Abstract
We performed a systematic evaluation of how variations in sequencing depth and other parameters influence interpretation of Chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) experiments. Using Drosophila S2 cells, we generated ChIP-seq datasets for a site-specific transcription factor (Suppressor of Hairy-wing) and a histone modification (H3K36me3). We detected a chromatin state bias, open chromatin regions yielded higher coverage, which led to false positives if not corrected and had a greater effect on detection specificity than any base-composition bias. Paired-end sequencing revealed that single-end data underestimated ChIP library complexity at high coverage. The removal of reads originating at the same base reduced false-positives while having little effect on detection sensitivity. Even at a depth of ~1 read/bp coverage of mappable genome, ~1% of the narrow peaks detected on a tiling array were missed by ChIP-seq. Evaluation of widely-used ChIP-seq analysis tools suggests that adjustments or algorithm improvements are required to handle datasets with deep coverage.
Introduction
ChIP-seq has become the predominant technique for profiling in vivo DNA-protein interactions1, 2 and histone marks3, 4 on a genome-wide scale. Multiple factors in the experimental design and data analysis influence the final interpretation of a ChIP-seq experiment. One important factor is the potential bias in the genomic coverage of sequencing reads, which can confound the true signal of interest. A second factor is whether the DNA libraries are prepared for paired-end (PE) or single-end (SE) sequencing. PE libraries are well suited to characterize genomic rearrangements and identify novel chimeric transcripts or alternative splice isoforms. However, the benefits of PE libraries for a standard ChIP-seq experiment are unclear. A third factor is the absolute and relative sequencing depth of the ChIP and chromatin input samples used as control for background signal. Chromatin input samples are generated by fragmentation or enzymatic digestion of chromatin extracts. (Supplementary Note). ChIP-seq is presumed to have many advantages over ChIP followed by array hybridization (ChIP-chip)5; some, such as greater resolution and better genome coverage are proven 6,7, others such as higher sensitivity, , and larger dynamic range, remain to be tested in a direct comparison between ChIP-chip data and ChIP-seq data at a deep coverage from the same samples. A fourth factor is the computational algorithm that is used for ChIP-seq peak calling. In an earlier systematic study of ChIP-chip performance, the choice of the analysis algorithm and parameters had a larger effect on the accuracy of the final results than any other single experimental factor5. The most popular ChIP-seq peak callers were developed and evaluated based on early low-coverage ChIP-seq8,9 or simulated datasets ((http://seqanswers.com/forums/showthread.php?t=1039; http://sourceforge.net/projects/useq/files/CommunityChIPSeqChallenge/)).
To evaluate the aforementioned factors, we generated a high-quality ChIP-seq datasets (Supplementary Note) from Drosophila melanogaster S2 cells with a depth of ~1 read/bp of mappable fly genome (corresponding to ~2.4 billion reads in human) 10 enriching for the site-specific transcription factor (TF) Suppressor of Hairy-wing (Su(Hw)) 13, yielding narrow peaks, and the broadly distributed histone mark H3K36me311, 12, 14., .
Results
The effect of DNA base composition and chromatin state
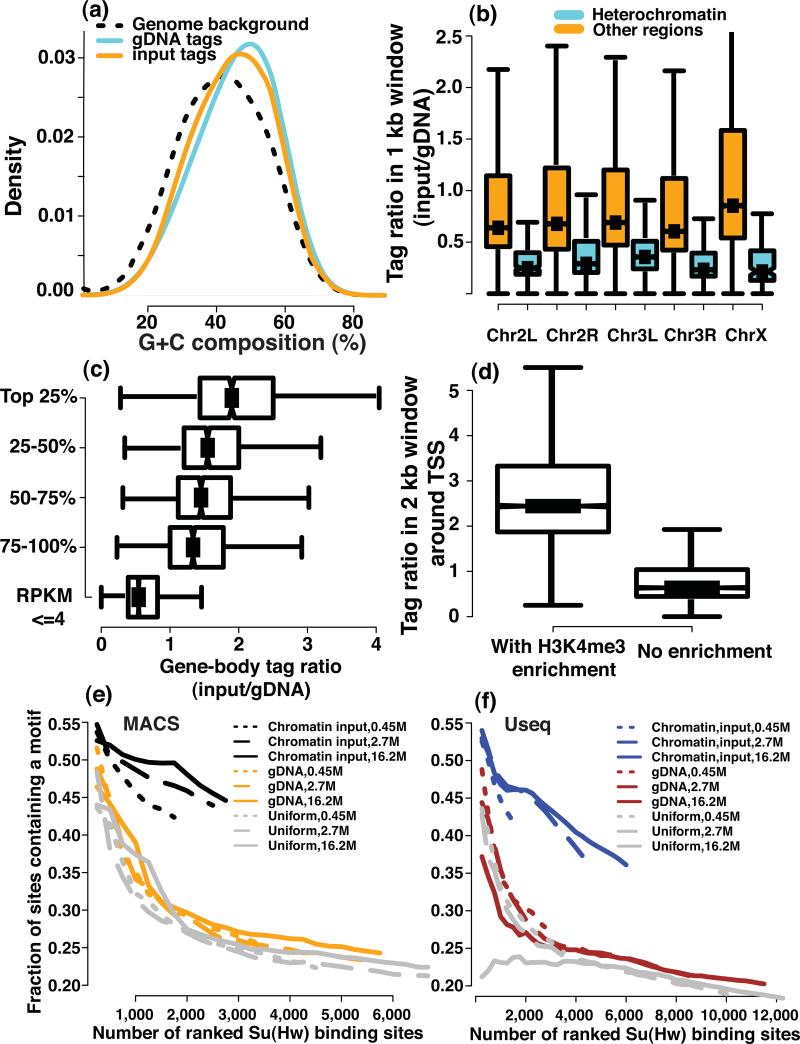
In a ChIP-seq experiment, biases could be introduced during the processing, for example PCR amplification and library preparation, and sequencing of DNA fragments. Consistent with earlier results15, 16, sequencing reads from our gDNA samples have a higher G+C content than the whole genome background (Online Methods) (Fig. 1a). We also observed that the sequencing reads of the chromatin input sample have a G+C composition distribution that is different from that of the gDNA sample (Fig. 1a, gDNA-GC-median=47%, Chromatin-GC-median=44%, Mann-Whitney (MW) test, P < 2.2 × 10-16) –, suggesting that chromatin may affect sequencing coverage.
