.4 min (M + 32: 3259.2). The fraction eluting at 20.6 min (Figure 2A) contains the product, which has a mass (M − 2: 3235.2) that is smaller by 2 D than the molar mass M of the starting oligonucleotide sequence. This product is identical to the 5’-d(CCATCG*CT*ACC oligonucleotide (Figure 2B) that has the same mass and contains the intrastrand cross-link between the G and T bases as described in our earlier work.[16]
Figure 2.
Anion-exchange HPLC elution profile of the end-products derived from the oxidation of the single-stranded oligonucleotide, 5’-d(CCATCGCTACC) by CO3•− radicals. A) The 5’-d(CCATCGCTACC) sequence (0.01 mM) was irradiated for 20 s in air-equilibrated buffer solution (pH 7.5) containing 2 mM [Co(NH3)4CO3]+. B) The 5’-d(CCATCGCTACC) sequence (0.01 mM) was irradiated for 10 s in air-equilibrated buffer solution (pH 7.5) containing 300 mM NaHCO3 and 10 mM Na2S2O8. A 100 W Xe arc continuous light source was used in both cases (300 – 340 nm). HPLC elution conditions (detection at 260 nm): 10 – 90% linear gradient of solvent B (10% acetonitrile and 90% 1.5 M ammonium acetate) in solvent A (10% acetonitrile and 90% water) for 30 min at a flow rate of 1 mL/min. The fractions containing the unmodified oligonucleotide (labeled oligo), the cyclic cross-linked adduct (G*CT*), the oligonucleotides with the single G residue converted to the guanidinohydantoin lesion (Gh), and spiroiminodihydantoin lesions with (+)-R-Sp and (−)-S-Sp configurations eluting at 21.7 min, and 22.4 min, respectively, were identified as discussed in the text (the absolute configurations of the Sp lesions were determined as described by Durandin et al.;[32] note: an alternate R and S assignment was proposed by Cadet and co-workers[33]).
To confirm that CO3•− radicals generated by the photolysis of [Co(NH3)4CO3]+ induce the formation of a covalent bond between G and T, we generated the cross-linked product by oxidation of the 5’-d(GpCpT) trinucleotide. The positive ion spectra (MS/MS) of the cross-linked product obtained (Figure S2, Supporting Information) showed a molecular ion [M + H]+ at m/z 859.1, which is smaller by 2 Da than the mass of the unmodified 5’-d(GpCpT) observed at m/z 861.2. Furthermore, multi-step fragmentation of the parent ion (m/z 859.1) gives rise to the product ion at m/z 276.0 that is characteristic of the ion derived from the G*-T* base-base dimer. Exactly the same fragmentation patterns and ions were observed when the 5’-d(G*CT*) product was obtained via oxidation of 5’-d(GpCpT) by CO3•− radicals derived from the photolysis of S2O82−/HCO3−.[16] Thus, CO3•− radicals generated by the photolysis of [Co(NH3)4CO3]+ complexes produce the same cyclic cross-linked products as the photolysis of S2O82−/HCO3−.
Localization of the G*CT* intrastrand cross-link in the 5’-d(CCATCG*CTACC) sequence
The oxidatively modified 5’-d(CCATCG*CT*ACC) oligonucleotide with mass M – 2 obtained in the [Co(NH3)4CO3]+ photolysis experiment (Figure 2A) was subjected to enzymatic digestion with snake venom phosphodiesterase I and bovine spleen phosphodiesterase II. These two exonucleases digest single-stranded DNA from the 3’- and 5’-ends, respectively. Detailed analysis of the enzymatic digestion patterns and masses of partially digested oligonucleotide fragments by MALDI-TOF/MS methods showed that the enzyme stalling patterns are consistent with the formation of exonuclease-resistant 5’-d(CCATCG*CT* and 5’-CG*CT*ACC fragments (Figure S3, Supporting information). The localization of the cyclic G*CT* cross-link within the 11-mer M – 2 sequence was also confirmed by standard hot piperidine treatment,[34] followed by analysis of the cleavage products by high resolution denaturing polyacrylamide gel electrophoresis assays (Figure S4 in Supporting Information) and MALDI-TOF/MS (Figure S5). The cross-linked sequences are more sensitive to cleavage induced by the standard hot piperidine treatment (90 °C, 30 – 60 min) (Figures S4 and S6). Collectively these results showed that the same intrastrand cross-linked products are formed when the carbonate radical ions are produced by the photolytic decomposition of the [Co(NH3)4CO3]+ complex or by the oxidation of HCO3− by SO4•− radicals.
Effect of pH on the distributions of different guanine lesions
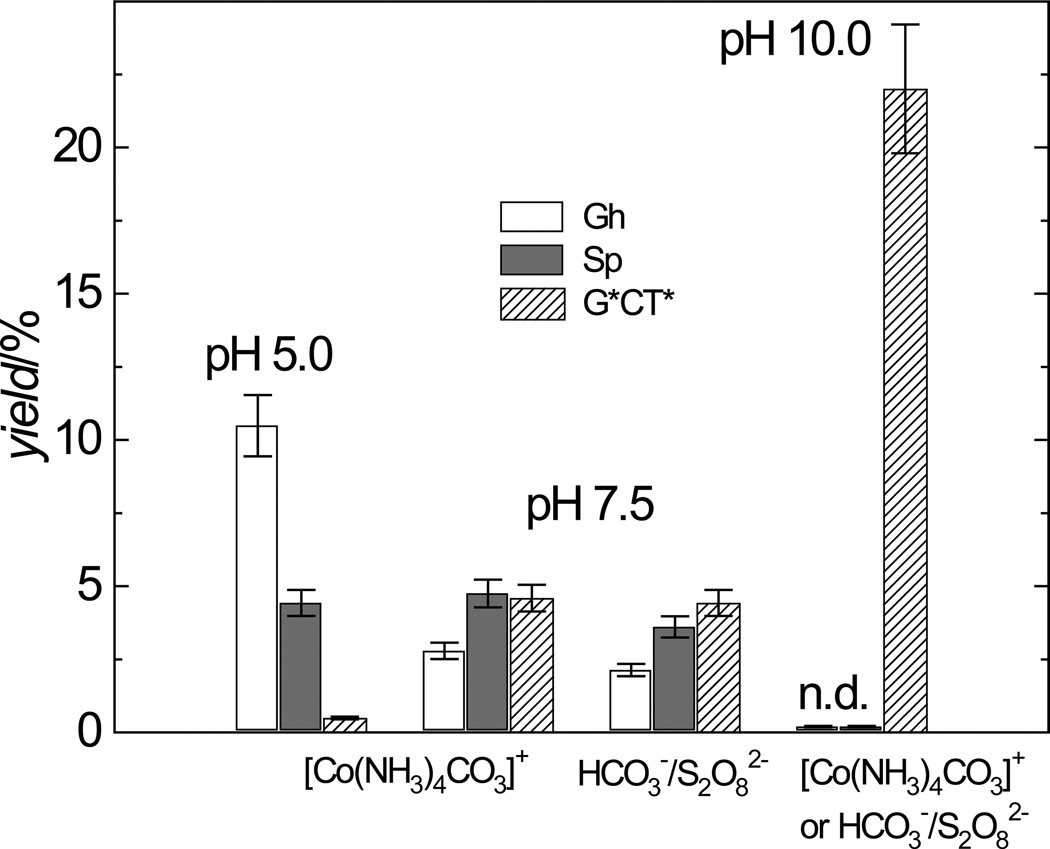
In the neutral solutions (pH 7.5) the yields of any given adduct, Gh, Sp, or G*CT*, does not depend significantly on the method of generating the CO3•− radicals by either the photolysis of [Co(NH3)4CO3]+ complexes or by the oxidation of HCO3− by photochemically generated SO4•− radicals (Figure 3). At pH 7.5 the yields of the Sp adducts and the G*CT* intrastrand cross-linked products are similar, while the yields of the Gh adduct are smaller by a factor of ~1.5 (Figure 3). In our previous experiments, employing reversed-phase HPLC rather than the anion exchange column used in this work, the yields of the Gh adducts were underestimated due to overlap of the Gh and Sp elution fractions.[12] The generation of carbonate radical anions by the photolysis of [Co(NH3)4CO3]+ complexes allows us to investigate the oxidation of the oligonucleotides at lower pH values, as already discussed. Decreasing the pH from 7.5 to 5 enhances the formation of the Gh adducts by a factor of 3.7, and strongly diminishes the yields of the G*CT* cross-linked products (by a factor of ~ 9). However, the yields of the Sp lesions remain practically unchanged, but the ratio of the Gh/Sp yields increases from 0.6 at pH 7.5 to 2.4 at pH 5.0 (Figure 3). In turn, at pH 10.0, the Gh and Sp lesions are not detectable, but the formation of the G*CT* cross-linked products is enhanced by a factor of 4.8 relative to the pH 7.5 value, and by a factor of 110 relative to the pH 5.0 value (Figure 3).