Figure 1. The impact of genomic sequence composition and chromatin state on read coverage.
(a) The histograms of GC composition for reads from gDNA and chromatin input samples are compared with the genomic background. Boxplots of the read count ratio of chromatin input to a gDNA sample are shown for (b) non-overlapping 1 kb windows in annotated heterochromatin and euchromatin regions of the corresponding chromosomes, (c) for the 2 kb windows centered at TSS that are with or without H3K4me3 enrichment, and (d) for the coding regions of genes with different expression levels (e,f) The fraction of computationally identified Su(Hw) peaks that contains a Su(Hw) binding motif is plotted as a function of the number of top-ranked binding sites for different types of controls (chromatin input, genomic DNA and a uniform background) and for two algorithms (e) MACS and (f) Useq. The ranking is based on the statistical significance of each peak that is assigned by individual algorithms.
We compared the gDNA-normalized coverage of the chromatin input sample in different genomic regions using ratios of the chromatin input to the gDNA sample in non-overlapping 1 kb windows. We first compared heterochromatin and euchromatin based on the annotation from UCSC dm3 (Online Methods). –. Read ratios in heterochromatin regions were significantly lower than those in euchromatin (Fig. 1b, MW test, P < 2.2 × 10-16). Comparison with 15 histone marks17-19 (Online Methods), confirmed that the normalized chromatin input coverage had a positive correlation with active histone marks and a negative correlation with repressive histone marks (Supplementary Fig. 1). We also observed higher coverage in euchromatin on the X chromosome than euchromatin of autosomes in the male-derived S2 lines (Fig. 1b). This is consistent with the dosage compensation mechanism in Drosophila20.
We further observed that genes with higher expression levels had higher read ratios in gene bodies (Fig. 1c, MW test, P < 7.2 × 10-7), and that the promoter regions with H3K4me3 enrichment exhibited higher read ratios than those without H3K4me3 (Fig. 1d, MW test, P < 2.2 × 10-16). These observations agree with results in Saccharomyces cerevisiae21 and indicate that coverage was higher in regions with more open chromatin states both at a chromosomal scale and at the level of individual genes.
To characterize the impact of GC bias and chromatin-state bias on the identification of ChIP-enriched regions, we identified Su (Hw) peaks using two different algorithms, the same ChIP data, but with “control” data from either chromatin input, genomic DNA, or generated from a uniform background model across the genome that ignores GC bias and chromatin-state bias. The genomic DNA data does not contain any information on the chromatin-state, and serves only to correct the GC bias. The chromatin input control corrects for both the GC bias and the chromatin-state bias. Peaks identified using chromatin input as a control showed much better enrichment of the Su(Hw) binding motif than those identified by other controls (Fig. 1e, f).
If we consider the fraction of the peaks that do not contain a motif as a crude proxy of false discovery rate (FDR) for peak-calling, then at a fixed FDR using chromatin input control resulted in more discovered binding sites than using other controls (Fig. 1e,f). Four to ten percent of ChIP-enriched regions identified using chromatin inputs were missed by using other controls, indicating that ignoring the GC bias and the chromatin-state bias also had a negative effect on detection sensitivity.
Single-end versus Paired-end reads for ChIP-seq
PE sequencing has been widely utilized in DNA- and RNA-seq experiments to uncover fusion transcripts, genomic structural variations, rearrangements and novel splice junctions, but the benefits of PE sequencing for regular ChIP-seq experiments are less clear. We first compared the percentage of the uniquely mapped PE reads that were also uniquely mapped when the PE reads were treated as if they were independent SE reads at different read lengths. The percentage of uniquely mapped SE reads was below 10% at a read length of 18 bp and was over 80% when the read length exceeds 22 bp (Supplementary Fig. 2a and Supplementary Note ).
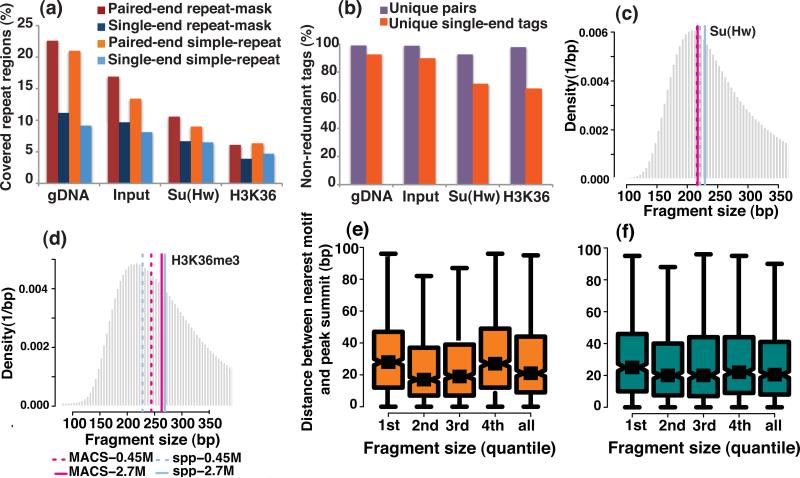
Next, we observed that the difference in sequencing coverage of repeat regions by uniquely mapped PE reads when they were mapped as either PE or SE reads (36 bp) at a sequencing depth of 16.2 M reads was approximately twice that of the SE reads for the gDNA sample. This sequencing depth approximately corresponds to 327 M reads for the mappable human genome10. In contrast, for the chromatin input sample and for the ChIP samples of Su(Hw) and H3K36me3, the difference in sequencing coverage of the repeat regions between PE and SE reads is less pronounced (Fig. 2a). The gain from PE data in discovering Su(Hw) or H3K36me3-enriched regions in repeat regions was typically less than 15% (Supplementary Fig. 2b).