Figure 3.
Effects of pH on the yields of the G*CT*, Gh, and Sp adducts derived from the oxidation of 5’-d(CCATCGCTACC) by CO3•− radicals generated by the photolysis of [Co(NH3)4CO3]+ and separately, by the oxidation of HCO3− by photochemically generated SO4•− radicals using continuous irradiation (300 – 340 nm) from a 100 W Xe arc lamp. The reaction conditions were identical to those used for generating the results shown in Figure 2. Neither Sp nor Gh adducts were detected (n.d.) at pH 10.0.
Mechanistic considerations
The one-electron oxidation of guanine by CO3•− radicals in the 5’-d(CCATCGCTACC) sequences triggers a cascade of chemical reactions that result in the formation of the stable oxidation end-products observed. The formation of these final products can be formally considered as the result of a two-electron (G*CT*), or a four-electron (Sp/Gh) oxidation mechanism (Scheme 1). Since the pKa of the guanine radical cation, G•+ radical is 3.9 [30], at pH≥ 5 this radical exists mostly in its neutral form, G(-H)• (Figure 1A), in agreement with the results of our previous experiments.[11, 12]. The G(-H)• radicals are usually considered to be O-centered radicals with the unpaired electron positioned on the O6 atom,[36, 37] which explains the low reactivity of the G(-H)• radical with molecular oxygen.[11, 12] The formation of stable products from this radical typically occurs via the addition of free radicals or nucleophiles to the C5 or C8 positions.[38, 39] Our results suggest that the C8-centered G(-H)• radical can react with thymine which is a weak nucleophile. The deprotonation of thymine greatly enhances its nucleophilicity, a conclusion that correlates well with the remarkable enhancement of the G*CT* yield at pH 10; under these conditions, the thymine exists mostly in the deprotonated form since its pKa is 9.67.[40] The radical adduct arising from the nucleophilic addition of the N3-atom of T to the C8 position of the G(-H)• radical is oxidized by O2 to form the G*CT* cross-linked product (Scheme 1). Indeed, the yields of the cross-linked products are negligible in the absence of O2.[16] Similar mechanisms have been proposed by Perrier et al. for the Nε-(guanin-8-yl)-lysine cross-link formation.[41] In our case, the generation of the G*CT* cross-linked product requires the abstraction of only one-electron (“single hit”), because the second electron is most likely abstracted by the O2 molecule[16] which is present in air-saturated solutions under physiological conditions.
Other types of intrastrand cross-linked lesions have been found between adjacent G and T bases (the so-called tandem lesions) that are produced when DNA is exposed to ionizing radiation.[42–44] These cross-linked products involve a covalent bond between the C8 atom of guanine and the C-atom of the methyl group of thymine. The formation of these cross-linked G*C8-T*(CH3) products is initiated by the hydrogen atom abstraction from the CH3-group of thymine, and O2 was found to suppress the formation of these products because it reacts readily with the 5-(2’-deoxyuridinyl)methyl radical.[45, 46]
In contrast, the formation of the Sp/Gh lesions from the G(-H)• radical requires three additional oxidizing equivalents (Scheme 1). Our previous experiments have demonstrated that the reactions resulting in the formation of Sp/Gh lesions include the formation of 8-oxoGua lesions as an intermediate.[12] We found that the maximum yield of 8-oxodGuo enzymatically excised from the 5’-d(CCATCGCTACC) sequence oxidized by CO3•− radicals and determined by the HPLC-amperometric detection method, does not exceed ~ 2%. The low yield of 8-oxodGuo is a clear indication that the oxidation of 5’-d(CCATC[8-oxoGua]CTACC) is much faster than that of the parent 5’-d(CCATCGCTACC) sequence. Indeed, the value of k4 for the oxidation of 5’-d(CCATC[8-oxoGua]CTACC) is by factor of ~ 13 greater than the value of the analogous rate constant k3 for the oxidation of 5’-d(CCATCGCTACC) (Table 1). Here, we found that the further oxidation of 5’-d(CCATC[8-oxoGua]CTACC) by CO3•− radicals results in the formation of the Sp/Gh lesions only (crosslinked adducts were not detectable) and that, therefore, the sequence 5’-d(CCATC[8-oxoGua]CTACC) is likely to be an intermediate in the oxidation of 5’-d(CCATCGCTACC) by CO3•− radicals with G transformed to either Sp or Gh.
A deeper understanding of the mechanistic aspects of the successive oxidation of the G(-H)• radicals by CO3•− radicals was gained by performing oxidation of DNA in H218O buffer solutions (see, Figure S7 in Supporting Information). These experiments showed that two oxygen atoms are added to the C5 and C8 positions of the oxidized guanine intermediates during the course of their stepwise reactions, and that both oxygen atoms originate from the CO3•− radicals in agreement with our previous results for the oxidation of 2’,3’,5’-tri-O-acetylguanosine and 2’,3’,5’-tri-O-acetyl-8-oxo-7,8-dihydroguanosine mononucleosides.[14] The 5-HO-8-oxoGua intermediate is a precursor of the Sp and Gh products[47, 48] that arises from a consecutive three-electron oxidation of the G(-H)• radical involving the transfer of two O− anions from CO3•− radicals to the C5 and C8 positions of guanine. The subsequent transformation of the 5-HO-8-oxoGua depends on the solution pH and results in the formation of either spiroiminodihydantoin or guanidinohydantoin (Gh) lesions.[47–49] Decreasing the pH favors pyrimidine ring opening, followed by the formation of the Gh lesions; in contrast, increasing the pH facilitates the acyl shift leading to the Sp lesions,[49] in agreement with our observations (Figure 3). This pH dependence qualitatively agrees with the enhancement of the yield of Gh products that has been reported for the oxidation of 8-oxoGuo by either peroxynitrite,[49] photoexcited riboflavin, IrCl62−,[50, 51] or Cr(VI) complexes.[52]
The formation of the Sp and Gh end-products involves several consecutive reactions of intermediates with CO3•− radicals, while the formation of the G*CT* cross-linked products involves only one carbonate radical. In the laser pulse excitation experiments described previously,[11–14] the transient carbonate radical concentrations were significantly higher than in the steady-state irradiation experiments[16] described in this work. Therefore, in laser pulse excitation experiments, the formation of Gh and Sp products is favored over the formation of the cross-linked products, which explains why we overlooked the G*CT* products in earlier laser pulse irradiation experiments at neutral pH.[12] In contrast, the steady-state, continuous irradiation method used here, as well as earlier,[16] enhances the relative yields of G*CT* products, particularly under basic conditions.
Conclusion
The CO3•− radicals oxidize guanine bases in DNA by a one-electron transfer reaction that ultimately results in the formation of stable guanine oxidation products. Here, we demonstrate that the generation of CO3•− radicals by the photolysis of carbonatotetramminecobalt(III) complexes in aqueous solution stimulated by either laser pulses or steady-state irradiation from a xenon arc lamp is a convenient method for generating carbonate radical anions. Using this method, the pH dependence of the distributions of the guanine oxidation products Gh, Sp, and the cross-linked G*CT* product in the 2’-deoxyribonucleotide sequence 5’-d(CCATCGCTACC) can be conveniently investigated. Variations in the relative yields of these three oxidation products at acidic, neutral, and basic pH values can provide valuable insights into the mechanistic aspects of these oxidative reactions.
Experimental Section
Materials
All chemicals (analytical grade) were used as received. The oligonucleotides were purchased from Integrated DNA Technologies (Coraville, IA), purified, and desalted using reversed-phase HPLC. The integrity of the oligonucleotides and nucleosides was confirmed by MALDI-TOF/MS and LC/MS/MS methods. The [Co(NH3)4CO3]ClO4 complex was a gift of Dr. Carol Creutz (Brookhaven National Laboratory, Upton, NY).