Figure 2. A comparison of several features between the PE and SE reads, and an evaluation of the effect of DNA fragment size.
The features include (a) genomic coverage in repeat regions and (b) the estimated library complexity for PE and SE reads. The repeat-mask refers to the DNA sequences of interspersed repeats and of low-complexity DNA that were identified by the RepeatMasker program (Online Methods). The simple-repeat refers to the simple tandem repeats (possibly imperfect repeats) that were located by the Tandem Repeats Finding program (Online Methods). . Fragment size that was estimated from the SE reads by MACS and spp was compared with the mode of the fragment size histogram that was derived from the PE reads for the (c) Su(Hw) and (d) H3K36me3 ChIP samples. The pink solid and dashed lines represent the fragment size that was estimated from the SE reads by MACS at the sequencing depth of 2.7M and 0.45M reads, respectively. The blue solid and dashed lines represent the fragment size that was estimated from the SE reads by spp at the sequencing depth of 2.7M and 0.45M reads. A box-plot comparison of the summit resolution of the peaks identified by (e) MACS and (f) spp is shown for the cases in which PE reads from DNA fragments with different sizes were used.
A common quality measure for ChIP-seq libraries is library complexity (Online Methods). There are many factors that can lead to poor library quality, such as poor antibody quality, over-crosslinking, an insufficient amount of starting material of ChIP-DNA, inappropriate sonication and over-amplification by PCR. We observed a major discrepancy between the PE- and SE-based estimates of the library complexity for the Su(Hw) and H3K36me3 ChIP samples, but not for the gDNA and chromatin input samples at a sequencing depth of 16.2 M PE reads (Fig. 2b). Therefore, caution is warranted when using SE ChIP-seq data to model library complexity.
Choosing ChIP-seq data analysis algorithms for evaluation
There are more than 30 published algorithms for identifying peaks from ChIP-seq data, with more being published continuously27, 28. In this study, we selected 7 algorithms7, 10, 22-26, that are capable of using chromatin input data12, that are not restricted to analysis of only TFs or histone marks, that directly support analyzing ChIP-seq data from Drosophila, and that are among the most highly cited. We evaluated their performance at different sequencing depths (Online Methods).
The size of the sequenced fragments and peak calling
MACS7 and spp24 explicitly report the estimated sizes of the DNA fragments in the library from the SE data. For Su(Hw) ChIP-seq data, MACS and spp gave notably accurate size estimations that deviated from the PE-inferred fragment size by only 10-20bp (Fig. 2c). However, both algorithms were less accurate in the H3K36me3 dataset (Fig. 2d). To characterize the influence of the fragment size on the spatial resolution of the narrow peaks, we next utilized PE reads from different size fragments in the same library for peak calling. For both MACS and spp, the larger the fragment size, the wider the peak (Supplementary Fig. 3). In contrast, the peak-summit resolution did not depend strongly on the size of the sequencing fragments. Thus, the use of smaller fragments did not necessarily improve the peak-summit resolution (Fig. 2e, f).
Sensitivity and specificity
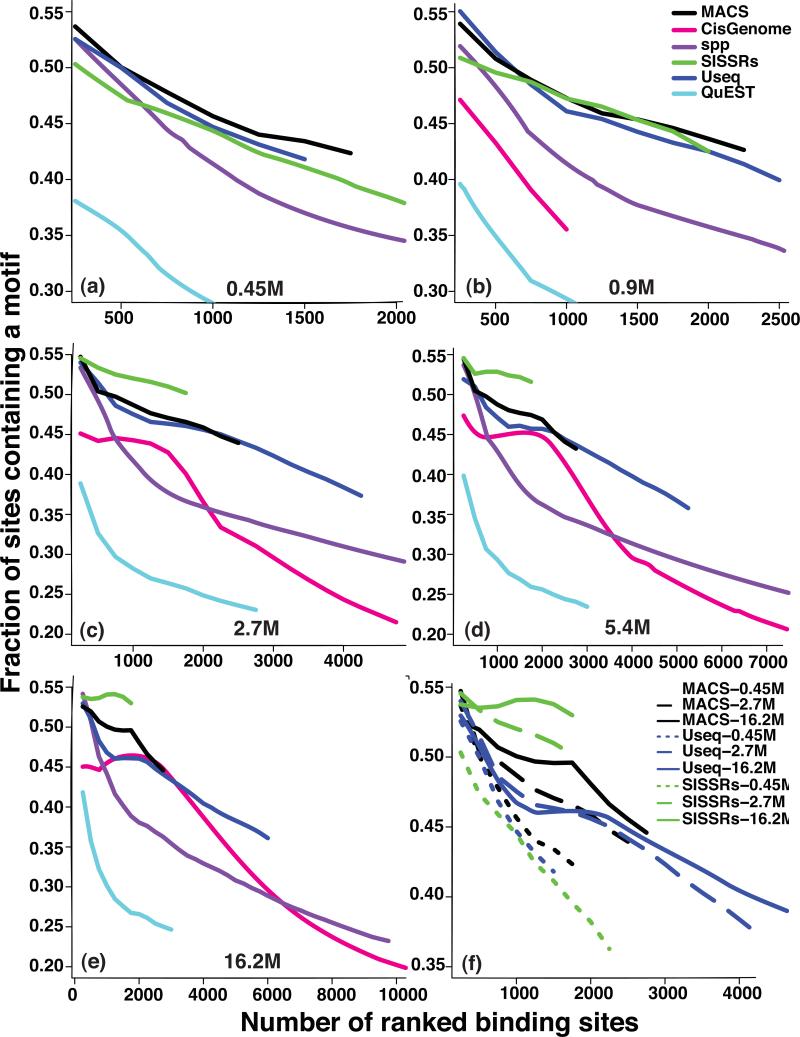
We used the enrichment of the Su(Hw) binding motif within peaks to evaluate the specificity of different algorithms. SISSRs, MACS and Useq were the three best-performing algorithms in terms of specificity (Fig. 3a, b, c, d, e), and they showed a notable improvement with an increasing sequencing depth (Fig. 3f). At most sequencing depths, SISSRs had the best overall specificity for all of the identified peaks but has fewer peaks than the other algorithms. To evaluate the overall sensitivity of the different algorithms for all of the identified peaks, we used the confidently enriched regions that were identified from ChIP-chip analysis as a proxy for true positives (Online Methods). Useq and spp had the highest overall sensitivity (Supplementary Fig. 4).