Laser kinetic spectroscopy
The kinetics of oxidative reactions initiated by CO3•− radicals were monitored directly using a fully-computerized kinetic spectrometer system (~7 ns response time) described elsewhere.[53] The transient absorbance was probed along a 1 cm optical path by a light beam (75 W xenon arc lamp) oriented perpendicular to the laser beam. The signal was detected with a Hamamtsu 928 photomultiplier tube and recorded by a Tektronix TDS 5052 oscilloscope operating in its high resolution mode that provided a satisfactory signal/noise ratio after a single laser shot. The rate constants were determined by least squares fits of the appropriate kinetic equations to the experimentally measured transient absorption profiles as described in detail elsewhere.[12, 54] The values reported are averages of five independent measurements.
Oxidation of oligonucleotides by CO3•− radicals
The oligonucleotides (10 nmol) were dissolved in air-equilibrated buffer solutions (pH 7.5, 1 mL) containing: either (i) 1 mM [Co(NH3)4CO3]+ (1 mM) and NaCl (300 mM) or (ii) Na2S2O8 (10 mM) and NaHCO3 (300 mM). Continuous light in the 300 – 340 nm spectral range from a 100 W xenon arc lamp was reflected from a dichroic mirror onto the sample. The energy incident on the sample was ~ 100 mWcm−2 and the irradiation time was varied from 10 to 30 s. After irradiation, the sample was immediately desalted by reversed-phase HPLC, concentrated, and subjected to anion-exchange HPLC analysis.
Synthesis of the authentic standards
The diastereomeric 5’-d(CCATC[Sp]CTACC adducts were synthesized by oxidation of the guanine in 5’-d(CCATCGCTACC) by CO3•− radicals derived from HCO3− oxidation by photochemically generated SO4•− radicals.[12] The 5’-d(CCATC[Gh]CTACC adduct was prepared by oxidation of 5’-d(CCATC[8-oxoGua]CTACC by IrCl62−.[51] All standards were isolated, purified, and desalted by HPLC methods and their identities were confirmed by MALDI-TOF/MS and LC/MS/MS methods.
HPLC isolation of oxidation products
The oxidatively modified oligonucleotides were isolated by anion-exchange HPLC employing an analytical (250 × 4 mm i.d.) DNAPac PA-100 column (Dionex, Sunnyvale, CA), using a 10 – 90% linear gradient of solvent B (10% acetonitrile and 90% 1.5 M ammonium acetate) in solvent A (10% acetonitrile and 90% water) for 30 min at a flow rate of 1 mL/min. The solutions were desalted by an analytical (250 mm × 4.6 mm i.d.) Microsorb-MV C18 column (Varian, Walnut Creek, CA) using the following mobile phases: 5 mM ammonium acetate (10 min), deionized water (10 min), and an isocratic 50 : 50 acetonitrile and H2O mixture (15 min).
Mass spectrometry
LC-MS/MS analysis of the photoproducts was performed with an Agilent 1100 Series capillary LC/MSD Ion Trap XCT mass spectrometer equipped with an electrospray ion source as described elsewhere.[16] The MALDI-TOF mass spectra were recorded in the negative mode using a Bruker OmniFLEX instrument.[12]
Supplementary Material
Acknowledgements
We are grateful to Dr. M. Greenberg (Johns Hopkins University, Baltimore, MD) for pointing out the relative stabilities of different guanine radicals and for insightful comments and discussions. We thank Dr. Carol Creutz (Brookhaven National Laboratory, Upton, NY) for providing the [Co(NH3)4CO3]ClO4 compound. This work was supported by the National Institutes of Health (5 RO1 ES 011589-07), and by the Kresge Foundation. Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from a Research Facilities Improvement Grant (C06 RR-16572) from the National Center for Research Resources, National Institutes of Health. The acquisition of the ion trap mass spectrometer was supported by the National Science Foundation (CHE-0234863).
References
- 1.Michelson AM, Maral J. Biochimie. 1983;65:95–104. doi: 10.1016/s0300-9084(83)80179-5. [DOI] [PubMed] [Google Scholar]
- 2.Pacher P, Beckman JS, Liaudet L. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balkwill F, Mantovani A. Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 4.Coussens LM, Werb Z. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huie RE, Padmaja S. Free Radic. Res. Commun. 1993;18:195–199. doi: 10.3109/10715769309145868. [DOI] [PubMed] [Google Scholar]
- 7.Davis KL, Martin E, Turko IV, Murad F. Annu. Rev. Pharmacol. Toxicol. 2001;41:203–236. doi: 10.1146/annurev.pharmtox.41.1.203. [DOI] [PubMed] [Google Scholar]
- 8.Ischiropoulos H, Beckman JS. J. Clin. Invest. 2003;111:163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lymar SV, Hurst JK. J. Am. Chem. Soc. 1995;117:8867–8868. [Google Scholar]
- 10.Merenyi G, Lind J. Chem. Res. Toxicol. 1997;10:1216–1220. doi: 10.1021/tx970101j. [DOI] [PubMed] [Google Scholar]
- 11.Shafirovich V, Dourandin A, Huang W, Geacintov NE. J. Biol. Chem. 2001;276:24621–24626. doi: 10.1074/jbc.M101131200. [DOI] [PubMed] [Google Scholar]
- 12.Joffe A, Geacintov NE, Shafirovich V. Chem. Res. Toxicol. 2003;16:1528–1538. doi: 10.1021/tx034142t. [DOI] [PubMed] [Google Scholar]
- 13.Joffe A, Mock S, Yun BH, Kolbanovskiy A, Geacintov NE, Shafirovich V. Chem. Res. Toxicol. 2003;16:966–973. doi: 10.1021/tx025578w. [DOI] [PubMed] [Google Scholar]
- 14.Crean C, Geacintov NE, Shafirovich V. Angew. Chem., Int. Ed. Engl. 2005;44:5057–5060. doi: 10.1002/anie.200500991. [DOI] [PubMed] [Google Scholar]
- 15.Shafirovich V, Mock S, Kolbanovskiy A, Geacintov NE. Chem. Res. Toxicol. 2002;15:591–597. doi: 10.1021/tx015593l. [DOI] [PubMed] [Google Scholar]
- 16.Crean C, Uvaydov Y, Geacintov NG, Shafirovich V. Nucleic Acids Res. 2008;36:742–755. doi: 10.1093/nar/gkm1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neta P, Huie RE, Ross AB. J. Phys. Chem. Ref. Data. 1988;17:1027–1284. [Google Scholar]
- 18.Cope VW, Hoffman MZ. J. Chem. Soc. Chem. Commun. 1972:227–228. [Google Scholar]
- 19.Cope VW, Chen S-N, Hoffman MZ. J. Am. Chem. Soc. 1973;95:3116–3121. [Google Scholar]
- 20.Chen S-N, Cope VW, Hoffman MZ. J. Phys. Chem. 1973;77:1111–1116. [Google Scholar]
- 21.Chen S-N, Hoffman MZ. Radiat. Res. 1973;56:40–47. [PubMed] [Google Scholar]
- 22.Chen S-N, Hoffman MZ. J. Phys. Chem. 1974;78:2099–2102. [Google Scholar]
- 23.Chen S-N, Hoffman MZ, Parsons GH., Jr J. Phys. Chem. 1975;79:1911–1912. [Google Scholar]
- 24.Cope VW, Hoffman MZ, Chen S-N. J. Phys. Chem. 1978;82:2665–2669. [Google Scholar]
- 25.Ferraudi G, Perkovic M. Inorg. Chem. 1993;32:2587–2590. [Google Scholar]
- 26.Kumar CV, Thota J. Inorg. Chem. 2005;44:825–827. doi: 10.1021/ic0488233. [DOI] [PubMed] [Google Scholar]
- 27.Kuimova MK, Cowan AJ, Matousek P, Parker AW, Sun XZ, Towrie M, George MW. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2150–2153. doi: 10.1073/pnas.0506860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huie RE, Clifton CL, Neta P. Radiat. Phys. Chem. 1991;38:477–481. [Google Scholar]
- 29.Czapski G, Lymar SV, Schwarz HA. J. Phys. Chem. A. 1999;103:3447–3450. [Google Scholar]