Figure 3. Quality of the Su(Hw) peaks.
The fraction Su(Hw) peaks, identified by the indicated peak callers, that contains a Su(Hw) binding motif is plotted as a function of the number of top-ranked binding sites at the sequencing depths of 0.45 M (a), 0.9 M (b) 2.7 M (c), 5.4 M (d), and 16.2 M (e) reads. The ranking is based on the statistical significance of each peak that is assigned by an individual algorithm. The evaluation results for the top 3 best-performing peak-callers at sequencing depths of 0.45 M, 2.7 M, and 16.2 M are shown in (f).
The effect of imbalanced coverage between ChIP and input
We evaluated how imbalanced sequencing coverage between ChIP and chromatin input samples influences peak calling. We focused on the MACS and Useq,. SISSRs was excluded from this evaluation because the number of identified peaks differed substantially between replicates when the sequencing coverage is unbalanced. For the same sequencing depth of the ChIP sample, deeper sequencing of the chromatin input sample gave rise to better detection specificity (Supplementary Fig. 5a-b). This observation holds at both small and large sequencing depths of the ChIP sample. Therefore it is beneficial to sequence the chromatin input sample to a depth at least equal to the ChIP sample, if not deeper.
The effect of redundant reads on narrow peak calling
Redundant reads in ChIP-seq datasets often indicate poor library complexity, and as a result, many peak callers remove redundant reads, that is, reads with the same 5’ genomic location, during peak calling. However, with very deep sequencing, redundant SE reads from ChIP samples may also result from ChIP-enrichment signal. We therefore evaluated the effect of retaining or removing redundant reads on both the sensitivity and specificity at large sequencing depths. We first compared the enrichment of the Su(Hw) motif within peaks that were identified by SISSRs, MACS and Useq between two conditions: one in which we kept only one read at each genomic location and another in which we retained the redundant reads completely (SISSRs and Useq) or partially (MACS). In general, removing redundant reads improved the specificity of the identified peaks for MACS and Useq. For SISSRs, removing redundant reads only improved the specificity at a high sequencing depth (Supplementary Fig. 5c-e).
The PE data allow us to differentiate the source of redundant reads in peak regions because the redundant reads that result from experimental artifacts, such as PCR amplification bias, should be identical at both ends. The data indicated that redundant reads from duplicate fragments represented fewer than 10% of all reads, whereas in most peak regions, the proportion of redundant reads was 20-40% (Supplementary Fig. 6). Thus, most redundant reads in peak regions represented true signal. Nonetheless, for two different algorithms, MACS and Useq, the removal of redundant reads had little (Supplementary Fig. 7a) or no effect (Supplementary Fig. 7b) on sensitivity. Overall, the removal of redundant reads was usually beneficial because it removes noise in the non-enriched regions while having little effect on detection sensitivity.
The detection dependence on sequencing depth
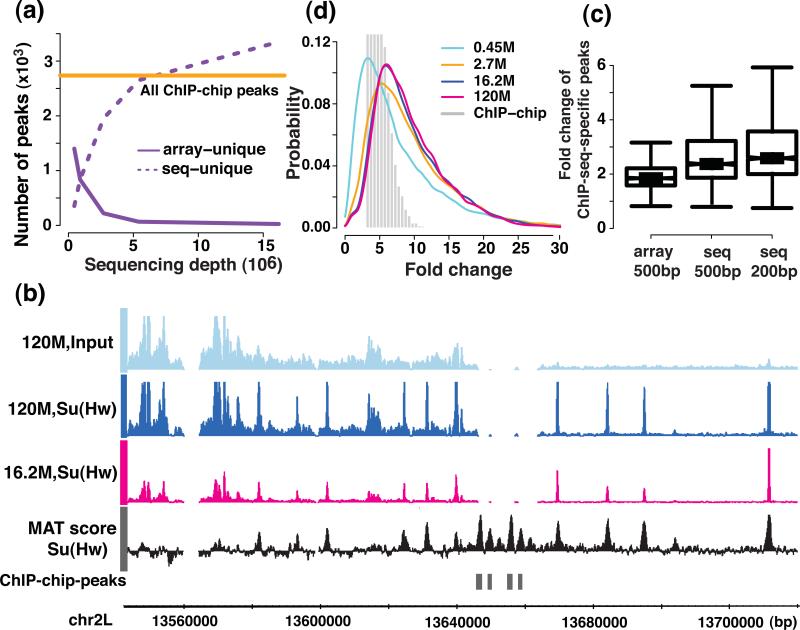
One indication that a sufficient sequencing depth has been reached is when the number of binding sites plateaus with an increasing read count. Interestingly, different algorithms had distinct saturation profiles. The number of binding sites identified by MACS, SISSRs, and QuEST started to plateau or plateaued at approximately 16.2 M reads (corresponding to ~327 M in human) (Supplementary Figs. 8a, d, f, and 9a), whereas the number of peaks identified by CisGenome, spp, and Useq steadily increased with depth (Supplementary Figs. 8b, c, e, and 9a). We then compared the enriched regions discovered at a given depth to those identified from the complete set of 120M reads. More than half of the Su(Hw)-enriched regions identified from the complete data were detected by most of the algorithms at a sequencing depth of 5.4 M reads (corresponding to ~110 M in human). More than 60% of the Su(Hw)-enriched regions (3-fold enrichment or greater) identified from the complete data, were identified by MACS and Useq at a depth of 2.7 M reads (corresponding to ~55 M in human; Supplementary Fig. 10a).