- 30.Candeias LP, Steenken S. J. Am. Chem. Soc. 1989;111:1094–1099. [Google Scholar]
- 31.Steenken S, Jovanovic SV, Bietti M, Bernhard K. J. Am. Chem. Soc. 2000;122:2373–2374. [Google Scholar]
- 32.Durandin A, Jia L, Crean C, Kolbanovskiy A, Ding S, Shafirovich V, Broyde S, Geacintov NE. Chem. Res. Toxicol. 2006;19:908–913. doi: 10.1021/tx060078e. [DOI] [PubMed] [Google Scholar]
- 33.Karwowski B, Dupeyrat F, Bardet M, Ravanat JL, Krajewski P, Cadet J. Chem. Res. Toxicol. 2006;19:1357–1365. doi: 10.1021/tx060088f. [DOI] [PubMed] [Google Scholar]
- 34.Burrows CJ, Muller JG. Chem. Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 35.Chatgilialoglu C, Caminal C, Altieri A, Vougioukalakis GC, Mulazzani QG, Gimisis T, Guerra M. J. Am. Chem. Soc. 2006;128:13796–13805. doi: 10.1021/ja062636h. [DOI] [PubMed] [Google Scholar]
- 36.Hildenbrand K, Schulte-Frohlinde D. Free Radic. Res. Commun. 1990;11:195–206. doi: 10.3109/10715769009088916. [DOI] [PubMed] [Google Scholar]
- 37.Bachler V, Hildenbrand K. Radiat. Phys. Chem. 1992;40:59–68. [Google Scholar]
- 38.Cadet J, Douki T, Ravanat JL. Nat. Chem. Biol. 2006;2:348–349. doi: 10.1038/nchembio0706-348. [DOI] [PubMed] [Google Scholar]
- 39.Pratviel G, Meunier B. Chem. Eur. J. 2006;12:6018–6030. doi: 10.1002/chem.200600539. [DOI] [PubMed] [Google Scholar]
- 40.Knobloch B, Linert W, Sigel H. Proc Natl Acad Sci U.S.A. 2005;102:7459–7464. doi: 10.1073/pnas.0501446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrier S, Hau J, Gasparutto D, Cadet J, Favier A, Ravanat JL. J. Am. Chem. Soc. 2006;128:5703–5710. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 42.Box HC, Budzinski EE, Dawidzik JD, Wallace JC, Evans MS, Gobey JS. Radiat. Res. 1996;145:641–643. [PubMed] [Google Scholar]
- 43.Box HC, Dawidzik JB, Budzinski EE. Free Radic. Biol. Med. 2001;31:856–868. doi: 10.1016/s0891-5849(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 44.Greenberg MM. Org. Biomol. Chem. 2007;5:18–30. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 45.Delatour T, Douki T, Gasparutto D, Brochier MC, Cadet J. Chem. Res. Toxicol. 1998;11:1005–1013. doi: 10.1021/tx980066w. [DOI] [PubMed] [Google Scholar]
- 46.Romieu A, Bellon S, Gasparutto D, Cadet J. Org. Lett. 2000;2:1085–1088. doi: 10.1021/ol005643y. [DOI] [PubMed] [Google Scholar]
- 47.Luo W, Muller JG, Rachlin EM, Burrows CJ. Org. Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 48.McCallum JE, Kuniyoshi CY, Foote CS. J. Am. Chem. Soc. 2004;126:16777–16782. doi: 10.1021/ja030678p. [DOI] [PubMed] [Google Scholar]
- 49.Niles JC, Wishnok JS, Tannenbaum SR. Chem. Res. Toxicol. 2004;17:1510–1519. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- 50.Leipold MD, Muller JG, Burrows CJ, David SS. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 51.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 52.Sugden KD, Campo CK, Martin BD. Chem. Res. Toxicol. 2001;14:1315–1322. doi: 10.1021/tx010088+. [DOI] [PubMed] [Google Scholar]
- 53.Shafirovich V, Dourandin A, Huang W, Luneva NP, Geacintov NE. J. Phys. Chem. B. 1999;103:10924–10933. [Google Scholar]
- 54.Kuzmin VA, Dourandin A, Shafirovich V, Geacintov NE. Phys. Chem. Chem. Phys. 2000;2:1531–1535. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.