Narrow peak differences between sequencing and array data
We compared ChIP-enriched regions identified by tiling arrays (Affymetrix) and sequencing platforms on the same set of Su(Hw) samples using Useq and MAT5, 29, respectively. At low sequencing depths (≤0.90 M, corresponding to ≤~18 M in human), ~30-50% of the ChIP-chip peaks were missed by ChIP-seq. When the sequencing depth reaches 2.7 M reads (corresponding to ~55 M in human), over 90% of ChIP-chip peaks were identified by ChIP-seq (Fig. 4a). Surprisingly, even when the sequencing depth reaches 16.2 M reads (corresponding to ~327 M in human), ~1% of the ChIP-chip peaks were not detected in the sequencing data. These peaks had sparse or no sequencing coverage, even using all reads in our dataset (Fig. 4b), mostly due to low mappability (Supplementary note)10, 28. The Su(Hw) peaks specific to ChIP-chip were enriched of the Su(Hw) binding motif, suggesting that they are genuine Su(Hw) binding sites. We performed ChIP-qPCR experiments and validated seven randomly selected ChIP-chip peaks that were missed in the sequencing data (Supplementary Fig. 11). Either a lack of probe coverage on the array or higher sensitivity of ChIP-seq relative to array5(Fig. 4c) contributed to those ChIP-seq specific peaks. The sequencing platform showed a larger dynamic range of fold change than the array and increased depth improves both the sensitivity and the quantification of regions with low fold enrichment (Fig. 4d).
Figure 4. Comparison of the identified narrow peaks and the dynamic range between the sequencing and the tiling array platform.
(a) the number of identified peaks on different platforms and (b) examples of ChIP-chip peaks that were missed in the sequencing platform, the MAT score for ChIP-chip data, and the ChIP-seq signal coverage at the sequencing depths of 16.2 M and 120 M are shown. (c) The fold change difference between sequencing and tiling arrays in 200 bp and 500 bp windows centered on the peaks that were unique to the sequencing platform at a sequencing depth of 16.2 M (d) the dynamic range of the signal (ChIP versus the chromatin input fold change) are shown for the sequencing and the tiling array platform.
Algorithm reproducibility for narrow peaks across replicates
We quantified the reproducibility of peak calling across replicates using the “irreproducible discovery rate (IDR)” 30, which assesses the consistency between the ranks of the peaks that were commonly identified on a pair of replicates. We found that the relative reproducibility of different algorithms depended on the sequencing depth. While MACS and spp were more reproducible across replicates than any other algorithms at shallow sequencing depths (fewer than 0.90 M, corresponding to fewer than 18 M in human), CisGenome and Useq became the most “reproducible” across replicates at or above 2.7 M reads (corresponding to ~55 M in human) (Fig. 5).
Figure 5. An evaluation of the reproducibility across replicates of six peak-callers.
The number of reproducible peaks at various IDR levels is plotted for sequencing depths of 0.45 M (a), 0.9 M (b), 2.7 M (c), 5.4 M (d), and 16.2 M (e) reads. In (f), the number of reproducible peaks identified at an IDR of 5% is plotted as a function of sequencing depth.
Detecting broad enriched regions at different depths
We evaluated sensitivity and specificity of different algorithms in detecting broad patterns of enrichment. As a “gold standard”, for H3K36me3-positive regions, we used all exonic regions from the genes with the top 4000 expression levels (Supplementary Fig. 12a-e; results were similar with the top 1000 or 2000 genes, Supplementary Figs. 13 and 14). We called the gene bodies of unexpressed genes H3K36me3-negative regions (Online Methods). To control for the width differences of the enriched regions that were identified by different algorithms, we used the coverage of the “true positives” normalized by the total width of all predicted enriched- regions as a proxy for the sensitivity and the width-normalized coverage of the “true negatives” as a proxy for the false positive rate. QuEST had the highest specificity, with PeakSeq second. Spp was among the algorithms with the lowest sensitivity and specificity (Supplementary Fig. 12a, b, f, g) at a shallower sequencing depth (fewer than 0.90 M, corresponding to fewer than ~18 M in human), but showed distinct improvement at larger sequencing depths (≥2.7 M reads, corresponding to ≥~55 M in human; Supplementary Fig. 12c, d, e, h, i, j).
Similar to the case of Su(Hw), different algorithms showed distinct saturation profiles in the broad enrichment data (Supplementary Figs. 15 and 9b). MACS, QuEST, spp and Useq showed a faster saturation in identifying ChIP-enriched regions of H3K36me3 than Su(Hw) (Supplementary Fig. 10b). Unlike the case of Su(Hw), the number of identified H3K36me3-enriched regions did not increase monotonically with the sequencing depth for many algorithms, including MACS, spp and QuEST, because neighboring regions started to merge at high sequencing depths.
Algorithm reproducibility for broad regions across replicates
We performed IDR analysis tailored to the broad regions (Online Methods) to evaluate the algorithm reproducibility across replicates. Again, we found that this depended on the sequencing depth. QuEST and Useq produced a greater number of reproducible regions across replicates than did other algorithms at or below 2.7 M (corresponding to ~55 M in human) reads (Supplementary Fig. 16a, b, c, f), whereas spp and Useq did so when the sequencing depth was above 2.7 M (Supplementary Fig. 16d, e, f).
Discussion
Sequencing depth had a profound impact on several aspects of ChIP-seq results, including some that were unexpected. Our study suggests that for such TFs as Su(Hw) and such histone marks as H3K36me3, the regularly-adopted sequencing depth of 15-20 M reads in humans may be insufficient for identifying vast majority of the enriched regions.
Our finding that the removal of redundant reads helped to reduce false positives and had little effect on the detection sensitivity is seemingly incompatible with the fact that at a high sequencing depth, most redundant reads in narrow peak regions represent true signals. The probable explanation is that most of the regions containing redundant reads were among the more highly enriched, such that even after the removal of redundant reads, the vast majority of those regions still show significant enrichment in ChIP versus chromatin input samples. Additionally, the removal of redundant reads in the non-enriched regions differentially reduces reads that originate from experimental bias in PCR amplification and library preparation. Because removing redundant reads influences quantitative information associated with enriched regions, for high-quality libraries, it may be most appropriate to identify peaks in the absence of redundant reads, but then to include all reads in downstream analyses.
There were notable variations in sensitivity and specificity between the algorithms under evaluation. Some algorithms exhibiting unexpected behavior at high sequencing depths, indicating the importance of improving algorithms for use at a high sequencing depth, including a more effective handling of reads mapped to multiple genomic locations. In practice, it is beneficial to use more than one algorithm to ensure the robustness of the analysis results for the deep sequencing data.
One important factor that was not assessed here is the choice of sequencing platform. We chose the Illumina platform for this study because the vast majority of publicly available ChIP-seq datasets were generated on this platform, including those from the ENCODE and modENCODE projects (http://www.genome.gov/10005107). There are also important open questions specifically regarding identifying broadly enriched regions that were not addressed in this study, such as how to determine the boundaries of broadly enriched regions and how sequencing depth influences the determination of the boundaries. We anticipate that our dataset will be a valuable resource (GEO Data accession code: GSE27679) for the ChIP-seq community to address these and other technical questions related to deep sequencing.
Online Methods
Overall experimental design
We amplified S2 cells from the modENCODE batch before transferring them to the plates. We put a total of 15 plates in culture until the cells reached the appropriate concentration. We harvested cells from 2 plates to extract the genomic DNA. We subsequently treated the remaining 13 plates with formaldehyde at the same time, and then extracted the chromatin. Next, we used 3 plates to produce a chromatin input sample corresponding to the chromatin DNA of the treated cells. Each plate corresponded to an Eppendorf tube of chromatin and we performed ChIP experiments on 5 tubes for H3K36me3 and on 5 tubes for Su(Hw). We performed ChIP and DNA extraction procedures independently on each tube. After the purification of the DNA, we pooled together the tubes corresponding to the same sample (Su(Hw), H3K36me3, chromatin input and gDNA). After mixing, we again aliquoted each sample into 5 tubes. Next, we used 1 aliquot of each for ChIP-chip on Affymetrix tiling arrays for quality control, 1 aliquot for SE sequencing at the high-throughput genomic analysis core of the University of Chicago, 1 aliquot for SE sequencing at the high-throughput sequencing facility of the University of North Carolina at Chapel Hill (UNC), 1 aliquot for PE sequencing at UNC, and the remaining aliquot to back up the original samples.
ChIP experiment
After the expansion of the S2 cell line, we transferred cells to a plate. Once confluent, we added 1.8% formaldehyde to the cell culture. We harvested cells in the presence of the formaldehyde with the help of a cell scraper. After 15 minutes of incubation at room temperature, we quenched the crosslinking reaction with glycine for 5 minutes. We subsequently washed cell pellets 3 times with a lysis buffer. We performed regular chromatin extraction before sonication. We then used sonicated chromatin for ChIP experiments or directly for DNA extraction for the chromatin input samples. We performed ChIP as previously described1, 2. The anti-H3K36me3 antibody was from Abcam (catalogue # ab9050/lot # 927884). The anti-Su(Hw) antibody was from Pamela K. Geyer's lab. Both are rabbit polyclonal antibodies.
Library preparation and sequencing
At the University of Chicago, we prepared the libraries according to Illumina's instructions accompanying the DNA Sample Kit (Part# 0801-0303). Briefly, we end-repaired DNA using a combination of T4 DNA polymerase, E. coli DNA Pol I large fragment (Klenow polymerase) and T4 polynucleotide kinase. We treated the blunt, phosphorylated ends with Klenow fragment (3’ to 5’ exo minus) and dATP to yield a protruding 3- ‘A’ base for ligation of Illumina's adapters, which have a single ‘T’ base overhang at the 3' end. After adapter ligation, we PCR amplified DNA with Illumina primers for 15 cycles, and band isolated library fragments of ~250 bp from a 2% agarose gel. We captured the purified DNA on an Illumina flow cell for cluster generation. We sequenced libraries on the Genome Analyzer IIx following the manufacturer's protocols. At UNC, we used a slightly different protocol of library preparation in which we band isolated the library fragments of ~150-500 bps immediately after the ligation of Illumina's adapters followed by 18 cycles of PCR amplification.
The definition of library complexity for PE and SE data
The library complexity of SE data is defined as the number of non-redundant SE reads divided by the total number of reads, where redundant SE reads are those that are mapped to the same location with the same orientation in the genome. The library complexity of PE data is defined as a non-redundant pair of reads divided by a total pair of reads, where redundant PE reads are those that have identical genomic locations on both ends.
The mapping of sequencing reads
We used ELAND to align the SE sequencing reads to the flybase BDGPv5 reference genome. We pooled together the uniquely mapped SE reads with no more than 2 mismatches from different runs up to 120 M for the ChIP-sample of Su(Hw) and H3K36me3. For chromatin input and gDNA samples, we added uniquely mapped PE reads to constitute the total 120 M reads, thereby compensating for the failed runs of SE sequencing of these two samples (Supplementary Table 1). For a comparison of differences in the read mappability and coverage of the repeats region, we performed an estimation of the library complexity for the PE and SE reads. We first used Bowtie-0.12.5 to map the PE reads with almost all of the default settings (-chunkmbs 120), and we constrained the fragment size between 80 and 600 bp. Next, we re-aligned the uniquely mapped PE reads in the SE mode using the same parameter settings.
The mappability, the heterochromatin and the repeat regions of the Drosophila genome
We obtained the mappability data of Drosophila genome from an early study3 and the details of how the mappability was calculated was described previously4. The interspersed repeats and of low-complexity DNA that were identified by the RepeatMasker program (http://www.repeatmasker.org). The simple-repeat refers to the simple tandem repeats (possibly imperfect repeats) that were identified by the Tandem Repeats Finding program5. Both the repeat region and the heterochromatin region annotation were based on UCSC dm3, and were downloaded from the UCSC genome browser.
ChIP-seq data analysis algorithms
We chose 7 algorithms that are capable of using chromatin input data, are not restricted to only TF or histone marks, are supportive for analyzing ChIP-seq data from Drosophila, and are among the most cited algorithms. These algorithms are CisGenome (v1.2), MACS (v1.40beta), spp (v1.8), QuEST (v2.4), Useq (v6.9), SISSRS (v1.4) and PeakSeq (v1.01). We did not include E-RANGE6 and F-Seq7, two highly cited algorithms, in the evaluation because the parameters of E-RANGE were optimized for the mammalian genome and F-Seq does not provide good support for peak finding in invertebrates. For the evaluation of the algorithm performance on the ChIP-seq data of Su(Hw), we did not include PeakSeq because it does not provide the information of the peak summit of each peak, whereas the evaluation of peak quality requires the information of the peak summit. When we used the middle point of each peak identified by PeakSeq as a surrogate of the peak summit, PeakSeq exhibited a poor performance, thereby making the comparison unfair. For the evaluation of the algorithm performance on the ChIP-seq data of H3k36me3, we did not include SISSRs because the width of the regions identified by SISSRs is too small to be consistent with that of the broad regions.
ChIP-seq data analysis at different sequencing depths
We randomly sampled reads at sequencing depths of 0.45 M, 0.9 M, 2.7 M, 5.4 M and 16.2 M reads from a pool of 120 M reads for both ChIP and chromatin input samples. These sequencing depths approximately correspond to 9 M, 18 M, 55 M, 109 M and 327 M reads in a human ChIP-seq experiment4. At each sampling depth, we generated five independent ”replicates” of the sequencing data. We averaged the analysis results of each algorithm over the five replicates before comparison.
ChIP-chip peak calling for Su(Hw) and the fold change calculation for the sequencing and tiling array platform
We performed peak calling for Su(Hw) using the MAT8 algorithm, which is among the best peak-calling algorithms for ChIP-chip data from Affymetrix data9 with a band width of 250 bp, a p-value cutoff of 10-5 and a false discovery rate (FDR) cutoff of 5%. We only considered the peaks with fold changes of no less than 3-fold (the detection limit of the Affymetrix tiling array9) for further comparison with ChIP-seq peaks. All 500 bp windows that centered on the summit of these ChIP-chip peaks of Su(Hw) were used as reference ChIP-chip peaks and as a proxy for the true positives to evaluate the sensitivity of different algorithms for identifying ChIP-seq peaks. The fold change of the signal (ChIP versus chromatin input) in each 500 bp-scanning window centered on the probes was calculated using Tiling Analysis Software (Affymetrix). The fold change in the 200 bp and 500 bp windows that were centered on the summit of ChIP-seq peaks was calculated as follows: (number of covered fragments+1)ChIP/(number of covered fragments+1)input.
The use of IDR to quantify the reproducibility of peak calling between a pair of replicates
The reproducibility across replicates is essential to ChIP experiments not only at the level of read count data but also at the level of peak calling because the identified peaks usually are the primary substrates for downstream analysis. IDR (irreproducible discovery rate) is a statistical measure that assesses the consistency of the rank orders between a pair of rank lists10. Unlike the usual scalar measures of reproducibility (e.g., the rank correlation), this measure describes reproducibility in terms of the extent to which the ranks of the entries on the lists are no longer consistent across replicates that are ordered in descending significance. Based on a copula mixture model, this measure provides a “score” that estimates the probability that each pair of peaks is reproducible, and it reports the expected rate of irreproducible discoveries (IDR) in the selected peaks in a fashion analogous to that of FDR. The number of reproducible peaks across replicates at given IDR levels can be used to compare the relative reproducibility of different peak calling algorithms.
IDR is independent of the threshold choice that is used for peak calling, and it emphasizes implicitly the consistency between the top-ranked peaks, rather than treating all of the ranks equally. Therefore, this method overcomes many limitations in traditional ways of measuring reproducibility and is suitable for our purposes. Detailed descriptions of the methodology and the implementation of IDR for narrow peak can be found in Ref. 10. For broad peaks, often one peak overlaps with multiple (small) peaks on the other replicate. When this occurs, all these small peaks are lumped as one peak and the most substantial significance of these small peaks is used as the significance of the lumped peak (unpublished results, personal communication with Dr. Kundaje).
Because the number of identified peaks of some algorithms is much larger than others, we took the top 3000 significant peaks from all of the identified peaks for the evaluation. We used the R package in Ref. 10 for all of the IDR analyses.
The RNA-seq and H3K4me3 ChIP-chip data
We calculated the gene-expression level summarized as the RPKM value based on the RNA-seq data from a previous study11. The H3K4me3 ChIP-chip data was generated, and the ChIP-enriched regions were identified, by the White lab. We considered the 2kb TSS-centered promoter to have H3K4me3 enrichment if it overlaps with the H3K4me3 peaks.
The data for enriched and depleted regions of various histone marks
The enriched and depleted regions of 15 histone marks that were identified from ChIP-chip data were obtained from modENCODE1, 12-14. These histone marks include H3K18ac, H3K27ac, H3K27me3, H3K36me1, H3K36me3, H3K4Me3, H3K4me1, H3K4me2, H3K79Me1, H3K79me2, H3K9ac, H3K9me3, H4K16ac, H4K5ac, and H4K8ac.
Motif enrichment analysis
We mapped the position-specific weight matrix (PSWM) of Su(Hw) onto the genome of Drosophila (dm3) using CisGenome with a 3rd-order Markov background model. We calculated the distance between the nearest mapped motif and the peak summit using a custom Perl script.
The definition of positive and negative regions for H3K36me3 and the measurement of sensitivity and the false positive rate
H3K36me3 is highly enriched in exonic regions but not in intronic regions of actively transcribed genes15, which allows us to approximate the positive and negative regions of H3K36me3 in the genome and to estimate the sensitivity and specificity of different algorithms using these predefined regions. We defined the positive regions of H3K36me3 as the exons of the top 4000 expressed genes (the evaluation results were similar for the top 1000 or top 2000 genes) and the negative regions as the gene body of the non-expressed genes. We estimated the expression level of each annotated gene based on the RNA-seq data from the S2 cells as the total number of reads of all of the unique exons per kb of total length of unique exons per million mapped reads (RPKM)16. We averaged the RPKM value of each gene over two biological replicates. We used the same criterion to define the non-expressed genes as in the previous study, where the non-expressed genes were those with the number of unique mapped reads per kb per million mapped reads (RKPM) smaller than or equal to 411. This cutoff was chosen based on the distribution of RKPM values in intergenic regions, where the probability of observing a RKPM value greater than or equal to 4 is approximately 5%. To control for the difference in peak width among algorithms, we used the coverage of the positive regions normalized by the total width of all predicted enriched-regions as a proxy for the sensitivity and the width-normalized coverage of the negative regions as a proxy for the false positive rate.
The calculation of the GC composition, the read-count ratio, and the coverage over different genomic features
We calculated the window-based GC composition (window-size 36bp, the same as read length) across the genome using the hgGcPercent program from Jim Kent (UCSC). We calculated the GC composition of sequencing reads of different samples using a custom Perl script. We calculated the window-based read-count ratio and the read coverage over different genomic features, including exons, gene bodies and repeat regions using the combination of custom Perl scripts and BEDTools17. We calculated the genomic distribution of most hyper/hyposequenced 1-kb windows between the chromatin input and the gDNA samples by the stand-alone Cis-regulatory Element Annotation System (CEAS) package18.
Statistical analysis
We performed all of the statistical analyses were in R, and showed all of the p-values smaller than 2.2 × 10-16 as P < 2.2 × 10-16, which is the default cutoff in R.
Supplementary Material
Acknowledgments
We thank the authors of all of the algorithms that were evaluated in this study: H. Ji, R. Jothi, P. Kharchenko, W. Li, D. Nix, J. Rozowsky and A. Valouev. We are also grateful for help from N. Bild, D. Roqueiro and M. Sabala in performing PeakSeq on the Bionimbus Cloud. Furthermore, we would like to thank D. Schmidt and D. Odom for sharing their sequencing data of the ENCODE spike-in sample, A. Kundaje for sharing his unpublished results on IDR analysis of H3K36me3 in humans, N. Rashid for sharing the mappability data of Drosophila genome and M. Greenberg's kind support in the early stage of this project. Finally, we are grateful for helpful discussions with E. Birney, M. Snyder, J. Ahringer, M. Gerstein, M. Kellis, P. Park, and other members of modENCODE consortium. This work was partially funded by NIH grant HG4069 to X.S.L, 3U01 HG004270-03S1 to X.S.L. and J.D.L., and U01HG004264 to K.P.W.
Footnotes
Author contributions
Y.C. performed bioinformatic analysis. N. N. performed the cell culture, ChIP experiments, and library preparation with the help from J. Z.. J. O. M. performed library preparation and sequencing experiments. Q. L. and P. J. B. contributed the code for IDR method. Q. L. also participated in writing the description of IDR method and interpretation of the IDR analysis result. M. S. performed ChIP-qPCR validation of the selected array-specific Su(Hw) peaks and analyzed the ChIP-qPCR data. T. L., Y. Z., T.K.K., H. H. H., Y. R., R. M. M. and B. J. W. contributed to the early development of the project. B. J. W., K. P. W, J. D. L and X. S. L. conceived the project. T.K.K., H. H. H., Y. R., and R. M. M. performed the pilot experiments. Y.C., J. D. L., and X. S. L. wrote the manuscript with the help from other co-authors.
References
- 1.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 2.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 3.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson DS, et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho JW, et al. ChIP-chip versus ChIP-seq: lessons for experimental design and data analysis. BMC Genomics. 2011;12:134. doi: 10.1186/1471-2164-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laajala TD, et al. A practical comparison of methods for detecting transcription factor binding sites in ChIP-seq experiments. BMC Genomics. 2009;10:618. doi: 10.1186/1471-2164-10-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIPseq peak detection. PLoS One. 2010;5:e11471. doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers RM, et al. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Methods. 2009;6:S22–32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Negre N, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Negre N, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy S, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larschan E, et al. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teytelman L, et al. Impact of chromatin structures on DNA processing for genomic analyses. PLoS One. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIPseq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng X, Grossman R, Stein L. PeakRanger: a cloud-enabled peak caller for ChIP-seq data. BMC Bioinformatics. 2011;12:139. doi: 10.1186/1471-2105-12-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. Annals of Applied Statistics. 2011;5:1752–1779. [Google Scholar]
- 31.Negre N, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negre N, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 37.Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson DS, et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of highthroughput experiments. Annals of Applied Statistics. 2011;5:1752–1779. [Google Scholar]
- 41.Zhang Y, et al. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy S, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 47.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.